Abstract
To increase the affinities of aptamers for their targets, we designed an aptamer dimer for thrombin and VEGF. This design is based on the avidity of the antibody, which enables the aptamer to connect easily since it is a single-strand nucleic acid. In this study, we connected a 15-mer thrombin-binding aptamer with a 29-mer thrombin-binding aptamer. Each aptamer recognizes a different part of the thrombin molecule, and the aptamer dimer has a Kd value which is 1/10 of that of the monomers from which it is composed. Also, the designed aptamer dimer has higher inhibitory activity than the reported (15-mer) thrombin-inhibiting aptamer. Additionally, we connected together two identical aptamers against vascular endothelial growth factor (VEGF165), which is a homodimeric protein. As in the case of the anti-thrombin aptamer, the dimeric anti-VEGF aptamer had a much lower Kd value than that of the monomer. This study demonstrated that the dimerization of aptamers effectively improves the affinities of those aptamers for their targets.
Keywords: Aptamer, dimerization, avidity
1. Introduction
Protein detection and molecule quantification play an essential role in clinical diagnosis. Aptamers are drawing attention as novel molecular recognition elements for protein detection [1]. Aptamers are single-strand nucleic acid ligands that bind to various target molecules with high affinity and specificity [2-4]. They have many advantages over the antibodies as molecular recognition elements, especially for diagnosis; for example, they are easy to design. By utilizing these advantages, simple, rapid and highly sensitive protein detection systems have been constructed [5-7]. Although the highly sensitive detection of target molecules has been reported, the lower detection limit is generally determined by the affinity of the aptamers used. Therefore, it is necessary for highly sensitive biosensing to use aptamers with high affinity for their targets.
It is indicated that bispecific antibody fragments that recognize adjacent and non-overlapping epitopes simultaneously have higher affinity [8-10]. By connecting two antibodies that recognize different parts of the target molecule, the Kd value of the antibody can be lowered. Because the kon value increases and the koff value decreases when binding together two recognition sites for the same target, the Kd value may be lowered by connecting two antibodies. Thus, the affinity of the antibody can be increased by connecting together two antibodies that have non-overlapping epitopes to the target.
Since it is easy to connect two aptamers, we tried to improve the affinity of aptamers by dimerization. In this study, as a model, we connected two thrombin-binding aptamers that have non-overlapping binding sites for thrombin and investigated the aptamer dimer's affinity and its thrombin-inhibitory activity. Furthermore, we have already reported a DNA aptamer against vascular endothelial growth factor (VEGF165) [11], which is the marker protein in some diseases [12, 13]. Because VEGF165 is a homodimeric protein, we tried to connect two identical aptamers and investigate the affinity of the aptamer dimer for the target.
2. Materials and Methods
2.1. Materials
Biotinylated and non-labeled oligonucleotides were purchased from Invitrogen and Operon Technologies, Inc. Thrombin and fibrinogen were obtained from Wako Chemicals. Recombinant human VEGF165 was purchased from R&D Systems as carrier-free lyophilized powders.
2.2. Binding assay measurements
The binding affinities of the aptamers for their targets were analyzed by surface plasmon resonance at 25 °C using a Biacore X instrument (Biacore AB).
The linked thrombin-binding aptamer
To immobilize the biotinylated aptamers to the SA sensor chip, the aptamers (20 nM) were injected after being equilibrated with binding buffer (50 mM Tris-HCl, 300 mM NaCl, 5 mM KCl, pH 8.0). After immobilization of the aptamers, various concentrations of thrombin were injected to measure the Kd values of the aptamers in running buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM KCl, pH 8.0). After injecting the thrombin, the sensor chip was regenerated with 0.5 % SDS for 30 seconds. To analyze the binding kinetics of the thrombin aptamers, we used the BIA evaluation software, version 3.1 with a local fit of the kinetic simultaneous Ka/Kd model, assuming a 1:2 heterogeneous binding.
