Abstract
The Staphylococcus aureus chromosome harbors two homologues of the YefM-YoeB toxin-antitoxin (TA) system. The toxins YoeBSa1 and YoeBSa2 possess ribosome-dependent ribonuclease (RNase) activity in Escherichia coli. This activity is similar to that of the E. coli toxin YoeBEc, an enzyme that, in addition to ribosome-dependent RNase activity, possesses ribosome-independent RNase activity in vitro. To investigate whether YoeBSa1 is also a ribosome-independent RNase, YoeBSa1 was expressed using a novel strategy, and its in vitro RNase activity, sequence specificity, and kinetics were characterized. Y88 of YoeBSa1 was critical for in vitro activity and cell culture toxicity. This residue was mutated to ortho-nitrobenzyl tyrosine (ONBY) via unnatural amino acid mutagenesis. YoeBSa1-Y88ONBY could be expressed in the absence of the antitoxin YefMSa1 in E. coli. Photocaged YoeBSa1-Y88ONBY displayed UV light-dependent RNase activity toward free mRNA in vitro. The in vitro ribosome-independent RNase activity of YoeBSa1-Y88ONBY, YoeBSa1-Y88F, and YoeBSa1-Y88TAG was significantly reduced or abolished. In contrast to YoeBEc, which cleaves RNA at both adenosine and guanosine with a preference for adenosine, YoeBSa1 cleaved mRNA specifically at guanosine. Using this information, a fluorometric assay was developed and used to determine the kinetic parameters for ribosome-independent RNA cleavage by YoeBSa1.
Toxin-antitoxin (TA) systems are genetic modules that are found in almost all free-living prokaryotes.1 TA loci encode a protein toxin and a protein or RNA antitoxin and are grouped into five types based on the mechanism by which the antitoxin counteracts the activity of the toxin.2–4 In Type II TA systems, the antitoxin is a protein that binds to the toxin and prevents it from interacting with its cellular target. The activity of Type II toxins is regulated by the differential susceptibility of the toxin and the antitoxin to proteolysis. The antitoxin is more labile than the toxin and must be continuously expressed to maintain cellular levels capable of inhibiting the toxin. Toxin inhibition is relieved by the activity of cellular proteases, which degrade the labile antitoxin and shift the antitoxin-to-toxin ratio to favor the free toxin, which can then act on its cellular target.5 The targets of TA system toxins include DNA replication, protein translation, and cell wall biosynthesis. Many toxins are ribonucleases (RNases) that inhibit translation by cleaving RNA in a ribosome-dependent or -independent fashion.2
TA systems were originally discovered on plasmids, where they function as a post-segregational killing mechanism to maintain a plasmid in a bacterial population. If a daughter cell inherits a plasmid containing a TA locus during replication, both the antitoxin and the toxin will be expressed to form a stable, innocuous complex, and the cell will survive. However, if the plasmid is not inherited, rapid proteolytic degradation of the antitoxin will release the toxin to act on its cellular target and kill the cell, thereby eliminating plasmid-free cells from the population.6 TA systems were subsequently discovered on the chromosomes of many bacteria and archaea.1, 7 The function of chromosomally-encoded TA systems is somewhat controversial, with at least thirteen proposed roles, including junk DNA, selfish genes, stabilization of mobile genetic elements, anti-addiction elements, gene regulation, growth control/stress response, persistence, growth arrest, programmed cell death, phage defense, biofilm formation, virulence, and phenotypic bistability.8, 9 However, it is generally accepted that cellular stress modulates transcription at the TA locus and stimulates degradation of the antitoxin, releasing the toxin to act on its cellular target and arrest growth until conditions become more favorable.5 Upon cessation of stress, the antitoxin is replenished, inactivating the toxin and allowing the cell to resume normal growth.10 The growth arrest and eventual cell death resulting from toxin overexpression have led to the proposal that artificial toxin activation could provide an effective antibacterial strategy.11–14
The Gram-positive pathogen Staphylococcus aureus is the leading cause of bloodstream, lower respiratory tract, and skin and soft tissue infections worldwide.15 A recent comparative genomic analysis identified between one and seven Type II TA loci in the sequenced genomes of fourteen S. aureus strains, including YefM-YoeBSa1 and YefM-YoeBSa2 (previously identified as Axe1-Txe1 and Axe2-Txe2)16, 17 which are homologues of the YefM-YoeBEc TA system from Escherichia coli.18 The toxin YoeBEc possesses ribosome-independent RNase activity in vitro as well as ribosome-dependent RNase activity in vitro and in cells.19, 20 YoeBEc binds in the A site of the 50S ribosomal subunit, causing cleavage of mRNA transcripts three bases downstream of the start codon.20 It is unknown whether YoeBEc-mediated mRNA cleavage on the ribosome results from direct catalysis by YoeBEc, from enhancement of the latent ribonucleolytic activity of the ribosome upon binding of YoeBEc, or from some combination of the two.20
A significant challenge when studying toxic proteins is the difficulty associated with their expression and purification in E. coli. Herein a novel solution to this problem was devised by incorporating a non-canonical amino acid, a photocaged tyrosine derivative, to replace a tyrosine that is critical for and toxicity of YoeBSa1. Using protein produced through this method, the substrate the activity specificity of YoeBSa1 was unveiled, enabling the creation of a fluorogenic substrate that can be used to monitor YoeBSa1 activity.
EXPERIMENTAL PROCEDURES
Bacterial Strains
MRSA clinical isolates were from a previously published collection.21 E. coli DH5α and NiCo21(DE3) were used for cloning and protein expression, respectively.
Primers
All primers used for PCR, RT-PCR, cloning, and site-directed mutagenesis were synthesized by Integrated DNA Technologies (IDT) and are listed in Supplementary Table S1.
PCR Analysis of Clinical Isolates
Total DNA was previously purified from a diverse collection of 78 clinical isolates of MRSA.21 Primers YefM-YoeBSa1-F and YefM-YoeBSa1-R or YefM-YoeBSa2-F and YefM-YoeBSa2-R were used to amplify the yefM-yoeBSa1 or yefM-yoeBSa2 loci, respectively, from the total DNA. Primers YefMSa1-NdeI-F, YefMSa1-XhoI-R, YoeBSa1-NdeI-F, YoeBSa1-HindIII-R, YefMSa2-NdeI-F, YefMSa2-HindIII-R, YoeBSa2-NdeI-F, and YoeBSa2-HindIII-R were used to amplify the yefMSa1, yoeBSa1, yefM-yoeBSa1, yefMSa2, yoeBSa2, and yefM-yoeBSa2 operons from the total DNA of strains NRS27 and NRS76. PCR amplification was performed on a DNA thermocycler using previously described reaction conditions.21, 22 PCR products were separated by electrophoresis on 1% agarose gels and stained with ethidium bromide.
Sequencing Analysis
Approximately 10% of the PCR products generated from PCR amplification of the yefM-yoeBSa1 and yefM-yoeBSa2 loci were submitted for DNA sequencing by the University of Illinois W. M. Keck Center for Comparative and Functional Genomics. Sequence data were analyzed using BioEdit 7.0.5.3 software. The sequences were aligned using CLUSTAL W23 and used as query sequences to search the BLAST database to verify the identity of the PCR products and sequence homology to known genes.24
Reverse Transcription (RT)-PCR Analysis of Clinical Isolates
MRSA isolates were streaked from glycerol stocks on Brain Heart Infusion (BHI) agar. Single colonies from freshly streaked plates were inoculated into 10 ml BHI medium and incubated aerobically at 37°C with shaking at 250 rpm overnight (14–16 h). Overnight cultures were diluted 1:100 in 10 ml BHI medium and incubated aerobically at 37°C with shaking at 250 rpm until the A600 reached 0.6 to 1.0. Logarithmically growing cultures were harvested by centrifugation at 3220 × g for 10 min at 4°C. Total RNA was purified using the FastRNA Pro Blue Kit (Qbiogene) according to the manufacturer’s instructions with the modification that lysis was performed by vortexing resuspended cells at maximum speed for 5 × 1 min pulses at room temperature with 1–5 min recovery on ice between pulses. Isolated RNA was treated with DNase I and purified with reagents from the Total RNA Kit I (Omega Bio-Tek) according to the RNA Cleanup protocol from the RNeasy Mini Handbook (Qiagen). Purified total RNA (10 ng) was used in RT-PCR and in PCR with Platinum Taq DNA Polymerase (Invitrogen) to detect DNA contamination. RT-PCR was performed with primers YefM-YoeBSa1-F and YefM-YoeBSa1-R or YefM-YoeBSa2-F and YefM-YoeBSa2-R using the Superscript III One-Step RT-PCR System with Platinum Taq (Invitrogen) as previously reported22 with the following modifications: the annealing temperature was raised to 55°C, and the number of cycles was reduced to 35. PCR products were separated by electrophoresis on 1% agarose gels and stained with ethidium bromide.
Construction of Plasmids
The yoeBSa1 ORF was PCR-amplified from the total DNA of MRSA S421 with primers YoeBSa1-NdeI-F and YoeBSa1-Y88F-HindIII-R or YoeBSa1-Y88TAG-HindIII-R and cloned into the corresponding sites of pET-28a (Novagen) to create pET-28a-yoeBSa1-Y88F and pET-28a-yoeBSa1-Y88TAG, respectively. The yefM-yoeBSa1 ORF was PCR-amplified from the total DNA of MRSA S421 with primers YefMSa1-NdeI-F and YoeBSa1-HindIII-R and cloned into the corresponding sites of pET-28a. Because a single non-silent point mutation was found in the sequence for YefMSa1, Quikchange site-directed mutagenesis was performed with primers YefMSa1-QC-F and YefMSa1-QC-R to create pET-28a-yefM-yoeBSa1. Site-directed mutagenesis was carried out with the Quikchange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions with the modification that E. coli DH5α was used as the host strain. The yefMSa1 ORF was PCR-amplified from the total DNA of MRSA NRS3 (Network on Antimicrobial Resistance in S. aureus) with primers YefMSa1-NdeI-F and YefMSa1-XhoI-R and cloned into the corresponding sites of pET-28a to create pET-28a-yefMSa1. All clones were confirmed by sequencing. Antibiotics were used at concentrations of 50 μg/ml kanamycin (pET-28a), 35 μg/ml chloramphenicol (pEVOL-ONBY), and 100 μg/ml ampicillin (Pentaprobes).
