Abstract
The functional repertoire of a cell is largely embodied in its proteome, the collection of proteins encoded in the genome of an organism. The molecular functions of proteins are the direct consequence of their structure and structure can be inferred from sequence using hidden Markov models of structural recognition. Here we analyze the functional annotation of protein domain structures in almost a thousand sequenced genomes, exploring the functional and structural diversity of proteomes. We find there is a remarkable conservation in the distribution of domains with respect to the molecular functions they perform in the three superkingdoms of life. In general, most of the protein repertoire is spent in functions related to metabolic processes but there are significant differences in the usage of domains for regulatory and extra-cellular processes both within and between superkingdoms. Our results support the hypotheses that the proteomes of superkingdom Eukarya evolved via genome expansion mechanisms that were directed towards innovating new domain architectures for regulatory and extra/intracellular process functions needed for example to maintain the integrity of multicellular structure or to interact with environmental biotic and abiotic factors (e.g., cell signaling and adhesion, immune responses, and toxin production). Proteomes of microbial superkingdoms Archaea and Bacteria retained fewer numbers of domains and maintained simple and smaller protein repertoires. Viruses appear to play an important role in the evolution of superkingdoms. We finally identify few genomic outliers that deviate significantly from the conserved functional design. These include Nanoarchaeum equitans, proteobacterial symbionts of insects with extremely reduced genomes, Tenericutes and Guillardia theta. These organisms spend most of their domains on information functions, including translation and transcription, rather than on metabolism and harbor a domain repertoire characteristic of parasitic organisms. In contrast, the functional repertoire of the proteomes of the Planctomycetes-Verrucomicrobia-Chlamydiae superphylum was no different than the rest of bacteria, failing to support claims of them representing a separate superkingdom. In turn, Protista and Bacteria shared similar functional distribution patterns suggesting an ancestral evolutionary link between these groups.
Keywords: functional annotation, fold superfamily, molecular function, protein domain, SCOP, structure, superkingdom
1. Introduction
Proteins are active components of molecular machinery that perform vital functions for cellular and organismal life [1,2]. Information in the DNA is copied into messenger RNA that is generally translated into proteins by the ribosome. Nascent polypeptide chains are unfolded random coils but quickly undergo conformational changes to produce characteristic and functional folds. These folds are three-dimensional (3D) structures that define the native state of proteins [3,4]. Biologically active proteins are made up of well-packed structural and functional units referred to as domains. Domains appear either singly or in combination with other domains in a protein and act as modules by engaging in combinatorial interplays that enhance the functional repertoires of cells [5]. While molecular interactions between domains in mutidomain proteins play important roles in the evolution of protein repertoires [6], it is the domain structure that is maintained in proteins for long periods of evolutionary time [7–9]. This is in sharp contrast to amino acid sequence, which is highly variable. For this reason, protein domains are also considered evolutionary units [7,10–12].
1.1. Classification of Domains
Domains that are evolutionarily related can be grouped together in hierarchical classifications [1,10,13]. One scheme of classifying protein domains is the well-established “Structural Classification of Proteins” (SCOP). The SCOP database groups domains that have sequence conservation (generally with >30% pairwise amino acid residue identities) into fold families (FFs), FFs with structural and functional evidence of common ancestry into fold superfamilies (FSFs), FSFs with common 3D structural topologies into folds (Fs), and Fs sharing a same general architecture into protein classes [10,14]. SCOP identifies protein domains using concise classification strings (css) (e.g., c.26.1.2, where c represents the protein class, 26 the F, 1 the FSF and 2 the FF). The 97,178 domains indexed in SCOP 1.73 (corresponding to 34,494 PDB entries) are classified into 1,086 F, 1,777 FSFs, and 3,464 FFs. Compared to the number of protein entries in UniProt (531,473 total entries as of July 27, 2011) the number of domain structural designs at these different levels of structural abstraction is quite limited. Their relatively small number suggests that fold space is finite and is evolutionarily highly conserved [1,7,15].
1.2. Assigning FSF Structures to Proteomes
Genome-encoded proteins can be scanned against advanced linear hidden Markov models (HMMs) of structural recognition in SUPERFAMILY [16,17]. HMM libraries are generated using the iterative Sequence Alignment and Modeling (SAM) method. SAM is considered one of the most powerful algorithms for detecting remote homologies [18]. The SUPERFAMILY database currently provides FSF structural assignments for a total of 1,245 model organisms including 96 Archaea, 861 Bacteria and 288 Eukarya.
1.3. Assigning Functional Categories to Protein Domains
Assigning molecular functions to FSFs is a difficult task since approximately 80% of the FSFs defined in SCOP are multi-functional and highly diverse [19]. For example, most of the ancient FSFs, such as the P-loop-containing NTP hydrolase FSF (c.37.1), are highly abundant in nature and include many FFs (20 in case of c.37.1). Each of those families may have functions that impinge on multiple and distinct pathways or networks. The functional annotation scheme introduced by Vogel and Chothia in SUPERFAMILY is a one-to-one mapping scheme that is based on information from various resources, including the Cluster of Orthologus Groups (COG) and Gene Ontology (GO) databases and manual surveys [20–23]. When a FSF is involved in multiple functions, the most predominant function is assigned to that multi-functional FSF under the assumption that the most dominant function is the most ancient and predominantly present in all proteomes. The error rate in assignments is estimated to be <10% for large FSFs and <20% for all FSFs [23].
The SUPERFAMILY functional classification maps seven general functional categories to 50 detailed functional categories in a two-tier hierarchy (Table 1). The seven general categories include Metabolism, Information, Intracellular processes (ICP), Extracellular processes (ECP), Regulation, General, and Other (we will refer to them as “categories” and “functional repertoires” interchangeably). In this study, we take advantage of this coarse-grained functional annotation scheme to assign individual functional categories to FSFs. We are aware that this one-to-one mapping may not provide a complete profile for multi-functional domains [19]. Dissection of such detailed functions and their comparison across organisms is a difficult problem that we will not address in this study. In contrast, we focus on domains defined at FSF level and use the coarse-grained functional annotation scheme to explore the functional diversity of the proteomes encoded in genomes that have been completely sequenced. Our results yield a global picture of the functional organization of proteomes that is only possible with this classification scheme. Results suggest that the functional structure of proteomes is remarkably conserved across all organisms, ranging from small bacteria to complex eukaryotes. There is also evidence for the existence of few outliers that deviate from global trends. Here we explore what makes these proteomes distinct.
Table 1.
Mapping between the general and minor functional categories for 1,781 protein domains defined in structural classification of proteins (SCOP) 1.73 and the number of fold superfamilies (FSFs) corresponding to each minor category in our dataset of 965 organisms. A total of 135 FSFs could not be annotated. m/tr, metabolism and transport.
| Functional category | Minor categories | No. of FSF domains |
|---|---|---|
| Metabolism (533 FSFs) | Energy | 54 |
| Photosynthesis | 20 | |
| E- transfer | 31 | |
| Amino acids m/tr | 20 | |
| Nitrogen m/tr | 1 | |
| Nucleotide m/tr | 30 | |
| Carbohydrate m/tr | 30 | |
| Polysaccharide m/tr | 21 | |
| Storage | 0 | |
| Coenzyme m/tr | 50 | |
| Lipid m/tr | 17 | |
| Cell envelope m/tr | 8 | |
| Secondary metabolism | 11 | |
| Redox | 55 | |
| Transferases | 29 | |
| Other enzymes | 156 | |
| General (131 FSFs) | Small molecule binding | 27 |
| Ion binding | 13 | |
| Lipid/membrane binding | 4 | |
| Ligand binding | 3 | |
| General | 28 | |
| Protein interaction | 49 | |
| Structural protein | 7 | |
| Information (201 FSFs) | Chromatin structure | 7 |
| Translation | 92 | |
| Transcription | 24 | |
| DNA replication/repair | 68 | |
| RNA processing | 10 | |
| Nuclear structure | 0 | |
| Other (273 FSFs) | Unknown function | 200 |
| Viral proteins | 73 | |
| Extracellular processes (95 FSFs) | Cell adhesion | 31 |
| Immune response | 19 | |
| Blood clotting | 5 | |
| Toxins/defense | 40 | |
| Intracellular processes (208 FSFs) | Cell cycle, Apoptosis | 20 |
| Phospholipid m/tr | 6 | |
| Cell motility | 20 | |
| Trafficking/secretion | 0 | |
| Protein modification | 35 | |
| Proteases | 52 | |
| Ion m/tr | 21 | |
| Transport | 54 | |
| Regulation (205 FSFs) | RNA binding, m/tr | 19 |
| DNA-binding | 66 | |
| Kinases/phosphatases | 15 | |
| Signal transduction | 53 | |
| Other regulatory function | 34 | |
| Receptor activity | 18 |
2. Results and Discussion
2.1. General Patterns in the Distribution of FSF Domain Functions
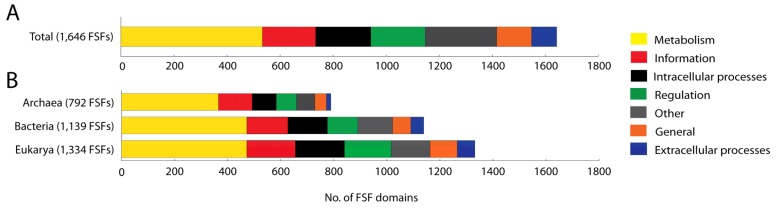
We studied the molecular functions of 1,646 domains defined at the FSF level of structural abstraction (SCOP 1.73) that are present in the proteomes of a total of 965 organisms spanning the three superkingdoms. A total of 135 FSFs that could not be annotated were excluded from analysis. For these FSFs, the functional annotation is not available. Out of the 1,646 FSFs studied, approximately one-third (32.38%) performs molecular functions related to Metabolism. Categories Other (16.58%), ICP (12.63%), Regulation (12.45%), and Information (12.21%) are uniformly distributed within proteomes. In contrast, General (7.96%) and ECP (5.77%) are significantly underrepresented compared to the rest (Figure 1(A)). The total number of FSFs in each category exhibits the following decreasing trend: Metabolism > Other > ICP > Regulation > Information > General > ECP. These patterns of FSF number and relative proteome content are for the most part maintained when studying the functional annotation of FSFs belonging to each superkingdom (Figure 1(B)). However, the number of FSFs in each superkingdom varies considerably and increases in the order Archaea, Bacteria and Eukarya, as we have shown in earlier studies [7].
Figure 1.
Number of protein FSFs annotated for each functional category defined in SCOP 1.73 (A) and in the three superkingdoms (B). The functional distributions show that coarse-grained functions are conserved across cellular proteomes and Metabolism is the most dominant functional category. Numbers in parentheses indicate the total number of FSFs annotated in each dataset. The number of FSFs increases in the order Archaea, Bacteria and Eukarya.
The significantly higher number of FSFs devoted to Metabolism is an anticipated result given the central importance of metabolic networks. However, the much larger number of FSFs corresponding to Other is quite unexpected. The 273 FSFs belonging to this category include 200 and 73 FSFs in sub-categories unknown functions and viral proteins, respectively. The sub-category unknown function includes FSFs for which the functions are either unknown or are unclassifiable. Viruses are defined as simple biological entities that are considered to be “gene poor” relatives of cellular organisms [24]. However, the number of domains belonging to viral proteins that are present in cellular organisms makes a noteworthy contribution to the total pool of FSFs (4.43%). Thus, viruses have a much more rich and diverse repertoire of domain structures than previously thought and their association with cellular life has contributed considerable structural diversity to the proteomic make up (A. Nasir, K.M. Kim and G. Caetano-Anollés, ms. in preparation).
The numbers of FSFs belonging to categories Regulation, Information, and ICP are uniformly distributed in proteomes. However, the ECP category is the least represented, perhaps because this category is the last to appear in evolution [7,15]. Extra cellular processes are more important to multicellular organisms (mainly eukaryotes) than to unicellular organisms. Multicellular organisms need efficient communication, such as signaling and cell adhesion. They also trigger immune responses and produce toxins when defending from parasites and pathogens. These ECP processes, which are depicted in the minor categories of cell adhesion, immune response, blood clotting and toxins/defense, are needed when interacting with environmental biotic and abiotic factors and for maintaining the integrity of multicellular structure. These categories are also present in the microbial superkingdoms but their functional role may be different than in Eukarya.
We note that current genomic research is highly shifted towards the sequencing of microbial genomes, especially those that hold parasitic lifestyles and are of bacterial origin. In fact, 67% of proteomes in our dataset belong to Bacteria. This bias can affect conclusions drawn from global trends such as those in Figure 1(A), including the under-representation of ECP FFs, because of their decreased representation in microbial proteomes.
