Summary
Mef2 transcription factors have been strongly linked with early heart development. D-mef2 is required for heart formation in Drosophila, but whether Mef2 is essential for vertebrate cardiomyocyte (CM) differentiation is unclear. In mice, although Mef2c is expressed in all CMs, targeted deletion of Mef2c causes lethal loss of second heart field (SHF) derivatives and failure of cardiac looping, but first heart field CMs can differentiate. Here we examine Mef2 function in early heart development in zebrafish. Two Mef2c genes exist in zebrafish, mef2ca and mef2cb. Both are expressed similarly in the bilateral heart fields but mef2cb is strongly expressed in the heart poles at the primitive heart tube stage. By using fish mutants for mef2ca and mef2cb and antisense morpholinos to knock down either or both Mef2cs, we show that Mef2ca and Mef2cb have essential but redundant roles in myocardial differentiation. Loss of both Mef2ca and Mef2cb function does not interfere with early cardiogenic markers such as nkx2.5, gata4 and hand2 but results in a dramatic loss of expression of sarcomeric genes and myocardial markers such as bmp4, nppa, smyd1b and late nkx2.5 mRNA. Rare residual CMs observed in mef2ca;mef2cb double mutants are ablated by a morpholino capable of knocking down other Mef2s. Mef2cb over-expression activates bmp4 within the cardiogenic region, but no ectopic CMs are formed. Surprisingly, anterior mesoderm and other tissues become skeletal muscle. Mef2ca single mutants have delayed heart development, but form an apparently normal heart. Mef2cb single mutants have a functional heart and are viable adults. Our results show that the key role of Mef2c in myocardial differentiation is conserved throughout the vertebrate heart.
Keywords: Second heart field, mef2c, mef2ca, mef2cb, mef2a, Heart, Hand2, Myl7, bulbus arteriosus, outflow tract, cardiomyocyte, differentiation
Introduction
Congenital heart defects occur in almost 1% of human births, which highlights the complexity of building the heart (Bruneau, 2008). In recent years, it has become clear that mammalian and avian hearts are built from two pools of progenitor cells: first heart field (FHF) cells generate the early cardiac tube and contribute to the left ventricle, atrioventricular canal (AVC) and atria, whereas the second heart field (SHF), also referred to as the anterior heart field, contributes to the outflow tract (OFT), right ventricle, and inflow region (Buckingham et al., 2005; Rochais et al., 2009). The SHF is the main source of myocardial progenitors added at a later stage to the arterial and venous poles of the heart (Cai et al., 2003; Kelly et al., 2001; Mjaatvedt et al., 2001; Waldo et al., 2001). Recent studies have suggested that, like the amniote heart, zebrafish heart is built from FHF and SHF progenitor cell pools. Whereas the FHF is the source of cells contributing to the primitive heart tube, the putative SHF provides cells that are added to the two poles of the heart tube, and build the structures at the inflow and outflow tract (de Pater et al., 2009; Grimes et al., 2010; Grimes et al., 2006; Hami et al., 2011; Lazic and Scott, 2011; Zhou et al., 2011). This provides a new opportunity to increase understanding of the molecular regulation of cardiomyogenesis.
Myocyte enhancer factor 2 (Mef2) transcription factors are key cardiomyogenic regulators, but their role in vertebrate heart development remains uncertain. In Drosophila, the single Mef2, D-mef2 is essential for the formation of all muscle types, including the heart. In D-mef2 mutants, cardiomyocyte (CM) precursors are properly specified and positioned, but fail to differentiate (Lilly et al., 1995; Ranganayakulu et al., 1995). In vertebrates, the Mef2 family of transcription factors has four members Mef2A, B, C and D (Breitbart et al., 1993; McDermott et al., 1993; Pollock and Treisman, 1991). Cell culture analyses implicate Mef2 activity, and particularly Mef2c, in CM differentiation (Ieda et al., 2010; Karamboulas et al., 2006). Although Mef2c is expressed during development of all murine CMs, FHF CMs differentiate in Mef2c mutants, which die at E9.5 from gross heart defects. These mutants have only a single ventricular chamber, abnormalities in the inflow and outflow tracts and defective cardiac looping reminiscent of SHF defects (Bi et al., 1999; Edmondson et al., 1994; Lin et al., 1998; Lin et al., 1997b; Verzi et al., 2005; Vong et al., 2006). Thus, in mice, there is a clear discrepancy between the strong SHF phenotype of Mef2c loss of function and the fact that Mef2c is expressed throughout the myocardium. It has been suggested that other Mef2 proteins may help drive FHF CM differentiation (Lin et al., 1997b; Vong et al., 2006). Other Mef2 genes are expressed in murine CMs, but are not, individually, essential for their formation. Mice mutant in Mef2d appear normal, whereas mutants lacking Mef2a form a myocardium but exhibit perinatal lethality from late cardiovascular defects (Arnold et al., 2007; Naya et al., 2002). Mef2b is expressed in early heart development (Molkentin et al., 1996b), but mice null for Mef2b have not been described. Thus, studies in mice have so far failed to determine whether Mef2 activity is essential for all CM differentiation.
Zebrafish have the advantages of surviving for several days without a heart and having two Mef2c genes, mef2ca and mef2cb (Hinits and Hughes, 2007; Miller et al., 2007; Ticho et al., 1996). Mef2ca is expressed in zebrafish heart (Ticho et al., 1996). Mef2cb is expressed in both FHF and SHF myocardium and, like loss of Mef2c function in mouse, mef2cb knockdown with morpholinos (MOs) has been shown to eliminate a subset of SHF CMs at the arterial pole (Lazic and Scott, 2011). A combinatorial loss of function analysis is therefore needed to establish the role Mef2 in the early steps of CM differentiation. Here we show that Mef2c activity controls an essential step in CM differentiation throughout the heart, in both FHF and SHF. By using loss- and gain-of-function models, we show that the two zebrafish Mef2c paralogues, Mef2ca and Mef2cb, control the expression of myocardial sarcomeric genes and other markers of CM maturation, such as nppa, smyd1b and bmp4, reminiscent of the function of the single Drosophila Mef2. Without Mef2ca and Mef2cb, the heart fails to form, and CMs are specified but developmentally arrested. Embryos lacking either Mef2ca or Mef2cb alone develop a normal heart. Together, our data reveal the essential role of Mef2 factors in the differentiation of FHF and SHF cardiomyocytes.
Materials and methods
Zebrafish lines and maintenance
Mutant and transgenic lines: mef2catn213 (Piotrowski et al., 1996), mef2cab1086(Miller et al., 2007), Tg(fli1a:EGFP)y1(Lawson and Weinstein, 2002), Tg(myl7:EGFP)twu26 (Huang et al., 2003), Tg(−5.1myl7:nDsRed2)f2 (Mably et al., 2003) and mef2cab1086;Tg(myl7:EGFP)twu26 were maintained on King’s wild type background. Mef2cbfh288 mutant allele was identified by TILLING (Draper et al., 2004) in the *AB background (http://labs.fhcrc.org/moens/Tilling_Mutants/index.html), and was further cleaned of linked contaminating mutations. Further crossing created the double mutant lines mef2cab1086;mef2cbfh288 and mef2cab1086;mef2cbfh288;Tg(myl7:EGFP)twu26. Staging and husbandry were as described (Westerfield, 1995). Genotyping was performed by sequencing of PCR products amplified from fin clip or embryo genomic DNA using primers 5’-AAAGCAGGCAAATAGAAAAACACT-3’ and 5’-AAAAGGCCAAACTCAACAGGAACT-3’ for b1086 allele, 5’-GGAAGAAGCGCTGTATTTAGGAC-3’ and 5’-ATATCTGTGCTGGCGTACTGG-3’ for fh288 allele and other methods in http://labs.fhcrc.org/moens/Tilling_Mutants/index.html).
Cloning of over-expression and other plasmids
pHS-mef2cb-IRES-GFP was made by cloning the full-length coding sequence of mef2cb (with four introduced mismatches) generated by PCR using the primers 5’-CGCTCTAGAATGGGACGAAAGAAAATTCAGATCACACGG-3’ and 5’-GTCGACTCATGTGGCCCACCCTTCCGAGA-3’ into the XbaI and SalI sites of hsp70-4-MCS-IRES-mGFP6 plasmid (Hinits and Hughes, 2007). DNA sequence was verified. Mef2cb mRNA was made with mMESSAGE mMACHINE kit from a linear DNA fragment containing the full CDS of mef2cb flanked by β-globin UTRs, which was made by a two-step PCR. Firstly, two PCR products were amplified by using the following primers: T3 and 5’-AATTTTCTTTCGTCCCATGAGCTCGATATCTCTCT-3’; T7U and 5’-TCTCGGAAGGGTGGGCCACATGAGTCGACGGATCCAGATCTGG-3’ on pβUT3 template (Hinits et al., 2009). Secondly, by using T3 and T7U primers on a template mix of pHS-mef2cb-IRES- GFP plus the two PCR products generated at the first step, a linear template with T7 and T3U ends was generated and confirmed by sequencing. pCMV:mef2cb-GFP was constructed by amplifying a fragment of the mef2cb gene 5’UTR including 166 bp immediately upstream and 25 bp downstream from the start codon with primers: 5’-AGATCTAAGCTTGTACAGCTACTGGAATCTTTGAAC-3’ and 5’-AGATCTGAATTCTGATCTGAATCTTTTTTCTCCCCAT-3’ and cloning into EcoRI/HindIII digested pEGFP-N1 (Clontech) in frame with EGFP and was sequence verified.
mRNA in situ hybridisation
In situ mRNA hybridization was performed as described previously (Hinits et al., 2009). To avoid cross-reactivity between mef2ca and mef2cb probes, for each gene we used two different probes, one containing only 3’UTR sequences and the other having part of the coding sequence. The two mef2ca probes gave identical patterns of expression (data not shown). We therefore used the longer, original probe used by Ticho and colleagues (Ticho et al., 1996). The mef2cb probe containing CDS as well as 3’UTR (IMAGE: 6519749) hybridized strongly to all of the domains detected by the 3’UTR only probe and additionally marked expression in the cephalic vascular system and was therefore preferred. For mef2aa, a plasmid containing the full CDS, MGC:55208, was used for PCR with the following primers (reverse primer contains T3 site) 5’-TGTACGGAAGTGTTACTTCTGCTC-3’ and 5’-GGATCCATTAACCCTCACTAAAGGGAAGGCCGCGACCTGCAGCTC-3’ (Thisse and Thisse, 2004). Other probes used were: myl7, vmhc (Yelon et al., 1999), bmp4 (Walsh and Stainier, 2001), hand2 (eu880, Thisse et al., 2005), nkx2.5 (Lee et al., 1996), nppa (Berdougo et al., 2003), tbx20 (Ahn et al., 2000), gata4, gata5, gata6 (Reiter et al., 1999) smbpc, tnnc, (Xu et al., 2000), egr2b (Oxtoby and Jowett, 1993), ntl (Schulte-Merker et al., 1992), kdrl (Thompson et al., 1998) and cdh5 (Larson et al., 2004). Embryos were photographed as wholemounts on Olympus DP70 or dissected and flatmounted in glycerol and photographed on a Zeiss Axiophot with Axiocam using Openlab software.