ONBY Synthesis
o-Nitrobenzyl tyrosine (ONBY) was synthesized according to a previously published method25 with some modifications. 2.0 g (11 mmol) L-tyrosine was stirred with 1.9 g (7.6 mmol) CuSO4·5H2O in 20 ml 1 M NaOH at 60°C for 20 min. The mixture was cooled to room temperature, quenched with hydrochloric acid, filtered, and washed with water. The resulting solid was stirred with 1.5 g (11 mmol) K2CO3 and 1.8 g (8.3 mmol) o-nitrobenzyl bromide in 60 ml 75% aqueous DMF at room temperature in the dark for three days. The resulting solid was filtered; washed with 75% aqueous DMF, water, 75% aqueous acetone, and ice-cold acetone; stirred in 100 ml 1 M HCl for 2 h; filtered; stirred in 100 ml 1 M HCl for 1 h; filtered; and washed with water and acetone. Yield: 1.8 g (5.7 mmol, 69%).
Purification of YoeBSa1-Y88F and YoeBSa1-Y88TAG
pET-28a-yoeBSa1-Y88F or pET-28a-yoeBSa1-Y88TAG was introduced into E. coli NiCo21(DE3). Overnight cultures were grown in LB/kanamycin with single colonies from freshly streaked plates, diluted 1:100 in LB/kanamycin, and grown at 37°C. When the A600 of the culture reached 0.4 to 0.6, protein expression was induced with 1 mM IPTG at 37°C for 4 h. Cultures were harvested by centrifugation at 6000 × g for 5 min at 4°C. Cell pellets were frozen at −20°C, thawed on ice for 30 min, and resuspended in 10 ml cold lysis buffer (20 mM Tris, 500 mM NaCl, 60 mM imidazole, pH 7.9). Cells were lysed by sonication on ice at 40% amplitude for 5 min with a 1 s pulse. The lysate was cleared by centrifugation at 35,000 × g for 30 min at 4°C. The supernatant was batch-loaded onto 1 ml 1:1 Ni-NTA agarose (Qiagen) at 4°C for 30 min with inversion. The resin was washed with 20 ml cold lysis buffer and eluted with 10 ml cold elution buffer A (50 mM Tris, 500 mM NaCl, 500 mM imidazole, 1 mM TCEP, 10% glycerol, pH 7.9). The eluted fraction was concentrated to ~0.5 ml using an Ultra-15 Centrifugal Filter Unit with Ultracel-3 membrane (Amicon), 0.2 μm filtered, and further purified on a HiLoad 16/60 Superdex 75 PG column (GE Healthcare) using FPLC buffer A (50 mM Tris, 200 mM NaCl, 1 mM TCEP, 10% glycerol, pH 7.9). Fractions containing pure protein were pooled and concentrated to ~0.5–1 ml. Purity was assessed by SDS-PAGE using 4–20% TGX Mini-PROTEAN gels (Bio-Rad). Concentration was determined by densitometry and by BCA assay (Pierce) performed according to the manufacturer’s instructions with the modification that lysozyme standards were used instead of bovine serum albumin (BSA) standards.
Purification of YoeBSa1-Y88ONBY
pET-28a-yoeBSa1-Y88TAG and pEVOL-ONBY were introduced into E. coli NiCo21(DE3). Overnight cultures were grown in LB/kanamycin/chloramphenicol with single colonies from a freshly streaked plate and diluted 1:100 in LB/kanamycin/chloramphenicol. 100 mM ONBY dissolved in 1 M NaOH was added to a final concentration of 1 mM, and the culture was grown at 37°C in the dark. When the A600 of the culture reached 0.5 to 0.6, arabinose was added to a final concentration of 0.2% to induce expression of ONBY-aaRS. When the A600 of the culture reached 1.0 to 1.2, IPTG was added to a final concentration of 1 mM to induce expression of YoeBSa1-Y88ONBY. Expression was allowed to proceed at 37°C for 15 h. Cultures were harvested and YoeBSa1-Y88ONBY was purified in the dark as described above for YoeBSa1-Y88F and YoeBSa1-Y88TAG.
Purification of YefMSa1
pET-28a-yefMSa1 was introduced into E. coli NiCo21(DE3), and protein expression and purification were performed as described for YoeBSa1-Y88F and YoeBSa1-Y88TAG, with the modification that YefMSa1 was eluted with 10 ml cold elution buffer B (20 mM Tris, 500 mM NaCl, 500 mM imidazole, pH 7.9). Gel filtration was performed twice using FPLC buffer B (20 mM Tris, 500 mM NaCl, pH 7.9) in order to remove RNase contamination. Fractions corresponding to the second major peak from the first gel filtration step were pooled, concentrated, and subjected to a second gel filtration step. Fractions corresponding to the second major peak from the second gel filtration step were pooled, concentrated, and used in experiments. Purity was assessed by SDS-PAGE using 4–20% TGX Mini-PROTEAN gels (Bio-Rad). Concentration was determined by BCA assay (Pierce) performed according to the manufacturer’s instructions using BSA standards.
ESI-MS
5 μg YefMSa1, YoeBSa1-Y88F, YoeBSa1-Y88TAG, or YoeBSa1-Y88ONBY was precipitated according to a previously published method.26 YoeBSa1-Y88ONBY was exposed to 312 nm UV light on a UV transilluminator for 0–600 s prior to precipitation. Precipitated, air-dried protein was analyzed by electrospray ionization mass spectrometry (ESI-MS) at the University of Illinois Mass Spectrometry Laboratory.
Agarose Gel RNase Activity Assay
pET-28a-yefM-yoeBSa1 was digested with XhoI (NEB) at 37°C for 3 h. The fully linearized plasmid was extracted once with 24.5:24.5:1 phenol:chloroform:isoamyl alcohol and twice with chloroform, precipitated with 3 M NaOAc, pH 5.3, and 100% EtOH, and resuspended in nuclease-free water. 1 μg linearized plasmid was used as template for standard RNA synthesis with the T7 High Yield RNA Synthesis Kit (NEB) according to the manufacturer’s instructions. RNA was purified with reagents from the Total RNA Kit I (Omega Bio-Tek) according to the RNA Cleanup protocol from the RNeasy Mini Handbook (Qiagen). RNase assays were performed according to a modified published method.19 For light-dependence of decaging, 10 pmol YoeBSa1-Y88ONBY in 50 mM Tris, 5% glycerol, pH 8.0, was exposed to UV light for 0–5 min. 320 ng yefM-yoeBSa1 RNA was then added with Human Placental RNase Inhibitor (NEB) at a final concentration of 1 unit/μl. Reactions were incubated at 37°C for 2 h and stopped by adding 1 μg proteinase K (Invitrogen) and incubating at 37°C for 15 min. 11 μl RNA loading dye I (95% formamide, 18 mM EDTA, 0.025% SDS, 0.025% bromophenol blue) was then added, and samples were incubated at 95°C for 5 min immediately prior to loading on 1.2% agarose, 0.5X TBE (45 mM Tris-borate, 1 mM EDTA, pH 8.3), 0.1 μg/ml EtBr gels for analysis. For inhibition of YoeBSa1 by YefMSa1, YoeBSa1 (10, 20, or 30 pmol) in 50 mM Tris, 5% glycerol, pH 8.0, was exposed to UV light for 3 min. YefMSa1 (0, 10, 20, or 30 pmol) was then added. Following incubation at 37°C for 30 min to allow complex formation, 320 ng yefM-yoeBSa1 RNA was added with Human Placental RNase Inhibitor at a final concentration of 1 unit/μl. Reactions were incubated at 37°C for 1 h, stopped, and analyzed as described above. For the time course of YoeBSa1 and YefMSa1 RNase activity, 10 pmol YoeBSa1-Y88ONBY, YoeBSa1-Y88F, or YoeBSa1-Y88TAG, or YefMSa1 was incubated with 320 ng yefM-yoeBSa1 RNA in the presence of 1 unit/μl Human Placental RNase inhibitor in 50 mM Tris, 5% glycerol, pH 8.0. One set of reactions containing YoeBSa1-Y88ONBY were exposed to UV light for 3 min prior to adding RNA. Reactions were incubated at 37°C for 1–20 h, stopped, and analyzed as described above.
Polyacrylamide Gel RNase Activity Assay
Quikchange site-directed mutagenesis was performed with primers PP1QC1-F and PP1QC1-R to insert a G into Pentaprobe 1 and with primers PP1QC2-F and PP1QC2-R, PP2QC-F and PP2QC-R, PP4QC-F and PP4QC-R, PP6QC-F and PP6QC-R, PP7QC-F and PP7QC-R, PP8QC-F and PP8QC-R, PP9QC-F and PP9QC-R, PP10QC-F and PP10QC-R, and PP12QC-F and PP12QC-R (Supplementary Table S1) to introduce XbaI sites into Pentaprobes 1, 2, 4, 6, 7, 8, 9, 10, and 12, respectively. Site-directed mutagenesis was carried out with the Quikchange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions, with the modification that E. coli DH5α was used as the host strain. Pentaprobe plasmids were digested with XbaI (NEB) at 37°C for 3 h. Fully linearized plasmids were purified by phenol-chloroform extraction as described above. RNA was synthesized and purified as described above. RNase assays were performed according to a modified published method.27 1 μg YoeBSa1-Y88ONBY was incubated with 1 μg Pentaprobe RNA in 50 mM Tris, pH 8.0, for 0.5–2 h at 37°C. Reactions were quenched by adding 1 μg proteinase K and incubating at 37°C for 15 min. An equal volume of RNA loading dye II (95% formamide, 5 mM EDTA, 0.025% bromophenol blue) was then added, reactions were incubated at 95°C for 5 min to denature the RNA, and the products were analyzed by electrophoresis on 8% polyacrylamide TBE-urea gels (89 mM Tris-borate, 2 mM EDTA, 8 M urea, pH 8.3) and post-stained with EtBr.
MALDI RNase Activity Assay
PP7-1, PP7-2, and PP7-3 were synthesized by Genscript. 10 μg YoeBSa1-Y88ONBY was incubated with 2 μg oligonucleotide for 1 or 2 h at 37°C. Reactions were quenched by adding 10 μg proteinase K (Invitrogen) and incubating at 37°C for 30 min followed by precipitation with 5 M NH4OAc (final concentration 2 M) and 3 volumes 100% EtOH at −80°C. Pellets were washed with ice-cold 70% EtOH, resuspended in 1 μl H2O, and submitted for matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) at the University of Illinois Mass Spectrometry Laboratory.
HPLC RNase Activity Assay
The fluorogenic chimeric oligonucleotide substrate 5′-6-FAM-AACrArArArArGrArArAAATT-IABkFQ-3′ (6-FAM, 6-carboxyfluorescein fluorophore; IABkFQ, Iowa Black fluorophore quencher) and the cleavage products 5-6-FAM-AACrArArArArG-3′ and 5′-rArArAAATT-IABkFQ-3′ were synthesized by IDT. YoeBSa1 (20 μM) was incubated with oligonucleotide (30 μM) in buffer at 25°C for 5 h. High-performance liquid chromatography (HPLC) was performed using an Alliance HPLC System (e2695 Separations Module, Waters) with detection at 260 nm (2489 UV/Visible Detector, Waters). A YMCbasic S5 column (4.6 × 150 mm, 5 μm, Waters) was used to separate the full-length oligonucleotide from the cleavage products with a linear gradient from 100 mM triethylammonium acetate (TEAA), pH 7.0, to 50 mM TEAA/50% acetonitrile, pH 7.0, over 25 min. Peak fractions were collected, concentrated to 5–10 μl, and submitted for MALDI-MS at the University of Illinois Mass Spectrometry Laboratory.