2.2. Distribution of FSF Domain Functions in the Three Superkingdoms of Life
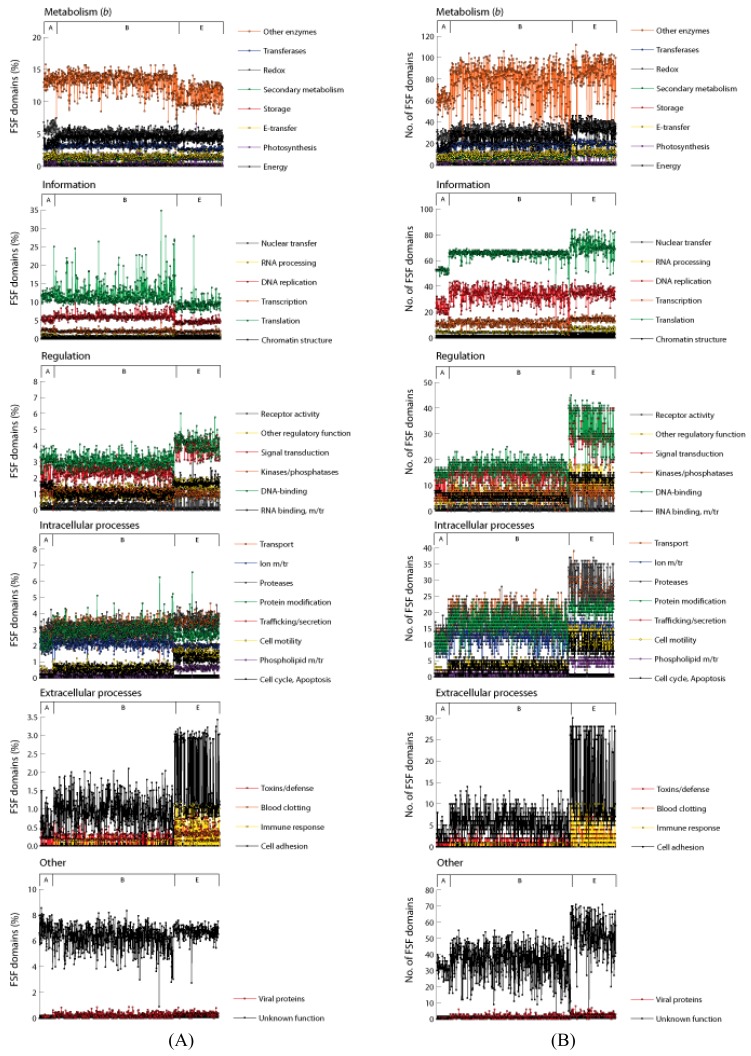
In order to explore whether the overall distribution of general functional categories differs in organisms belonging to the three superkingdoms, we analyzed proteomes at the species level and calculated both the percentage and actual number of FSFs corresponding to different functional repertoires (Figure 2).
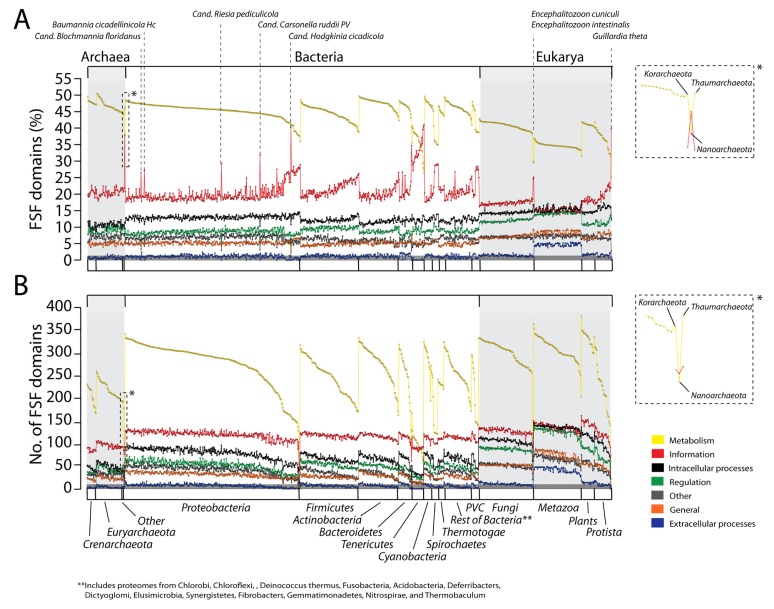
Figure 2.
The functional distribution of FSFs in individual proteomes of the three superkingdoms. Both the percentage (A) and actual FSF numbers (B) indicate conservation of functional distributions in proteomes and the existence of considerable functional flexibility between superkingdoms. Dotted vertical lines indicate genomic outliers. Insets highlight the interplay between Metabolism (yellow trend lines) and Information (red trend lines) in N. equitans.
FSF domains follow the following decreasing trend in both the percentage and actual counts of FSFs, and do so consistently for the three superkingdoms: Metabolism > Information > ICP > Regulation > Other > General > ECP. Note that trend lines across proteomes seldom overlap and cross in Figure 2. It is noteworthy however that this trend differs from the decreasing total numbers of FSFs we described above (Figure 1). Thus, no correlation should be expected between the numbers of FSFs for individual proteomes and the total set for each category. This suggests that variation in functional assignments across proteomes of superkingdoms may not necessarily match overall functional patterns.
Proteomes in microbial superkingdoms Archaea and Bacteria exhibit remarkably similar functional distributions of FSFs (Figure 2(A)). The only exception appears to be the slight overrepresentation of Regulation FSFs (green trend lines) and underrepresentation of ICP (black trend lines) in Archaea compared to Bacteria (especially Proteobacteria). These distributions are clearly distinct from those in Eukarya. Proteomic representations of FSFs corresponding to Metabolism and Information are decreased while those of all other five functional categories are significantly and consistently increased (Figure 2(A)). There is also more variation evident in Eukarya; large groups of proteomes exhibit different patterns of functional use (clearly evident in Information; red trend lines in Figure 2(A)).
On the whole, the relative functional make up of the proteomes of individual superkingdoms appear highly conserved (Figure 2(A)). There is however considerable variation in the metabolic functional repertoire of organisms, especially in Bacteria, where Metabolism ranges 30–50% of proteomic content (100–350 FSFs, Table S1 and Table S2). This variation is not present in other functional repertoires.
Consequently, tendencies of reduction in the metabolic repertoire are generally offset by small increases in the representation of the other six repertoires, with the notable exception of Information. In this particular case, when Metabolism goes down Information goes up. For example, bacterial proteomes with metabolic FSF repertoires of <45% offset their decrease by a corresponding increase in Information FSFs (generally from ∼20% to ∼35%, Figure 2(A)). In all superkingdoms, we identify groups of proteomes or few outliers that deviate from the global trends (vertical dotted lines in Figure 2(A)). As we will discuss below this is generally a consequence of reductive evolution imposed by the lifestyle of organisms (discussed in detail below). Outliers are particularly evident in Bacteria and harbor sharp increases in Information repertoires, not always with corresponding decreases in Metabolism. In Archaea, decreases of Metabolism are generally offset by increases of the Regulation category, with an exception in Nanoarchaeum equitans (see below). In Eukarya, decreases in Metabolism go in hand with decreases in Information, and are correspondingly offset mostly by increases in Regulation and ECP. Apparently, the advantages of regulatory control (e.g., signal transduction and transcriptional and posttranscriptional regulation) and multicellularity counteract the interplay of Metabolism and Information in eukaryotes.
When we look at the actual number of FSFs within each functional repertoire (Figure 2(B)), we observe a clear trend in domain use that matches the total trend for superkingdoms described above (Figure 1). In most cases, the functional repertoires of Archaea are smaller than those of Bacteria, and bacterial repertoires are generally smaller than those of Eukarya (Figure 2(B)). This holds true for all functional categories. However, the numbers of metabolic FSFs vary 1.5–4 fold in proteomes of superkingdoms, the change being maximal in Bacteria. While both proteomes in Eukarya and Bacteria show similar ranges of metabolic FSFs, the repertoire of Archaea is more constrained. Furthermore, FSFs belonging to categories Other and ECP are significantly higher in Eukarya than in the microbial superkingdoms. These remarkable observations suggest high conservation in the make up of proteomes of superkingdoms and at the same time considerable levels of flexibility in the metabolic make-up of organisms. Results also support the evolution of the protein complements of Archaea and Bacteria via reductive evolutionary processes and Eukarya by genome expansion mechanisms [7,25]. Reductive tendencies in microbial superkingdoms do not show bias in favor of any functional category. Furthermore, enrichment of eukaryal proteomes with viral proteins supports theories, which state that viruses have played an important role in the evolution of Eukarya [26].
2.3. Distribution of FSF Domain Functions in Individual Phyla/Kingdoms
Figure 2 also describes the functional distribution of FSFs at the phyla/kingdom level for each superkingdom. Plots describing the percentages (Figure 2(A)) and actual number of FSFs in proteomes (Figure 2(B)) highlight the existence of “outliers” (vertical dotted lines in Figure 2(A)) that deviate from the global functional trends that are typical of each superkingdom.
In Archaea, the functional repertoires of the proteomes of Euryarachaeota, Crenarchaeota, Korarcheota and Thaumarchaeota were remarkably conserved and consistent with each other. Only N. equitans could be considered an outlier (insets of Figure 2). Its proteome deviates from the global archaeal signature by reducing its proteomic make up (it has only 200 distinct FSFs) and by exchanging Information for metabolic FSFs. N. equitans is an obligate intracellular parasite [27] that is part of a new phylum of Archaea, the Nanoarchaeota [28]. N. equitans has many atypical features, including the almost complete absence of operons and presence of split genes [29], tRNA genes that code for only half of the tRNA molecule [30], and the complete absence of the nucleic acid processing enzyme RNAse P [31]. Some of these features were used to propose that N. equitans is a living fossil [32], represents the root of superkingdom Archaea and the tree of life [33], and is part of a very ancient and yet to be described superkingdom (M. Di Giulio, personal communication). Phylogenomic analyses of domain structures in proteomes suggest Archaea is the most ancient superkingdom [19,34] and has placed N. equitans at the base of the tree of life together with other archaeal species. Its ancestral nature is therefore in line with the evolutionary and functional uniqueness of N. equitans and the very distinct functional repertoire we here report.
In Bacteria, the functional repertoires of bacterial phyla were also remarkably conserved. Only Information and Metabolism showed significantly distinct patterns and considerable variation in the use of FSFs. Again, decreases in representation of metabolic FSFs were generally offset by increases in informational FSFs (Figure 2(A)). Notable outliers include the Tenericutes and the Spirochetes. As groups, they have the highest relative usage of Information FSFs, which are clearly offset by a decrease in metabolic FSFs. The Tenericutes is a phylum of bacteria that includes class Mollicutes. Members of the Mollicutes are typical obligate parasites of animals and plants (some of medical significance such as Mycoplasma) that lack cell walls and have gliding motility. These organisms are characterized by small genome sizes [35] considered to have evolved via reductive evolutionary processes [36]. Because of its unique properties and history, mycoplasmas have been used recently to produce a completely synthetic genome [37]. There were also clear outliers in the Proteobacteria. These included Candidatus Blochmannia floridanus (symbiont of ants), Baumannia cicadellinicola (symbiont of sharpshooter insect), Candidatus Riesia pediculicola, Candidatus Carsonella ruddii (symbiont of sap-feeding insects) and Candidatus Hodgkinia cicadicola (symbiont of cicadas). These bacteria are generally endosymbionts of insects (e.g., ants, sharpshooters, psyllids, cicadas) that have undergone irreversible specialization to an intracellular lifestyle. Candidatus Carsonella ruddii has the smallest genome of any bacteria [38]. There were also bacterial proteome groups that were expected to be outliers but were no different than the rest. Bacteria belonging to the superphylum Planctomycetes-Verrucomicrobia-Chlamydiae (PVC) are different from other bacterial phyla because they have an “eukaryotic touch” [39]. Indeed, PVC bacteria display genetic and cellular features that are characteristics of Eukarya and Archaea, including the presence of Histone H1, condensed DNA surrounded by membrane, α-helical repeat domains and β-propeller folds that make up eukaryotic-like membrane coats, reproduction by budding, ether lipids and lack of cell walls [40–42]. Due to the unique nature of the PVC superphylum, it was proposed that these organisms be identified as a separate superkingdom that contributed to the evolution of Eukarya and Archaea [40]. However, trees of life generated from domain structures in hundreds of proteomes did not dissect the PVC superphylum into a separate group [7,19,34]. Functional distributions of FSFs now show PVC proteomes appear no different from bacteria (Figure 2). These results do not support PVC-inspired theories that explain the diversification of the three cellular superkingdoms of life.
In contrast to the functional repertoires of bacterial and archaeal phyla, proteomes belonging to individual kingdoms in Eukarya had functional signatures that were highly conserved (Figure 2(A)). However, these signatures differed between groups. Plants and fungi had functional representations that were very similar and showed little diversity. In contrast, Metazoa functional distributions increased the representation of ECP and Regulation FSFs in exchange of FSFs in Metabolism and Information. Protista had patterns that resemble those of Plants and Fungi but had widely varying metabolic repertoires, very much like Bacteria. This possible link between basal eukaryotes and bacteria revealed by our comparative analysis is consistent with the existence of an ancestor of Bacteria and Eukarya and the early rise of Archaea [34]. Only few outliers belonging to kingdoms Fungi (Encephalitozoon cuniculi and Encephalitozoon intestinalis) and Protista (Guillardia theta) were identified. E. cuniculi and E. intestinalis are eukaryotic parasites with highly reduced genomes [43,44]. Similarly, Guillardia theta is a nucleomorph that has a highly compact and reduced genome with loss of nearly all metabolic genes [45].
When we look at the actual number of FSFs in proteomes of phyla and kingdoms (Figure 2(B)) we observe that while the overall patterns match those of FSF representation (Figure 2(A)), FSF number revealed considerable variation in the metabolic repertoire of Protista and Bacteria. FSFs in these groups typically ranged 130–340, with PVC and Spirochetes exhibiting the smallest range (130–300 FSFs). In contrast, metabolic repertoires of Archaea and the other eukaryotic kingdoms typically ranged 200–260 FSFs and 270–350 FSFs, respectively. This observation is significant. It provides comparative information to support a unique evolutionary link of phyla within superkingdoms Eukarya and Bacteria. Plots of FSF number also clarified functional patterns in outliers, revealing they did not have more numbers of FSFs in Information but rather have reduced metabolic repertoires. This shows that parasitic outliers get rid of metabolic domains and become more and more dependent on host cells.