Embryo staining
Anti-Mef2 (Santa Cruz) and Anti-Mef2c (McDermott et al., 1993) were used as described (Hinits and Hughes, 2007). Anti-Mef2 can detect Mef2cb protein, as high-level nuclear expression was detected in a mosaic fashion after injecting BAC CH211-202E12 DNA, containing the mef2cb locus into embryos (Fig. S1A-E). Anti-Mef2ca/cb (1:200, Anaspec) reacted similar to anti-Mef2c and did not cross-react with other Mef2s (Fig. S2A-C). It also detects Mef2cb, as injecting either hs-mef2cb-IRES-GFP plasmid DNA (followed by a heat shock) or mef2cb mRNA at 1-2 cell stage, resulted in many cells co-expressing strong nuclear Mef2c and GFP (Fig. S1F,G). Other primary antibodies used were against sarcomeric myosin heavy chain (MyHC; A4.1025 (Blagden et al., 1997) or MF20 (DSHB, Iowa)), slow MyHC (F59, (Devoto et al., 1996)), DM-Grasp (zn5, ZIRC), Elastin (Miao et al., 2007), GFP (rabbit, Torrey Pines or chicken, Abcam ab13970) or RFP (rabbit PM0005, Medical and Biological Laboratories). Secondary antibodies were either HRP-conjugated (Vector) or Alexa dye-conjugated (Invitrogen). Embryos for immunohistochemistry were fixed in 4% PFA for 30 min to 2 h (except embryos stained with DAF2DA were fixed in 2% PFA for 30 min) and stained as described (Hinits and Hughes, 2007). Embryos were mounted in Citifluor (Agar) or low melting point agarose. Confocal images collected on a Zeiss LSM510 and some processed using Volocity software. DAF-2 DA (Santa Cruz) was used as described (Grimes et al., 2006).
Embryo manipulation
All morpholinos, plasmid and BAC DNA were injected into 1-2 cell stage embryos. mef2d/c MO and mef2ca ATG MO were described (Hinits and Hughes, 2007). Mef2cb ATG MO (5’TGTCCCCGTCTTTTCGTCTCTCTCT3’, Gene-Tools, 0.25 ng) and mef2cb E1I1 MO (5’TTCCGGTCAGCGTGACTCACCTGTC3’, Gene-Tools, 1 ng) were used for Mef2cb knockdown. To evaluate the effectiveness of mef2cb ATG MO, we co-injected pCMV:mef2cb-GFP with mef2cb ATG MO or other control MOs (Fig. S3A,B). Mef2cb E1I1 MO was checked by RT-PCR using primers 5’-CACACGGATTATGGATGAACG-3’ and 5’-TCCTTTGACTCTGGGCTGTGG-3’ matching the first and the third exons of mef2cb, which produces the 321 bp normal splicing product and an additional PCR product of 403 bp. Sequencing indicated the use of a hidden splice donor site inside intron 1, and so creating an aberrant transcript with a premature stop codon (Fig. S3C). Hand2 MO 5’-CCTCCAACTAAACTCATGGCGACAG-3’) was used as described (Maves et al., 2009).
Results
Mef2ca and mef2cb are the only Mef2 orthologues expressed in the early phases of heart development in zebrafish
Developing zebrafish hearts were screened for expression of Mef2 family transcription factors. Four Mef2 orthologues are known in zebrafish: mef2aa (previously named mef2a), mef2ca (previously named mef2c), mef2cb and mef2d (Hinits and Hughes, 2007; Lazic and Scott; Miller et al., 2007; Ticho et al., 1996). Two more, mef2ab and mef2b, were identified bioinformatically as orthologues of mammalian Mef2a and Mef2b, respectively. Only mef2ca, mef2cb and mef2aa genes were detectably expressed in the developing zebrafish heart (Fig. 1 and data not shown). Mef2ca mRNA accumulated in cells of the anterior lateral plate mesoderm (ALPM) from around 6ss (6 somite stage, data not shown), and was strong at 10-13ss. Expression persisted as the heart fields fuse to form the primitive heart tube (Fig. 1A,D) (Hinits and Hughes, 2007; Ticho et al., 1996). At 24 hpf, mef2ca was still detected weakly throughout the heart but was downregulated thereafter (Fig. 1H and data not shown). Mef2cb mRNA accumulated weakly in differentiated somitic adaxial cells in the bilateral heart fields as early as 7-8ss (Fig. 1B,E and data not shown, see also (Lazic and Scott, 2011)). During the heart cone stage mef2cb mRNA and Mef2c protein had accumulated in the differentiating CMs (Fig. 1F,G). Around 24 hpf, mef2cb mRNA pattern became distinct from that of mef2ca mRNA. Mef2cb was detected in the arterial and venous poles of the heart, in addition to the overall weak signal in the rest of the heart tube (Fig. 1I). Mef2cb mRNA was present without detectable mef2ca mRNA in the telencephalon and vascular system (Fig. S4A). Conversely, mef2cb mRNA was absent in the branchial arches that express mef2ca highly (Miller et al., 2007; Ticho et al., 1996). Mef2aa mRNA was not apparent before FHF CMs differentiate, but was detected in the heart at later stages than mef2ca and mef2cb and remained until at least 48 hpf (Fig. 1C and data not shown). Thus, the main Mef2 genes expressed during early cardiac development are mef2ca and mef2cb.
Figure 1. Mef2ca and mef2cb expression during early cardiogenesis.

In situ mRNA hybridisation of indicated genes, or immunodetection of Mef2ca/cb protein (G) for wild type (A-E, G-I) and Tg(myl7:EGFP) (F) in dorsal (A,B) or lateral (C-E) views of wholemount embryos or in flatmounts of dorsal views of the cardiac region, anterior to top (F-I). A,B. Mef2ca and mef2cb mRNAs accumulate in the bilateral heart fields in ALPM (arrowheads) and adaxial cells (green arrowhead). C-E. Egr2b expression in rhombomeres 3 and 5, and ntl expression in the notochord positions the row of ventral cells in the ALPM (black arrowheads; D,E) that contain mef2ca and mef2cb, but not mef2aa mRNA (C). F. Confocal stack of Tg(myl7:EGFP) heart at 25ss showing co-localisation of mef2cb mRNA (Fast Red) and EGFP. G. Mef2c protein in nuclei of a similar crescent of CMs spanning the midline. H,I. By 24 hpf, both mef2ca (H) and mef2cb (I) mRNAs are detected weakly in the heart tube, but mef2cb also accumulates strongly in the venous (blue arrow) and arterial (pink arrow) poles of the heart. Scale = 100 μm (A-F), 20 μm (G-I).
Mef2ca and mef2cb are required for myocardial differentiation
To test the role of Mef2c during early cardiac development, we analysed a double mef2cab1086;mef2cbfh288 mutant made by crossing double heterozygotes for mef2ca+/b1086 (hoover) (Hinits and Hughes, 2007; Miller et al., 2007) and a novel mef2cbfh288 allele with an A to T single point mutation changing Arg at residue 24 of the Mef2cb protein to a stop codon early in the MADS domain, thereby generating a predicted null mutation (http://labs.fhcrc.org/moens/Tilling_Mutants/index.html; (Molkentin et al., 1996a; Yu et al., 1992)). The mef2cbfh288 mutation was cleaned of linked contaminating mutations. In wild type embryos, differentiation of myocardial precursors begins around 14ss in ALPM, and is marked by accumulation of mRNAs encoding sarcomeric protein genes such as myl7 and vmhc (Fig. 2A and (Yelon et al., 1999)). Close to 1/16th of embryos from a mef2ca+/b1086;mef2cb+/fh288 in-cross entirely lacked these markers at 15ss (Fig. 2F, quantification of all experiments is presented in Table S1). We confirmed this result in a mef2ca single mutant injected with mef2cb MO (either of two mef2ca null mutant alleles injected with either of two mef2cb MOs lacked markers in one quarter of embryos), the mef2cb null mutant injected with mef2ca MO, or dual mef2ca and mef2cb MOs (Figs. 2B, S5A,B). Hereafter, we refer to embryos lacking Mef2c activity as ‘dual loss of function’ irrespective of the combination of mutant allele and/or MOs employed; all lack the vast majority of differentiated CMs. We also injected wild type embryos with a single mef2d/c ATG MO which we previously showed ablates several Mef2 proteins due to the high degree of conservation between various Mef2 genes at the beginning of the coding region (Hinits and Hughes, 2007)). This MO also ablated all CMs differentiation at 15ss (Fig. S5A,B).
Figure 2. Early cardiomyocytes fail to differentiate after loss of Mef2c function.
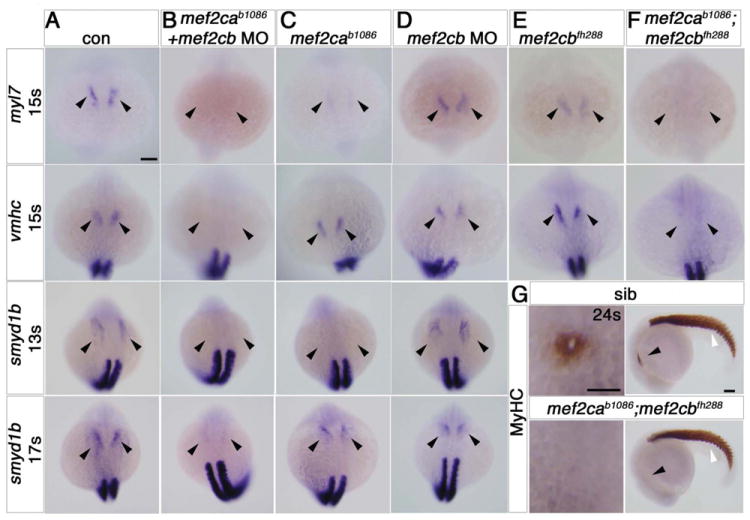
A-F. In situ mRNA hybridisation for myl7, vmhc and smyd1b in wild type control (A), mef2cab1086+mef2cb MO (B), mef2cab1086 (C) mef2cb MO (D), mef2cbfh288 (E) and mef2cab1086;mef2cbfh288 (F) embryos, shown in wholemounts in dorsal view, anterior to top. Loss of both mef2ca and mef2cb function greatly reduces myl7, vmhc and smyd1b mRNAs in the bilateral heart fields (arrowheads; A,B,F). Mef2cab1086 mutant embryos have weak myl7 and smyd1b mRNAs early, but recover later, and show no change in vmhc (C). Mef2cb single morphants or mef2cbfh288 mutant show no changes (D). G. Immunostaining for MyHC (A4.1025) in 24ss mef2cab1086;mef2cbfh288 embryos and their siblings, shown in wholemounts in dorsal view, anterior to top (left panel) and lateral view, anterior to left (right panel). No MyHC is detected in the heart, whereas somitic muscle appears normal (white arrowheads). Scale = 100 μm.
Zebrafish smyd1b mRNA normally marks differentiating CMs in the FHF from 13ss, and is an orthologue of mouse Smyd1/Bop, a known Mef2c target (Phan et al., 2005). Mef2ca;mef2cb dual loss of function embryos lacked smyd1b mRNA both before (13ss) and after (17ss) the time of CM differentiation (Fig. 2A,B and data not shown). At 24ss, such embryos had no sarcomeric myosin heavy chain (MyHC) protein in heart, whereas this protein was present in skeletal muscle (Fig. 2G). At 24 hpf, mef2ca;mef2cb double mutants and other dual loss of function embryos had no beating heart cells, and lacked almost all myl7 and vmhc mRNA and MyHC protein in heart; however, a few residual CMs were variably present (Fig. 3A,B,F and Fig. S6A). The residual cells had both ventricular and atrial character, having vmhc mRNA caudally and myl7 mRNA alone anteriorly. At this stage, mef2cbfh288 mutant embryos injected with mef2ca MO had somewhat more cells expressing myl7 mRNA than the double mef2ca;mef2cb mutants, and their heart had a thin string-like shape (Fig. S6B). Although mef2ca morphants phenocopied the jaw defects seen in the mutant at 5 dpf ((Miller et al., 2007) and data not shown), this finding suggests incomplete knockdown of Mef2ca protein by mef2ca MO. Conversely, using the mef2d/c MO we observed no cardiac cells expressing MyHC or a variety of other myocardial markers (Fig. 3G-I and data not shown). In addition, mRNAs encoding bmp4, smyd1b and atrial natriuretic factor (nppa), all markers of maturing CMs, were absent (Fig. 3B,F,J). Thus, lack of both Mef2ca and Mef2cb results in failure of CM differentiation and heart formation.