Fluorometric Assay
Wells of a black 384-well plate were filled with 15 μl 0.5–20 μM intact fluorogenic substrate or cleavage products diluted in assay buffer (200 mM sodium phosphate, pH 6.0, 5% glycerol) and allowed to equilibrate for 30 min at room temperature. YoeBSa1-Y88ONBY was diluted to 10 μM in assay buffer, exposed to UV light at 312 nm for 3 min, and added to wells containing intact substrate and cleavage products to a final concentration of 5 μM. Fluorescence was measured once every minute for 100 min using a Criterion Analyst AD (Molecular Devices) with 485 ± 15 nm excitation and 530 ± 15 nm emission filters and a 505 nm cutoff dichroic mirror. The fluorophore was excited with a 1000 W continuous xenon arc lamp with 10 reads per well. Three separately purified batches of YoeBSa1-Y88ONBY were each assayed in technical triplicate on two different days. Results are the average of the average rate for each batch of protein.
Construction of Calibration Plot
Fluorescence values for 1:1 molar mixtures of the cleavage products at 30 min after addition of YoeBSa1 were used to construct a calibration plot of pmol cleaved substrate versus relative fluorescence units (RFU). A separate calibration plot was constructed each time the fluorometric assay was performed.
Kinetic Analysis
In some of the progress curves, an increase in fluorescence was not observed until ~15 min after addition of YoeBSa1. However, the increase in measured fluorescence was linear over the majority of the remainder of the assay. Consequently, initial rates were calculated from the linear portion of the progress curves between 20 and 30 min. Fluorescence values were corrected by subtracting measured fluorescence at 20 min from subsequent time points. Corrected fluorescence values were converted to pmol cleaved substrate using the slope from the calibration plot, and linear regression was performed using Microsoft Excel to obtain initial velocities. Linear velocities were plotted versus substrate concentration, and the resulting data points were fit with the Hill equation using OriginPro 8.0 software.
RESULTS
Prevalence, Conservation, and Transcription of yefM-yoeBSa1 and yefM-yoeBSa2 in MRSA Clinical Isolates
The artificial activation of TA systems is of significant interest as an intriguing antibacterial strategy.11–14 YoeBSa1 and YoeBSa2 specifically present outstanding targets for this approach, as expression of YoeBSa1 and YoeBSa2 in the absence of their respective antitoxins YefMSa1 and YefMSa2 induces growth arrest in E. coli.28 In order for artificial activation of YoeBSa1 or YoeBSa2 to be an effective antibacterial strategy, YefM-YoeBSa1 and YefM-YoeBSa2 must be present and functional in clinical isolates of S. aureus. Thus the presence, conservation, and transcription of yefM-yoeBSa1 and yefM-yoeBSa2 were investigated in a collection of 78 clinical isolates of MRSA from 3 Illinois hospitals and the Network on Antimicrobial Resistance in S. aureus (NARSA). Multiple-locus variable number of tandem repeats analysis previously confirmed that these isolates were not clonal.21
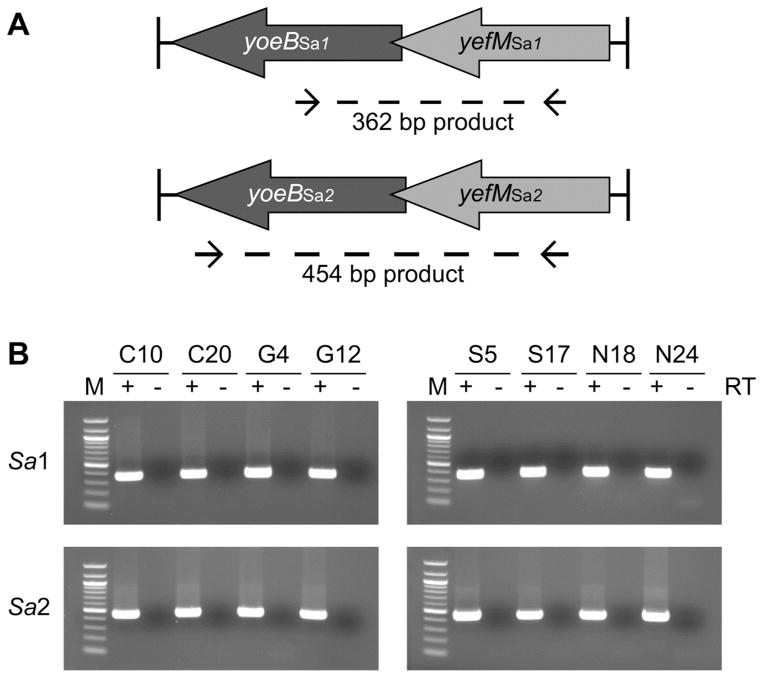
The total DNA of these isolates was probed for the presence of yefM-yoeBSa1 and yefM-yoeBSa2 using PCR with intragenic specific primers (Figure 1A). The yefM-yoeBSa1 and yefM-yoeBSa2 genes were present in 99% (77 out of 78) of the clinical isolates (Supplementary Table S2). Approximately 10% of the PCR products were subjected to DNA sequencing, and the sequences were aligned with the reference genome from the S. aureus COL strain using CLUSTAL W23 (Supplementary Figures S1 and S2) and compared with yefM-yoeBSa1 and yefM-yoeBSa2 loci from published S. aureus genomes using BLAST.24 The yefM-yoeBSa1 PCR products had greater than 97% identity with the yefM-yoeBSa1 genes in 29 of the 30 genomes containing that locus, while the yefM-yoeBSa2 PCR products had at least 96% identity with the yefM-yoeBSa2 genes in all 26 genomes containing that locus.
Figure 1. yefM-yoeBSa1 and yefM-yoeBSa2 are prevalent and transcribed in MRSA clinical isolates.
(A) Locations of homology for primers used in PCR and RT-PCR. Primer sequences were designed from the S. aureus COL genome. (B) RT-PCR analysis of yefM-yoeBSa1 (Sa1, upper two panels) and yefM-yoeBSa2 (Sa2, lower two panels) transcription in MRSA clinical isolates. Lane M: 100 bp DNA ladder (1517, 1200, 1000, 900, 800, 700, 600, 500, 400, 300, 200, and 100 bp, NEB). (+) and (−) denote the inclusion or exclusion, respectively, of reverse transcriptase. Clinical isolates are identified by strain number.
PCR products of ~1.3 kb and ~2.5 kb resulted from attempts to detect yefM-yoeBSa1 and yefM-yoeBSa2, respectively, in NRS27. DNA sequence analysis revealed that the product amplified by yefM-yoeBSa1-specific primers spanned a phage transcriptional repressor and DNA polymerase III alpha subunit, while the product amplified by yefM-yoeBSa2-specific primers spanned an ornithine cyclodeaminase, a siderophore biosynthesis protein, and a multidrug resistance efflux pump. Sites of partial complementarity were identified between the yefM-yoeBSa1- and yefM-yoeBSa2-specific primers and these genes, which could allow amplification in the absence of yefM-yoeBSa1 and yefM-yoeBSa2. To confirm the absence of yefM-yoeBSa1 and yefM-yoeBSa2 in NRS27, the yefMSa1, yoeBSa1, yefM-yoeBSa1, yefMSa2, yoeBSa2, and yefM-yoeBSa2 operons were individually amplified using primers designed to clone each of the full genes. PCR products were detected in the positive control strain NRS76, but not in NRS27. These results indicate that the yefM-yoeBSa1 and yefM-yoeBSa2 genes are not present in NRS27.
To investigate the transcription of yefM-yoeBSa1 and yefM-yoeBSa2 in MRSA, RT-PCR was performed on total RNA from eight isolates using the same primers used in the PCR screen. yefM-yoeBSa1 and yefM-yoeBSa2 were transcribed as bicistronic messages in each of the strains (Figure 1B). No PCR products were detected in the absence of reverse transcriptase, indicating the absence of DNA contamination. Taken together, these results suggest that yefM-yoeBSa1 and yefM-yoeBSa2 are widespread, conserved, and transcribed in clinical MRSA isolates. Thus an activator of YoeBSa1 or YoeBSa2 with antibacterial activity could have broad efficacy against S. aureus.
Photocaging Allows Expression of YoeBSa1 in E. coli
YoeBEc exhibits ribosome-dependent RNase activity in vitro and in E. coli.20 YoeBSa1 and YoeBSa2 were found to exhibit RNase activity similar to that of YoeBEc upon overexpression in E. coli, suggesting that they are ribosome-dependent RNases with the same mechanism of action as YoeBEc.28 Intriguingly, YoeBEc also possesses “residual” ribosome-independent RNase activity.19 Evaluation of the possibility that YoeBSa1 and YoeBSa2 might also possess such activity necessitates the expression and purification of full-length, functional YoeBSa1 and YoeBSa2. However, YoeBSa1 and YoeBSa2 inhibit translation initiation and induce growth arrest in E. coli,28 effectively preventing overexpression and characterization. As yefM-yoeBSa1 proved more amenable to manipulation using standard molecular biology techniques than yefM-yoeBSa2, a number of strategies were explored in efforts to obtain pure, functional YoeBSa1.
The most common method employed to obtain a functional TA system toxin for in vitro characterization involves co-expression of the antitoxin with the toxin, purification of the resulting TA complex, isolation of the toxin by denaturation of the complex, and subsequent refolding. Active YoeBEc has been obtained by dissociation from YefMEc in 6 M guanidine hydrochloride (GuHCl) followed by refolding via gradual dialysis into HEPES buffer.19 YoeBEc and YoeBSa1 share a high degree of sequence homology, suggesting that a similar method could enable purification of functional YoeBSa1. The yefM-yoeBSa1 operon was therefore cloned into pET-28a with a hexahistidine tag at the N-terminus of YefMSa1 to facilitate purification, and the complex was expressed in E. coli and purified using Ni-NTA resin. A variety of conditions were screened to identify an optimal method for denaturing, purifying, and refolding YoeBSa1. In each case, the YefM-YoeBSa1 complex was denatured by extended dialysis against either 6 M GuHCl or 8 M urea, YefMSa1 was extracted with Ni-NTA resin, and the presence and purity of the remaining YoeBSa1 was assessed using SDS-PAGE. If 6 M GuHCl was used as the denaturant, the recaptured YefMSa1 was highly pure, but YoeBSa1 was not recovered in the unbound fraction, probably due to the low initial ratio of YoeBSa1 to YefMSa1 and to the necessity of diluting and precipitating samples containing GuHCl for SDS-PAGE. In contrast, when 8 M urea was used as the denaturant, YoeBSa1 was recovered, but a significant amount of residual YefMSa1 was also present. Efforts to refold the impure toxin by dilution or gradual dialysis resulted in substantial precipitation. These unpromising results led to the abandonment of denaturing purification and refolding as a viable approach to obtain large quantities of pure, active YoeBSa1.