The linked VEGF-binding aptamer
VEGF was immobilized on the sensor surface and the aptamers were injected onto the surface. VEGF165 (10 μg/ml in 10 mM acetate, pH 6.0) was immobilized on one of the two flow cells (Fc-1) of the CM5 sensor chip (Biacore AB) by the amine coupling procedure. On another flow cell surface (Fc-2), VEGF165 was not immobilized in order to evaluate the nonspecific binding of aptamers to the sensor surface. For the interaction assay, the aptamer solutions were injected onto both flow cell surfaces. TBSE (10 mM Tris-HCl, pH 7.0, 100 mM NaCl, 0.05 mM EDTA) served as both the running buffer and the aptamer dilution buffer. NaOH at concentrations of 10∼50 mM or 0.5 % SDS was used as the regeneration solution. The differences in the response units Fc-1 and Fc-2 were used as the response signals, and these signals were used for the construction of a Scatchard plot. The dissociation constants of the aptamers were calculated from the Scatchard plot. In this evaluation, we assumed that the binding ratio of VEGF165 to the aptamer is 1:1 (one aptamer monomer binds to one VEGF165 monomer, and one aptamer dimer binds to one VEGF165 homodimer).
2.3. Thrombin activity inhibition assay using linked aptamers
In a buffer solution consisting of 50 mM Tris-HCl, 100 mM NaCl and 5 mM KCl at pH 8.0, linked aptamers and thrombin (final concentration, 54 nM) were mixed for 5 min at 37 °C. Next, fibrinogen (final concentration, 2 mg/ml) was added, and the clotting time was measured using an automated fibrometer (Amelung KC-4A Micro Coagulation Analyzer) at 37 °C.
3. Results and Discussion
3.1. The linked thrombin-binding aptamer
3.1.1. Design of the aptamer dimer
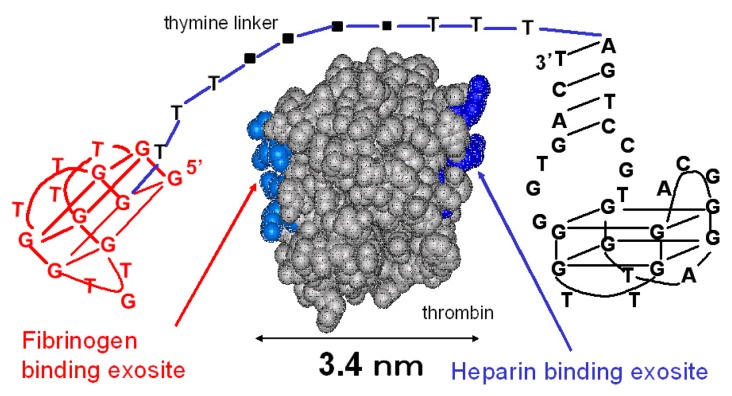
The sequences of the designed thrombin-binding aptamers are shown in Table 1. The 15-mer thrombin-binding aptamer having a G-quartet structure recognizes the fibrinogen-binding exosite of thrombin via a T-loop and inhibits the thrombin activity (Fig. 1) [14]. The 29-mer thrombin-binding aptamer recognizes the heparin-binding site of thrombin and has no thrombin-inhibitory activity (Fig. 1) [15]. We connected the 15-mer and the 29-mer aptamers using poly(dT) linkers of various lengths. The distance between the fibrinogen-binding exosite and the heparin-binding site is about 3.4 nm, and the total length of five sequential thymine bases is about 5 nm. Considering this, we varied the number of dTs inserted between the 15-mer and the 29-mer (0, 5, 10, 15 or 20 dTs).
Table 1.