Figure 3. Redundant and specific functions of Mef2ca and Mef2cb drive cardiomyogenesis and heart tube formation.
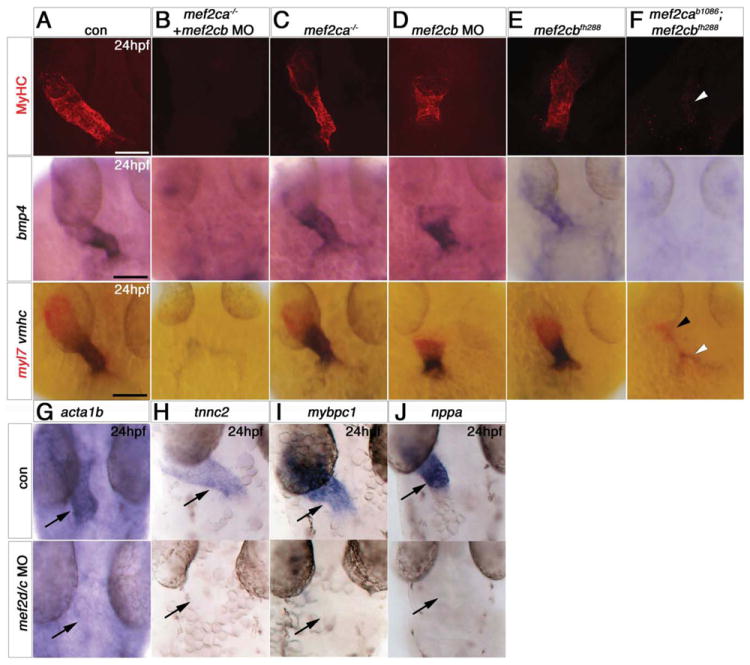
Immunodetection of MyHC (confocal stacks, top panels) or in situ mRNA hybridisation for indicated genes (A-F, lower panels and G-J) in hearts of 24 hpf zebrafish embryos shown in a dorsal view, anterior to top. A,B,F. Loss of both mef2ca (tn213 allele, in MyHC and b1086 allele, in bmp4 and myl7+vmhc) and mef2cb function (B,F) led to lack of all markers, compared with control (A). Note the few cells expressing myl7 only (black arrowhead) or both myl7 and vmhc (white arrowhead). C. Mef2ca mutants have a normal heart. D,E. Mef2cb morphants have a shortened heart with substantial loss of both atrial and ventricular volume, yet mef2cbfh288 mutants have a normal heart. G-I. Loss of Mef2c function with mef2d/c MO ablated all actin (acta1b, G), tnnc2 (H), mybpc1 (I) and nppa (J) mRNAs. Scale = 100 μm.
By 48 hpf, double mef2ca;mef2cb mutant embryos had pericardial oedema accompanied by lack of most myocardial cells (Fig. 4A, S6C). Around 30 residual CMs were present in small beating tube-shaped structure(s) forming either one or two zones (usually in the location of the venous pole, occasionally in both poles, and, more rarely, in a single string-like structure; Fig. 4B,C and data not shown). Wild type hearts had undergone looping by this stage and two chambers are readily distinguished. Double in situ hybridisation for myl7 and vmhc marks the ventricular cells with both mRNAs, whereas atrial cells express myl7 only. Each small residual tube in a mef2ca;mef2cb double mutant had either the ventricular or the atrial expression pattern appropriate to its position in the cardiac region (Fig. 4B). nppa and bmp4 mRNAs are missing or weakly restricted to residual regions (Fig. 4E,F). At 72 hpf, an incross of mef2ca+/b1086;mef2cb+/fh288;Tg(myl7:EGFP)twu26 had one or two residual GFP+ tube-shaped structures in double mutant embryos, of which ~80% (n=17) were beating (Fig. 4G and data not shown). Thus, lack of both Mef2ca and Mef2cb results in failure of most CM differentiation and heart formation, although a few cells showing either atrial or ventricular fates are able to differentiate after 24 hpf.
Figure 4. Loss of Mef2c function abolishes sarcomeric gene expression.
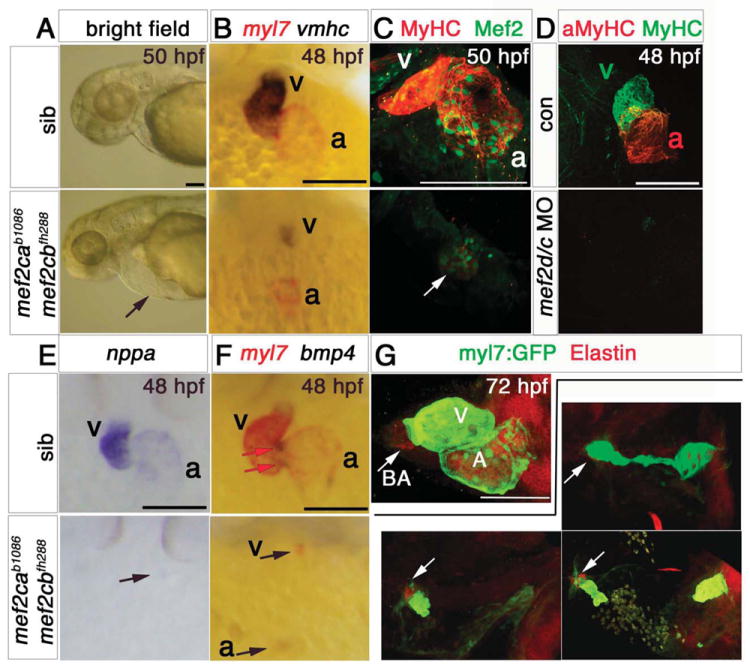
Hearts of 48-50 hpf (A-F) or 72 hpf (G) mef2cab1086;mef2cbfh288 mutant embryos and their siblings in bright field (A), after immunodetection (C,D,G) or in situ mRNA hybridisation (B,E,F) shown in lateral view, anterior to left (A,G) or ventral (B-F) view, anterior to top. A. mef2ca b1086;mef2cbfh288 embryos had a tiny residual heart (arrow) and cardiac chamber edema. B,E,F. Double mutant embryos lacked almost all myl7, vmhc, bmp4 and nppa mRNAs as well as MyHC and Mef2 proteins in ventricle (v), atrium (a) or AV canal (red arrows in F) except for one (arrows in C,E) or two small heart structures expressing these markers. C. Confocal stack showing genotyped double mutant expressing low levels MyHC and nuclear Mef2 in a residual myocardial tissue. Sibling presented is mef2ca+/b1086;mef2cb+/+. D. No MyHC or atrial MyHC is detected in mef2d/c morphants. G. Immunodetection for GFP (atrium and ventricle, green) and Elastin (bulbus arteriosus, ba) in hearts of three different genotyped mef2cab1086;mef2cbfh288;Tg(myl7:EGFP)twu26 showing variation in residual differentiated myocardial and bulbus tissue, compared to the normal heart of a mef2ca+/b1086;mef2cb+/fh288;Tg(myl7:EGFP)twu26 sibling. Scale = 100 μm.
In contrast, hearts of mef2d/c morphant embryos had a more severe phenotype; neither MyHC nor atrial-specific MyHC immunostaining was detected in mef2d/c morphants (Fig. 4D). No Mef2 immunoreactivity was observed in the cardiac region of mef2d/c morphants (data not shown). Lack of additional Mef2s in mef2d/c MO appears to prevent residual CM differentiation.
We also tested whether non-myocardial cells that form the bulbus arteriosus can differentiate when lacking Mef2ca and Mef2cb. Double mutant embryos had a smaller than normal bulbus arteriosus structure marked by Elastin immunostaining (Grimes et al.; Grimes et al., 2006; Hami et al.; Lazic and Scott; Miao et al., 2007), but the bulbus size was reduced in proportion to that of the residual CM tube structure in the arterial pole (Fig. 4G). Thus, Mef2c activity directly or indirectly controls bulbus arteriosus size.
Mef2ca and mef2cb are required for a late step in cardiomyocyte differentiation
To define the role of Mef2 proteins in the cascade of events leading to CM differentiation, we examined mef2ca;mef2cb dual loss of function embryos at early stages when myocardial precursors reside bilaterally within the ALPM. Specified CMs in the ALPM express a transcriptional program that includes several GATA, Tbx, Nkx2 and Hand2 factors (Begemann and Ingham, 2000; Reiter et al., 1999; Ruvinsky et al., 2000; Serbedzija et al., 1998; Yelon et al., 1999; Yelon et al., 2000). At 12ss, nkx2.5, gata4,5,6 and hand2 mRNAs appeared indistinguishable in dual loss of function embryos compared to controls (Fig. 5A-C and S5E). Congruently, all embryos from mef2cb+/b1086;mef2cb+/fh288 incross analysed at this stage showed similar expression of these mRNAs (data not shown). By 22ss, expression of various genes that are part of the myocardial program, such as tbx20 and gata6, began to show defects in dual loss of function embryos and mef2d/c MO by lacking the characteristic ring of expression around the endocardium and had a disorganised pattern at the midline (Fig. 5D,E). Paralleling this defect there was a failure of nkx2.5 mRNA maintenance. Whereas early nkx2.5 expression was unaffected by loss of Mef2ca and Mef2cb function, its mRNA was later lost, presumably due to the lack of differentiated CMs (Fig. 5A,F). Similarly, smyd1b and most bmp4 mRNA was lost (Figs 2B and S5C,D), indicating their role in the maturation of differentiated CMs that are lost after dual loss of function. Thus, Mef2cs function late, during sarcomeric differentiation of CMs.
Figure 5. Cardiomyocytes of mef2ca;mef2cb dual loss of function are specified but developmentally arrested.
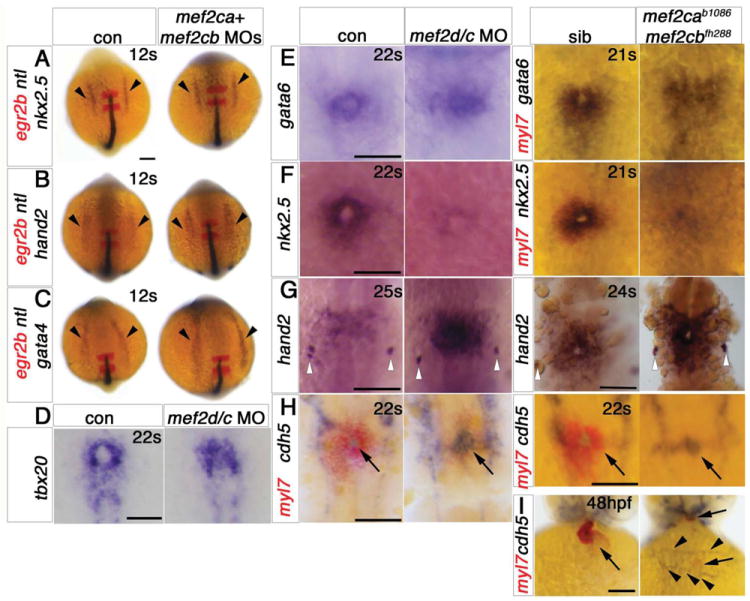
In situ mRNA hybridisation for indicated genes (wholemounts in dorsal view, anterior to top, except J, ventral view). A-C. ALPM expression (arrowheads) of nkx2.5 (A), hand2 (B) and gata4 (C) at 12ss is unaffected by lack of Mef2ca and Mef2cb. Notochord and rhombomeres 3 and 5 are marked by ntl (blue) and egr2b (red), respectively. D,E. Tbx20 and gata6 mRNAs are present but disorganised in the heart region of mef2d/c morphants and mef2cab1086;mef2cbfh288 mutant embryos (lacking myl7 expression in red, E, right panel) compared to the typical ring-shape in control and sibling embryos. F. During heart cone stage (22ss), nkx2.5 mRNA is abolished. G. hand2 mRNA is enhanced in a sheet of cells spanning the cardiac region but not elsewhere. Pharyngeal pouch expression is unchanged (white arrowheads). H, Whereas myocardial cells are undifferentiated in mef2d/c morphants and mef2cab1086;mef2cbfh288 mutants (myl7, red), the endocardium is expanded and cdh5 is up-regulated. I. Endocardium (cdh5, arrow) lines the myocardium (myl7, red) in the normal looped heart of a sibling embryo. In mef2ca;mef2cb mutant embryos, little endocardial makrer is co-localised with the residual myocardium (arrows). Extra cdh5-expressing tissue is detected in the cardiac region (arrowheads). Scale = 100 μm.