2.4. Effect of Organism Lifestyle
The analysis thus far revealed the existence of a small group of outliers within each superkingdom. Manual inspection of lifestyles of these organisms showed that all of these organisms are united by a parasitic or symbiotic lifestyle. For example, N. equitans is the smallest archaeal genome ever sequenced and represents a new phylum, the Nanoarchaeaota [28]. This organism interacts with Ignicoccus hospitalis, establishing the only known parasite/symbiont relationship of Archaea, and harbors a highly reduced genome [29]. Parasitic/symbiotic relationships with various plants and animals can be found in Tenericutes and in the endosymbionts of insects that belong to Proteobacteria. Similarly, the Encephalitozoon species are eukaryotic parasites that lack mitochondria and have highly reduced genomes [43,44]. E. cunniculi has even a chromosomal dispersion of its ribosomal genes, very much like N. equitans, and the rRNA of the large ribosomal subunit reduced to its universal core [46]. Similarly, Guillardia theta is a nucleomorph that has a highly compact and reduced genome with loss of nearly all metabolic genes [45]. Thus, all outliers exhibit extreme or unique cases of genome reduction.
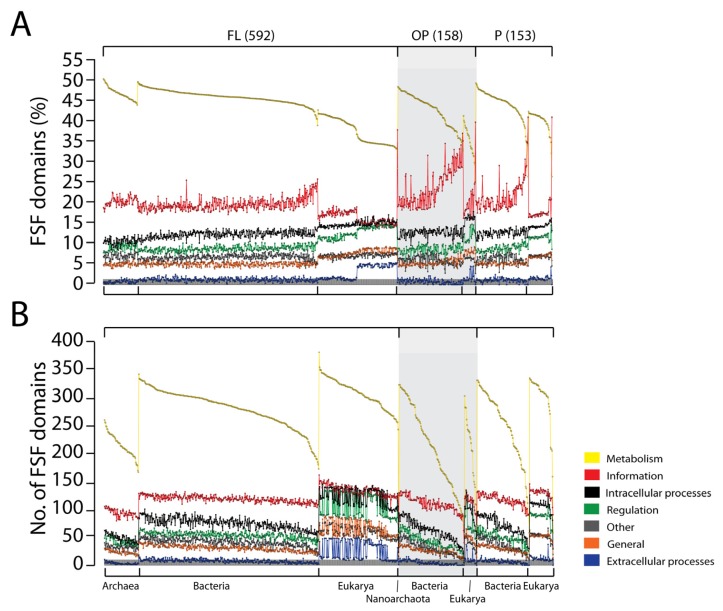
In order to explore whether organisms that engage in parasitic or symbiotic interactions have general tendencies that resemble those of the outliers, we classified organisms into three different lifestyles: free living (FL) (592 proteomes), facultative parasitic (P) (153 proteomes), and obligate parasitic (OP) (158 proteomes). Functional distributions for the seven general functional categories for these proteomic sets explained the role of parasitic life on proteomic constitution (Figure 3). Plots of percentages (Figure 3(A)) and actual number of FSFs in proteomes (Figure 3(B)) showed FSF distribution in FL organisms were remarkably homogenous and that the vast majority of variability within superkingdoms was ascribed to the P and OP lifestyles. This variability was for the most part explained by a sharp decline in the number of metabolic FSFs that are assigned to the Metabolism general category (Figure 3(B)). Plots also support the hypothesis that parasitic organisms have gone the route of massive genome reduction in a tendency to loose all of their metabolic genes. This tendency makes them more and more dependent on host cells for metabolic functions and survival [47,48].
Figure 3.
The functional distribution of FSFs with respect to organism lifestyle. Both the percentage (A) and actual FSF numbers (B) indicate that obligate parasitic (OP) and facultative parasitic (P) organisms exhibit considerable variability in their metabolic repertoires (yellow trend lines) that is offset by corresponding increases in the Information FSFs (red trend lines).
The number of domains corresponding to each general functional category in the proteomes of FL organisms increases in the order Archaea, Bacteria and Eukarya (Table S3). When compared to the total proteomic set (Figure 2), Metabolism remains the predominant functional category and a large number of domains in all the proteomes perform metabolic functions. Again, the proteomes of Eukarya have the richest FSF repertoires, and those of Archaea the most simple. Since maximum variability lies within the proteome repertoires of parasitic/symbiotic organisms (Figure 3) and parasitism/symbiosis in these organisms is the result of secondary adaptations, the analysis of proteomic diversity in FL organisms allows us to test if the functional repertoires of superkingdoms are indeed statistically significant. Analysis of variance showed that the number of FSFs for each functional repertoire was consistently different between superkingdoms (p < 0.0001; Table S3). This supports the conclusions drawn from earlier analyses that the microbial superkingdoms followed a genome reduction path while Eukarya expanded their genomic repertoires [7,25].
2.5. Analysis of Minor Functional Categories
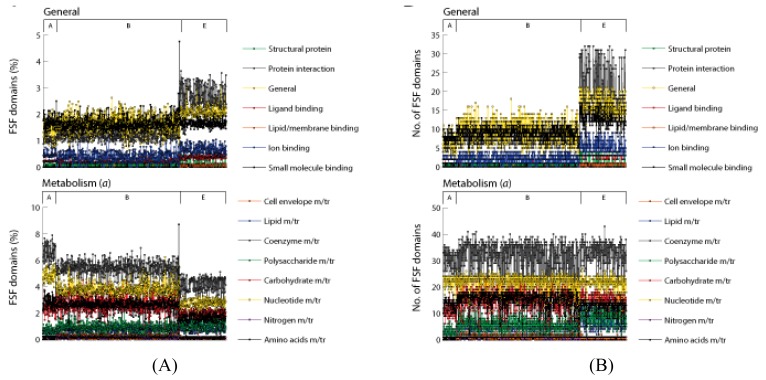
The seven general categories of molecular functions map to 50 minor categories (Table 1). We explored the distribution of FSFs corresponding to each minor category in superkingdoms (Figure 4). Only category “not annotated” (NONA) was excluded from analysis. In terms of percentage (Figure 4(A)), the overall functional signature is split into two components: prokaryotic and eukaryotic. Prokaryotes spend most of their domain repertoire on Metabolism and Information whereas Eukarya stand out in ECP (particularly cell adhesion, immune response), Regulation (DNA binding, signal transduction), and all the minor functional categories corresponding to ICP and General.
Figure 4.
The percentage (A) and number (B) of FSFs in minor functional categories across superkingdoms. Archaea (A) and Bacteria (B) spend most of their proteomes in functions related to Metabolism and Information whereas Eukarya (E) stand out in the minor categories of Regulation, General, Intracellular processes (ICP) and Extracellular processes (ECP). In turn, the number of FSFs increases in the order Archaea, Bacteria and Eukarya. Eukaryal proteomes have the richest functional repertoires for Regulation, Other, General, ICP and ECP.
In terms of domain counts (Figure 4(B)), proteomes of Eukarya have the richest functional repertoires with a significantly large number of FSFs devoted for each minor functional category. Bacteria and Archaea work with small number of domains. However, the number of FSFs in Bacteria is significantly higher compared to Archaea (supporting results of Figure 1, Figure 2 and Table S3). These results are consistent with the evolutionary trends in proteomes described previously [7,19,25]. Our results support the complex nature of the Last Universal Common Ancestor (LUCA) [19] and are consistent with the evolution of microbial superkingdoms via reductive evolutionary processes and the evolution of eukaryal proteomes by genome expansion [7,25]. It appears that Archaea went on the route of genome reduction very early in evolution and was followed by Bacteria and finally Eukarya. Late in evolution, the eukaryal superkingdom increased the representation of FSFs and developed a rich proteome. This can explain the relatively huge and diverse nature of eukaryal proteomes compared to prokaryotic proteomes. Finally, there appears to be no significant difference in the distributions of FSFs corresponding to Metabolism and Information between Bacteria and Eukarya except for minor category “Translation” (green trend lines in Figure 4(B, Information)) that is significantly higher in Eukarya compared to Bacteria. This shows that Bacteria exhibit incredible metabolic and informational diversity despite their reduced genomic complements. We conclude that the genome expansion in Eukarya occurred primarily for functions related to ECP, ICP, Regulation and General.
2.6. Reliability of Functional Annotations and Conclusions of this Study
Our analysis depends upon the accuracy of assigning structures to protein sequences and the SCOP protein classification and SUPERFAMILY functional annotation schemes. Databases such as SCOP and SUPERFAMILY are continuously updated with more and more genomes and new assignments. We therefore ask the reader to focus on the general trends in the data as opposed to the specifics such as the exact percentage or numbers of FSFs in each functional repertoire. Trends related to the number of domains in Archaea relative to Bacteria and Eukarya and the reduction of metabolic repertoires in parasitic organisms should be considered robust since these have been reliably observed in previous studies with more limited datasets [1,7,15,19,34]. Biases in sampling of proteomes in the three superkingdoms is not expected to over or underestimate the remarkably conserved nature of the functional makeup. We show that the conservation of molecular functions in proteomes is only broken in genomic outliers that are united by parasitic lifestyles. Thus equal sampling will not significantly alter the global trends described for individual superkingdoms. In light of our results, organism lifestyle is the only factor affecting the conserved nature of proteomes. Finally, we propose that lower or higher than expected numbers of FSFs in any category (subcategory) can be explained either by possible limitations of the scheme used to annotate molecular functions of FSFs or the simple nature of the functional repertoire. For example, the number of FSFs in subcategory structural proteins (main category General) is 7 (Table 1) despite the importance of structural proteins in cellular organization. Table S4 lists the description of these FSFs and shows that indeed these FSF domains play important structural roles. Their limited number indicates that the structural and functional organization is quite limited and very few folds play important structural roles. Another possibility is the “hidden” overlap between FSFs and molecular functions due to the one-to-one mapping limitations of the SUPERFAMILY functional annotation scheme. Most of the large FSFs include many FFs and participate in multiple pathways; for few FSFs a complete functional profile may not be intuitively obvious. This may be one of the shortcomings of using this functional annotation scheme but dissection of such detailed functions and pathways is a difficult task and is not described in this study. In summary, we do not believe that the classification or annotation schemes, despite their limitations, would undergo serious revisions or weaken our findings.
3. Experimental Section
3.1. Data Retrieval
We downloaded the protein architecture assignments for a total of 965 organisms including 70 Archaea, 651 Bacteria and 244 Eukarya (Table S5) from SUPERFAMILY ver. 1.73 MySQL [16,17] at an E-value cutoff of 10−4. This cutoff is considered a stringent threshold to eliminate the rate of false positives in HMM assignments [19]. Classification of organisms according to their lifestyles was done manually and resulted in 592 FL, 153 P, and 158 OP organisms.
3.2. Assigning Functional Categories to Protein Domains
The most recent domain functional annotation file for SCOP 1.73 was downloaded from the SUPERFAMILY webserver [23]. For each genome we extracted the set of unique FSFs present and then mapped them to the 7 general and 50 detailed functional categories. We calculated both the percentage and actual number of domains using programming implementations in Python 3.1 (http://www.python.org/download/).
3.3. Statistical Analysis
The statistical significance between the numbers of functional FSFs in FL organisms of superkingdoms was evaluated by Welch's ANOVA in SAS (http://www.sas.com/software/sas9), which is the appropriate test to detect differences between means for groups having unequal variances [49]. We excluded organisms with P and OP lifestyles in order to remove noise from the data. Additionally, in order to meet asymptotic normality, we used the Log10 transformation and rescaled the data to 0–7 using the following formula,
where Nxy is the count of a FSF in x functional category in y superkingdom; Nmax is the largest value in the matrix and Nnormal is the normalized and scaled score for FSF x in y superkingdom.
4. Conclusions
Our analysis revealed a remarkable conservation in the functional distribution of protein domains in superkingdoms for proteomes for which we have structural assignments. Figure S1 showcases average distribution of FSFs in phyla, kingdoms, and superkingdoms. The biggest proportion of each proteome is devoted in all cases to functions related to Metabolism. Phylogenomic analysis has shown that Metabolism appeared earlier than other functional groups and their structures were the first to spread in life [1,50]. This would explain the relative large representation of Metabolism in the functional toolkit of cells. Usage of domains related to ECP and Regulation is significantly higher in Metazoa compared to the rest. This showcases the importance of regulation signal transduction mechanisms for eukaryotic organisms [51,52]. Our results support the view that prokaryotes evolved via reductive evolutionary processes whereas genome expansion was the route taken by eukaryotic organisms. Genome expansion in Eukarya seems to be directed towards innovation of FSF architectures, especially those linked to Regulation, ECP and General. Finally, viral structures make up a substantial proportion of cellular proteomes and appear to have played an important role in the evolution of cellular life.
Organisms with parasitic lifestyles have simple and reduced proteomes and rely on host cells for metabolic functions. Tenericutes are unique in this regard. They spend most of their proteomic resources in functions linked to Information (e.g., translation, replication). Remarkably, we find that the conservation of molecular functions in proteomes is only broken in “outliers” with parasitic lifestyles that do not obey the global trends. We conclude that organism lifestyle is a crucial factor in shaping the nature of proteomes.
Acknowledgments
This study began as a class project in CPSC 567, a course in bioinformatics and systems biology taught by G.C.-A. at the University of Illinois in spring 2011. We thank Kyung Mo Kim and Liudmila Yafremava for information about lifestyles. A.N., A.Na., M.J.K. and H.D.L.-N. conceived the experiments and analyzed the data. G.C.-A. supervised the project and edited the manuscript. Research was supported by the National Science Foundation (MCB-0749836), CREES-USDA and the Soybean Disease Biotechnology Center (to G.C.-A.). Any opinions, findings, and conclusions and recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the funding agencies.
Supplementary Materials
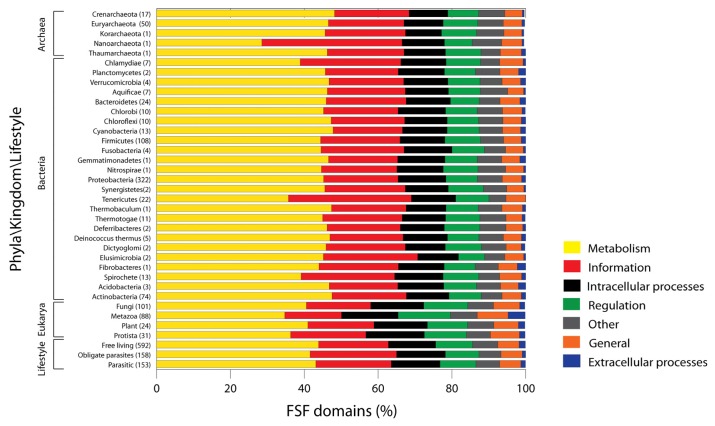
Figure S1.