An alternative to denaturing purification of the toxin from the TA complex is to use a commercially available protease to selectively degrade the labile antitoxin from the TA complex, leaving the toxin unscathed.27 The YefM-YoeBSa1 complex was expressed and purified as described above and subjected to digestion with trypsin. Analysis of the digest products by SDS-PAGE suggested that YoeBSa1 remained intact while YefMSa1 was rapidly and selectively degraded. However, the YoeBSa1 protein obtained from trypsin digestion of YefM-YoeBSa1 did not bind to YefMSa1 in pulldown experiments. Native PAGE of the digestion timecourse suggested that a fragment of YefMSa1 remained bound to YoeBSa1, prohibiting complex formation and indicating that this approach could not be used to obtain pure YoeBSa1.
The yoeBSa1 gene was then cloned at the C-terminus of glutathione S-transferase (GST) in hopes that a larger fusion partner would reduce the toxicity of YoeBSa1 by preventing its ability to bind to the ribosome. However, all sequenced clones contained frameshift mutations introducing premature stop codons in the yoeBSa1 gene. Two rounds of site-directed mutagenesis were carried out to correct these mutations. Additional premature stop codons or inactivating mutations were introduced in each round, suggesting that low levels of full-length GST-YoeBSa1, produced by leaky expression from the T7 promoter even in the absence of T7 RNA polymerase, retained the ability to bind to the ribosome, inhibit translation, induce growth arrest, and prevent colony formation. Only mutant clones encoding premature stop codons were not toxic. These results hinted that the C-terminal region of YoeBSa1 is necessary for activity and toxicity. As the C-terminal residues H83 and Y84 of YoeBEc are required for both in vitro RNase activity and toxicity in E. coli,19 the inability to clone the full-length yoeBSa1 gene at the C-terminus of GST suggested that the analogous H87 and Y88 in YoeBSa1 have similar roles.
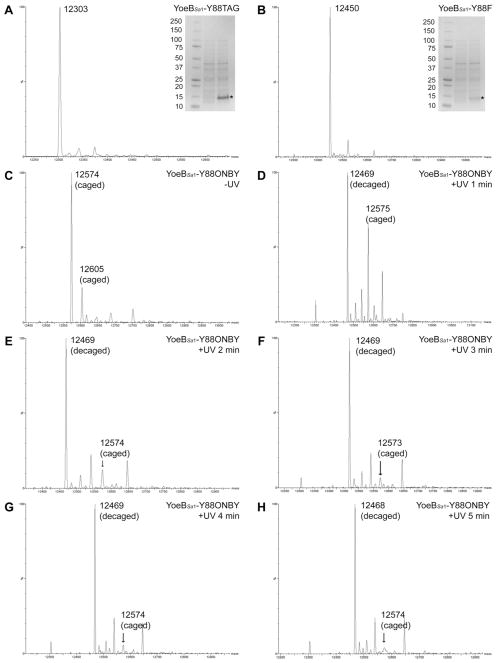
Mutation of the C-terminal Y84 of YoeBEc to phenylalanine or alanine was previously found to diminish both in vitro RNase activity and toxicity in E. coli,19 suggesting that similar mutations to the homologous C-terminal Y88 of YoeBSa1 might sufficiently reduce toxicity to allow expression in E. coli. To investigate the contribution of Y88 to the toxicity of YoeBSa1, the TAT codon for Y88 in YoeBSa1 was changed to TAG for the amber stop codon (Y88TAG) or to TTT for phenylalanine (Y88F). The genes encoding these mutants were cloned into pET-28a and expressed in E. coli. The resulting proteins were purified with yields of 5 mg of YoeBSa1-Y88TAG and 2.5 mg of YoeBSa1-Y88F per liter of culture. ESI mass spectrometry (ESI-MS) showed that the molecular weights of these mutants matched the predicted values (Figure 2A: YoeBSa1-Y88TAG, expected molecular weight: 12,307 Da, observed: 12,303 Da; Figure 2B: YoeBSa1-Y88F, expected molecular weight: 12,454 Da, observed: 12,450 Da). These results indicated that Y88 contributes to YoeBSa1 toxicity in E. coli and suggested that a non-permanent modification of the structure of Y88 might sufficiently alleviate the toxicity of YoeBSa1 to allow expression of the full-length protein in E. coli.
Figure 2. ESI-MS analysis of YoeBSa1 mutants.
(A) YoeBSa1-Y88TAG (calculated MW: 12,307 Da; observed MW: 12,303 Da). (B) YoeBSa1-Y88F (calculated MW: 12,454 Da; observed MW: 12,450 Da). (A) and (B) Gel inset lanes (L–R): Kaleidoscope prestained standards (Bio-Rad), expression culture lysate immediately prior to induction with IPTG, and expression culture lysate 4 h post-induction. * indicates the band corresponding to YoeBSa1-Y88TAG in (A) and YoeBSa1-Y88F in (B). (C) YoeBSa1-Y88ONBY prior to exposure to UV light (calculated MW: 12,606 Da; observed MW: 12,573 Da). (D–H) YoeBSa1-Y88ONBY exposed to UV light for 1, 2, 3, 4, or 5 min (calculated MW: 12,470 Da; observed MW: 12,468–12,469 Da).
Unnatural amino acid (UAA) mutagenesis enables the incorporation of a non-canonical amino acid site-specifically into a protein via the use of an aminoacyl-tRNA synthetase (aaRS)/tRNA pair that is orthogonal to the host organism’s translational machinery.29–31 The photocaged UAA o-nitrobenzyl tyrosine (ONBY) has been incorporated into a number of proteins in which a tyrosine is critical for enzymatic activity.32–36 Mutation of tyrosine to ONBY “cages” the enzyme by inhibiting catalysis, substrate binding, or both. Irradiation with UV light between 300 and 365 nm releases the photocaging o-nitrobenzyl group,37 revealing the free tyrosine and activating the enzyme for catalysis. As Y88 contributes to the toxicity and, presumably, the activity of YoeBSa1, it was hypothesized that mutation of Y88 to ONBY would allow expression of an inactive, non-toxic YoeBSa1 variant that could be activated by UV light following expression and purification.
The pEVOL plasmids for UAA mutagenesis encode an optimized amber (TAG) stop codon suppressor tRNA and two copies of an evolved UAA-specific Methanocaldococcus jannaschii aaRS, one under the control of a constitutive glnS’ promoter and the other under the control of an arabinose-inducible araBAD promoter, which allows the expression level of the aaRS to be finely tuned.38 As Y88 is the last residue in YoeBSa1, it would be challenging to purify ONBY-containing YoeBSa1 from prematurely truncated YoeBSa1. A variety of expression conditions were therefore screened by ESI-MS for maximal incorporation of ONBY into YoeBSa1. Maximum incorporation was achieved when 1 mM ONBY was present in the culture medium from the beginning of the expression, when aaRS expression was induced in early logarithmic growth phase, and when YoeBSa1 expression was induced in late logarithmic or early stationary phase and allowed to proceed overnight at 37°C. Using this method, 1 mg of pure YoeBSa1-Y88ONBY was obtained per liter of culture. ESI-MS revealed that the mass of the full-length caged protein was ~30 Da less than predicted (Figure 2C; YoeBSa1-Y88ONBY expected molecular weight: 12,606 Da, observed: 12,573 Da). This discrepancy is attributed to reduction of the nitro group of ONBY to an amino group, a phenomenon that has been observed when ESI-MS is carried out in protic solvents, resulting in a loss of 30 Da that corresponds to the observed mass shift.39 Importantly, very little truncated YoeBSa1-Y88TAG is present (expected molecular weight: 12,307 Da, observed: 12,305 Da). UV light-induced loss of the o-nitrobenzyl moiety was followed by ESI-MS (Figure 2D–H; YoeBSa1-Y88 expected molecular weight: 12,470). The extent of decaging increased as UV exposure was increased up to 3 minutes, after which no further decaging was observed (Figure 2F–H).
Decaged YoeBSa1 Is a Ribosome-Independent RNase
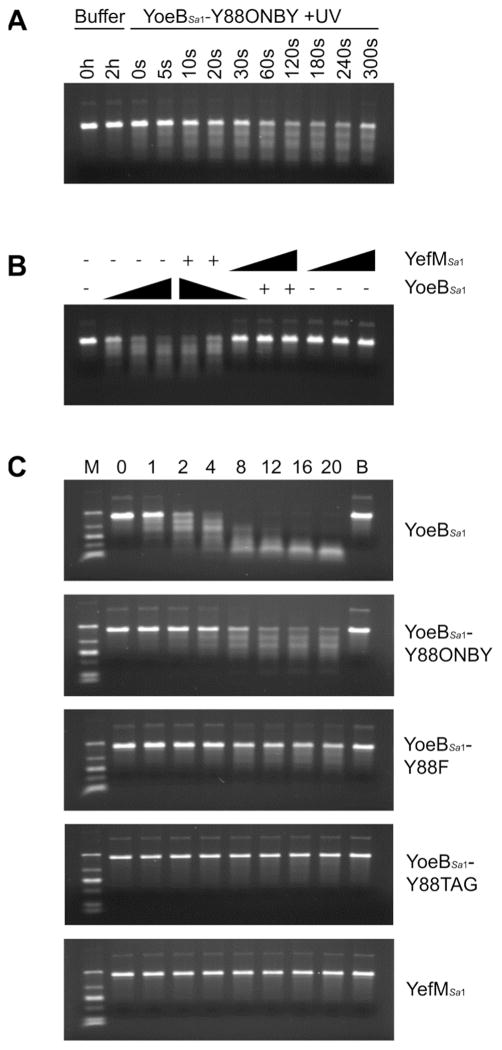
The ribosome-independent RNase activity of YoeBSa1 was assessed toward yefM-yoeBSa1 RNA in vitro. Decaging of YoeBSa1-Y88ONBY resulted in UV light-dependent degradation of the RNA (Figure 3A). In agreement with ESI-MS data showing that the extent of decaging did not increase following three minutes of UV exposure (Figure 2F), the RNase activity of YoeBSa1-Y88ONBY did not increase with UV exposure greater than three minutes (Figure 3A). YoeBSa1 activity was inhibited by the addition of an equimolar amount of YefMSa1 (Figure 3B), demonstrating that the observed RNase activity was due specifically to YoeBSa1 and not to RNase contamination. This result also suggests that YefMSa1 inhibits YoeBSa1 at a one-to-one ratio, which is identical to the ratio of YefMEc to YoeBEc required to inhibit YoeBEc in a similar in vitro assay.19 Comparison of the activity of decaged YoeBSa1 with that of YoeBSa1 mutants and YefMSa1 reveals that YoeBSa1-Y88ONBY retains weak in vitro RNase activity, while YoeBSa1-Y88F degrades RNA very slowly, and no detectable RNase activity is observed for YoeBSa1-Y88TAG or YefMSa1 (Figure 3C).