In order to place the function of Mef2cs in relation to other factors implicated in CM differentiation, we examined the bHLH transcription factor hand2, which promotes CM differentiation, especially in ventricle (Yelon et al., 2000). In mice, it has been suggested that Mef2c is upstream of Hand2, as expression of Hand2 was reduced in Mef2c null mice (Lin et al., 1997b). However, we found that, at 20-25ss, hand2 mRNA levels appear up-regulated after mef2ca;mef2cb dual loss of function in a wide sheet cells in the prospective cardiac region (Fig. 5G). Although hand2 mRNA normally persists in the heart tube, its levels are down-regulated in the pre-cardiac region as CMs differentiate (Yelon et al., 2000). These data indicate that CM development was arrested in the absence of Mef2 function. Although hand2 mRNA expression precedes that of mef2ca and mef2cb in the ALPM, both are expressed in hand2 mutants and morphants, ((Yelon et al., 2000), and Fig. S7A,B). Thus, it is not likely that mef2ca/mef2cb are targets for Hand2 in zebrafish.
Endothelial expression of Mef2c is not required for endocardium formation
The lack of a clear cardiac cone revealed by tbx20 and gata6 mRNA in dual loss of function embryos suggested that embryos lacking Mef2ca and Mef2cb may have defects in the endocardium or, more generally, in endothelium (Fig. 5D,E). Indeed, mef2cb mRNA is clearly detected in vasculature (Fig. S4A), consistent with the presence in mef2ca, mef2cb and mouse Mef2c upstream regulatory regions of a FOX:ETS binding motif capable of driving GFP in zebrafish endothelium (De Val et al., 2008). However, endothelial markers fli-1, kdr (flk-1) and cdh5 are relatively unperturbed in vasculature of double mef2ca;mef2cb mutants and in mef2d/c MO embryos (Fig. S4B-E). Endocardial markers were also present in dual loss of function embryos. Whereas in 22ss control embryos a ring of myl7+ CMs surrounds a small circle of cdh5+ endocardial cells, in dual loss of function and mef2d/c MO embryos lacking heart myl7 mRNA, cdh5 mRNA was not only present but even expanded and up-regulated (Fig. 5H). Intriguingly, by 48 hpf, endothelial cells marked with cdh5 mRNA were abnormally organised in the cardiac region where the heart had failed to form (Fig. 5I). Thus, lack of myocardial differentiation leads to the disorganisation of other layers of the heart.
Over-expression of mef2cb dominantly changes cell fate
Dual loss of function shows Mef2c factors are necessary for myocardial differentiation. To test whether Mef2 is also sufficient for CM differentiation, we used a gain of function approach. Embryos injected at the 1-2 cell stage with RNA encoding mef2cb had widespread ectopic muscle in the head region (Figs 6A and S1G). Most ectopic muscle was composed of elongated cells expressing high levels of MyHC and smyd1b mRNA and lacking bmp4 mRNA, and therefore may differ from CMs (Fig. 6A-C). Interestingly, mef2cb mRNA injection did up-regulate bmp4 mRNA in many cells in a rectangle within the cardiogenic region, but not elsewhere in the embryo (Fig. 6C). Nevertheless, few cells in the rectangle appeared to form CMs, and hearts in such embryos were not bigger than normal (Fig. 6A). Taken together, these data indicate that although Mef2cb can induce bmp4 in the cardiogenic region, Mef2cb alone is insufficient to drive widespread ectopic CM differentiation.
Figure 6. Mef2cb overexpression induces skeletal muscle at the expense of myocardial and endothelial cells.
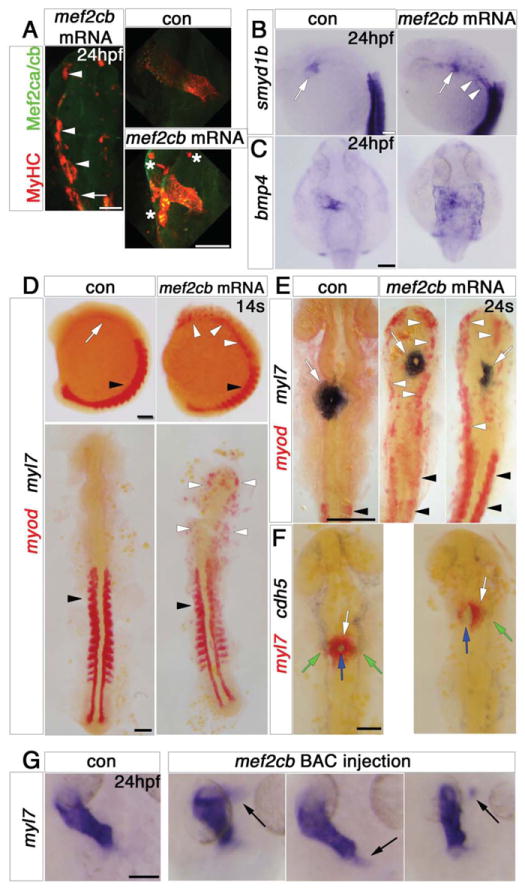
In situ mRNA hybridisation (or immunodetection, A) for indicated genes, shown as wholemounts in dorsal (G,C) or lateral (B, D, top panel) views, or as flatmounts in dorsal view (A, D, bottom panel, E,F). A. Injection of mef2cb RNA results in many ectopic muscle cells expressing strong Mef2ca/cb and MyHC at 24 hpf (white arrowheads) anterior to the first somite (white arrow). In the heart, MyHC and Mef2ca/cb are mosaically stronger than in control embryos. Asterisks = ectopic muscle. B. Smyd1b is upregulated in CMs (white arrow), and in ectopic muscle in the head region (white arrowheads). C. Expression of bmp4 is upregulated in much of a sheet of cardiogenic cells but not elsewhere in the embryo. D. In 14ss control embryos, myod mRNA is expressed in somites (black arrowheads) and myl7 is expressed weakly in CMs (white arrow). Embryos injected with mef2cb RNA express no ectopic myl7, but have ectopic myod mRNA in the head region (white arrowheads). E. By 24ss, high levels of ectopic myod correlated with reduction or lack of myl7-expressing CMs (right panel) and defective brain development. F. At 24ss, mef2cb RNA-injected embryos that had fewer myl7-expressing CMs (white arrows) also had less cdh5 expression in vascular endothelium (green arrows) and disorganised endocardium (blue arrows). G. Compared to wild type control (leftmost panel), three examples of embryos injected with mef2cb BAC DNA show ectopic myl7 mRNA either contiguous with (left panels) or detached from (right panel) the arterial (flanking panels) or venous (middle panel) poles. Scale = 100 μm.
The elongated nature of ectopic muscle cells induced outside the cardiogenic region by Mef2cb over-expression was reminiscent of skeletal muscle. To test this possibility, we used double in situ hybridisation for myod and myl7, which exclusively mark skeletal muscle or myocardial cells, respectively. Injection of 20 pg of mef2cb RNA caused high levels of ectopic myogenesis accompanied by a dramatic failure of brain morphogenesis (Fig. 6D,E). At both 14ss and 23ss, no ectopic myl7 expression was found in embryos injected with mef2cb RNA, whereas many cells in the head region expressed ectopic myod (Fig. 6D,E). Moreover, at 23ss, fewer CMs expressed myl7 in Mef2cb-overexpressing embryos (Fig. 6E). In addition, cdh5+ endothelial cells were also reduced in both endocardium and vasculature in such embryos (Fig. 6G). The ectopic expression of the skeletal muscle marker myod in mesodermal and probably other territories, together with the depletion of cardiac and endothelial markers, indicates that high levels of Mef2cb can dramatically alter cell fate in the early embryo.
In order to restrict our overexpression analysis to cells normally expressing mef2cb, we drove mosaic over-expression of mef2cb by injection of a BAC CH211-202E12 DNA, containing the mef2cb locus. This approach leads to greatly increased expression of genes contained within the BAC, but in normal locations (Minchin and Hughes, 2008). The BAC also contains Dre-mir-9-5 microRNA. mir-9 is highly expressed in the brain, and is suggested to regulate neurogenesis, but is not expressed in zebrafish heart (Leucht et al., 2008; Wienholds et al., 2005), and thus is not predicted to effect the heart. Injection of the BAC increased levels of mef2cb mRNA and Mef2 protein in scattered somitic muscle, heart, telencephalon and vasculature, regions were mef2cb is normally detected (Fig. S1A-D). Within the cardiogenic region, patches of ectopic myl7 mRNA appeared around the heart (Fig. 6G), and could reflect either ectopic induction of CMs or their aberrant migration. Ectopic skeletal muscle was not observed elsewhere (data not shown). Thus, the skeletal myogenic effect of Mef2cb over-expression appears to be restricted to certain cells in early embryos.
Lack of Mef2ca delays cardiomyocyte maturation
Dual loss of function of both mef2ca and mef2cb blocked CM differentiation, but loss of function of either gene alone did not prevent heart formation (Figs 2,3). Analysis of two mutant alleles of mef2ca, b1086 and tn213, revealed weaker myl7 mRNA accumulation in FHF CMs at 15ss, suggesting a decreased rate of differentiation in the absence of Mef2ca (Fig. 2C and data not shown). However, vmhc expression was indistinguishable from that in sibling and control wild type embryos (Fig. 2C and data not shown). Consistent with the view that FHF differentiation is retarded, smyd1b mRNA was not detected in the bilateral heart of a quarter of mef2ca+/- incross progeny at 13ss, but appeared normal by 17ss (Fig. 2C). By 24 hpf, mef2ca mutant hearts had normal shape and size as revealed by various sarcomeric genes and bmp4 mRNA accumulation (Fig. 3C), even though Mef2 protein was greatly reduced (Fig. S2B and (Hinits and Hughes, 2007)). At 48 hpf, mef2ca mutant embryos had normal gene expression and had undergone normal looping (Fig. S8A,B). Mutant inflow and outflow tracts (IFT and OFT) were well-formed and addition of new CMs in these locations occurred during the 2-3 dpf. At 5 dpf, mef2ca mutants had indistinguishable hearts from their siblings (data not shown). Moreover, we found no significant difference in heart rate between mef2ca mutants, mef2ca morphants and their respective control embryos (Fig. S8E). Taken together with mef2ca and mef2cb expression data and the dual loss of function results, these data suggest that either mef2cb is the main functional Mef2 gene in the heart, or that redundancy between Mef2ca and Mef2cb is sufficient to support a functional heart.