The sequences of aptamers and aptamer dimmers.
| aptamers | Sequences (5'-3') |
|---|---|
| Thrombin-binding aptamer | |
| 15-mer | GGTTGGTGTGGTTGG |
| 29-mei | AGTCCGTGGTAGGGCAGGTTGGGGTGACT |
| Linker 0 | GGTTGGTGTGGTTGG--------------------AGTCCGTGGTAGGGCAGGTTGGGGTGACT |
| Linker 5 | GGTTGGTGTGGTTGG-------TTTTT--------AGTCCGTGGTAGGGCAGGTTGGGGTGACT |
| Linker 10 | GGTTGGTGTGGTTGG-----TTTTTTTTTT-----AGTCCGTGGTAGGGCAGGTTGGGGTGACT |
| Linker 20 | GGTTGGTGTGGTTGGTTTTTTTTTTTTTTTTTTTTAGTCCGTGGTAGGGCAGGTTGGGGTGACT |
| VEGF-binding aptamer | |
| VEa5 | ATACCAGTCTATTCAATTGGGCCCGTCCGTATGGTGGGTGTGCTGGCCAGATAGTATGTGCAATCA |
| VEa5-VEa5 | [VEa5]--------------------[VEa5] |
| VEa5-T10-VEa5 | [VEa5]----- TTTTTTTTTT-----[VEa5] |
| VEa5-T20-VEa5 | [VEa5] TTTTTTTTTTTTTTTTTTTT[VEa5] |
| del5-1 | ATACCAGTCTATTCAATTGGGCCCGTCCGTATGGTGGGTGTGCTGGCCAG |
| del5-1-del5-1 | [del5-1]--------------------[del5-1] |
| del5-1-T10-del5-1 | [del5-1]----- TTTTTTTTTT-----[del5-1] |
| del5-1-T20-del5-1 | [del5-1] TTTTTTTTTTTTTTTTTTTT[del5-1] |
Figure 1.
The linked thrombin-binding aptamer.
3.1.2. Binding assay using SPR measurement
We investigated the affinities of the designed aptamers by surface plasmon resonance (SPR) measurement on a Biacore X instrument. For this purpose, biotinylated aptamers were immobilized on the SA chip modified with streptavidin. The injection of the biotinylated aptamers onto the SA chip caused the RU value to increase (data not shown). Increased RU values were also obtained upon injection of various concentrations of thrombin (Fig. 2). The Kd value of the designed aptamers were calculated using the BIA evaluation software, version 3.1. As a result, the Kd value of the 5-linker aptamer became subnanomolar, i.e. 1/10 of that of the 29-mer (Table 2). This was caused mainly by the much smaller koff value. This finding indicates that the concatenated aptamer might recognize two different sites on the thrombin molecule owing to the flexible dT linkers. Thus, by connecting two thrombin-binding aptamers that recognize different parts of the thrombin molecule, avidity is obtained. The 5-linker aptamer had the highest affinity for thrombin among the concatenated aptamers. The linker we used is a single-strand DNA with a length of approximately 5 nm. It is expected that the flexibility and length of the 5-linker aptamer will enable it to bind to two different recognition sites of thrombin simultaneously.
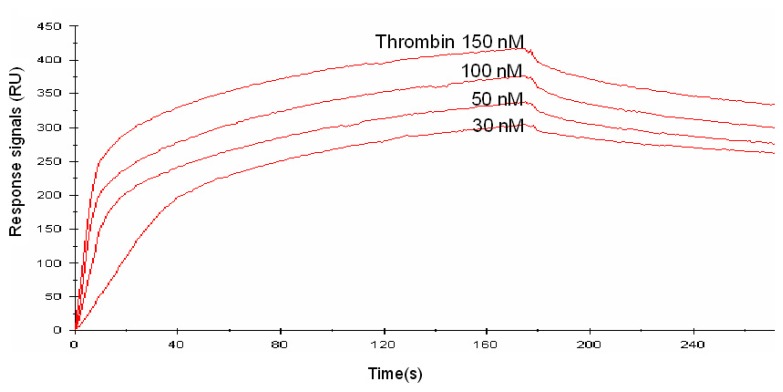
Figure 2.
A sensorgram of the linked aptamer (10-linker), as determined by SPR. Thrombin (30, 50, 100, 150 nM) was injected onto the linked aptamer-immobilized SA chip under running buffer at a flow rate of 20 μl/min.
Table 2.