Figure 3. RNase activity of YoeBSa1 mutants.
(A) RNase activity of YoeBSa1-Y88ONBY exposed to UV light for 0, 5, 10, 20, 30, 60, 120, 180, 240, or 300 s and incubated with yefM-yoeBSa1 RNA for 2 h. Buffer controls were incubated with yefM-yoeBSa1 RNA for 0 or 2 h. (B) Inhibition of YoeBSa1 by YefMSa1. YoeBSa1-Y88ONBY was exposed to UV light for 3 min. YefMSa1 and YoeBSa1 were mixed and incubated at 37°C for 30 min to allow complex formation prior to incubation with yefM-yoeBSa1 RNA at 37°C for 1 h. “−” designates 0 pmol protein, “+” designates 10 pmol protein, and right triangles designate increasing protein from 10–30 pmol. (C) RNase activity of YoeBSa1, YoeBSa1-Y88ONBY, YoeBSa1-Y88F, YoeBSa1-Y88TAG, and YefMSa1. 10 pmol protein was incubated with yefM-yoeBSa1 RNA for 0, 1, 2, 4, 8, 12, 16, or 20 h. Lane M: Low-Range ssRNA Ladder (1000, 500, 300, 150, 80, and 50 nt; NEB). Lane B: buffer + RNA at 20 h. Top panel only: YoeBSa1-Y88ONBY was exposed to UV light for 3 min.
YoeBSa1 Cleaves RNA After Guanosine Residues
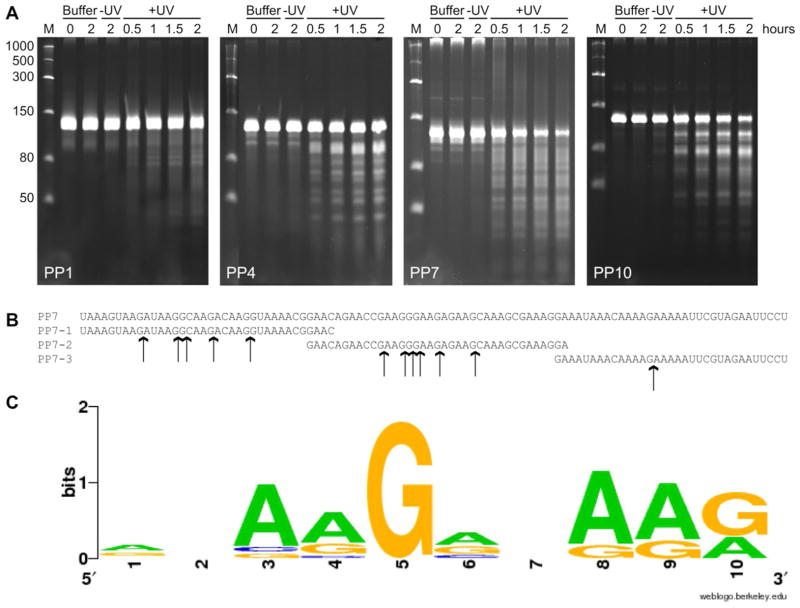
The Pentaprobes are a set of twelve plasmids that together encode every possible combination of 5 nucleotides in sequences of ~100 nucleotides per plasmid.40 The Pentaprobes have been used to determine the sequence specificity of RNases with recognition sequences of five or fewer nucleotides. Cleavage of each Pentaprobe by the RNase of interest is assessed by PAGE. Once cleaved Pentaprobes are identified, deconvolution occurs via MALDI-MS analysis of cleavage of small oligonucleotides.27 The RNase activity of YoeBSa1 toward each of the twelve Pentaprobe RNA transcripts was assessed by denaturing PAGE. YoeBSa1 was least active toward Pentaprobe 1 and most active toward Pentaprobes 4, 7, and 10 (Figure 4A, other Pentaprobes not shown). Cleavage of Pentaprobe 7 was the most dramatic, with rapid and significant degradation of the full-length parent band and appearance of a number of smaller discrete product bands, indicating that YoeBSa1 cleaves this Pentaprobe at multiple sites. The sequence of Pentaprobe 7 is rich in purine residues (Figure 4B), suggesting that the sequence specificity of YoeBSa1 might be similar to that of YoeBEc, which cleaves RNA after purine residues with a preference for adenosine.19
Figure 4. YoeBSa1 cleaves RNA at guanosine residues.
(A) RNase activity of YoeBSa1 toward Pentaprobes 1, 4, 7, and 10. Lane M: Low-Range ssRNA Ladder (NEB). Buffer lanes: RNA was incubated in buffer alone for 0 or 2 h. −UV lanes: YoeBSa1-Y88ONBY was incubated with RNA for 2 h. +UV lanes: YoeBSa1-Y88ONBY was exposed to UV light for 2 min and incubated with RNA for 0.5, 1, 1.5, or 2 h. (B) Primary sequences of Pentaprobe 7 and Pentaprobe 7 oligonucleotides PP7-1, PP7-2, and PP7-3. YoeBSa1 cleavage sites identified by MALDI-MS are indicated with arrows. (C) YoeBSa1 consensus cleavage sequence calculated by WebLogo.74
To identify the specific sequence recognized and cleaved by YoeBSa1 within Pentaprobe 7, cleavage of three overlapping oligonucleotides spanning Pentaprobe 7 (Figure 4B) was assessed by MALDI-MS. YoeBSa1 cleaved each oligonucleotide selectively after guanosine, leaving a 3′-cyclic phosphate on the 5′ RNA product. This suggests that YoeBSa1 is a guanosine-specific RNase in vitro and furthermore that the mechanism of YoeBSa1 RNA cleavage involves activation of the 2′-OH for nucleophilic attack of the 3′ phosphodiester bond. Fragments corresponding to both the 5′ and 3′ products were observed for five of the nine guanosines in PP7-1, six of thirteen in PP7-2, and one of four in PP7-3. Most of the non-cleaved guanosines were near the termini of the oligonucleotides, which reduces the likelihood of observing cleavage at these sites by MALDI due to the 2 kDa lower mass limit of the detector and a reduction in MALDI sensitivity with increasing oligonucleotide length. YoeBSa1 may also require a minimum number of residues 5′ or 3′ from the cleavage site in order to bind and cleave RNA and thus may skip guanosines near the termini of an oligonucleotide.
In the consensus sequence cleaved by YoeBSa1, adenosine residues precede and follow the guanosine at the cleavage site (Figure 4C). This may be an artifact of oligonucleotide design and selection, as most of the guanosines in Pentaprobe 7 are preceded by one or more adenosines. However, it is also possible that YoeBSa1 prefers to cleave after guanosines in the midst of purine-rich sequences. The fact that YoeBSa1 cleaved PP7-3 at only one of the three internal guanosines suggests that the ability of YoeBSa1 to cleave free mRNA may be inhibited by secondary structure, as PP7-3 had more predicted secondary structure than PP7-1 and PP7-2, which may have limited the ability of YoeBSa1 to access other potential cleavage sites in PP7-3.
Kinetic Characterization of YoeBSa1 RNase Activity
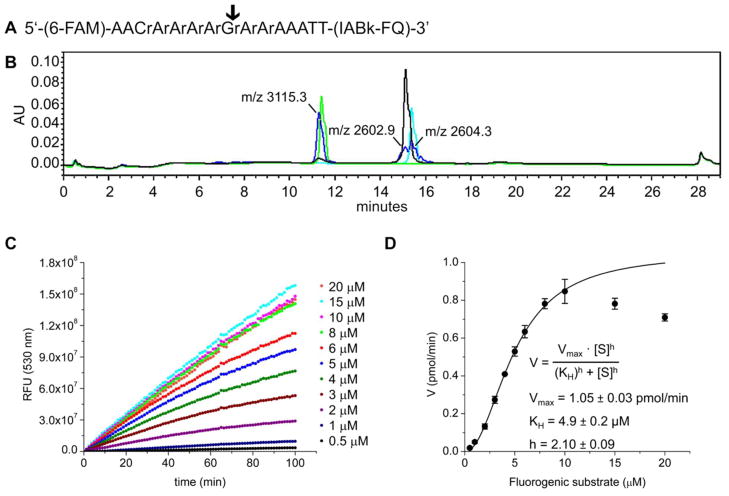
With the objective of developing a YoeBSa1 substrate that could be used to assess the activation of this RNase in a high-throughput setting, a fluorogenic oligonucleotide substrate was designed based on the data presented in Figure 4. An analogous design was previously used for the creation of a substrate for the ribosome-independent RNase MazF.41, 42 One of the sequences from PP7 that was cleaved by YoeBSa1 was converted into a 15-mer chimeric oligonucleotide with eight internal RNA residues surrounded by DNA nucleotides for stability (Figure 5A). Based on the specificity of YoeBSa1 identified using the Pentaprobes, cleavage of this substrate was predicted to occur after the central guanosine residue. A 6-carboxyfluorescein fluorophore (excitation: 495 nm; emission 520 nm) and an Iowa Black FQ quencher (absorbance maximum: 530 nm) were appended to the 5′ and 3′ ends, respectively, of the oligonucleotide. In the intact substrate, proximity enables the quencher to absorb fluorescence emitted by the fluorophore. Cleavage of the substrate increases the distance between the fluorophore and the quencher, producing an increase in fluorescence that can be monitored spectrophotometrically.
Figure 5. Kinetic analysis of YoeBSa1 using fluorometric assay.
(A) Fluorogenic substrate design. “r” designates RNA nucleotides. (B) HPLC traces of products of YoeBSa1 activity. Black: 30 μM intact substrate. Green: 30 μM 5′ cleavage product. Cyan: 30 μM 3′ cleavage product. Blue: 30 μM substrate cleaved by 20 μM YoeBSa1 after 5 h at 25°C. Peak fractions from the blue trace were analyzed by MALDI-MS and are labeled with the observed molecular weights (calculated MW, 5′ cleavage product: 3115.0 Da; calculated MW, 3′ cleavage product: 2602.8 Da). (C) Representative set of progress curves from one experiment. (D) Initial slopes of fluorometric assay curves (filled circles) with Hill fit (solid curve). Error bars represent standard deviation (n = 3 separately purified batches of YoeBSa1, each assayed in technical triplicate on two different days; plotted data are the average of the average for each batch).