Average distribution of FSFs in phyla, kingdom, and superkingdoms suggest conservation of functional design in proteomes. Numbers in parentheses indicate total number of proteomes analyzed for each phyla/kingdom.
Table S1.
Average number of FSF domains in each phyla/kingdom corresponding to the seven general functional categories. Numbers were rounded up when the decimal value exceeded 0.5 and rounded down otherwise. Nanoarchaeota and Tenericutes have the least number of metabolic domains and are highlighted in bold. Eukaryal kingdoms (Fungi, Metazoa, Plants and Protista) have the richest FSF repertoires compared to the prokaryotes.
| Superkingdom | Phyla/Kingdom | Metabolism | Information | ICP | Regulation | Other | General | ECP |
|---|---|---|---|---|---|---|---|---|
| Archaea | Crenarchaeota | 204 | 85 | 44 | 35 | 30 | 20 | 2 |
| Euryarchaeota | 219 | 96 | 50 | 44 | 32 | 24 | 4 | |
| Korarchaeota | 178 | 85 | 38 | 37 | 29 | 19 | 2 | |
| Nanoarchaeota | 57 | 76 | 23 | 15 | 16 | 11 | 1 | |
| Thaumarchaeota | 202 | 91 | 49 | 42 | 23 | 25 | 5 | |
| Bacteria | Proteobacteria | 274 | 119 | 78 | 52 | 42 | 31 | 7 |
| Firmicutes | 246 | 117 | 67 | 53 | 35 | 26 | 7 | |
| Actinobacteria | 275 | 115 | 66 | 50 | 33 | 30 | 7 | |
| Bacteroidetes | 251 | 113 | 65 | 43 | 32 | 29 | 9 | |
| Tenericutes | 99 | 90 | 33 | 25 | 13 | 14 | 0 | |
| Cyanobacteria | 289 | 112 | 73 | 52 | 39 | 30 | 8 | |
| Spirochaetes | 171 | 104 | 56 | 41 | 24 | 25 | 5 | |
| Thermotogae | 231 | 110 | 60 | 48 | 36 | 22 | 4 | |
| Rest of Bacteria * | 255 | 113 | 67 | 48 | 37 | 27 | 6 | |
| PVC | 206 | 110 | 58 | 43 | 28 | 27 | 6 | |
| Eukarya | Fungi | 298 | 127 | 105 | 87 | 51 | 52 | 10 |
| Metazoa | 307 | 135 | 136 | 126 | 65 | 75 | 42 | |
| Plants | 332 | 145 | 117 | 87 | 58 | 54 | 14 | |
| Protista | 220 | 117 | 94 | 67 | 39 | 46 | 9 |
Includes proteomes from Chlorobi, Chloroflexi, Aquificae, Deinococcus thermus, Fusobacteria, Acidobacteria, Deferribacters, Dictyoglomi, Elusimicrobia, Synergistetes, Fibrobacters, Gemmatimonadetes, Nitrospirae, and Thermobaculum.
Table S2.
Average percentage of FSF domains in each phyla/kingdom corresponding to the seven general functional categories. Numbers were rounded up when the decimal value exceeded 0.5 and rounded down otherwise. Nanoarchaeota (highlighted in bold) is an outlier considering it has the smallest percentage for metabolic domains compared to the rest and this decrease is offset by an increase in the informational FSFs.
| Superkingdom | Phyla/Kingdom | Metabolism | Information | ICP | Regulation | Other | General | ECP |
|---|---|---|---|---|---|---|---|---|
| Archaea | Crenarchaeota | 48 | 21 | 10 | 9 | 7 | 5 | 1 |
| Euryarchaeota | 47 | 20 | 11 | 9 | 7 | 5 | 1 | |
| Korarchaeota | 46 | 22 | 10 | 9 | 7 | 5 | 1 | |
| Nanoarchaeota | 29 | 38 | 12 | 8 | 8 | 6 | 1 | |
| Thaumarchaeota | 46 | 21 | 11 | 10 | 5 | 6 | 1 | |
| Bacteria | Proteobacteria | 45 | 20 | 13 | 8 | 7 | 5 | 1 |
| Firmicutes | 44 | 21 | 12 | 10 | 6 | 5 | 1 | |
| Actinobacteria | 48 | 20 | 12 | 9 | 6 | 5 | 1 | |
| Bacteroidetes | 46 | 22 | 12 | 8 | 6 | 5 | 2 | |
| Tenericutes | 36 | 33 | 12 | 9 | 5 | 5 | 0 | |
| Bacteria | Cyanobacteria | 48 | 19 | 12 | 9 | 6 | 5 | 1 |
| Spirochaetes | 39 | 25 | 13 | 10 | 6 | 6 | 1 | |
| Thermotogae | 45 | 22 | 12 | 9 | 7 | 4 | 1 | |
| Rest of Bacteria * | 46 | 21 | 12 | 9 | 7 | 5 | 1 | |
| PVC | 42 | 24 | 12 | 9 | 6 | 6 | 1 | |
| Eukarya | Fungi | 41 | 17 | 14 | 12 | 7 | 7 | 1 |
| Metazoa | 35 | 15 | 15 | 14 | 7 | 8 | 5 | |
| Plants | 41 | 18 | 14 | 11 | 7 | 7 | 2 | |
| Protista | 36 | 20 | 16 | 11 | 6 | 8 | 2 |
Includes proteomes from Chlorobi, Chloroflexi, Aquificae, Deinococcus thermus, Fusobacteria, Acidobacteria, Deferribacters, Dictyoglomi, Elusimicrobia, Synergistetes, Fibrobacters, Gemmatimonadetes, Nitrospirae, and Thermobaculum
Table S3.
Comparison of functional categories across superkingdoms using Welch's ANOVA.
| Functional category | F-ratio | DF | P-value * |
|---|---|---|---|
| Metabolism | 350.21 | 2 | <0.0001 |
| Information | 582.28 | 2 | <0.0001 |
| ICP | 1271.32 | 2 | <0.0001 |
| Regulation | 966.75 | 2 | <0.0001 |
| Other | 520.97 | 2 | <0.0001 |
| General | 1043.76 | 2 | <0.0001 |
| ECP | 263.44 | 2 | <0.0001 |
All the P-values are statistically significant at 0.05.
Table S4.
Names and description of FSF domains corresponding to subcategory structural proteins in the main category General.
| No. | SCOP Id | FSF Id | Description |
|---|---|---|---|
| 1 | 103589 | g.71.1 | Mini-collagen I, C-terminal domain |
| 2 | 49695 | b.11.1 | Gamma-crystallin-like |
| 3 | 51269 | b.85.1 | Anti-freeze protein (AFP) III-like domain |
| 4 | 56558 | d.182.1 | Baseplate structural protein gp11 |
| 5 | 58002 | h.1.6 | Chicken cartilage matrix protein |
| 6 | 58006 | h.1.7 | Assembly domain of catrillage oligomeric matrix protein |
| 7 | 75404 | d.213.1 | Vesiculovirus (VSV) matrix proteins |
Table S5.
List of organisms analyzed with their taxonomic classifications.
| No. | Genome Name | Phyla/Kingdom | Superkingdom |
|---|---|---|---|
| 1 | Malassezia globosa CBS 7966 | Fungi | Eukaryota |
| 2 | Ustilago maydis | Fungi | Eukaryota |
| 3 | Puccinia graminis f. sp. tritici CRL 75-36-700-3 | Fungi | Eukaryota |
| 4 | Melampsora laricis-populina | Fungi | Eukaryota |
| 5 | Sporobolomyces roseus IAM 13481 | Fungi | Eukaryota |
| 6 | Serpula lacrymans var. lacrymans S7.9 | Fungi | Eukaryota |
| 7 | Coprinopsis cinerea okayama7 130 v3 | Fungi | Eukaryota |
| 8 | Pleurotus ostreatus | Fungi | Eukaryota |
| 9 | Laccaria bicolor S238N-H82 | Fungi | Eukaryota |
| 10 | Agaricus bisporus var. bisporus | Fungi | Eukaryota |
| 11 | Schizophyllum commune | Fungi | Eukaryota |
| 12 | Heterobasidion annosum | Fungi | Eukaryota |
| 13 | Phanerochaete chrysosporium RP-78 2.1 | Fungi | Eukaryota |
| 14 | Postia placenta | Fungi | Eukaryota |
| 15 | Tremella mesenterica | Fungi | Eukaryota |
| 16 | Cryptococcus neoformans JEC21 | Fungi | Eukaryota |
| 17 | Magnaporthe grisea 70-15 | Fungi | Eukaryota |
| 18 | Podospora anserina | Fungi | Eukaryota |
| 19 | Sporotrichum thermophile ATCC 42464 | Fungi | Eukaryota |
| 20 | Thielavia terrestris NRRL 8126 | Fungi | Eukaryota |
| 21 | Chaetomium globosum CBS 148.51 | Fungi | Eukaryota |
| 22 | Neurospora tetrasperma | Fungi | Eukaryota |
| 23 | Neurospora discreta FGSC 8579 | Fungi | Eukaryota |
| 24 | Neurospora crassa OR74A | Fungi | Eukaryota |
| 25 | Cryphonectria parasitica | Fungi | Eukaryota |
| 26 | Verticillium dahliae VdLs.17 | Fungi | Eukaryota |
| 27 | Verticillium albo-atrum VaMs.102 | Fungi | Eukaryota |
| 28 | Fusarium oxysporum f. sp. lycopersici 4286 | Fungi | Eukaryota |
| 29 | Nectria haematococca mpVI | Fungi | Eukaryota |
| 30 | Fusarium verticillioides 7600 | Fungi | Eukaryota |
| 31 | Fusarium graminearum | Fungi | Eukaryota |
| 32 | Trichoderma atroviride | Fungi | Eukaryota |
| 33 | Trichoderma reesei 1.2 | Fungi | Eukaryota |
| 34 | Trichoderma virens Gv29-8 | Fungi | Eukaryota |
| 35 | Botrytis cinerea B05.10 | Fungi | Eukaryota |
| 36 | Sclerotinia sclerotiorum | Fungi | Eukaryota |
| 37 | Alternaria brassicicola | Fungi | Eukaryota |
| 38 | Pyrenophora tritici-repentis | Fungi | Eukaryota |
| 39 | Cochliobolus heterostrophus | Fungi | Eukaryota |
| 40 | Stagonospora nodorum | Fungi | Eukaryota |
| 41 | Mycosphaerella fijiensis CIRAD86 | Fungi | Eukaryota |
| 42 | Mycosphaerella graminicola IPO323 | Fungi | Eukaryota |
| 43 | Ajellomyces dermatitidis SLH14081 | Fungi | Eukaryota |
| 44 | Histoplasma capsulatum class NAmI strain WU24 | Fungi | Eukaryota |
| 45 | Microsporum canis CBS 113480 | Fungi | Eukaryota |
| 46 | Microsporum gypseum | Fungi | Eukaryota |
| 47 | Arthroderma benhamiae CBS 112371 | Fungi | Eukaryota |
| 48 | Trichophyton equinum CBS 127.97 | Fungi | Eukaryota |
| 49 | Trichophyton verrucosum HKI 0517 | Fungi | Eukaryota |
| 50 | Trichophyton tonsurans CBS 112818 | Fungi | Eukaryota |
| 51 | Trichophyton rubrum CBS 118892 | Fungi | Eukaryota |
| 52 | Paracoccidioides brasiliensis Pb18 | Fungi | Eukaryota |
| 53 | Coccidioides posadasii RMSCC 3488 | Fungi | Eukaryota |
| 54 | Coccidioides immitis RS | Fungi | Eukaryota |
| 55 | Uncinocarpus reesii 1704 | Fungi | Eukaryota |
| 56 | Aspergillus fumigatus Af293 | Fungi | Eukaryota |
| 57 | Neosartorya fischeri NRRL 181 | Fungi | Eukaryota |
| 58 | Penicillium chrysogenum Wisconsin 54-1255 | Fungi | Eukaryota |
| 59 | Penicillium marneffei ATCC 18224 | Fungi | Eukaryota |
| 60 | Aspergillus carbonarius ITEM 5010 | Fungi | Eukaryota |
| 61 | Aspergillus terreus NIH2624 | Fungi | Eukaryota |
| 62 | Aspergillus oryzae RIB40 | Fungi | Eukaryota |
| 63 | Aspergillus niger ATCC 1015 | Fungi | Eukaryota |
| 64 | Aspergillus flavus NRRL3357 | Fungi | Eukaryota |
| 65 | Aspergillus clavatus NRRL 1 | Fungi | Eukaryota |
| 66 | Aspergillus nidulans FGSC A4 | Fungi | Eukaryota |
| 67 | Tuber melanosporum Vittad | Fungi | Eukaryota |
| 68 | Pichia stipitis CBS 6054 | Fungi | Eukaryota |
| 69 | Candida guilliermondii ATCC 6260 | Fungi | Eukaryota |
| 70 | Lodderomyces elongisporus NRRL YB-4239 | Fungi | Eukaryota |
| 71 | Debaromyces hansenii | Fungi | Eukaryota |
| 72 | Candida dubliniensis CD36 | Fungi | Eukaryota |
| 73 | Candida tropicalis MYA-3404 | Fungi | Eukaryota |
| 74 | Candida parapsilosis | Fungi | Eukaryota |
| 75 | Candida albicans SC5314 | Fungi | Eukaryota |
| 76 | Yarrowia lipolytica CLIB122 | Fungi | Eukaryota |
| 77 | Candida lusitaniae ATCC 42720 | Fungi | Eukaryota |
| 78 | Vanderwaltozyma polyspora DSM 70294 | Fungi | Eukaryota |
| 79 | Candida glabrata CBS138 | Fungi | Eukaryota |
| 80 | Kluyveromyces thermotolerans CBS 6340 | Fungi | Eukaryota |
| 81 | Lachancea kluyveri | Fungi | Eukaryota |
| 82 | Kluyveromyces waltii | Fungi | Eukaryota |
| 83 | Ashbya gossypii ATCC 10895 | Fungi | Eukaryota |
| 84 | Zygosaccharomyces rouxii | Fungi | Eukaryota |
| 85 | Saccharomyces mikatae MIT | Fungi | Eukaryota |