The binding parameters of thrombin-binding aptamers investigated by BIA evaluation 3.1.
| kon1 (1/M-s) | kon2 (1/M-s) | koff1 (1/s) | koff2 (1/s) | Kd1 (nM) | Kd2 (nM) | |
|---|---|---|---|---|---|---|
| 15-mer | 1.89 × 105 | 3.83× 10-3 | 20.2 | |||
| 29-mer | 4.40 × 10s | 1.54 × 10-3 | 3.5 | |||
| Linker 0 | 5.96 × 104 | 5.80 × 104 | 8.57 × 10-6 | 8.62 × 10-6 | 0.14 | 0.15 |
| Linker 5 | 4.89 × 105 | 6.11 × 104 | 2.91 × 10-6 | 1.54 × 10-5 | 0.06 | 0.25 |
| Linker 10 | 5.51 × 104 | 3.69 × 105 | 4.09 × 10-5 | 1.91 × 10-4 | 0.74 | 0.52 |
| Linker 20 | 4.91 × 104 | 5.02 × 104 | 1.74 × 10-5 | 1.76 × 10-5 | 0.35 | 0.35 |
3.1.3. The inhibitory activity of the linked aptamers
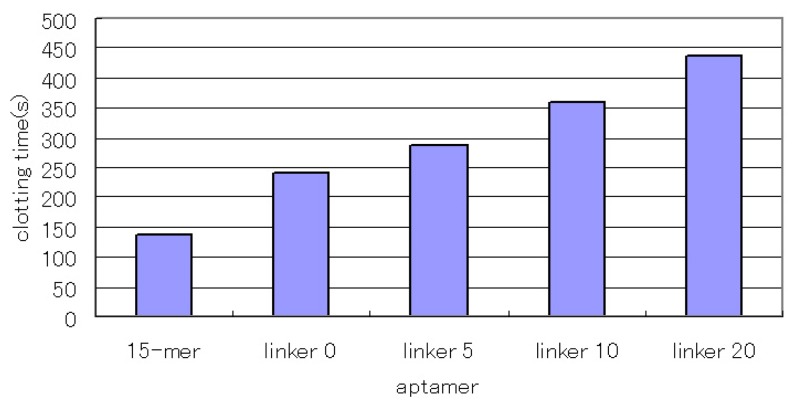
We investigated the inhibitory activity of the linked aptamers. The 15-mer aptamer recognizes the fibrinogen-binding site of thrombin and exerts an anticoagulant effect by inhibiting the reaction of thrombin and fibrinogen. We investigated the inhibitory activity of the concatenated aptamers by comparing them with the 15-mer aptamer. As a result, the concatenated aptamers showed increased inhibitory activity (Fig. 3), and the aptamers with the longer linkers showed higher inhibitory activity. This result does not seem to be consistent with the affinity values. However, all the concatenated aptamers showed subnanomolar Kd values, and their affinities were not significantly different. In this case, excess amounts of concatenated aptamers for thrombin were used, and most of the thrombin seemed to bind to the aptamers. In that situation, larger concatenated aptamers might hinder the approach of fibrinogen to a greater degree, which might result in higher inhibitory activity as the linkers become longer.
Figure 3.
The inhibitory activity of the linked aptamer for thrombin. We compared it with the inhibitory activity of the 15-mer thrombin-inhibiting aptamer. After the aptamer (1 μM) and thrombin (54 nM) were incubated for 5 min at 37 °C, we added fibrinogen (2 mg/ml) and measured the clotting time.
Muller et al. have reported a fusion aptamer consisting of two distinct aptamers that target thrombin [16]. But they aimed to improve not the affinity but the inhibitory activity, and the observed affinities were increased only two- to three-fold compared to those of the individual precursor aptamers. On the other hand, the Kd values of the linked aptamers designed by us in the present study were 1/10 of those of the monomer aptamers. The reason for this difference resides in the linkers used to connect the aptamers. Muller's group used poly(dA) as the linker, but we used poly(dT). The pyrimidine nucleosides are smaller than the purine nucleotides, so dT linkers are more flexible than dA linkers and the aptamers connected with dT linkers fit both binding sites of thrombin.