The ability of YoeBSa1 to cleave this substrate was assessed by HPLC. The retention times of the products from YoeBSa1 activity overlapped with those of the independently synthesized standards (Figure 5B), suggesting that cleavage occurred primarily at guanosine, as predicted. MALDI-MS analysis of fractions collected from the elution peaks confirmed that the majority of cleavage events occurred at guanosine (Supplementary Figure S3). These results corroborate the guanosine-specificity of YoeBSa1 ribosome-independent RNase activity.19
To determine the kinetic parameters of YoeBSa1 as a baseline for future high-throughput screening efforts, the RNase activity of YoeBSa1 toward the fluorogenic substrate was assessed in 384-well plate format. YoeBSa1 (5 μM) was incubated with a range of concentrations of the fluorogenic substrate (0.5–20 μM) to produce a set of progress curves (representative data from one replicate of one experiment shown in Figure 5C). For each experiment, a calibration curve was constructed by mixing the two chimeric products resulting from YoeBSa1 cleavage at a 1:1 molar ratio in the presence of YoeBSa1 (5 μM). The relationship between fluorescence and concentration of the cleavage products was linear up to 1.5 μM. This calibration curve was used to convert relative fluorescence units (RFU) at 530 nm to pmol of cleaved substrate. Ideally, initial rates would be measured immediately following addition of YoeBSa1 to the substrate. However, in some experiments, a significant increase in fluorescence was not observed until ~15 min after addition of YoeBSa1. Consequently, the rates plotted in Figure 5D were determined from the linear portion of the progress curves between 20 and 30 min.
Traditional Michaelis-Menten enzyme kinetics are characterized by a hyperbolic rate, V, versus substrate concentration, [S], curve that can be fit by the equation
| (1) |
where Vmax is the maximum reaction velocity and Km is the Michaelis constant, equal to the substrate concentration at which half-maximal activity is observed. This equation describes a hyperbolic rate that increases linearly when [S] is low and asymptotically approaches Vmax at high [S].43, 44 The rate data in Figure 5D reveals that the kinetics of YoeBSa1 RNase activity toward the fluorogenic substrate are complex, with a sigmoidal increase in rate up to 10 μM substrate, followed by a marked decrease in rate at 15 and 20 μM substrate. The shape of the rate data suggests that the activity of YoeBSa1 is influenced by positive cooperativity at low substrate concentrations and by substrate inhibition at high substrate concentrations. However, careful consideration of the assay conditions and the nature of the fluorogenic substrate points to alternative explanations for the apparent cooperativity and substrate inhibition.
In traditional Michaelis-Menten enzyme kinetics, the substrate must be present in significant excess relative to the enzyme so that the substrate concentration is not substantially reduced by formation of the enzyme-substrate complex.45 In the fluorometric assay, however, YoeBSa1 (5 μM) is present at a concentration similar to that of the fluorogenic substrate (0.5–20 μM) in each reaction. Rather than Michaelis-Menten conditions, these enzyme and substrate concentrations resemble those of mutual depletion systems, in which formation of the enzyme-substrate complex significantly reduces the concentrations of both enzyme and substrate.46 Consequently, the classical Michaelis-Menten equation (1) cannot be used to fit the YoeBSa1kinetic data, but this does not necessarily mean that YoeBSa1 does not follow Michaelis-Menten kinetics. Interestingly, it has been shown that when the concentration of enzyme, [E]T, is equal to or greater than the Km, enzymes that follow Michaelis-Menten kinetics appear to have cooperative activity. The magnitude of the apparent cooperativity increases as [E]T increases relative to Km, causing a concomitant increase in the substrate concentration ([S]T)0.5 at which half-maximal velocity is observed.47 This increase is given by equation (2):
| (2) |
Equation (2) can be used to calculate Km from a plot of reaction velocity versus substrate concentration for enzymes operating under mutual depletion conditions.46, 47
However, the maximal velocity Vmax must be known in order to estimate ([S]T)0.5 and calculate Km using equation (2). The reduction in rate observed at 15 and 20 μM substrate precludes extrapolation to Vmax from the kinetic plot and suggests the influence of substrate inhibition. However, this behavior is likely an artifact of the substrate design, in which a central guanosine RNA residue is flanked by three or four adenosine RNA residues (Figure 5A). These RNA residues could not be removed without abolishing the activity of YoeBSa1 toward the substrate. Although YoeBSa1 cleaves primarily after guanosine, minor cleavage is also observed after the neighboring adenosines (Figure S3). As a result, the activity measured in the fluorometric assay is the sum of the rates at which YoeBSa1 cleaves after all of the RNA residues in the substrate. The preference for guanosine over adenosine suggests that cleavage after guanosine is faster than cleavage after adenosine. Thus at non-saturating substrate concentrations, the observed rate will be influenced primarily by cleavage after guanosine. As the substrate concentration increases, the proportion of cleavage events that are catalyzed after adenosine rather than guanosine will also increase. Eventually, at super-saturating substrate concentrations, the proportion of cleavage events at adenosine will increase to the degree that it will cause a measurable decrease in the overall rate, as observed in Figure 5D.
Consequently, Vmax for guanosine-specific substrate cleavage cannot be determined from the kinetic data. However, assuming that cleavage after adenosine makes a limited contribution to the measured rate at low substrate concentrations, fitting the kinetic data at substrate concentrations below those at which a measurable decrease in rate occurs will allow extrapolation to an estimate for Vmax. As it is possible that cleavage after adenosine makes a non-negligible contribution to the overall rate even at low substrate concentrations, this approach will provide minimal estimates only for Vmax and Km.
The Hill equation (3) was originally derived to describe the cooperative binding of ligands to a protein containing multiple binding sites when one or more ligands is already bound:
| (3) |
where θ is the ratio of occupied binding sites to total binding sites, [L] is the ligand concentration, KA is the association constant, and h is the Hill coefficient, which quantifies the degree of cooperativity.48 The maximum possible value of h for a perfectly cooperative protein is equal to the number of ligand-binding sites. h > 1 indicates positive cooperativity, where the rate of ligand binding is enhanced by already bound ligands; h < 1 indicates negative cooperativity, where the rate of ligand binding is reduced by already bound ligands; and h = 1 indicates no cooperativity, where the rate of ligand binding is unaffected by already bound ligands. If h = 1, the Hill equation (3) reduces to a form similar to that of the Michaelis-Menten equation (1).44
In addition to ligand-binding proteins, many enzymes exhibit cooperative enhancements or reductions in rate that result from conformational changes induced by enzyme oligomerization or the binding of allosteric effectors. Another smaller but growing class of monomeric enzymes with single ligand-binding sites also exhibits cooperative behavior caused by conformational transformations between two or more enzyme forms with differing substrate affinities. If the rate of conformational interconversion is slower than the rate of the reaction, substrate binding cannot reach equilibrium, resulting in non-hyperbolic, non-Michaelis-Menten kinetics.49
Therefore an alternative to mutual depletion as an explanation for the sigmoidal kinetics of YoeBSa1 is that its activity is cooperative, which would require oligomerization, allosteric effector binding, or a slow conformational change. Comparison of the gel filtration elution volume of YoeBSa1 with those of known standards indicates that YoeBSa1 is monomeric under the conditions employed in this assay. Furthermore, the assay buffer contains no known allosteric effectors, indicating that oligomerization and effector binding are unlikely to be responsible for sigmoidal shape of the kinetic data. On the other hand, the crystal structures of YefM-YoeBEc and YoeBEc reveal that YefMEc induces a conformational change in the active site of YoeBEc,19 which suggests that this area may be conformationally flexible. Moreover, as the primary cellular function of YoeB homologues appears to be ribosome-dependent RNase activity,20, 28, 50–52 the conformation of YoeBSa1 that cleaves RNA in a ribosome-dependent fashion may be distinct from the conformations that exist free in solution and cleave free mRNA. Slow conformational change is therefore a plausible explanation for the observed cooperativity in the kinetics of YoeBSa1 activity toward the fluorogenic substrate.
Regardless of whether the sigmoidal shape of the kinetic data results from mutual depletion or monomeric cooperativity, a modified version of the Hill equation can be used to provide a minimal estimate for Vmax. The simplest and most common method applied to analyze the kinetics of cooperative monomeric enzymes with single ligand-binding sites is a combination of the Hill and Michaelis-Menten equations:
| (4) |
where Vmax is the maximum reaction velocity, [S] is the substrate concentration, KH is the substrate concentration at which half-maximal activity is observed, and h is the Hill coefficient.49 If YoeBSa1 is cooperative, h quantifies the degree of cooperativity, and KH is equivalent to Km. On the other hand, if mutual depletion occurs, h quantifies the deviation from hyperbolic Michaelis-Menten kinetics, and Km can be calculated using equation (2) with KH = ([S]T)0.5. Equation (4) was therefore used to fit the kinetic data for substrate concentrations between 0.5 and 10 μM to allow extrapolation to a minimal estimate for Vmax. This gave Vmax = 1.05 ± 0.03 pmol/min and KH = 4.9 ± 0.2 μM, with h = 2.10 ± 0.09 (Figure 5D). Using KH = ([S]T)0.5 in equation (2) gives Km = 2.4 ± 0.2 μM.
DISCUSSION
The YoeBSa1 and YoeBSa2 toxins from S. aureus were previously found to induce growth arrest and to exhibit cellular RNase activity identical to that of YoeBEc upon overexpression in E. coli.28 As YoeBEc possesses weak ribosome-independent RNase activity with a preference for purine nucleotides in vitro,19 the possibility that YoeBSa1 also has ribosome-independent RNase activity was investigated.
One of the most significant obstacles to characterizing the activities and determining the structures of toxic proteins is the difficulty of overexpression in heterologous hosts such as E. coli. Many TA system toxins cannot be obtained in sufficient quantities for characterization because their overexpression arrests growth and/or kills the cells. After the failure of multiple strategies to provide pure, full-length, functional YoeBSa1, the discovery that Y88 is required for YoeBSa1 toxicity in E. coli led to the utilization of UAA mutagenesis to replace this residue with the photocaged amino acid ONBY, which reduced the toxicity of YoeBSa1 to a sufficient degree to allow overexpression in E. coli.