| 86 | Saccharomyces paradoxus MIT | Fungi | Eukaryota |
| 87 | Saccharomyces cerevisiae SGD | Fungi | Eukaryota |
| 88 | Saccharomyces bayanus MIT | Fungi | Eukaryota |
| 89 | Pichia pastoris GS115 | Fungi | Eukaryota |
| 90 | Kluyveromyces lactis | Fungi | Eukaryota |
| 91 | Schizosaccharomyces octosporus yFS286 | Fungi | Eukaryota |
| 92 | Schizosaccharomyces japonicus yFS275 | Fungi | Eukaryota |
| 93 | Schizosaccharomyces pombe | Fungi | Eukaryota |
| 94 | Allomyces macrogynus ATCC 38327 | Fungi | Eukaryota |
| 95 | Rhizopus oryzae RA 99-880 | Fungi | Eukaryota |
| 96 | Phycomyces blakesleeanus | Fungi | Eukaryota |
| 97 | Mucor circinelloides | Fungi | Eukaryota |
| 98 | Spizellomyces punctatus DAOM BR117 | Fungi | Eukaryota |
| 99 | Batrachochytrium dendrobatidis JEL423 | Fungi | Eukaryota |
| 100 | Encephalitozoon cuniculi | Fungi | Eukaryota |
| 101 | Encephalitozoon intestinalis | Fungi | Eukaryota |
| 102 | Homo sapiens 59_37d (all transcripts) | Metazoa | Eukaryota |
| 103 | Pan troglodytes 59_21n (all transcripts) | Metazoa | Eukaryota |
| 104 | Gorilla gorilla 59_3b (all transcripts) | Metazoa | Eukaryota |
| 105 | Pongo pygmaeus 59_1e (all transcripts) | Metazoa | Eukaryota |
| 106 | Macaca mulatta 59_10n (all transcripts) | Metazoa | Eukaryota |
| 107 | Callithrix jacchus 59_321a (all transcripts) | Metazoa | Eukaryota |
| 108 | Otolemur garnettii 59_1g (all transcripts) | Metazoa | Eukaryota |
| 109 | Microcebus murinus 59_1d (all transcripts) | Metazoa | Eukaryota |
| 110 | Tarsius syrichta 59_1e (all transcripts) | Metazoa | Eukaryota |
| 111 | Rattus norvegicus 59_34a (all transcripts) | Metazoa | Eukaryota |
| 112 | Mus musculus 59_37l (all transcripts) | Metazoa | Eukaryota |
| 113 | Spermophilus tridecemlineatus 59_1i (all transcripts) | Metazoa | Eukaryota |
| 114 | Dipodomys ordii 59_1e (all transcripts) | Metazoa | Eukaryota |
| 115 | Cavia porcellus 59_3c (all transcripts) | Metazoa | Eukaryota |
| 116 | Oryctolagus cuniculus 59_2b (all transcripts) | Metazoa | Eukaryota |
| 117 | Ochotona princeps 59_1e (all transcripts) | Metazoa | Eukaryota |
| 118 | Tupaia belangeri 59_1h (all transcripts) | Metazoa | Eukaryota |
| 119 | Sus scrofa 59_9c (all transcripts) | Metazoa | Eukaryota |
| 120 | Bos taurus 59_4h (all transcripts) | Metazoa | Eukaryota |
| 121 | Vicugna pacos 59_1e (all transcripts) | Metazoa | Eukaryota |
| 122 | Tursiops truncatus 59_1e (all transcripts) | Metazoa | Eukaryota |
| 123 | Canis familiaris 59_2o (all transcripts) | Metazoa | Eukaryota |
| 124 | Felis catus 59_1h (all transcripts) | Metazoa | Eukaryota |
| 125 | Equus caballus 59_2f (all transcripts) | Metazoa | Eukaryota |
| 126 | Myotis lucifugus 59_1i (all transcripts) | Metazoa | Eukaryota |
| 127 | Pteropus vampyrus 59_1e (all transcripts) | Metazoa | Eukaryota |
| 128 | Sorex araneus 59_1g (all transcripts) | Metazoa | Eukaryota |
| 129 | Erinaceus europaeus 59_1g (all transcripts) | Metazoa | Eukaryota |
| 130 | Procavia capensis 59_1e (all transcripts) | Metazoa | Eukaryota |
| 131 | Loxodonta africana 59_3b (all transcripts) | Metazoa | Eukaryota |
| 132 | Echinops telfairi 59_1i (all transcripts) | Metazoa | Eukaryota |
| 133 | Dasypus novemcinctus 59_2c (all transcripts) | Metazoa | Eukaryota |
| 134 | Macropus eugenii 59_1b (all transcripts) | Metazoa | Eukaryota |
| 135 | Monodelphis domestica 59_5k (all transcripts) | Metazoa | Eukaryota |
| 136 | Ornithorhynchus anatinus 59_1m (all transcripts) | Metazoa | Eukaryota |
| 137 | Anolis carolinensis 59_1c (all transcripts) | Metazoa | Eukaryota |
| 138 | Taeniopygia guttata 59_1e (all transcripts) | Metazoa | Eukaryota |
| 139 | Meleagris gallopavo 57_2 (all transcripts) | Metazoa | Eukaryota |
| 140 | Gallus gallus 59_2o (all transcripts) | Metazoa | Eukaryota |
| 141 | Xenopus laevis | Metazoa | Eukaryota |
| 142 | Xenopus tropicalis 59_41p (all transcripts) | Metazoa | Eukaryota |
| 143 | Danio rerio 59_8e (all transcripts) | Metazoa | Eukaryota |
| 144 | Gasterosteus aculeatus 59_1l (all transcripts) | Metazoa | Eukaryota |
| 145 | Oryzias latipes 59_1k (all transcripts) | Metazoa | Eukaryota |
| 146 | Tetraodon nigroviridis 59_8d (all transcripts) | Metazoa | Eukaryota |
| 147 | Takifugu rubripes 59_4m (all transcripts) | Metazoa | Eukaryota |
| 148 | Branchiostoma floridae 1.0 | Metazoa | Eukaryota |
| 149 | Ciona savignyi 59_2j (all transcripts) | Metazoa | Eukaryota |
| 150 | Ciona intestinalis 59_2o (all transcripts) | Metazoa | Eukaryota |
| 151 | Strongylocentrotus purpuratus | Metazoa | Eukaryota |
| 152 | Helobdella robusta | Metazoa | Eukaryota |
| 153 | Capitella sp. I | Metazoa | Eukaryota |
| 154 | Bombyx mori | Metazoa | Eukaryota |
| 155 | Nasonia vitripennis | Metazoa | Eukaryota |
| 156 | Apis mellifera 38.2d (all transcripts) | Metazoa | Eukaryota |
| 157 | Drosophila grimshawi 1.3 | Metazoa | Eukaryota |
| 158 | Drosophila willistoni 1.3 | Metazoa | Eukaryota |
| 159 | Drosophila pseudoobscura 2.13 | Metazoa | Eukaryota |
| 160 | Drosophila persimilis 1.3 | Metazoa | Eukaryota |
| 161 | Drosophila yakuba 1.3 | Metazoa | Eukaryota |
| 162 | Drosophila simulans 1.3 | Metazoa | Eukaryota |
| 163 | Drosophila sechellia 1.3 | Metazoa | Eukaryota |
| 164 | Drosophila melanogaster 59_525a (all transcripts) | Metazoa | Eukaryota |
| 165 | Drosophila erecta 1.3 | Metazoa | Eukaryota |
| 166 | Drosophila ananassae 1.3 | Metazoa | Eukaryota |
| 167 | Drosophila virilis 1.2 | Metazoa | Eukaryota |
| 168 | Drosophila mojavensis 1.3 | Metazoa | Eukaryota |
| 169 | Aedes aegypti 55 (all transcripts) | Metazoa | Eukaryota |
| 170 | Culex pipiens quinquefasciatus | Metazoa | Eukaryota |
| 171 | Anopheles gambiae 49_3j (all transcripts) | Metazoa | Eukaryota |
| 172 | Tribolium castaneum 3.0 | Metazoa | Eukaryota |
| 173 | Pediculus humanus corporis | Metazoa | Eukaryota |
| 174 | Acyrthosiphon pisum | Metazoa | Eukaryota |
| 175 | Daphnia pulex | Metazoa | Eukaryota |
| 176 | Ixodes scapularis | Metazoa | Eukaryota |
| 177 | Lottia gigantea | Metazoa | Eukaryota |
| 178 | Pristionchus pacificus | Metazoa | Eukaryota |
| 179 | Meloidogyne incognita | Metazoa | Eukaryota |
| 180 | Brugia malayi WS218 | Metazoa | Eukaryota |
| 181 | Caenorhabditis japonica | Metazoa | Eukaryota |
| 182 | Caenorhabditis brenneri | Metazoa | Eukaryota |
| 183 | Caenorhabditis remanei | Metazoa | Eukaryota |
| 184 | Caenorhabditis elegans 59_210a (all transcripts) | Metazoa | Eukaryota |
| 185 | Caenorhabditis briggsae 2 | Metazoa | Eukaryota |
| 186 | Schistosoma mansoni | Metazoa | Eukaryota |
| 187 | Nematostella vectensis 1.0 | Metazoa | Eukaryota |
| 188 | Hydra magnipapillata | Metazoa | Eukaryota |
| 189 | Trichoplax adhaerens | Metazoa | Eukaryota |
| 190 | Giardia lamblia 2.3 | Protista | Eukaryota |
| 191 | Trypanosoma cruzi strain CL Brener | Protista | Eukaryota |
| 192 | Trypanosoma brucei | Protista | Eukaryota |
| 193 | Leishmania mexicana 2.4 | Protista | Eukaryota |
| 194 | Leishmania major strain Friedlin | Protista | Eukaryota |
| 195 | Leishmania infantum JPCM5 2.4 | Protista | Eukaryota |
| 196 | Leishmania braziliensis MHOM/BR/75/M2904 2.4 | Protista | Eukaryota |
| 197 | Aureococcus anophagefferens | Protista | Eukaryota |
| 198 | Phytophthora ramorum 1.1 | Protista | Eukaryota |
| 199 | Phytophthora sojae 1.1 | Protista | Eukaryota |
| 200 | Phytophthora infestans T30-4 | Protista | Eukaryota |
| 201 | Phytophthora capsici | Protista | Eukaryota |
| 202 | Paramecium tetraurelia | Protista | Eukaryota |
| 203 | Tetrahymena thermophila SB210 1 | Protista | Eukaryota |
| 204 | Babesia bovis T2Bo | Protista | Eukaryota |
| 205 | Theileria parva | Protista | Eukaryota |
| 206 | Theileria annulata | Protista | Eukaryota |
| 207 | Plasmodium falciparum 3D7 | Protista | Eukaryota |
| 208 | Plasmodium vivax SaI-1 7.0 | Protista | Eukaryota |
| 209 | Plasmodium knowlesi strain H | Protista | Eukaryota |
| 210 | Plasmodium yoelii ssp. yoelii 1 | Protista | Eukaryota |
| 211 | Plasmodium chabaudi | Protista | Eukaryota |
| 212 | Plasmodium berghei ANKA | Protista | Eukaryota |
| 213 | Cryptosporidium hominis | Protista | Eukaryota |
| 214 | Cryptosporidium muris | Protista | Eukaryota |
| 215 | Cryptosporidium parvum Iowa II | Protista | Eukaryota |
| 216 | Neospora caninum Nc-Liverpool 6.2 | Protista | Eukaryota |
| 217 | Neospora caninum | Protista | Eukaryota |
| 218 | Toxoplasma gondii ME49 | Protista | Eukaryota |
| 219 | Naegleria gruberi | Protista | Eukaryota |
| 220 | Guillardia theta | Protista | Eukaryota |
| 221 | Arabidopsis lyrata | Plantae | Eukaryota |
| 222 | Arabidopsis thaliana 10 (all transcripts) | Plantae | Eukaryota |
| 223 | Carica papaya | Plantae | Eukaryota |
| 224 | Medicago truncatula | Plantae | Eukaryota |
| 225 | Glycine max | Plantae | Eukaryota |
| 226 | Cucumis sativus | Plantae | Eukaryota |
| 227 | Populus trichocarpa 6.0 | Plantae | Eukaryota |
| 228 | Vitis vinifera | Plantae | Eukaryota |
| 229 | Brachypodium distachyon | Plantae | Eukaryota |
| 230 | Oryza sativa ssp. japonica 5.0 | Plantae | Eukaryota |
| 231 | Zea mays subsp. mays | Plantae | Eukaryota |
| 232 | Sorghum bicolor | Plantae | Eukaryota |
| 233 | Selaginella moellendorffii | Plantae | Eukaryota |
| 234 | Physcomitrella patens subsp. patens | Plantae | Eukaryota |
| 235 | Ostreococcus sp. RCC809 | Plantae | Eukaryota |
| 236 | Ostreococcus lucimarinus CCE9901 | Plantae | Eukaryota |
| 237 | Ostreococcus tauri | Plantae | Eukaryota |
| 238 | Micromonas sp. RCC299 | Plantae | Eukaryota |
| 239 | Micromonas pusilla CCMP1545 | Plantae | Eukaryota |
| 240 | Coccomyxa sp. C-169 | Plantae | Eukaryota |
| 241 | Chlorella sp. NC64A | Plantae | Eukaryota |
| 242 | Chlorella vulgaris | Plantae | Eukaryota |
| 243 | Volvox carteri f. nagariensis | Plantae | Eukaryota |
| 244 | Chlamydomonas reinhardtii 4.0 | Plantae | Eukaryota |
| 245 | Candidatus Koribacter versatilis Ellin345 | Acidobacteria | Bacteria |