The approach of connecting two aptamers with different epitopes is very simple and easily improves aptamer affinity. However, to obtain such aptamers, we needed to construct two aptamers which recognize distinct regions of the target protein. So, we designed our aptamer dimer using only the aptamer against the homodimeric protein VEGF165.
3.2. The linked VEGF-binding aptamer
3.2.1. Design of the aptamer dimer
We had isolated the DNA aptamer against vascular endothelial growth factor (VEGF165), named “VEa5,” and investigated the properties of this aptamer in a previous study [11]. Since VEGF165 is a homodimeric protein, we attempted the dimerization of VEa5 and that of the deletion mutant of VEa5, del5-1, to improve their affinities for the target (Table 1). VEGF165 contains two domains, a receptor-binding domain and a heparin-binding domain [17], and VEa5 recognizes the heparin-binding domain [11]. Although the 3D structures of each domain of VEGF165 have already been determined [17, 18], the structure of the full length of the VEGF165 has not been reported. Therefore, the distance between the heparin-binding domains of each monomer in homodimer is still unclear. So, we connected the aptamers together via 0-, 10- or 20-mer dT linkers to determine the suitable length between the linked aptamers.
3.2.2. Binding assay using SPR measurement
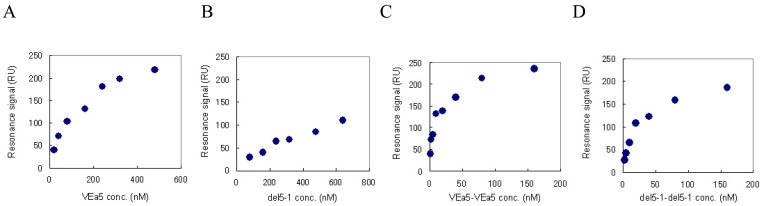
We have already confirmed by SPR measurement that VEa5 binds VEGF165 with a Kd value of 120 nM (Fig. 4A) [11]. The affinities of del5-1 and the aptamer dimers against VEGF165 were also evaluated by SPR measurement. Aptamer solutions of different concentrations were assayed on VEGF165-immobilized sensor chips and the corresponding signals were obtained. Figure 4 shows the signal at equilibrium plotted as a function of the aptamer concentration. The Scatchard plot analysis showed that del5-1 had a Kd value of 500 nM (Table 3). In both VEa5 and del5-1, the constants of dissociation with VEGF165 of the aptamer dimers without linkers were less than 1/10 of those of the monomer aptamers (Table 3). Meanwhile, when dimer aptamers with dT linkers were injected, the equilibrium response values were not even partially dependent on the aptamer concentrations, and the Scatchard plots were not linear (data not shown). This might be caused by the low stabilities of aptamer dimers with dT linkers.
Figure 4.
The equilibrium responses are plotted as a function of the aptamer concentration. VEGF165 was immobilized on a sensor chip and aptamers were injected for the SPR measurement. (A) VEa5, (B) del5-1, (C) VEa5-VEa5, (D) del5-1-del5-1 were assayed.
Table 3.
The dissociation constants of VEGF-binding aptamers calculated by scachard plots.
| Aptamers | kd (nM) |
|---|---|
| VEa5 | 116.28 |
| VEa5-VEa5 | 6.24 |
| del5-1 | 476.19 |
| del5-1-del5-1 | 17.15 |
From these results, the dimerization of aptamers which bind to a homodimeric protein would be effective for improving their affinities. Furthermore, VEGF is a marker protein of some diseases [12, 13], so aptamer dimers having higher affinities would be good sensor elements for highly sensitive VEGF detection in disease diagnosis.
4. Conclusion
We connected two thrombin-binding aptamers which recognize different sites of the thrombin molecule via flexible dT linkers, and the concatenated aptamer showed a higher affinity than the individual monomers (15-mer and 29-mer). Also, the concatenated aptamer showed a higher thrombin-inhibitory activity. Furthermore, the dimerization of two identical aptamers against the homodimeric protein VEGF165 notably improved their affinities. We demonstrated here a simple approach to the improvement of aptamer affinity.