To our knowledge, this is the first reported use of in vivo UAA mutagenesis to facilitate the isolation and characterization of a toxin from a TA system. Incorporation of ONBY has also been used to overexpress a toxic zinc-finger nuclease in an inactive, non-toxic form in E. coli,36 suggesting that incorporation of a photocaged version of a critical catalytic residue via UAA mutagenesis may provide a general strategy to allow expression and characterization of toxic enzymes, provided that an appropriate aaRS/tRNA pair is available. For example, Y87 in YoeBSa2 is homologous to Y88 in YoeBSa1 and appears to be important for YoeBSa2 toxicity in E. coli (unpublished data). Consequently, incorporation of ONBY into YoeBSa2 using UAA mutagenesis may allow YoeBSa2 expression in E. coli for in vitro characterization as well. Furthermore, aaRS/tRNA pairs have also been developed for photocaged cysteine, serine, and lysine.31 Photocaged versions of aspartate, glutamate, glycine, alanine, and histidine have been synthesized,53–55 but corresponding aaRS/tRNA pairs do not yet exist. A list of residues identified by mutagenesis and structural data to be responsible for the activity and/or toxicity of a number of TA system toxins is provided in Table 1. Notably, each characterized toxin possesses one or more residues corresponding to an accessible photocaged amino acid. Introduction of aaRS/tRNA pairs specific for photocaged aspartate and histidine could allow for the general application of the photocaging strategy to express these proteins and study their activities and cellular roles in vitro and in vivo.
Table 1.
Residues responsible for the activity and/or toxicity of various TA system toxins.
| Toxin | Source(s) | Residues Implicated in Activity and/or Toxicity |
|---|---|---|
| CcdB 57, 58 | V. fischeri, Plasmid F | Trp99, Gly100, Ile101 |
| Doc 59 | Plasmid P1 | His66, Asp70 |
| FitB 60 | N. gonorrhoeae | Asp5, Glu42, Asp104, Asp122 |
| HigB 61 | Proteus spp. | His92 |
| HipA 62 | E. coli | Ser150, Asp209, Asp332 |
| Kid 63 | E. coli | Asp75, Arg73, His17 |
| MazF 64 | E. coli | Glu24, His28 |
| MqsR 65 | E. coli | Lys56, Gln68, Tyr81, Lys96 |
| PemK 66 | B. anthracis | Glu24, His59 |
| PezT 67, 68 | S. pneumoniae | Lys45, Asp66, Thr 118, Arg157, Arg170 |
| RelE 56 | E. coli | Arg61, Arg81, Tyr87 |
| VapC-3 69 | M. tuberculosis | Asp9, Glu43, Asp99, Asp117 |
| VapC-5 70 | M. tuberculosis | Asp26, Glu57, Asp115, Asp135 |
| VapC 71 | S. flexneri | Asp7, Glu42, Asp98 |
| YafQ 72 | E. coli | His50, His63, Asp67, Trp68, Arg83, His87, Phe91 |
| YoeB 19 | E. coli | Glu46, Arg65, His83, Tyr88 |
| ζ73 | S. pyogenes | Lys46, Asp67 |
YoeBEc belongs to the RelE structural family. RelE is a ribosome-dependent RNase toxin that is active only in the presence of the ribosome. In the ribosome-bound structure of RelE, Y87 of RelE stacks with the residue upstream of the cleavage site, and its hydroxyl is the closest functional group to the 2′-OH of the cleaved residue, suggesting a dual role in substrate binding and catalysis. Y87 is hypothesized to act as a general base through hydrogen bonding to a water molecule, causing the deprotonation and activation of the 2′-OH for attack of the 3′-phosphodiester bond.56 The role of Y88 in the catalytic mechanism of YoeBSa1 is currently unknown and may be different in the presence and absence of the ribosome. Mutation of Y88 in YoeBSa1-Y88TAG, YoeBSa1-Y88F, and YoeBSa1-Y88ONBY sufficiently reduced cellular toxicity to allow overexpression in E. coli (Figure 2). The relative yields of YoeBSa1-Y88TAG (5 mg/L) and YoeBSa1-Y88F (2.5 mg/L) under identical expression conditions indicate that the Y88TAG mutation renders YoeBSa1 less toxic than the Y88F mutation, which suggests that both the aromatic ring and hydroxyl functional group of Y88 contribute to cellular RNase activity. Similarly, YoeBSa1-Y88F and YoeBSa1-Y88ONBY retained the capacity to cleave free mRNA in vitro, albeit at a significantly reduced rate and to a much lesser extent than decaged YoeBSa1, while the RNase activity of YoeBSa1-Y88TAG was almost completely abolished (Figure 3C). These results suggest that both the aromatic ring and hydroxyl functional group of Y88 also contribute to the in vitro activity of YoeBSa1, although the exact role played by each of these moieties in ribosome-independent RNase activity remains unknown.
YoeBEc was previously found to possess purine-specific ribosome-independent RNase activity with a preference for adenosine over guanosine.19 In contrast, YoeBSa1 preferentially cleaves free mRNA after guanosine and to a much lesser extent after adenosine (Figures 4B, 5B, and S3). This specificity was used to design a chimeric fluorogenic substrate to study the kinetics of YoeBSa1 activity. The resulting kinetic data shows a sigmoidal increase in rate at low substrate concentrations followed by a decrease at high substrate concentrations. There are several plausible explanations for these deviations from standard hyperbolic Michaelis-Menten kinetics. First, the sigmoidal shape of the kinetic data may be an artifact of the high enzyme concentration used in these experiments,47 as it was necessary for the concentrations of YoeBSa1 and the fluorogenic substrate to be roughly equal (5 μM and 0.5–20 μM, respectively) in order to observe YoeBSa1 activity using the fluorometric assay. Alternatively, a slow transition between inactive and active enzyme conformations may be required for catalysis, resulting in failure to achieve a steady-state of bound substrate due to the faster speed of enzymatic catalysis. The seeming substrate inhibition is likely an artifact resulting from the unavoidable inclusion of adenosine RNA residues bordering guanosine in the substrate. Cleavage at these adenosine residues is hypothesized to cause a measurable reduction in rate at high substrate concentrations, necessitating the exclusion of these points from the kinetic analysis. It should be noted that these explanations are not mutually exclusive, and two or all three could conceivably be involved in the complex kinetics of YoeBSa1 activity toward the fluorogenic substrate. The Hill equation was used to fit the sigmoidal portion of the kinetic data in order to provide minimum estimates for Vmax and Km. Fitting the kinetic data with the Hill equation (4) for cooperative enzymatic activity gave Vmax = 1.05 ± 0.03 pmol/min, KH = 4.9 ± 0.2 μM, and Hill coefficient h = 2.10 ± 0.09 (Figure 5D). Using equation (2) to convert KH to Km gives Km = 2.4 ± 0.2 μM.
The relatively low affinity and slow rate observed for YoeBSa1 cleavage of free mRNA suggest that ribosome-independent RNase activity is unlikely to make a significant contribution to the activity of YoeBSa1 in cells. This is consistent with prior evidence for only ribosome-dependent RNase activity upon overexpression of YoeBSa1 in E. coli.28 YoeBEc is structurally homologous to RNase Sa from Streptomyces aureofaciens and Barnase from Bacillus amyloliquefaciens, both of which are guanosine-specific RNases,19 and the catalytic Glu-Arg-His triad found in these and other microbial guanosine-specific RNases is conserved in YoeBEc, YoeBSa1, and YoeBSa2. However, YoeB homologues lack a C-terminal extension that orients the nucleotide downstream of the cleavage site via stacking interactions with aromatic side chains. Ribosomal nucleotides may supply these interactions to facilitate proper orientation and cleavage of bound mRNA when YoeB binds in the ribosome, as in the crystal structure of the RelE-ribosome complex.56 The absence of the C-terminal extension and ribosomal interactions may explain the low affinity and slow rate of the RNase activity of free YoeBSa1.
Although YoeBEc was originally found to have ribosome-independent RNase activity in in vitro experiments,19 the only cellular RNase activity observed for YoeB homologues has been ribosome-dependent.20, 28, 50–52 This suggests that the ribosome-independent RNase activity of YoeBSa1 described here may have little relevance in cells apart from the discovery of an artificial activator capable of stabilizing a ribosome-independent RNase conformation of YoeBSa1. Then activation of YoeBSa1 from the YefM-YoeBSa1 complex in S. aureus could lead to global non-specific RNA degradation via the guanosine-specific ribosome-independent RNase activity reported here, in addition to translation inhibition via the ribosome-dependent RNase activity described previously,28 causing growth arrest and possibly eventual death. The fluorometric assay developed for YoeBSa1 ribosome-independent RNase activity will facilitate high-throughput screens of chemical and peptide libraries to identify molecules capable of activating YoeBSa1 via prevention of YefM-YoeBSa1 complex formation or disruption of the complex.
Supplementary Material
Acknowledgments
Funding sources: This work was supported by NIH grant 2R01-GM068385 to PJH. ASL was partially supported by the NIH Chemistry-Biology Interface Training Grant (NIGMS-NIH grant 5T32-GM070421).
We thank Prof. Peter Schultz at the Scripps Research Institute for providing pEVOL-ONBY and Prof. Joel Mackay at the University of Sydney, Australia, for providing the Pentaprobes. We also thank Prof. Alex Deiters and Jessica Torres Kolbus at North Carolina State University for advice regarding the synthesis of ONBY.