| 246 | Candidatus Solibacter usitatus Ellin6076 | Acidobacteria | Bacteria |
| 247 | Acidobacterium capsulatum ATCC 51196 | Acidobacteria | Bacteria |
| 248 | Gardnerella vaginalis 409-05 | Actinobacteria | Bacteria |
| 249 | Bifidobacterium longum NCC2705 | Actinobacteria | Bacteria |
| 250 | Bifidobacterium animalis ssp. lactis AD011 | Actinobacteria | Bacteria |
| 251 | Bifidobacterium dentium Bd1 | Actinobacteria | Bacteria |
| 252 | Bifidobacterium adolescentis ATCC 15703 | Actinobacteria | Bacteria |
| 253 | Kineococcus radiotolerans SRS30216 | Actinobacteria | Bacteria |
| 254 | Catenulispora acidiphila DSM 44928 | Actinobacteria | Bacteria |
| 255 | Stackebrandtia nassauensis DSM 44728 | Actinobacteria | Bacteria |
| 256 | Acidothermus cellulolyticus 11B | Actinobacteria | Bacteria |
| 257 | Nakamurella multipartita DSM 44233 | Actinobacteria | Bacteria |
| 258 | Geodermatophilus obscurus DSM 43160 | Actinobacteria | Bacteria |
| 259 | Frankia sp. CcI3 | Actinobacteria | Bacteria |
| 260 | Frankia alni ACN14a | Actinobacteria | Bacteria |
| 261 | Thermobifida fusca YX | Actinobacteria | Bacteria |
| 262 | Thermomonospora curvata DSM 43183 | Actinobacteria | Bacteria |
| 263 | Streptosporangium roseum DSM 43021 | Actinobacteria | Bacteria |
| 264 | Streptomyces griseus ssp. griseus NBRC 13350 | Actinobacteria | Bacteria |
| 265 | Streptomyces avermitilis MA-4680 | Actinobacteria | Bacteria |
| 266 | Streptomyces scabiei 87.22 | Actinobacteria | Bacteria |
| 267 | Streptomyces coelicolor | Actinobacteria | Bacteria |
| 268 | Actinosynnema mirum DSM 43827 | Actinobacteria | Bacteria |
| 269 | Saccharomonospora viridis DSM 43017 | Actinobacteria | Bacteria |
| 270 | Saccharopolyspora erythraea NRRL 2338 | Actinobacteria | Bacteria |
| 271 | Kribbella flavida DSM 17836 | Actinobacteria | Bacteria |
| 272 | Nocardioides sp. JS614 | Actinobacteria | Bacteria |
| 273 | Propionibacterium acnes KPA171202 | Actinobacteria | Bacteria |
| 274 | Salinispora arenicola CNS-205 | Actinobacteria | Bacteria |
| 275 | Salinispora tropica CNB-440 | Actinobacteria | Bacteria |
| 276 | Gordonia bronchialis DSM 43247 | Actinobacteria | Bacteria |
| 277 | Rhodococcus jostii RHA1 | Actinobacteria | Bacteria |
| 278 | Rhodococcus opacus B4 | Actinobacteria | Bacteria |
| 279 | Rhodococcus erythropolis PR4 | Actinobacteria | Bacteria |
| 280 | Nocardia farcinica IFM 10152 | Actinobacteria | Bacteria |
| 281 | Mycobacterium abscessus ATCC 19977 | Actinobacteria | Bacteria |
| 282 | Mycobacterium sp. MCS | Actinobacteria | Bacteria |
| 283 | Mycobacterium avium ssp. paratuberculosis K-10 | Actinobacteria | Bacteria |
| 284 | Mycobacterium vanbaalenii PYR-1 | Actinobacteria | Bacteria |
| 285 | Mycobacterium tuberculosis H37Rv | Actinobacteria | Bacteria |
| 286 | Mycobacterium bovis AF2122/97 | Actinobacteria | Bacteria |
| 287 | Mycobacterium ulcerans Agy99 | Actinobacteria | Bacteria |
| 288 | Mycobacterium gilvum PYR-GCK | Actinobacteria | Bacteria |
| 289 | Mycobacterium marinum M | Actinobacteria | Bacteria |
| 290 | Mycobacterium smegmatis MC2 155 | Actinobacteria | Bacteria |
| 291 | Mycobacterium leprae TN | Actinobacteria | Bacteria |
| 292 | Corynebacterium aurimucosum ATCC 700975 | Actinobacteria | Bacteria |
| 293 | Corynebacterium kroppenstedtii DSM 44385 | Actinobacteria | Bacteria |
| 294 | Corynebacterium efficiens YS-314 | Actinobacteria | Bacteria |
| 295 | Corynebacterium urealyticum DSM 7109 | Actinobacteria | Bacteria |
| 296 | Corynebacterium jeikeium K411 | Actinobacteria | Bacteria |
| 297 | Corynebacterium glutamicum ATCC 13032 Kitasato | Actinobacteria | Bacteria |
| 298 | Corynebacterium diphtheriae NCTC 13129 | Actinobacteria | Bacteria |
| 299 | Tropheryma whipplei Twist | Actinobacteria | Bacteria |
| 300 | Sanguibacter keddieii DSM 10542 | Actinobacteria | Bacteria |
| 301 | Kytococcus sedentarius DSM 20547 | Actinobacteria | Bacteria |
| 302 | Beutenbergia cavernae DSM 12333 | Actinobacteria | Bacteria |
| 303 | Leifsonia xyli ssp. xyli CTCB07 | Actinobacteria | Bacteria |
| 304 | Clavibacter michiganensis ssp. michiganensis NCPPB 382 | Actinobacteria | Bacteria |
| 305 | Jonesia denitrificans DSM 20603 | Actinobacteria | Bacteria |
| 306 | Brachybacterium faecium DSM 4810 | Actinobacteria | Bacteria |
| 307 | Xylanimonas cellulosilytica DSM 15894 | Actinobacteria | Bacteria |
| 308 | Kocuria rhizophila DC2201 | Actinobacteria | Bacteria |
| 309 | Rothia mucilaginosa DY-18 | Actinobacteria | Bacteria |
| 310 | Arthrobacter sp. FB24 | Actinobacteria | Bacteria |
| 311 | Arthrobacter chlorophenolicus A6 | Actinobacteria | Bacteria |
| 312 | Arthrobacter aurescens TC1 | Actinobacteria | Bacteria |
| 313 | Renibacterium salmoninarum ATCC 33209 | Actinobacteria | Bacteria |
| 314 | Micrococcus luteus NCTC 2665 | Actinobacteria | Bacteria |
| 315 | Cryptobacterium curtum DSM 15641 | Actinobacteria | Bacteria |
| 316 | Eggerthella lenta DSM 2243 | Actinobacteria | Bacteria |
| 317 | Slackia heliotrinireducens DSM 20476 | Actinobacteria | Bacteria |
| 318 | Atopobium parvulum DSM 20469 | Actinobacteria | Bacteria |
| 319 | Conexibacter woesei DSM 14684 | Actinobacteria | Bacteria |
| 320 | Rubrobacter xylanophilus DSM 9941 | Actinobacteria | Bacteria |
| 321 | Acidimicrobium ferrooxidans DSM 10331 | Actinobacteria | Bacteria |
| 322 | Sulfurihydrogenibium sp. YO3AOP1 | Aquificae | Bacteria |
| 323 | Sulfurihydrogenibium azorense Az-Fu1 | Aquificae | Bacteria |
| 324 | Persephonella marina EX-H1 | Aquificae | Bacteria |
| 325 | Hydrogenobaculum sp. Y04AAS1 | Aquificae | Bacteria |
| 326 | Thermocrinis albus DSM 14484 | Aquificae | Bacteria |
| 327 | Aquifex aeolicus VF5 | Aquificae | Bacteria |
| 328 | Hydrogenobacter thermophilus TK-6 | Aquificae | Bacteria |
| 329 | Dyadobacter fermentans DSM 18053 | Bacteroidetes | Bacteria |
| 330 | Cytophaga hutchinsonii ATCC 33406 | Bacteroidetes | Bacteria |
| 331 | Spirosoma linguale DSM 74 | Bacteroidetes | Bacteria |
| 332 | Candidatus Azobacteroides pseudotrichonymphae genomovar. | Bacteroidetes | Bacteria |
| 333 | Prevotella ruminicola 23 | Bacteroidetes | Bacteria |
| 334 | Parabacteroides distasonis ATCC 8503 | Bacteroidetes | Bacteria |
| 335 | Porphyromonas gingivalis W83 | Bacteroidetes | Bacteria |
| 336 | Bacteroides vulgatus ATCC 8482 | Bacteroidetes | Bacteria |
| 337 | Bacteroides thetaiotaomicron VPI-5482 | Bacteroidetes | Bacteria |
| 338 | Bacteroides fragilis NCTC 9343 | Bacteroidetes | Bacteria |
| 339 | Candidatus Amoebophilus asiaticus 5a2 | Bacteroidetes | Bacteria |
| 340 | Salinibacter ruber DSM 13855 | Bacteroidetes | Bacteria |
| 341 | Rhodothermus marinus DSM 4252 | Bacteroidetes | Bacteria |
| 342 | Chitinophaga pinensis DSM 2588 | Bacteroidetes | Bacteria |
| 343 | Pedobacter heparinus DSM 2366 | Bacteroidetes | Bacteria |
| 344 | Candidatus Sulcia muelleri GWSS | Bacteroidetes | Bacteria |
| 345 | Zunongwangia profunda SM-A87 | Bacteroidetes | Bacteria |
| 346 | Gramella forsetii KT0803 | Bacteroidetes | Bacteria |
| 347 | Robiginitalea biformata HTCC2501 | Bacteroidetes | Bacteria |
| 348 | Flavobacteriaceae bacterium 3519-10 | Bacteroidetes | Bacteria |
| 349 | Capnocytophaga ochracea DSM 7271 | Bacteroidetes | Bacteria |
| 350 | Flavobacterium psychrophilum JIP02/86 | Bacteroidetes | Bacteria |
| 351 | Flavobacterium johnsoniae UW101 | Bacteroidetes | Bacteria |
| 352 | Blattabacterium sp. Bge | Bacteroidetes | Bacteria |
| 353 | Candidatus Protochlamydia amoebophila UWE25 | Chlamydiae | Bacteria |
| 354 | Chlamydophila pneumoniae TW-183 | Chlamydiae | Bacteria |
| 355 | Chlamydophila caviae GPIC | Chlamydiae | Bacteria |
| 356 | Chlamydophila felis Fe/C-56 | Chlamydiae | Bacteria |
| 357 | Chlamydophila abortus S26/3 | Chlamydiae | Bacteria |
| 358 | Chlamydia muridarum Nigg | Chlamydiae | Bacteria |
| 359 | Chlamydia trachomatis D/UW-3/CX | Chlamydiae | Bacteria |
| 360 | Pelodictyon phaeoclathratiforme BU-1 | Chlorobi | Bacteria |
| 361 | Chlorobium luteolum DSM 273 | Chlorobi | Bacteria |
| 362 | Chlorobium chlorochromatii CaD3 | Chlorobi | Bacteria |
| 363 | Chlorobium phaeobacteroides DSM 266 | Chlorobi | Bacteria |
| 364 | Chlorobium phaeovibrioides DSM 265 | Chlorobi | Bacteria |
| 365 | Chlorobium limicola DSM 245 | Chlorobi | Bacteria |
| 366 | Chlorobaculum parvum NCIB 8327 | Chlorobi | Bacteria |
| 367 | Chlorobium tepidum TLS | Chlorobi | Bacteria |
| 368 | Chloroherpeton thalassium ATCC 35110 | Chlorobi | Bacteria |
| 369 | Prosthecochloris aestuarii DSM 271 | Chlorobi | Bacteria |
| 370 | Dehalococcoides sp. CBDB1 | Chloroflexi | Bacteria |
| 371 | Dehalococcoides ethenogenes 195 | Chloroflexi | Bacteria |
| 372 | Thermomicrobium roseum DSM 5159 | Chloroflexi | Bacteria |
| 373 | Sphaerobacter thermophilus DSM 20745 | Chloroflexi | Bacteria |
| 374 | Herpetosiphon aurantiacus ATCC 23779 | Chloroflexi | Bacteria |
| 375 | Roseiflexus sp. RS-1 | Chloroflexi | Bacteria |
| 376 | Roseiflexus castenholzii DSM 13941 | Chloroflexi | Bacteria |
| 377 | Chloroflexus sp. Y-400-fl | Chloroflexi | Bacteria |
| 378 | Chloroflexus aggregans DSM 9485 | Chloroflexi | Bacteria |
| 379 | Chloroflexus aurantiacus J-10-fl | Chloroflexi | Bacteria |
| 380 | Gloeobacter violaceus PCC 7421 | Cyanobacteria | Bacteria |
| 381 | Acaryochloris marina MBIC11017 | Cyanobacteria | Bacteria |
| 382 | Prochlorococcus marinus MIT 9313 | Cyanobacteria | Bacteria |
| 383 | Nostoc punctiforme PCC 73102 | Cyanobacteria | Bacteria |
| 384 | Nostoc sp. PCC 7120 | Cyanobacteria | Bacteria |
| 385 | Anabaena variabilis ATCC 29413 | Cyanobacteria | Bacteria |
| 386 | Trichodesmium erythraeum IMS101 | Cyanobacteria | Bacteria |
| 387 | Thermosynechococcus elongatus BP-1 | Cyanobacteria | Bacteria |
| 388 | cyanobacterium UCYN-A | Cyanobacteria | Bacteria |
| 389 | Cyanothece sp. ATCC 51142 | Cyanobacteria | Bacteria |
| 390 | Synechocystis sp. PCC 6803 | Cyanobacteria | Bacteria |