Mef2cb mutant is viable and shows no heart defects
We next tested the function of Mef2cb in zebrafish development. Unlike mef2ca mutants, at 15ss, mef2cb morphants had no change of myl7 or vmhc mRNA accumulation (Fig. 2D). Expression of smyd1b was also normal at both 13ss and 17ss (Fig. 2D). Considering mef2cb mRNA accumulation at both poles of 24 hpf hearts (Fig. 1I), we hypothesized an effect of lack of Mef2cb function on structures added to both poles. Indeed, Lazic and Scott, 2011 using an ATG-MO that has only a 4 bp overlap with ours, have reported that Mef2cb is necessary for ventricular development and for late CM addition at the arterial pole (Lazic and Scott, 2011). We also find that mef2cb morphants had smaller hearts at 24 hpf with both cardiac chambers appear defective (Fig. 3D). By 48 hpf, Mef2cb morphant hearts did not undergo looping, and their linear hearts showed mis-regulation various genes (Fig. S9A-C and data not shown). Other, non-myocardial derivatives, such as bulbus arteriosus that is marked by DAF-2 DA and Elastin (Grimes et al.; Grimes et al., 2006; Hami et al.; Lazic and Scott; Miao et al., 2007) were missing at 72 hpf (Fig. S9C,D). In order to test these results we analysed the TILLING mutant mef2cbfh288 (see earlier section). We did not observe any heart phenotype in embryos from various incrosses of mef2cb+/fh288 fish. Moreover, progeny from incrosses that were allowed to grow to adulthood revealed close to expected numbers of adult homozygous mutants mef2cbfh288/fh288 (5/14, 35% compared with 25% expected). These adults are healthy and able to breed. In agreement with these facts, genotyped mef2cbfh288/fh288 homozygous mutants had no change in myl7, vmhc or bmp4 mRNAs or MyHC protein accumulation at 15ss or 24 hpf (Figs 2E, 3E). Hearts were looped and expressed normal levels and pattern of myl7, vmhc and bmp4 mRNAs (Fig. 7A,B). The bulbus arteriosus also formed correctly and expressed Elastin (Fig. 7C). Thus, mef2cbfh288 mutants have normal hearts, at least under normal growing conditions
Figure 7. Loss of mef2cb function results in no heart phenotype.
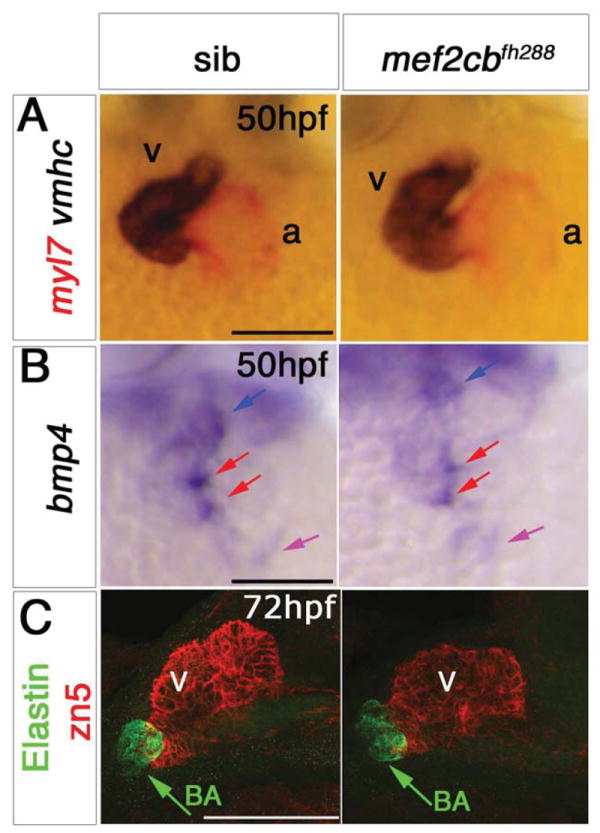
In situ mRNA hybridisation for myl7, vmhc and bmp4 (A,B, ventral view, anterior to top) and immunodetection of DM-GRASP (zn5) and Elastin (C, lateral view, anterior to left) of hearts at indicated stage of genotyped mef2cbfh288 mutants and their siblings. A,B. mef2cbfh288 mutants had a normal looped heart and normal expression of chamber markers, and bmp4 mRNA in OFT (blue arrow), IFT (purple arrow) and AV canal (red arrows). C. Confocal stacks of genotyped mef2cbfh288 mutants and siblings with a normal looped heart with developed chambers and bulbus arteriosus. Scale = 100 μm.
Discussion
The current work makes five major points. First, Mef2c homologues are essential for heart formation in zebrafish. Second, early Mef2 function during heart development is in a specific step of CM differentiation that follows specification. Third, Mef2ca and Mef2cb function redundantly to drive differentiation of both early and late added CMs. Fourth, both mef2ca and mef2cb single mutants have a seemingly normal heart. Fifth, Mef2cb, if mis-expressed outside the cardiogenic region during early embryonic development, has the ability to override cell fates and convert cells to skeletal muscle. These findings provide experimental confirmation of the long-speculated view that Mef2 activity is essential for vertebrate CM differentiation.
Mef2 role in sarcomeric muscle differentiation
Our data show that Mef2 activity is required for differentiation of all CMs of both FHF and the putative SHF of zebrafish. Sarcomeric gene expression was lost when the two Mef2c paralogues in zebrafish, mef2ca and mef2cb, were knocked down by morpholinos or in genetic null mutants. Mef2c does not act alone; sarcomeric genes such as myl7 and vmhc also require other transcription factors for their expression in CMs (Peterkin et al., 2007; Yelon et al., 2000). Nevertheless, to date, we have found no cardiac sarcomeric gene that is normally expressed without Mef2 activity. Expression of many other genes, including smyd1b, nppa and bmp4, that are involved in the maturation of CMs were also ablated after Mef2 loss of function. Mammalian orthologues of several of these genes are direct Mef2c targets, including Bop (Smyd1) and Anf (Nppa) (Morin et al., 2000; Phan et al., 2005; Zang et al., 2004). Bmp4 is suggested to function by increasing myofibrillar gene expression, turning cardioblasts into beating CMs (Tirosh-Finkel et al., 2010). Our findings place Mef2 function in CM differentiation at a step that precedes Bmp4 expression, although it is not clear if bmp4 is a direct Mef2c target.
Mef2c is clearly required at a later step than Nkx2.5, GATA and Hand2 transcription factors. Cardiac precursors lacking both Mef2ca and Mef2cb express nkx2.5 until the point of differentiation into contractile CMs. When differentiation fails, nkx2.5 expression is lost, suggesting that CM precursors do not simply remain in an immature state. Other manipulations, that prevent differentiation of CMs, such as ectopic expression of dominant negative BMP receptors or gata6 MO, also specifically block late nkx2.5 expression (Peterkin et al., 2003; Shi et al., 2000). Combined loss of function of GATA factors prevents CM specification prior to mef2ca expression, whereas lack of GATA5 or GATA6 alone causes cardia bifida (Holtzinger and Evans, 2007; Peterkin et al., 2003, 2007). Loss of function of hand2, the only Hand factor in the current zebrafish genome, does not prevent mef2ca and mef2cb expression but results in cardia bifida with reduced CM number (Yelon et al., 2000). In contrast, heart cells lacking Mef2 are specified and migrate normally to the midline, but fail to differentiate. Mef2ca;mef2cb dual loss of function embryos retain high levels of hand2 mRNA in the cardiogenic region, suggesting a developmental arrest at a pre-differentiation step involving Hand2 expression. Loss of Mef2c in the mouse, on the other hand, leads to a mild delay in hand1 and absence of late hand2 expression (Lin et al., 1997b; Vong et al., 2006). As far as we are aware, early Hand2 expression remains to be investigated in Mef2c null mice. The up-regulation of both hand2 and the endocardial marker cdh5 in Mef2 loss of function zebrafish embryos suggests that failure of CM differentiation has effects beyond myocardium.
The mef2d/c morpholino, which targets mef2d, both mef2cs and possibly mef2aa and mef2b (Hinits and Hughes, 2007), causes a complete loss of cardiomyogenesis, showing that Mef2 is essential for CM differentiation. The stronger effect of mef2d/c MO when compared with other dual loss of function mutant or morphant combinations, which ablate most but not all CMs, strongly suggests that other Mef2 proteins are responsible for the residual cardiomyogenesis in mef2ca;mef2cb double mutants. Strong evidence supporting this view is that the residual CMs in double mutants have Mef2, but not Mef2c, immunoreactivity. Indeed, mef2aa mRNA is detected in the residual myocardial regions of double mutants (data not shown). We conclude that mef2d/c MO is likely to knock down Mef2aa activity sufficiently to prevent CM differentiation, despite six bases of mismatch. Moreover, Mef2aa appears, like the Mef2cs, to be able to drive CM differentiation.
Mef2-dependency of zebrafish myocardium is remarkably similar to that in Drosophila. The single Mef2 factor in Drosophila, D-Mef2, is essential for expression of sarcomeric genes in the fly heart in the dorsal vessel, but is not required for expression of specification genes. Moreover, the dorsal vessel is more sensitive to the lack of D-Mef2 than is body wall (i.e., skeletal) muscle (Lilly et al., 1995; Ranganayakulu et al., 1995). In zebrafish skeletal muscle, Myogenic Regulatory Factors (MRFs) drive expression of some sarcomeric genes after knockdown of Mef2 activity (Hinits and Hughes, 2007; Hinits et al., 2009; Hinits et al., 2011). Thus, cardiac and skeletal myocytes may differ in Mef2 dependency for terminal differentiation. Nevertheless, both in skeletal and cardiac muscle, Mef2c expression parallels differentiation of the contractile phenotype (Hinits and Hughes, 2007 ; Potthoff et al., 2007). Our findings suggest a deep conservation of function of Mef2 in heart development through animal evolution and contrast with the divergence of function of MRFs between vertebrates and Drosophila.
Redundant roles for Mef2ca and Mef2cb during cardiac development
The FHF had early maturation defects in mef2ca mutants that were not observed in mef2cb morphants. Both mef2ca probable null alleles (Miller et al., 2007) and morphants showed a short developmental delay affecting some sarcomeric and maturation genes in the FHF. Mef2ca deficient hearts recovered well, but whether they are normal in all respects, for example under stress, remains to be determined.
The recovery of FHF cardiomyogenesis suggests that Mef2ca and Mef2cb are redundant during FHF development, but indicates a more prominent role for Mef2ca in FHF development. At 24 hpf, we detect strong mef2cb mRNA at both the arterial and venous poles, A similar pattern was observed in Atlantic cod, Gadus morhua, that has two Mef2c paralogues, each expressed strongly in a single cardiac pole at heart tube stage (Torgersen et al.). This is where SHF precursors reside (de Pater et al., 2009; Zhou et al., 2011). The mef2cb expression pattern suggests a more prominent role at the cardiac poles. Lazic and Scott (2011) described an arterial pole defect using mef2cb MO; we found the same result with our distinct mef2cb MO. However, the likely null mef2cbfh288 mutant has an apparently normal heart and grows well to a viable and fertile adult. This shows that although distinct regulatory elements cause distinct expression of each gene, a genetic redundancy of Mef2ca and Mef2cb function exist for both early/central FHF and later-added/polar SHF CMs.
How can one explain the cardiac pole-specific defects in mef2cb morphants? Clearly, the MOs work to knockdown Mef2cb protein, as Mef2c immunreactivity is ablated when they are used in combination with the mef2ca mutant. Moreover, the absence of Mef2c protein and the early stop codon in the mef2cb mutant make it highly likely to be null. We hypothesise that the cardiac pole defects arise from a non-specific effect of the MO added to a loss of Mef2cb function that preferentially sensitizes CMs at the cardiac poles.