ABBREVIATIONS
- TA system
toxin-antitoxin system
- RNase
ribonuclease
- ONBY
ortho-nitrobenzyl tyrosine
Footnotes
SUPPORTING INFORMATION AVAILABLE
Includes two tables listing the primers used in this study (Table S1) and the results of the PCR screen for yefM-yoeBSa1 and yefM-yoeBSa2 in MRSA clinical isolates (Table S2) as well as three figures showing the sequence alignments of yefM-yoeBSa1 (Figure S1) and yefM-yoeBSa2 (Figure S2) PCR products and the MALDI spectra of the fluorogenic substrate cleavage products (Figure S3). This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 3.Masuda H, Tan Q, Awano N, Wu KP, Inouye M. YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol Microbiol. 2012;84:979–989. doi: 10.1111/j.1365-2958.2012.08068.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, Benedik MJ, Page R, Wood TK. A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat Chem Biol. 2012;8:855–861. doi: 10.1038/nchembio.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes K, Rasmussen PB, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, Van Melderen L. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 2011;39:5513–5525. doi: 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magnuson RD. Hypothetical functions of toxin-antitoxin systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett. 2013;340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen K, Christensen SK, Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 11.Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12:66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 12.DeNap JC, Hergenrother PJ. Bacterial death comes full circle: targeting plasmid replication in drug-resistant bacteria. Org Biomol Chem. 2005;3:959–966. doi: 10.1039/b500182j. [DOI] [PubMed] [Google Scholar]
- 13.Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles’ heel of drug-resistant bacteria. Curr Opin Chem Biol. 2008;12:389–399. doi: 10.1016/j.cbpa.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams JJ, Hergenrother PJ. Artificial activation of toxin-antitoxin systems as an antibacterial strategy. Trends Microbiol. 2012;20:291–298. doi: 10.1016/j.tim.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32(Suppl 2):S114–132. doi: 10.1086/320184. [DOI] [PubMed] [Google Scholar]
- 16.Donegan NP, Cheung AL. Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J Bacteriol. 2009;191:2795–2805. doi: 10.1128/JB.01713-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donegan NP, Thompson ET, Fu Z, Cheung AL. Proteolytic regulation of toxin-antitoxin systems by ClpPC in Staphylococcus aureus. J Bacteriol. 2010;192:1416–1422. doi: 10.1128/JB.00233-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct. 2009;4:19. doi: 10.1186/1745-6150-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamada K, Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell. 2005;19:497–509. doi: 10.1016/j.molcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Inouye M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J Biol Chem. 2009;284:6627–6638. doi: 10.1074/jbc.M808779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams JJ, Halvorsen EM, Dwyer EM, DiFazio RM, Hergenrother PJ. Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2011;322:41–50. doi: 10.1111/j.1574-6968.2011.02330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moritz EM, Hergenrother PJ. Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci U S A. 2007;104:311–316. doi: 10.1073/pnas.0601168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Miller JC, Silverman SK, England PM, Dougherty DA, Lester HA. Flash decaging of tyrosine sidechains in an ion channel. Neuron. 1998;20:619–624. doi: 10.1016/s0896-6273(00)81001-6. [DOI] [PubMed] [Google Scholar]
- 26.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie JL, Duyvestyn JM, Smith T, Bendak K, Mackay J, Cursons R, Cook GM, Arcus VL. Determination of ribonuclease sequence-specificity using Pentaprobes and mass spectrometry. Rna. 2012;18:1267–1278. doi: 10.1261/rna.031229.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshizumi S, Zhang Y, Yamaguchi Y, Chen L, Kreiswirth BN, Inouye M. Staphylococcus aureus YoeB homologues inhibit translation initiation. J Bacteriol. 2009;191:5868–5872. doi: 10.1128/JB.00623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 30.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 31.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 32.Deiters A, Groff D, Ryu Y, Xie J, Schultz PG. A genetically encoded photocaged tyrosine. Angew Chem Int Ed. 2006;45:2728–2731. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]
- 33.Chou C, Young DD, Deiters A. A light-activated DNA polymerase. Angew Chem Int Ed. 2009;48:5950–5953. doi: 10.1002/anie.200901115. [DOI] [PubMed] [Google Scholar]
- 34.Edwards WF, Young DD, Deiters A. Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem Biol. 2009;4:441–445. doi: 10.1021/cb900041s. [DOI] [PubMed] [Google Scholar]
- 35.Chou C, Young DD, Deiters A. Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. Chembiochem. 2010;11:972–977. doi: 10.1002/cbic.201000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou C, Deiters A. Light-activated gene editing with a photocaged zinc-finger nuclease. Angew Chem Int Ed. 2011;50:6839–6842. doi: 10.1002/anie.201101157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao H, Sterner ES, Coughlin EB, Theato P. o-Nitrobenzyl alcohol derivatives: opportunities in polymer and materials science. Macromolecules. 2012;45:1723–1736. [Google Scholar]
- 38.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E coli. J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 39.Karancsi T, Slegel P. Reliable molecular mass determination of aromatic nitro compounds: elimination of gas-phase reduction occurring during atmospheric pressure chemical ionization. J Mass Spectrom. 1999;34:975–977. doi: 10.1002/(SICI)1096-9888(199909)34:9<975::AID-JMS852>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 40.Bendak K, Loughlin FE, Cheung V, O’Connell MR, Crossley M, Mackay JP. A rapid method for assessing the RNA-binding potential of a protein. Nucleic Acids Res. 2012;40:e105. doi: 10.1093/nar/gks285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang NR, Hergenrother PJ. A continuous fluorometric assay for the assessment of MazF ribonuclease activity. Anal Biochem. 2007;371:173–183. doi: 10.1016/j.ab.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rensburg JJ, Hergenrother PJ. Detection of endogenous MazF enzymatic activity in Staphylococcus aureus. Anal Biochem. 2013;443:81–87. doi: 10.1016/j.ab.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michaelis L, Menten ML, Johnson KA, Goody RS. The original Michaelis constant: translation of the 1913 Michaelis-Menten paper. Biochemistry. 2011;50:8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson DL, Lehninger AL, Cox MM. Lehninger Principles of Biochemistry. 5. W.H. Freeman; New York: 2008. [Google Scholar]
- 45.Schnell S, Maini PK. A century of enzyme kinetics: reliability of the Km and Vmax estimates. Comm Theor Biol. 2003;8:169–187. [Google Scholar]
- 46.Griffiths JR. Steady-state enzyme kinetics in mutual depletion systems. Biochem Soc Trans. 1979;7:429–439. doi: 10.1042/bst0070429. [DOI] [PubMed] [Google Scholar]
- 47.Laurent M, Kellershohn N. Apparent co-operativity for highly concentrated Michaelian and allosteric enzymes. J Mol Biol. 1984;174:543–555. doi: 10.1016/0022-2836(84)90335-8. [DOI] [PubMed] [Google Scholar]
- 48.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:iv–vii. [Google Scholar]
- 49.Porter CM, Miller BG. Cooperativity in monomeric enzymes with single ligand-binding sites. Bioorg Chem. 2012;43:44–50. doi: 10.1016/j.bioorg.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halvorsen EM, Williams JJ, Bhimani AJ, Billings EA, Hergenrother PJ. Txe, an endoribonuclease of the enterococcal Axe-Txe toxin-antitoxin system, cleaves mRNA and inhibits protein synthesis. Microbiology. 2011;157:387–397. doi: 10.1099/mic.0.045492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sevillano L, Diaz M, Yamaguchi Y, Inouye M, Santamaria RI. Identification of the first functional toxin-antitoxin system in Streptomyces. PLoS One. 2012;7:e32977. doi: 10.1371/journal.pone.0032977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nolle N, Schuster CF, Bertram R. Two paralogous yefM-yoeB loci from Staphylococcus equorum encode functional toxin-antitoxin systems. Microbiology. 2013 doi: 10.1099/mic.0.068049-0. [DOI] [PubMed] [Google Scholar]
- 53.Wilcox M, Viola RW, Johnson KW, Billington AP, Carpenter BK, Mccray JA, Guzikowski AP, Hess GP. Synthesis of photolabile precursors of amino-acid neurotransmitters. J Org Chem. 1990;55:1585–1589. [Google Scholar]
- 54.Zhang Z, Papageorgiou G, Corrie JET, Grewer C. Pre-steady-state currents in neutral amino acid transporters induced by photolysis of a new caged alanine derivative. Biochemistry. 2007;46:3872–3880. doi: 10.1021/bi0620860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakayama K, Heise I, Gorner H, Gartner W. Peptide release upon photoconversion of 2-nitrobenzyl compounds into nitroso derivatives. Photochem Photobiol. 2011;87:1031–1035. doi: 10.1111/j.1751-1097.2011.00957.x. [DOI] [PubMed] [Google Scholar]
- 56.Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, Brodersen DE. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bahassi EM, Salmon MA, Van Melderen L, Bernard P, Couturier M. F plasmid CcdB killer protein: ccdB gene mutants coding for non-cytotoxic proteins which retain their regulatory functions. Mol Microbiol. 1995;15:1031–1037. doi: 10.1111/j.1365-2958.1995.tb02278.x. [DOI] [PubMed] [Google Scholar]
- 58.Loris R, Dao-Thi MH, Bahassi EM, Van Melderen L, Poortmans F, Liddington R, Couturier M, Wyns L. Crystal structure of CcdB, a topoisomerase poison from E. coli. J Mol Biol. 1999;285:1667–1677. doi: 10.1006/jmbi.1998.2395. [DOI] [PubMed] [Google Scholar]
- 59.Magnuson R, Yarmolinsky MB. Corepression of the P1 addiction operon by Phd and Doc. J Bacteriol. 1998;180:6342–6351. doi: 10.1128/jb.180.23.6342-6351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mattison K, Wilbur JS, So M, Brennan RG. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J Biol Chem. 2006;281:37942–37951. doi: 10.1074/jbc.M605198200. [DOI] [PubMed] [Google Scholar]
- 61.Hurley JM, Woychik NA. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korch SB, Hill TM. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J Bacteriol. 2006;188:3826–3836. doi: 10.1128/JB.01740-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamphuis MB, Bonvin AM, Monti MC, Lemonnier M, Munoz-Gomez A, van den Heuvel RH, Diaz-Orejas R, Boelens R. Model for RNA binding and the catalytic site of the RNase Kid of the bacterial parD toxin-antitoxin system. J Mol Biol. 2006;357:115–126. doi: 10.1016/j.jmb.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 64.Li GY, Zhang Y, Chan MC, Mal TK, Hoeflich KP, Inouye M, Ikura M. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J Mol Biol. 2006;357:139–150. doi: 10.1016/j.jmb.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 65.Brown BL, Grigoriu S, Kim Y, Arruda JM, Davenport A, Wood TK, Peti W, Page R. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009;5:e1000706. doi: 10.1371/journal.ppat.1000706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Agarwal S, Mishra NK, Bhatnagar S, Bhatnagar R. PemK toxin of Bacillus anthracis is a ribonuclease: an insight into its active site, structure, and function. J Biol Chem. 2010;285:7254–7270. doi: 10.1074/jbc.M109.073387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khoo SK, Loll B, Chan WT, Shoeman RL, Ngoo L, Yeo CC, Meinhart A. Molecular and structural characterization of the PezAT chromosomal toxin-antitoxin system of the human pathogen Streptococcus pneumoniae. J Biol Chem. 2007;282:19606–19618. doi: 10.1074/jbc.M701703200. [DOI] [PubMed] [Google Scholar]
- 68.Sedwick C. PezT: a bacterial suicide gene. PLoS Biol. 2011;9:e1001036. doi: 10.1371/journal.pbio.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Min AB, Miallau L, Sawaya MR, Habel J, Cascio D, Eisenberg D. The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg2+ ion in the active site and a putative RNA-binding site. Protein Sci. 2012;21:1754–1767. doi: 10.1002/pro.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miallau L, Faller M, Chiang J, Arbing M, Guo F, Cascio D, Eisenberg D. Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J Biol Chem. 2009;284:276–283. doi: 10.1074/jbc.M805061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dienemann C, Boggild A, Winther KS, Gerdes K, Brodersen DE. Crystal structure of the VapBC toxin-antitoxin complex from Shigella flexneri reveals a hetero-octameric DNA-binding assembly. J Mol Biol. 2011;414:713–722. doi: 10.1016/j.jmb.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Armalyte J, Jurenaite M, Beinoraviciute G, Teiserskas J, Suziedeliene E. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J Bacteriol. 2012;194:1523–1532. doi: 10.1128/JB.06104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meinhart A, Alonso JC, Strater N, Saenger W. Crystal structure of the plasmid maintenance system ε/ζ: functional mechanism of toxin ζ and inactivation by ε2ζ2 complex formation. Proc Natl Acad Sci U S A. 2003;100:1661–1666. doi: 10.1073/pnas.0434325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.