| 391 | Synechococcus elongatus PCC 6301 | Cyanobacteria | Bacteria |
| 392 | Microcystis aeruginosa NIES-843 | Cyanobacteria | Bacteria |
| 393 | Denitrovibrio acetiphilus DSM 12809 | Deferribacteres | Bacteria |
| 394 | Deferribacter desulfuricans SSM1 | Deferribacteres | Bacteria |
| 395 | Deinococcus deserti VCD115 | Deinococcus-Thermus | Bacteria |
| 396 | Deinococcus geothermalis DSM 11300 | Deinococcus-Thermus | Bacteria |
| 397 | Deinococcus radiodurans R1 | Deinococcus-Thermus | Bacteria |
| 398 | Meiothermus ruber DSM 1279 | Deinococcus-Thermus | Bacteria |
| 399 | Thermus thermophilus HB27 | Deinococcus-Thermus | Bacteria |
| 400 | Dictyoglomus turgidum DSM 6724 | Dictyoglomi | Bacteria |
| 401 | Dictyoglomus thermophilum H-6-12 | Dictyoglomi | Bacteria |
| 402 | Elusimicrobium minutum Pei191 | Elusimicrobia | Bacteria |
| 403 | uncultured Termite group 1 bacterium phylotype Rs-D17 | Elusimicrobia | Bacteria |
| 404 | Fibrobacter succinogenes ssp. succinogenes S85 | Fibrobacteres | Bacteria |
| 405 | Acidaminococcus fermentans DSM 20731 | Firmicutes | Bacteria |
| 406 | Veillonella parvula DSM 2008 | Firmicutes | Bacteria |
| 407 | Natranaerobius thermophilus JW/NM-WN-LF | Firmicutes | Bacteria |
| 408 | Symbiobacterium thermophilum IAM 14863 | Firmicutes | Bacteria |
| 409 | Anaerococcus prevotii DSM 20548 | Firmicutes | Bacteria |
| 410 | Finegoldia magna ATCC 29328 | Firmicutes | Bacteria |
| 411 | Clostridiales genomosp. BVAB3 UPII9-5 | Firmicutes | Bacteria |
| 412 | Candidatus Desulforudis audaxviator MP104C | Firmicutes | Bacteria |
| 413 | Pelotomaculum thermopropionicum SI | Firmicutes | Bacteria |
| 414 | Desulfitobacterium hafniense Y51 | Firmicutes | Bacteria |
| 415 | Desulfotomaculum reducens MI-1 | Firmicutes | Bacteria |
| 416 | Desulfotomaculum acetoxidans DSM 771 | Firmicutes | Bacteria |
| 417 | Eubacterium rectale ATCC 33656 | Firmicutes | Bacteria |
| 418 | Eubacterium eligens ATCC 27750 | Firmicutes | Bacteria |
| 419 | Syntrophomonas wolfei ssp. wolfei Goettingen | Firmicutes | Bacteria |
| 420 | Heliobacterium modesticaldum Ice1 | Firmicutes | Bacteria |
| 421 | Alkaliphilus oremlandii OhILAs | Firmicutes | Bacteria |
| 422 | Alkaliphilus metalliredigens QYMF | Firmicutes | Bacteria |
| 423 | Clostridium phytofermentans ISDg | Firmicutes | Bacteria |
| 424 | Clostridium novyi NT | Firmicutes | Bacteria |
| 425 | Clostridium kluyveri DSM 555 | Firmicutes | Bacteria |
| 426 | Clostridium cellulolyticum H10 | Firmicutes | Bacteria |
| 427 | Clostridium beijerinckii NCIMB 8052 | Firmicutes | Bacteria |
| 428 | Clostridium thermocellum ATCC 27405 | Firmicutes | Bacteria |
| 429 | Clostridium tetani E88 | Firmicutes | Bacteria |
| 430 | Clostridium perfringens 13 | Firmicutes | Bacteria |
| 431 | Clostridium difficile 630 | Firmicutes | Bacteria |
| 432 | Clostridium botulinum A ATCC 3502 | Firmicutes | Bacteria |
| 433 | Clostridium acetobutylicum ATCC 824 | Firmicutes | Bacteria |
| 434 | Caldicellulosiruptor saccharolyticus DSM 8903 | Firmicutes | Bacteria |
| 435 | Anaerocellum thermophilum DSM 6725 | Firmicutes | Bacteria |
| 436 | Coprothermobacter proteolyticus DSM 5265 | Firmicutes | Bacteria |
| 437 | Thermoanaerobacter tengcongensis MB4 | Firmicutes | Bacteria |
| 438 | Carboxydothermus hydrogenoformans Z-2901 | Firmicutes | Bacteria |
| 439 | Moorella thermoacetica ATCC 39073 | Firmicutes | Bacteria |
| 440 | Ammonifex degensii KC4 | Firmicutes | Bacteria |
| 441 | Thermoanaerobacter pseudethanolicus ATCC 33223 | Firmicutes | Bacteria |
| 442 | Thermoanaerobacter sp. X514 | Firmicutes | Bacteria |
| 443 | Thermoanaerobacter italicus Ab9 | Firmicutes | Bacteria |
| 444 | Halothermothrix orenii H 168 | Firmicutes | Bacteria |
| 445 | Enterococcus faecalis V583 | Firmicutes | Bacteria |
| 446 | Oenococcus oeni PSU-1 | Firmicutes | Bacteria |
| 447 | Leuconostoc citreum KM20 | Firmicutes | Bacteria |
| 448 | Leuconostoc mesenteroides ssp. mesenteroides ATCC 8293 | Firmicutes | Bacteria |
| 449 | Lactobacillus casei ATCC 334 | Firmicutes | Bacteria |
| 450 | Lactobacillus crispatus ST1 | Firmicutes | Bacteria |
| 451 | Lactobacillus rhamnosus GG | Firmicutes | Bacteria |
| 452 | Lactobacillus johnsonii NCC 533 | Firmicutes | Bacteria |
| 453 | Lactobacillus salivarius UCC118 | Firmicutes | Bacteria |
| 454 | Lactobacillus fermentum IFO 3956 | Firmicutes | Bacteria |
| 455 | Lactobacillus sakei ssp. sakei 23K | Firmicutes | Bacteria |
| 456 | Lactobacillus reuteri DSM 20016 | Firmicutes | Bacteria |
| 457 | Lactobacillus gasseri ATCC 33323 | Firmicutes | Bacteria |
| 458 | Lactobacillus plantarum WCFS1 | Firmicutes | Bacteria |
| 459 | Lactobacillus helveticus DPC 4571 | Firmicutes | Bacteria |
| 460 | Lactobacillus delbrueckii ssp. bulgaricus ATCC 11842 | Firmicutes | Bacteria |
| 461 | Lactobacillus brevis ATCC 367 | Firmicutes | Bacteria |
| 462 | Lactobacillus acidophilus NCFM | Firmicutes | Bacteria |
| 463 | Pediococcus pentosaceus ATCC 25745 | Firmicutes | Bacteria |
| 464 | Lactococcus lactis ssp. lactis Il1403 | Firmicutes | Bacteria |
| 465 | Streptococcus gallolyticus UCN34 | Firmicutes | Bacteria |
| 466 | Streptococcus equi ssp. zooepidemicus MGCS10565 | Firmicutes | Bacteria |
| 467 | Streptococcus dysgalactiae ssp. equisimilis GGS_124 | Firmicutes | Bacteria |
| 468 | Streptococcus mitis B6 | Firmicutes | Bacteria |
| 469 | Streptococcus uberis 0140J | Firmicutes | Bacteria |
| 470 | Streptococcus pyogenes M1 GAS | Firmicutes | Bacteria |
| 471 | Streptococcus pneumoniae TIGR4 | Firmicutes | Bacteria |
| 472 | Streptococcus agalactiae NEM316 | Firmicutes | Bacteria |
| 473 | Streptococcus mutans UA159 | Firmicutes | Bacteria |
| 474 | Streptococcus thermophilus LMG 18311 | Firmicutes | Bacteria |
| 475 | Streptococcus suis 05ZYH33 | Firmicutes | Bacteria |
| 476 | Streptococcus sanguinis SK36 | Firmicutes | Bacteria |
| 477 | Streptococcus gordonii Challis subCH1 | Firmicutes | Bacteria |
| 478 | Exiguobacterium sp. AT1b | Firmicutes | Bacteria |
| 479 | Exiguobacterium sibiricum 255-15 | Firmicutes | Bacteria |
| 480 | Bacillus tusciae DSM 2912 | Firmicutes | Bacteria |
| 481 | Alicyclobacillus acidocaldarius ssp. acidocaldarius DSM 446 | Firmicutes | Bacteria |
| 482 | Brevibacillus brevis NBRC 100599 | Firmicutes | Bacteria |
| 483 | Paenibacillus sp. JDR-2 | Firmicutes | Bacteria |
| 484 | Listeria welshimeri ser. 6b SLCC5334 | Firmicutes | Bacteria |
| 485 | Listeria innocua Clip11262 | Firmicutes | Bacteria |
| 486 | Listeria seeligeri ser. 1/2b SLCC3954 | Firmicutes | Bacteria |
| 487 | Listeria monocytogenes EGD-e | Firmicutes | Bacteria |
| 488 | Lysinibacillus sphaericus C3-41 | Firmicutes | Bacteria |
| 489 | Oceanobacillus iheyensis HTE831 | Firmicutes | Bacteria |
| 490 | Anoxybacillus flavithermus WK1 | Firmicutes | Bacteria |
| 491 | Geobacillus sp. WCH70 | Firmicutes | Bacteria |
| 492 | Geobacillus thermodenitrificans NG80-2 | Firmicutes | Bacteria |
| 493 | Geobacillus kaustophilus HTA426 | Firmicutes | Bacteria |
| 494 | Bacillus subtilis ssp. subtilis 168 | Firmicutes | Bacteria |
| 495 | Bacillus licheniformis ATCC 14580 | Firmicutes | Bacteria |
| 496 | Bacillus amyloliquefaciens FZB42 | Firmicutes | Bacteria |
| 497 | Bacillus halodurans C-125 | Firmicutes | Bacteria |
| 498 | Bacillus weihenstephanensis KBAB4 | Firmicutes | Bacteria |
| 499 | Bacillus thuringiensis ser. konkukian 97-27 | Firmicutes | Bacteria |
| 500 | Bacillus cereus ATCC 14579 | Firmicutes | Bacteria |
| 501 | Bacillus anthracis Ames Ancestor | Firmicutes | Bacteria |
| 502 | Bacillus pseudofirmus OF4 | Firmicutes | Bacteria |
| 503 | Bacillus clausii KSM-K16 | Firmicutes | Bacteria |
| 504 | Bacillus pumilus SAFR-032 | Firmicutes | Bacteria |
| 505 | Bacillus megaterium QM B1551 | Firmicutes | Bacteria |
| 506 | Macrococcus caseolyticus JCSC5402 | Firmicutes | Bacteria |
| 507 | Staphylococcus saprophyticus ssp. saprophyticus ATCC 15305 | Firmicutes | Bacteria |
| 508 | Staphylococcus lugdunensis HKU09-01 | Firmicutes | Bacteria |
| 509 | Staphylococcus haemolyticus JCSC1435 | Firmicutes | Bacteria |
| 510 | Staphylococcus epidermidis RP62A | Firmicutes | Bacteria |
| 511 | Staphylococcus carnosus ssp. carnosus TM300 | Firmicutes | Bacteria |
| 512 | Staphylococcus aureus ssp. aureus NCTC 8325 | Firmicutes | Bacteria |
| 513 | Streptobacillus moniliformis DSM 12112 | Fusobacteria | Bacteria |
| 514 | Sebaldella termitidis ATCC 33386 | Fusobacteria | Bacteria |
| 515 | Leptotrichia buccalis C-1013-b | Fusobacteria | Bacteria |
| 516 | Fusobacterium nucleatum ssp. nucleatum ATCC 25586 | Fusobacteria | Bacteria |
| 517 | Gemmatimonas aurantiaca T-27 | Gemmatimonadetes | Bacteria |
| 518 | Thermodesulfovibrio yellowstonii DSM 11347 | Nitrospirae | Bacteria |
| 519 | Rhodopirellula baltica SH 1 | Planctomycetes | Bacteria |
| 520 | Pirellula staleyi DSM 6068 | Planctomycetes | Bacteria |
| 521 | Nautilia profundicola AmH | Proteobacteria | Bacteria |
| 522 | Sulfurospirillum deleyianum DSM 6946 | Proteobacteria | Bacteria |
| 523 | Arcobacter butzleri RM4018 | Proteobacteria | Bacteria |
| 524 | Campylobacter hominis ATCC BAA-381 | Proteobacteria | Bacteria |
| 525 | Campylobacter lari RM2100 | Proteobacteria | Bacteria |
| 526 | Campylobacter curvus 525.92 | Proteobacteria | Bacteria |
| 527 | Campylobacter concisus 13826 | Proteobacteria | Bacteria |
| 528 | Campylobacter jejuni ssp. jejuni NCTC 11168 | Proteobacteria | Bacteria |
| 529 | Campylobacter fetus ssp. fetus 82-40 | Proteobacteria | Bacteria |
| 530 | Sulfurimonas denitrificans DSM 1251 | Proteobacteria | Bacteria |
| 531 | Wolinella succinogenes DSM 1740 | Proteobacteria | Bacteria |
| 532 | Helicobacter hepaticus ATCC 51449 | Proteobacteria | Bacteria |
| 533 | Helicobacter mustelae 12198 | Proteobacteria | Bacteria |
| 534 | Helicobacter acinonychis Sheeba | Proteobacteria | Bacteria |
| 535 | Helicobacter pylori 26695 | Proteobacteria | Bacteria |
| 536 | Nitratiruptor sp. SB155-2 | Proteobacteria | Bacteria |