Acknowledgments
This work was supported by the 2006 Industrial Technology Research Grant Program of the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Contributor Information
Koji Sode, Email: sode@cc.tuat.ac.jp.
Kazunori Ikebukuro, Email: ikebu@cc.tuat.ac.jp.
References and Notes
- 1.Jayasena S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 2.Ellington A.D., Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 4.Noma T., Ikebukuro K. Aptamer selection based on inhibitory activity using an evolution-mimicking algorithm. Biochem. Biophys. Res. Commun. 2006;347:226–231. doi: 10.1016/j.bbrc.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 5.Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S.M., Ostman A., Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida W., Sode K., Ikebukuro K. Homogeneous DNA sensing using enzyme-inhibiting DNA aptamers. Biochem. Biophys. Res. Commun. 2006;348:245–252. doi: 10.1016/j.bbrc.2006.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida W., Sode K., Ikebukuro K. Aptameric enzyme subunit for biosensing based on enzymatic activity measurement. Anal. Chem. 2006;78:3296–3303. doi: 10.1021/ac060254o. [DOI] [PubMed] [Google Scholar]
- 8.Adams G.P., Schier R. Generating improved single-chain Fv molecules for tumor targeting. J. Immunol. Methods. 1999;231:249–260. doi: 10.1016/s0022-1759(99)00161-1. [DOI] [PubMed] [Google Scholar]
- 9.Viti F., Tarli L., Giovannoni L., Zardi L., Neri D. Increased binding affinity and valence of recombinant antibody fragments lead to improved targeting of tumoral angiogenesis. Cancer Res. 1999;59:347–352. [PubMed] [Google Scholar]
- 10.Neri D., Momo M., Prospero T., Winter G. High-affinity antigen binding by chelating recombinant antibodies (CRAbs) J. Mol. Biol. 1995;246:367–373. doi: 10.1006/jmbi.1994.0091. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa H., Sode K., Ikebukuro K. Selection of DNA aptamers against VEGF(165) using a protein competitor and the aptamer blotting method. Biotechnol. Lett. 2008 doi: 10.1007/s10529-007-9629-6. [DOI] [PubMed] [Google Scholar]
- 12.Nakahara H., Song J., Sugimoto M., Hagihara K., Kishimoto T., Yoshizaki K., Nishimoto N. Anti-interleukin-6 receptor antibody therapy reduces vascular endothelial growth factor production in rheumatoid arthritis. Arthritis Rheum. 2003;48:1521–1529. doi: 10.1002/art.11143. [DOI] [PubMed] [Google Scholar]
- 13.Voorzanger-Rousselot N., Garnero P. Biochemical markers in oncology. Part I: molecular basis. Part II: clinical uses. Cancer Treat. Rev. 2007;33:230–283. doi: 10.1016/j.ctrv.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Bock L.C., Griffin L. C., Latham J. A., Vermaas E. H., Toole J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 15.Tasset D.M., Kubik M. F., Steiner W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J Mol Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 16.Muller J., Wulffen B., Potzsch B., Mayer G. Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer. Chembiochem. 2007;8:2223–2226. doi: 10.1002/cbic.200700535. [DOI] [PubMed] [Google Scholar]
- 17.Stauffer M.E., Skelton N. J., Fairbrothe W. J. Refinement of the solution structure of the heparin-binding domain of vascular endothelial growth factor using residual dipolar couplings. J. Biomol. NMR. 2002;23:57–61. doi: 10.1023/a:1015346504499. [DOI] [PubMed] [Google Scholar]
- 18.Muller Y. A., Li B., Christinger H. W., Wells J. A., Cunningham B. C., de Vos A. M. Vascular endothelial growth factor: crystal structure and functional mapping of the kinase domain receptor binding site. Proc. Natl. Acad. Sci. USA. 1997;94:7192–7197. doi: 10.1073/pnas.94.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]