Given time, the residual CMs in double mef2ca;mef2cb mutants can reach their normal position, aggregate, take on ventricular or atrial character, form small tubular structures, beat and even recruit non-myocardial cells into bulbus arteriosus. As discussed above, Mef2aa may contribute towards this residual differentiation. Mef2aa is expressed later than the two Mef2cs in zebrafish heart, and we detected mef2aa mRNA only in differentiated CMs. Consistent with this, Mef2aa knockdown causes late cardiac defects relating to contractility (Wang et al., 2005). So late recovery of CMs in double mutants may reflect independent activation of mef2aa. Alternatively, other factors such as GATAs and Hand2 may be sufficient to slowly rescue a few cells to CM differentiation. In mouse, Mef2a (and Mef2d) expression begins only at the heart tube stage, whereas Mef2c (and Mef2b) are expressed in pre-cardiac mesoderm, (Edmondson et al., 1994). We speculate that redundancy between Mef2c and Mef2b may explain the ability of CMs in early heart development of Mef2c knockout mice. Mef2a null mice exhibit perinatal lethality from an array of cardiovascular defects (Ewen et al., 2011; Naya et al., 2002). Thus, expression and function of zebrafish Mef2c and Mef2a orthologues appear conserved between fish and mammals.
Mef2cb can drive ectopic skeletal myogenesis
Our data show that over-expression of Mef2cb leads to ectopic expression of both cardiac and skeletal muscle genes. Extra Mef2cb in its normal location, induced by BAC injection, leads to ectopic CMs within the heart field, but not elsewhere. Strikingly, however, the entire cardiogenic region appears able to up-regulate bmp4 expression in response to Mef2cb, when it is introduced early by mef2cb mRNA injection, but does not undergo extensive ectopic cardiomyogenesis. This finding suggests that the ability of Mef2 to drive CM differentiation is under tight additional controls.
Elsewhere in the embryo, particularly, but not exclusively, in head mesoderm, mef2cb mRNA over-expression causes high levels of conversion of cells to skeletal muscle. This effect is efficient, comparable to that of myod or mrf4 mRNA injection (Hinits et al., 2009; Osborn et al., 2011), and can lead to loss of myocardial markers, possibly through conversion of early cranial mesoderm to skeletal muscle before it attains the character of the cardiogenic region. The distinct action of Mef2cb over-expression in the cardiogenic and other regions of the embryo suggests that the activity of Mef2cb within a cell is strongly influenced by other signals/molecules involved in head mesoderm patterning ((Tzahor and Evans, 2011) and others).
Our findings indicate a potential role for Mef2 as a skeletal muscle determination factor, and is reminscent of the observation that MEF2A can initiate skeletal myogenesis in some circumstances (Kaushal et al., 1994). However, other studies have shown that Mef2c alone is unable to activate the myogenic program, but requires Myod or another MRF (Black et al., 1998; Molkentin et al., 1995). As no MRFs are expressed in the zebrafish head region prior to 24 hpf, it appears that Mef2cb can trigger the MRF expression required for efficient enhancement of terminal muscle differentiation by Mef2s (della Gaspera et al., 2009; Molkentin et al., 1995). Indeed, Xenopus Mef2a can activate the Myod promoter (Wong et al., 1994). In Drosophila, Dmef2 is a key regulator of body wall striated myogenesis and does not require on MRF (Gunthorpe et al., 1999; Lin et al., 1997a). In C. elegans, by contrast, Mef2 is dispensible for myogenesis (Dichoso et al., 2000). It remains to be established whether the ability of Mef2cb is shared by other Mef2 factors and/or plays a role during development of any vertebrate skeletal muscle in the wild type situation.
Supplementary Material
Figure S1. Mef2cb mRNA and protein are upregulated after mef2cb over-expression.
A-D. In situ hybridisation for mef2cb mRNA in controls or embryos injected with BAC DNA containing the mef2cb locus shown in flatmounts, lateral view, anterior to left (A), transverse section through the somites (B), wholemounts in dorsal view, anterior to top (C,D). Cells with high levels of mef2cb are indicated with arrows in muscle fibres (A,B), head vasculature (C), heart (D) and telencephalon (white arrows, C). E. Mef2 immunoreactivity is chimerically strong (yellow arrows) in somitic cells after injection of mef2cb BAC DNA. F. Mef2ca/cb and GFP immunoreactivity are ectopically strong and co-localise in fibres (white arrowhead) in the first somite (asterisk), and in cells outside the somite and in the head (white arrows) after injection of hs-mef2cb-IRES-GFP plasmid DNA followed by heat shock activation, shown in dorsal view, anterior to top. G. Ectopic muscle fibres are forming in the head region of embryos injected with 20pg mef2cb mRNA and immunoreact with Mef2ca/cb and MyHC. Scale = 100 μm (except in E,G= 20 μm).
Figure S2. Mef2c protein is downregulated in mef2ca mutants.
Immunodetection with antibodies to Mef2ca/cb (Anaspec, red in A-C, blue in D), general Mef2 (Santa Cruz, green, E), MyHC (red, D) and zn-5 (red, E) and DAPI (blue, A,C) in confocal stacks of wildtype, and mef2ca mutant embryos. A. Reaction with Mef2c antibody is strong in nuclei of differentiated slow muscle fibres in the somite (left panel), but is missing from the PSM (right panel), and from cranial ganglia indicating that the antibody is not cross-reacting with Mef2d and Mef2a protein, respectively. B,C. Anti-Mef2c antibody reacts weakly in CMs of mef2cab1086 mutant embryos (B), but is absent from nuclei in the myotome of the somites (C). Scale = 50μm (except in A=100 μm).
Figure S3. Mef2cb morpholinos are specific and efficient in targeting translation and splicing of mef2cb.
A,B. mef2cb ATG MO specifically blocked translation of mef2cb-GFP mRNA. Representative embryos (A) and quantification (B) of mosaic GFP accumulation in 24 hpf embryos injected with pCMV:mef2cb-GFP, with or without MOs. Embryos injected with plasmid alone or with plasmid plus control myog MO, have numerous cells with strong GFP. In contrast, embryos injected with plasmid plus mef2cb ATG MO had little if any GFP expression. C. Schematic diagram of 5’UTR (white) and ORF (black) of Exons 1-3 in mef2cb mRNA and RT-PCR strategy used to detect spliced mRNAs in control and mef2cb E1I1 MO-injected embryos (top left). Gel showing RT-PCR of mRNA from 24 hpf uninjected control and mef2cb E1I1 MO-injected embryos (two independent samples each, top right). Morphant cDNA had strong reduction of the normal splice form (nor; 321 bp) and the appearance of a large aberrant band (ab; 403 bp). The aberrant transcript (bottom) results in a premature stop codon. Exon (upper case) and intron sequences (lower case) are coloured (exon1, blue; intron1, orange; exon2, green; exon3, purple). Primer sequences are underlined. Amino acid sequence of the aberrant CDS is shown below the nucleotide sequence leading to a stop codon in the first half of the MADS domain. Scale = 100 μm.
Figure S4. Endothelial markers are little affected by Mef2 knockdown.
In situ mRNA hybridisation for mef2cb (A), kdrl (flk-1, B,C), myl7 (C, red) and cdh5 (E) or immunodetection of MyHC and GFP (D) in wild type or Tg(fli1:GFP) control or injected with mef2d/c MO (except C; mef2cab1086;mef2cbfh288 and sibling embryos). A.Mef2cb mRNA detected in heart (blue arrowhead), telencephalon (white arrowheads), and head vasculature (yellow arrows). B,C.Kdrl expression in mef2d/c morphant and double mef2ca;mef2cb mutant embryos at 21s is not changed compare with control and sibling embryos, respectively. D. Expression of endothelial marker fli1:GFP at 24 hpf is similar in control and mef2d/c morphants in the heart and head region (upper panels), but some defects in intersomitic vessels of morphants (lower panels) parallel the somitic muscle MyHC phenotype previously described (Hinits and Hughes, 2007). E. Mild upregulation of cdh5 mRNA in head and heart region (upper panels) and in the trunk region (lower panels) at 24 hpf. lda, lateral dorsal aorta; pmbc, primordial midbrain channel; da, dorsal aorta; pcv, posterior cardinal vein; se, intersegmental vessel. Scale = 100 μm.
Figure S5. Mef2ca and Mef2cb dual loss of function abolishes myocardial differentiation.
In situ mRNA hybridisation for myl7 (A), vmhc (B), bmp4 (C), smyd1b (D) and gata4,5,6 (E) in hearts of zebrafish embryos shown in a dorsal view, anterior to top (A-C,E) or in lateral view, anterior to left (D). A,B.Myl7 and vmhc expression in hearts (arrowheads) is lost after dual loss of function of Mef2ca and Mef2cb by various combinations of MOs and mutants. C.Bmp4 is dramatically reduced from heart of 22s mef2d/c morphants. D.Smyd1b mRNA is abolished from hearts (arrowheads) of 22s mef2d/c morphants. Expression in somites is slightly downregulated (arrow). E.Gata4, gata5 and gata6 appear unchange in 22s mef2d/c morphants. Scale = 100 μm.
Figure S6. Mef2ca and Mef2cb dual loss of function abolishes myocardial differentiation.
A,B. In situ mRNA hybridisation for myl7 in hearts of 24 hpf mef2cab1086;mef2cbfh288 mutant embryos and their siblings (A) or control and mef2cb+/fh288 incross embryos non-injected or injected with mef2ca MO shown in a dorsal view, anterior to top. Double mutants have almost no myl7 expression albeit few cells in the cardiac region (arrows, A). Mef2cbfh288 mutant embryos injected with mef2ca MO have a thinner than normal heart tube. All other embryos with only one or no Mef2s depleted have a normal heart. C. Bright field images of mef2cbfh288 mutant embryos injected with mef2ca MO showing cardiac edema, compared with mef2ca MO injected siblings with normal heart and no edema. Scale = 100 μm.
Figure S7. Mef2ca and Mef2cb are expressed in hand2 knockdown embryos.
In situ mRNA hybridisation for mef2ca (A) and mef2cb (B) in hearts of control, and hand2 MO zebrafish embryos shown in a dorsal view, anterior to top. A,B. Hearts of hand2 morphants show a cardia bifida phenotype (arrows). CMs on both sides of the midline express both mef2ca and mef2cb. Expression of mef2ca in the branchial arches is marked with asterisks. Scale = 100 μm.
Figure S8. Mef2ca mutant has a normal heart.
A,B. Double mRNA in situ hybridisation for myl7 (red, A,B), vmhc (blue, A) and bmp4 (blue, B) in genotyped 48 hpf mef2ca mutant embryos and their siblings shown in ventral view, anterior to top. Mutants have normal looping and gene expression is no different than siblings. OFT, blue arrow, IFT, purple arrow, AVC, red arrows. C. Confocal stacks of 72 hpf mef2cab1086;Tg(myl7:GFP) mutants and their siblings and of Tg(myl7:GFP) injected with mef2ca MO immunostained for Elastin showing no difference between morphants, mutants and siblings. D. GFP+dsRED- CMs can only be found in OFT and IFT of the heart (dotted line) of mef2cab1086;Tg(myl7:GFP;myl7;ndsRed) mutants and their siblings at 60 hpf. E. Heart rate (bpm) of control wild type, mef2ca morphants, mef2catn213 and their siblings at 25-48 hpf. Embryos were grown in 24-well plates and genotype was determined at 5 dpf according to jaw phenotype (see (Piotrowski et al., 1996)). No significant differences according to t-test statistics were found at any stage between morphants/mutants and control/siblings. Number of embryos per condition is shown on columns. Variation in heart rate between experiments is due to differences in temperature. Scale = 100 μm.
Figure S9. Mef2cb MO results in linear defective heart and loss of bulbus arteriosus.
In situ mRNA hybridisation for indicated genes (A,B, ventral view, anterior to top) and immunodetection of EGFP, MyHC (MF20), Elastin and DAF-2DA detection (C,D, lateral view, anterior to left) of hearts at indicated stage of mef2cb morphants and controls. A,B.Mef2cb morphants have a linear unlooped hearts and no obvious AV canal as marked by bmp4 expression (red arrows). C,D. Confocal stacks of control or mef2cb MO-injected Tg(myl7:EGFP) (E) or wild-type (F) embryos at 72 hpf. Hearts are unlooped and bulbus arteriosus markers (BA, arrows) are absent in mef2cb morphants. Insets in D show YZ sections at the OFT region, indicating to the area bordering myocardial and smooth muscle cells. sh, sternohyoideus; cl, cleithrum; V, ventricle; A, atrium, IFT, inflow tract; OFT, outflow tract; AVC, atrioventricular canal. Scale = 100μm, except for C,D=50 μm.
Table S1. Quantification of effects of loss- and gain-of-function.
Highlights.
Mef2c homologues are essential for heart formation in zebrafish.
Mef2ca and Mef2cb function redundantly to drive differentiation of FHF and SHF CMs.
mef2ca and mef2cb single mutants have a seemingly normal heart.
Mef2cb overexpression during early embryonic development converts cells into skeletal muscle.
Acknowledgments
SMH is a member of MRC scientific staff with Programme Grant support. Funding was from MRC and the British Heart Foundation. Identification of the mef2cbfh288 mutant was supported by NIH grant R01HG002995 to C.B. Moens and NIH grants DE13834 and HD22486 to C.B. Kimmel. We thank Charles B. Kimmel and members of his laboratory for mutant fish lines and communication of unpublished data. We thank R. Hampson, C. L. Hammond, D. Yelon, T. Evans, G. Burns, M. Miao, F.W. Keeley, A. Rodaway, R. Patient, P. Ingham, E. Ehler, S.J. Du, J.C McDermott and P. Riley for reagents and advice. We thank Massimo Ganassi and Susanna Molinari, University of Modena, Italy for their work in cloning the full length mef2cb.
Footnotes
Competing interests
Neither author has any competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn DG, Ruvinsky I, Oates AC, Silver LM, Ho RK. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95:253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Begemann G, Ingham PW. Developmental regulation of Tbx5 in zebrafish embryogenesis. Mech Dev. 2000;90:299–304. doi: 10.1016/s0925-4773(99)00246-4. [DOI] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Bi W, Drake CJ, Schwarz JJ. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol. 1999;211:255–267. doi: 10.1006/dbio.1999.9307. [DOI] [PubMed] [Google Scholar]
- Black BL, Molkentin JD, Olson EN. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Molecular & Cellular Biology. 1998;18:69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by Sonic Hedgehog. Genes Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart RE, Liang CS, Smoot LB, Laheru DA, Mahdavi V, Nadal-Ginard B. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095–1106. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater E, Clijsters L, Marques SR, Lin YF, Garavito-Aguilar ZV, Yelon D, Bakkers J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136:1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DY, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Gaspera B, Armand AS, Sequeira I, Lecolle S, Gallien CL, Charbonnier F, Chanoine C. The Xenopus MEF2 gene family: evidence of a role for XMEF2C in larval tendon development. Dev Biol. 2009;328:392–402. doi: 10.1016/j.ydbio.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Dichoso D, Brodigan T, Chwoe KY, Lee JS, Llacer R, Park M, Corsi AK, Kostas SA, Fire A, Ahnn J, Krause M. The MADS-Box factor CeMEF2 is not essential for Caenorhabditis elegans myogenesis and development. Dev Biol. 2000;223:431–440. doi: 10.1006/dbio.2000.9758. [DOI] [PubMed] [Google Scholar]
- Draper BW, McCallum CM, Stout JL, Slade AJ, Moens CB. A high-throughput method for identifying N-ethyl-N-nitrosourea (ENU)-induced point mutations in zebrafish. Methods in cell biology. 2004;77:91–112. doi: 10.1016/s0091-679x(04)77005-3. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- Ewen EP, Snyder CM, Wilson M, Desjardins D, Naya FJ. Mef2A coordinately regulates a costamere gene program in cardiac muscle. J Biol Chem. 2011;286:29644–53. doi: 10.1074/jbc.M111.268094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes AC, Duran AC, Sans-Coma V, Hami D, Santoro MM, Torres M. Phylogeny informs ontogeny: a proposed common theme in the arterial pole of the vertebrate heart. Evol Dev. 12:552–567. doi: 10.1111/j.1525-142X.2010.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes AC, Duran AC, Sans-Coma V, Hami D, Santoro MM, Torres M. Phylogeny informs ontogeny: a proposed common theme in the arterial pole of the vertebrate heart. Evolution & development. 2010;12:552–567. doi: 10.1111/j.1525-142X.2010.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes AC, Stadt HA, Shepherd IT, Kirby ML. Solving an enigma: arterial pole development in the zebrafish heart. Dev Biol. 2006;290:265–276. doi: 10.1016/j.ydbio.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Gunthorpe D, Beatty KE, Taylor MV. Different levels, but not different isoforms, of the Drosophila transcription factor DMEF2 affect distinct aspects of muscle differentiation. Developmental biology. 1999;215:130–145. doi: 10.1006/dbio.1999.9449. [DOI] [PubMed] [Google Scholar]
- Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 138:2389–2398. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hami D, Grimes AC, Tsai HJ, Kirby ML. Zebrafish cardiac development requires a conserved secondary heart field. Development. 2011;138:2389–2398. doi: 10.1242/dev.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134:2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Osborn DP, Hughes SM. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development. 2009;136:403–414. doi: 10.1242/dev.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Williams VC, Sweetman D, Donn TM, Ma TP, Moens CB, Hughes SM. Defective cranial skeletal development, larval lethality and haploinsufficiency in Myod mutant zebrafish. Developmental biology. 2011;358:102–112. doi: 10.1016/j.ydbio.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev Biol. 2007;312:613–622. doi: 10.1016/j.ydbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamboulas C, Dakubo GD, Liu J, De Repentigny Y, Yutzey K, Wallace VA, Kothary R, Skerjanc IS. Disruption of MEF2 activity in cardiomyoblasts inhibits cardiomyogenesis. J Cell Sci. 2006;119:4315–4321. doi: 10.1242/jcs.03186. [DOI] [PubMed] [Google Scholar]
- Kaushal S, Schneider JW, Nadal-Ginard B, Mahdavi V. Activation of the myogenic lineage by MEF2A, a factor that induces and cooperates with MyoD. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- Larson JD, Wadman SA, Chen E, Kerley L, Clark KJ, Eide M, Lippert S, Nasevicius A, Ekker SC, Hackett PB, Essner JJ. Expression of VE-cadherin in zebrafish embryos: a new tool to evaluate vascular development. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;231:204–213. doi: 10.1002/dvdy.20102. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Developmental biology. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- Lazic S, Scott IC. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Dev Biol. doi: 10.1016/j.ydbio.2011.03.028. [DOI] [PubMed] [Google Scholar]
- Lazic S, Scott IC. Mef2cb regulates late myocardial cell addition from a second heart field-like population of progenitors in zebrafish. Developmental biology. 2011;354:123–133. doi: 10.1016/j.ydbio.2011.03.028. [DOI] [PubMed] [Google Scholar]
- Lee KH, Xu Q, Breitbart RE. A new tinman-related gene, nkx2.7, anticipates the expression of nkx2.5 and nkx2.3 in zebrafish heart and pharyngeal endoderm. Dev Biol. 1996;180:722–731. doi: 10.1006/dbio.1996.0341. [DOI] [PubMed] [Google Scholar]
- Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin MH, Bour BA, Abmayr SM, Storti RV. Ectopic expression of MEF2 in the epidermis induces epidermal expression of muscle genes and abnormal muscle development in Drosophila. Developmental biology. 1997a;182:240–255. doi: 10.1006/dbio.1996.8484. [DOI] [PubMed] [Google Scholar]
- Lin Q, Lu J, Yanagisawa H, Webb R, Lyons GE, Richardson JA, Olson EN. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997b;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- Maves L, Tyler A, Moens CB, Tapscott SJ. Pbx acts with Hand2 in early myocardial differentiation. Developmental biology. 2009;333:409–418. doi: 10.1016/j.ydbio.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott JC, Cardoso MC, Yu YT, Andres V, Leifer D, Krainc D, Lipton SA, Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Molecular & Cellular Biology. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M, Bruce AE, Bhanji T, Davis EC, Keeley FW. Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 2007;26:115–124. doi: 10.1016/j.matbio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev Biol. 2007;308:144–157. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin JE, Hughes SM. Sequential actions of Pax3 and Pax7 drive xanthophore development in zebrafish neural crest. Dev Biol. 2008;317:508–522. doi: 10.1016/j.ydbio.2008.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt CH, Nakaoka T, Moreno-Rodriguez R, Norris RA, Kern MJ, Eisenberg CA, Turner D, Markwald RR. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238:97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Molecular and Cellular Biology. 1996a;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Firulli AB, Black BL, Martin JF, Hustad CM, Copeland N, Jenkins N, Lyons G, Olson EN. MEF2B is a potent transactivator expressed in early myogenic lineages. Molecular and Cellular Biology. 1996b;16:3814–3824. doi: 10.1128/mcb.16.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin S, Charron F, Robitaille L, Nemer M. GATA-dependent recruitment of MEF2 proteins to target promoters. Embo J. 2000;19:2046–2055. doi: 10.1093/emboj/19.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;15:15. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- Osborn DP, Li K, Hinits Y, Hughes SM. Cdkn1c drives muscle differentiation through a positive feedback loop with Myod. Developmental biology. 2011;350:464–475. doi: 10.1016/j.ydbio.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxtoby E, Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Research. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. Embo J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311:623–635. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan D, Rasmussen TL, Nakagawa O, McAnally J, Gottlieb PD, Tucker PW, Richardson JA, Bassel-Duby R, Olson EN. BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development. 2005;132:2669–2678. doi: 10.1242/dev.01849. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Schilling TF, Brand M, Jiang YJ, Heisenberg CP, Beuchle D, Grandel H, Van Eeden FJM, FurutaniSeiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Warga RM, NussleinVolhard C. Jaw and branchial arch mutants in zebrafish. II: Anterior arches and cartilage differentiation. Development. 1996;123:345–356. doi: 10.1242/dev.123.1.345. [DOI] [PubMed] [Google Scholar]
- Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Devel. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Arnold MA, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol Cell Biol. 2007;27:8143–8151. doi: 10.1128/MCB.01187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Developmental Biology. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104:933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Oates AC, Silver LM, Ho RK. The evolution of paired appendages in vertebrates: T-box genes in the zebrafish. Dev Genes Evol. 2000;210:82–91. doi: 10.1007/s004270050014. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Ho RK, Herrmann BG, Nusslein-Volhard C. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development. 1992;116:1021–1032. doi: 10.1242/dev.116.4.1021. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Chen JN, Fishman MC. Regulation in the heart field of zebrafish. Development. 1998;125:1095–1101. doi: 10.1242/dev.125.6.1095. [DOI] [PubMed] [Google Scholar]
- Shi Y, Katsev S, Cai C, Evans S. BMP signaling is required for heart formation in vertebrates. Dev Biol. 2000;224:226–237. doi: 10.1006/dbio.2000.9802. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Ransom DG, Pratt SJ, MacLennan H, Kieran MW, Detrich HW, 3rd, Vail B, Huber TL, Paw B, Brownlie AJ, Oates AC, Fritz A, Gates MA, Amores A, Bahary N, Talbot WS, Her H, Beier DR, Postlethwait JH, Zon LI. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Developmental biology. 1998;197:248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 http://zfin.org.
- Ticho BS, Stainier DY, Fishman MC, Breitbart RE. Three zebrafish MEF2 genes delineate somitic and cardiac muscle development in wild-type and mutant embryos. Mech Dev. 1996;59:205–218. doi: 10.1016/0925-4773(96)00601-6. [DOI] [PubMed] [Google Scholar]
- Tirosh-Finkel L, Zeisel A, Brodt-Ivenshitz M, Shamai A, Yao Z, Seger R, Domany E, Tzahor E. BMP-mediated inhibition of FGF signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development. 2010;137:2989–3000. doi: 10.1242/dev.051649. [DOI] [PubMed] [Google Scholar]
- Torgersen JS, Takle H, Andersen O. Differential spatial expression of mef2 paralogs during cardiac development in Atlantic cod (Gadus morhua) Comp Biochem Physiol B Biochem Mol Biol. 158:181–187. doi: 10.1016/j.cbpb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Tzahor E, Evans SM. Pharyngeal mesoderm development during embryogenesis: implications for both heart and head myogenesis. Cardiovascular research. 2011;91:196–202. doi: 10.1093/cvr/cvr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- Vong L, Bi W, O’Connor-Halligan KE, Li C, Cserjesi P, Schwarz JJ. MEF2C is required for the normal allocation of cells between the ventricular and sinoatrial precursors of the primary heart field. Dev Dyn. 2006;235:1809–1821. doi: 10.1002/dvdy.20828. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski DH, Wallis KT, Stadt HA, Hutson MR, Platt DH, Kirby ML. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128:3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- Walsh EC, Stainier DY. UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science. 2001;293:1670–1673. doi: 10.1126/science.293.5535.1670. [DOI] [PubMed] [Google Scholar]
- Wang YX, Qian LX, Yu Z, Jiang Q, Dong YX, Liu XF, Yang XY, Zhong TP, Song HY. Requirements of myocyte-specific enhancer factor 2A in zebrafish cardiac contractility. FEBS Lett. 2005;579:4843–4850. doi: 10.1016/j.febslet.2005.07.068. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book - a guide for the laboratory use of zebrafish (Danio rerio) 3. University of Oregon Press; 1995. [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wong MW, Pisegna M, Lu MF, Leibham D, Perry M. Activation of Xenopus MyoD transcription by members of the MEF2 protein family. Developmental Biology. 1994;166:683–695. doi: 10.1006/dbio.1994.1347. [DOI] [PubMed] [Google Scholar]
- Xu Y, He J, Wang X, Lim TM, Gong Z. Asynchronous activation of 10 muscle-specific protein (MSP) genes during zebrafish somitogenesis. Dev Dyn. 2000;219:201–215. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1043>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Yu YT, Breitbart RE, Smoot LB, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue- restricted MADS box transcription factors. Genes & Development. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- Zang MX, Li Y, Xue LX, Jia HT, Jing H. Cooperative activation of atrial naturetic peptide promoter by dHAND and MEF2C. J Cell Biochem. 2004;93:1255–1266. doi: 10.1002/jcb.20225. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, Gu A, Mosimann C, Sondalle S, Peterson RE, Heideman W, Burns CE, Burns CG. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474:645–648. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mef2cb mRNA and protein are upregulated after mef2cb over-expression.
A-D. In situ hybridisation for mef2cb mRNA in controls or embryos injected with BAC DNA containing the mef2cb locus shown in flatmounts, lateral view, anterior to left (A), transverse section through the somites (B), wholemounts in dorsal view, anterior to top (C,D). Cells with high levels of mef2cb are indicated with arrows in muscle fibres (A,B), head vasculature (C), heart (D) and telencephalon (white arrows, C). E. Mef2 immunoreactivity is chimerically strong (yellow arrows) in somitic cells after injection of mef2cb BAC DNA. F. Mef2ca/cb and GFP immunoreactivity are ectopically strong and co-localise in fibres (white arrowhead) in the first somite (asterisk), and in cells outside the somite and in the head (white arrows) after injection of hs-mef2cb-IRES-GFP plasmid DNA followed by heat shock activation, shown in dorsal view, anterior to top. G. Ectopic muscle fibres are forming in the head region of embryos injected with 20pg mef2cb mRNA and immunoreact with Mef2ca/cb and MyHC. Scale = 100 μm (except in E,G= 20 μm).
Figure S2. Mef2c protein is downregulated in mef2ca mutants.
Immunodetection with antibodies to Mef2ca/cb (Anaspec, red in A-C, blue in D), general Mef2 (Santa Cruz, green, E), MyHC (red, D) and zn-5 (red, E) and DAPI (blue, A,C) in confocal stacks of wildtype, and mef2ca mutant embryos. A. Reaction with Mef2c antibody is strong in nuclei of differentiated slow muscle fibres in the somite (left panel), but is missing from the PSM (right panel), and from cranial ganglia indicating that the antibody is not cross-reacting with Mef2d and Mef2a protein, respectively. B,C. Anti-Mef2c antibody reacts weakly in CMs of mef2cab1086 mutant embryos (B), but is absent from nuclei in the myotome of the somites (C). Scale = 50μm (except in A=100 μm).
Figure S3. Mef2cb morpholinos are specific and efficient in targeting translation and splicing of mef2cb.
A,B. mef2cb ATG MO specifically blocked translation of mef2cb-GFP mRNA. Representative embryos (A) and quantification (B) of mosaic GFP accumulation in 24 hpf embryos injected with pCMV:mef2cb-GFP, with or without MOs. Embryos injected with plasmid alone or with plasmid plus control myog MO, have numerous cells with strong GFP. In contrast, embryos injected with plasmid plus mef2cb ATG MO had little if any GFP expression. C. Schematic diagram of 5’UTR (white) and ORF (black) of Exons 1-3 in mef2cb mRNA and RT-PCR strategy used to detect spliced mRNAs in control and mef2cb E1I1 MO-injected embryos (top left). Gel showing RT-PCR of mRNA from 24 hpf uninjected control and mef2cb E1I1 MO-injected embryos (two independent samples each, top right). Morphant cDNA had strong reduction of the normal splice form (nor; 321 bp) and the appearance of a large aberrant band (ab; 403 bp). The aberrant transcript (bottom) results in a premature stop codon. Exon (upper case) and intron sequences (lower case) are coloured (exon1, blue; intron1, orange; exon2, green; exon3, purple). Primer sequences are underlined. Amino acid sequence of the aberrant CDS is shown below the nucleotide sequence leading to a stop codon in the first half of the MADS domain. Scale = 100 μm.
Figure S4. Endothelial markers are little affected by Mef2 knockdown.
In situ mRNA hybridisation for mef2cb (A), kdrl (flk-1, B,C), myl7 (C, red) and cdh5 (E) or immunodetection of MyHC and GFP (D) in wild type or Tg(fli1:GFP) control or injected with mef2d/c MO (except C; mef2cab1086;mef2cbfh288 and sibling embryos). A.Mef2cb mRNA detected in heart (blue arrowhead), telencephalon (white arrowheads), and head vasculature (yellow arrows). B,C.Kdrl expression in mef2d/c morphant and double mef2ca;mef2cb mutant embryos at 21s is not changed compare with control and sibling embryos, respectively. D. Expression of endothelial marker fli1:GFP at 24 hpf is similar in control and mef2d/c morphants in the heart and head region (upper panels), but some defects in intersomitic vessels of morphants (lower panels) parallel the somitic muscle MyHC phenotype previously described (Hinits and Hughes, 2007). E. Mild upregulation of cdh5 mRNA in head and heart region (upper panels) and in the trunk region (lower panels) at 24 hpf. lda, lateral dorsal aorta; pmbc, primordial midbrain channel; da, dorsal aorta; pcv, posterior cardinal vein; se, intersegmental vessel. Scale = 100 μm.
Figure S5. Mef2ca and Mef2cb dual loss of function abolishes myocardial differentiation.
In situ mRNA hybridisation for myl7 (A), vmhc (B), bmp4 (C), smyd1b (D) and gata4,5,6 (E) in hearts of zebrafish embryos shown in a dorsal view, anterior to top (A-C,E) or in lateral view, anterior to left (D). A,B.Myl7 and vmhc expression in hearts (arrowheads) is lost after dual loss of function of Mef2ca and Mef2cb by various combinations of MOs and mutants. C.Bmp4 is dramatically reduced from heart of 22s mef2d/c morphants. D.Smyd1b mRNA is abolished from hearts (arrowheads) of 22s mef2d/c morphants. Expression in somites is slightly downregulated (arrow). E.Gata4, gata5 and gata6 appear unchange in 22s mef2d/c morphants. Scale = 100 μm.
Figure S6. Mef2ca and Mef2cb dual loss of function abolishes myocardial differentiation.
A,B. In situ mRNA hybridisation for myl7 in hearts of 24 hpf mef2cab1086;mef2cbfh288 mutant embryos and their siblings (A) or control and mef2cb+/fh288 incross embryos non-injected or injected with mef2ca MO shown in a dorsal view, anterior to top. Double mutants have almost no myl7 expression albeit few cells in the cardiac region (arrows, A). Mef2cbfh288 mutant embryos injected with mef2ca MO have a thinner than normal heart tube. All other embryos with only one or no Mef2s depleted have a normal heart. C. Bright field images of mef2cbfh288 mutant embryos injected with mef2ca MO showing cardiac edema, compared with mef2ca MO injected siblings with normal heart and no edema. Scale = 100 μm.
Figure S7. Mef2ca and Mef2cb are expressed in hand2 knockdown embryos.
In situ mRNA hybridisation for mef2ca (A) and mef2cb (B) in hearts of control, and hand2 MO zebrafish embryos shown in a dorsal view, anterior to top. A,B. Hearts of hand2 morphants show a cardia bifida phenotype (arrows). CMs on both sides of the midline express both mef2ca and mef2cb. Expression of mef2ca in the branchial arches is marked with asterisks. Scale = 100 μm.
Figure S8. Mef2ca mutant has a normal heart.
A,B. Double mRNA in situ hybridisation for myl7 (red, A,B), vmhc (blue, A) and bmp4 (blue, B) in genotyped 48 hpf mef2ca mutant embryos and their siblings shown in ventral view, anterior to top. Mutants have normal looping and gene expression is no different than siblings. OFT, blue arrow, IFT, purple arrow, AVC, red arrows. C. Confocal stacks of 72 hpf mef2cab1086;Tg(myl7:GFP) mutants and their siblings and of Tg(myl7:GFP) injected with mef2ca MO immunostained for Elastin showing no difference between morphants, mutants and siblings. D. GFP+dsRED- CMs can only be found in OFT and IFT of the heart (dotted line) of mef2cab1086;Tg(myl7:GFP;myl7;ndsRed) mutants and their siblings at 60 hpf. E. Heart rate (bpm) of control wild type, mef2ca morphants, mef2catn213 and their siblings at 25-48 hpf. Embryos were grown in 24-well plates and genotype was determined at 5 dpf according to jaw phenotype (see (Piotrowski et al., 1996)). No significant differences according to t-test statistics were found at any stage between morphants/mutants and control/siblings. Number of embryos per condition is shown on columns. Variation in heart rate between experiments is due to differences in temperature. Scale = 100 μm.
Figure S9. Mef2cb MO results in linear defective heart and loss of bulbus arteriosus.
In situ mRNA hybridisation for indicated genes (A,B, ventral view, anterior to top) and immunodetection of EGFP, MyHC (MF20), Elastin and DAF-2DA detection (C,D, lateral view, anterior to left) of hearts at indicated stage of mef2cb morphants and controls. A,B.Mef2cb morphants have a linear unlooped hearts and no obvious AV canal as marked by bmp4 expression (red arrows). C,D. Confocal stacks of control or mef2cb MO-injected Tg(myl7:EGFP) (E) or wild-type (F) embryos at 72 hpf. Hearts are unlooped and bulbus arteriosus markers (BA, arrows) are absent in mef2cb morphants. Insets in D show YZ sections at the OFT region, indicating to the area bordering myocardial and smooth muscle cells. sh, sternohyoideus; cl, cleithrum; V, ventricle; A, atrium, IFT, inflow tract; OFT, outflow tract; AVC, atrioventricular canal. Scale = 100μm, except for C,D=50 μm.
Table S1. Quantification of effects of loss- and gain-of-function.