| 537 | Sulfurovum sp. NBC37-1 | Proteobacteria | Bacteria |
| 538 | Bdellovibrio bacteriovorus HD100 | Proteobacteria | Bacteria |
| 539 | Syntrophus aciditrophicus SB | Proteobacteria | Bacteria |
| 540 | Syntrophobacter fumaroxidans MPOB | Proteobacteria | Bacteria |
| 541 | Desulfotalea psychrophila LSv54 | Proteobacteria | Bacteria |
| 542 | Desulfatibacillum alkenivorans AK-01 | Proteobacteria | Bacteria |
| 543 | Desulfobacterium autotrophicum HRM2 | Proteobacteria | Bacteria |
| 544 | Desulfococcus oleovorans Hxd3 | Proteobacteria | Bacteria |
| 545 | Desulfohalobium retbaense DSM 5692 | Proteobacteria | Bacteria |
| 546 | Desulfomicrobium baculatum DSM 4028 | Proteobacteria | Bacteria |
| 547 | Lawsonia intracellularis PHE/MN1-00 | Proteobacteria | Bacteria |
| 548 | Desulfovibrio magneticus RS-1 | Proteobacteria | Bacteria |
| 549 | Desulfovibrio vulgaris Hildenborough | Proteobacteria | Bacteria |
| 550 | Desulfovibrio salexigens DSM 2638 | Proteobacteria | Bacteria |
| 551 | Desulfovibrio desulfuricans ssp. desulfuricans G20 | Proteobacteria | Bacteria |
| 552 | Pelobacter propionicus DSM 2379 | Proteobacteria | Bacteria |
| 553 | Pelobacter carbinolicus DSM 2380 | Proteobacteria | Bacteria |
| 554 | Geobacter uraniireducens Rf4 | Proteobacteria | Bacteria |
| 555 | Geobacter sp. FRC-32 | Proteobacteria | Bacteria |
| 556 | Geobacter lovleyi SZ | Proteobacteria | Bacteria |
| 557 | Geobacter bemidjiensis Bem | Proteobacteria | Bacteria |
| 558 | Geobacter sulfurreducens PCA | Proteobacteria | Bacteria |
| 559 | Geobacter metallireducens GS-15 | Proteobacteria | Bacteria |
| 560 | Haliangium ochraceum DSM 14365 | Proteobacteria | Bacteria |
| 561 | Sorangium cellulosum So ce 56 | Proteobacteria | Bacteria |
| 562 | Anaeromyxobacter sp. Fw109-5 | Proteobacteria | Bacteria |
| 563 | Anaeromyxobacter dehalogenans 2CP-C | Proteobacteria | Bacteria |
| 564 | Myxococcus xanthus DK 1622 | Proteobacteria | Bacteria |
| 565 | Magnetococcus sp. MC-1 | Proteobacteria | Bacteria |
| 566 | Sideroxydans lithotrophicus ES-1 | Proteobacteria | Bacteria |
| 567 | Aromatoleum aromaticum EbN1 | Proteobacteria | Bacteria |
| 568 | Dechloromonas aromatica RCB | Proteobacteria | Bacteria |
| 569 | Thauera sp. MZ1T | Proteobacteria | Bacteria |
| 570 | Laribacter hongkongensis HLHK9 | Proteobacteria | Bacteria |
| 571 | Chromobacterium violaceum ATCC 12472 | Proteobacteria | Bacteria |
| 572 | Neisseria meningitidis Z2491 | Proteobacteria | Bacteria |
| 573 | Neisseria gonorrhoeae FA 1090 | Proteobacteria | Bacteria |
| 574 | Methylotenera mobilis JLW8 | Proteobacteria | Bacteria |
| 575 | Methylovorus sp. SIP3-4 | Proteobacteria | Bacteria |
| 576 | Methylobacillus flagellatus KT | Proteobacteria | Bacteria |
| 577 | Thiobacillus denitrificans ATCC 25259 | Proteobacteria | Bacteria |
| 578 | Candidatus Accumulibacter phosphatis clade IIA UW-1 | Proteobacteria | Bacteria |
| 579 | Methylibium petroleiphilum PM1 | Proteobacteria | Bacteria |
| 580 | Leptothrix cholodnii SP-6 | Proteobacteria | Bacteria |
| 581 | Ralstonia eutropha JMP134 | Proteobacteria | Bacteria |
| 582 | Cupriavidus taiwanensis | Proteobacteria | Bacteria |
| 583 | Cupriavidus metallidurans CH34 | Proteobacteria | Bacteria |
| 584 | Ralstonia pickettii 12J | Proteobacteria | Bacteria |
| 585 | Ralstonia solanacearum GMI1000 | Proteobacteria | Bacteria |
| 586 | Polynucleobacter necessarius ssp. asymbioticus QLW-P1DMWA-1 | Proteobacteria | Bacteria |
| 587 | Burkholderia phytofirmans PsJN | Proteobacteria | Bacteria |
| 588 | Burkholderia phymatum STM815 | Proteobacteria | Bacteria |
| 589 | Burkholderia thailandensis E264 | Proteobacteria | Bacteria |
| 590 | Burkholderia pseudomallei K96243 | Proteobacteria | Bacteria |
| 591 | Burkholderia mallei ATCC 23344 | Proteobacteria | Bacteria |
| 592 | Burkholderia sp. 383 | Proteobacteria | Bacteria |
| 593 | Burkholderia ambifaria AMMD | Proteobacteria | Bacteria |
| 594 | Burkholderia cenocepacia AU 1054 | Proteobacteria | Bacteria |
| 595 | Burkholderia multivorans ATCC 17616 | Proteobacteria | Bacteria |
| 596 | Burkholderia vietnamiensis G4 | Proteobacteria | Bacteria |
| 597 | Burkholderia xenovorans LB400 | Proteobacteria | Bacteria |
| 598 | Burkholderia glumae BGR1 | Proteobacteria | Bacteria |
| 599 | Rhodoferax ferrireducens T118 | Proteobacteria | Bacteria |
| 600 | Verminephrobacter eiseniae EF01-2 | Proteobacteria | Bacteria |
| 601 | Delftia acidovorans SPH-1 | Proteobacteria | Bacteria |
| 602 | Polaromonas sp. JS666 | Proteobacteria | Bacteria |
| 603 | Polaromonas naphthalenivorans CJ2 | Proteobacteria | Bacteria |
| 604 | Variovorax paradoxus S110 | Proteobacteria | Bacteria |
| 605 | Acidovorax ebreus TPSY | Proteobacteria | Bacteria |
| 606 | Acidovorax sp. JS42 | Proteobacteria | Bacteria |
| 607 | Acidovorax citrulli AAC00-1 | Proteobacteria | Bacteria |
| 608 | Herminiimonas arsenicoxydans | Proteobacteria | Bacteria |
| 609 | Janthinobacterium sp. Marseille | Proteobacteria | Bacteria |
| 610 | Bordetella petrii DSM 12804 | Proteobacteria | Bacteria |
| 611 | Bordetella avium 197N | Proteobacteria | Bacteria |
| 612 | Bordetella pertussis Tohama I | Proteobacteria | Bacteria |
| 613 | Bordetella parapertussis 12822 | Proteobacteria | Bacteria |
| 614 | Bordetella bronchiseptica RB50 | Proteobacteria | Bacteria |
| 615 | Nitrosospira multiformis ATCC 25196 | Proteobacteria | Bacteria |
| 616 | Nitrosomonas eutropha C91 | Proteobacteria | Bacteria |
| 617 | Nitrosomonas europaea ATCC 19718 | Proteobacteria | Bacteria |
| 618 | Caulobacter sp. K31 | Proteobacteria | Bacteria |
| 619 | Caulobacter crescentus CB15 | Proteobacteria | Bacteria |
| 620 | Caulobacter segnis ATCC 21756 | Proteobacteria | Bacteria |
| 621 | Phenylobacterium zucineum HLK1 | Proteobacteria | Bacteria |
| 622 | Erythrobacter litoralis HTCC2594 | Proteobacteria | Bacteria |
| 623 | Sphingopyxis alaskensis RB2256 | Proteobacteria | Bacteria |
| 624 | Novosphingobium aromaticivorans DSM 12444 | Proteobacteria | Bacteria |
| 625 | Sphingobium japonicum UT26S | Proteobacteria | Bacteria |
| 626 | Sphingomonas wittichii RW1 | Proteobacteria | Bacteria |
| 627 | Zymomonas mobilis ssp. mobilis ZM4 | Proteobacteria | Bacteria |
| 628 | Maricaulis maris MCS10 | Proteobacteria | Bacteria |
| 629 | Hirschia baltica ATCC 49814 | Proteobacteria | Bacteria |
| 630 | Hyphomonas neptunium ATCC 15444 | Proteobacteria | Bacteria |
| 631 | Dinoroseobacter shibae DFL 12 | Proteobacteria | Bacteria |
| 632 | Jannaschia sp. CCS1 | Proteobacteria | Bacteria |
| 633 | Ruegeria sp. TM1040 | Proteobacteria | Bacteria |
| 634 | Ruegeria pomeroyi DSS-3 | Proteobacteria | Bacteria |
| 635 | Roseobacter denitrificans OCh 114 | Proteobacteria | Bacteria |
| 636 | Rhodobacter sphaeroides 2.4.1 | Proteobacteria | Bacteria |
| 637 | Rhodobacter capsulatus SB 1003 | Proteobacteria | Bacteria |
| 638 | Paracoccus denitrificans PD1222 | Proteobacteria | Bacteria |
| 639 | Magnetospirillum magneticum AMB-1 | Proteobacteria | Bacteria |
| 640 | Rhodospirillum centenum SW | Proteobacteria | Bacteria |
| 641 | Rhodospirillum rubrum ATCC 11170 | Proteobacteria | Bacteria |
| 642 | Azospirillum sp. B510 | Proteobacteria | Bacteria |
| 643 | Granulibacter bethesdensis CGDNIH1 | Proteobacteria | Bacteria |
| 644 | Gluconacetobacter diazotrophicus PAl 5 | Proteobacteria | Bacteria |
| 645 | Gluconobacter oxydans 621H | Proteobacteria | Bacteria |
| 646 | Acetobacter pasteurianus IFO 3283-01 | Proteobacteria | Bacteria |
| 647 | Candidatus Puniceispirillum marinum IMCC1322 | Proteobacteria | Bacteria |
| 648 | Candidatus Pelagibacter ubique HTCC1062 | Proteobacteria | Bacteria |
| 649 | Neorickettsia sennetsu Miyayama | Proteobacteria | Bacteria |
| 650 | Neorickettsia risticii Illinois | Proteobacteria | Bacteria |
| 651 | Wolbachia endosymbiont of Culex_quinquefasciatus Pel | Proteobacteria | Bacteria |
| 652 | Wolbachia endosymbiont of Drosophila melanogaster | Proteobacteria | Bacteria |
| 653 | Wolbachia endosymbiont TRS of Brugia malayi | Proteobacteria | Bacteria |
| 654 | Wolbachia sp. wRi | Proteobacteria | Bacteria |
| 655 | Ehrlichia chaffeensis Arkansas | Proteobacteria | Bacteria |
| 656 | Ehrlichia canis Jake | Proteobacteria | Bacteria |
| 657 | Ehrlichia ruminantium Welgevonden | Proteobacteria | Bacteria |
| 658 | Anaplasma phagocytophilum HZ | Proteobacteria | Bacteria |
| 659 | Anaplasma marginale St. Maries | Proteobacteria | Bacteria |
| 660 | Anaplasma centrale Israel | Proteobacteria | Bacteria |
| 661 | Orientia tsutsugamushi Boryong | Proteobacteria | Bacteria |
| 662 | Rickettsia bellii RML369-C | Proteobacteria | Bacteria |
| 663 | Rickettsia canadensis McKiel | Proteobacteria | Bacteria |
| 664 | Rickettsia typhi Wilmington | Proteobacteria | Bacteria |
| 665 | Rickettsia prowazekii Madrid E | Proteobacteria | Bacteria |
| 666 | Rickettsia peacockii Rustic | Proteobacteria | Bacteria |
| 667 | Rickettsia felis URRWXCal2 | Proteobacteria | Bacteria |
| 668 | Rickettsia massiliae MTU5 | Proteobacteria | Bacteria |
| 669 | Rickettsia africae ESF-5 | Proteobacteria | Bacteria |
| 670 | Rickettsia akari Hartford | Proteobacteria | Bacteria |
| 671 | Rickettsia rickettsii Sheila Smith | Proteobacteria | Bacteria |
| 672 | Rickettsia conorii Malish 7 | Proteobacteria | Bacteria |
| 673 | Xanthobacter autotrophicus Py2 | Proteobacteria | Bacteria |
| 674 | Azorhizobium caulinodans ORS 571 | Proteobacteria | Bacteria |
| 675 | Methylobacterium chloromethanicum CM4 | Proteobacteria | Bacteria |
| 676 | Methylobacterium extorquens PA1 | Proteobacteria | Bacteria |
| 677 | Methylobacterium sp. 4-46 | Proteobacteria | Bacteria |
| 678 | Methylobacterium populi BJ001 | Proteobacteria | Bacteria |
| 679 | Methylobacterium nodulans ORS 2060 | Proteobacteria | Bacteria |
| 680 | Methylobacterium radiotolerans JCM 2831 | Proteobacteria | Bacteria |
| 681 | Candidatus Hodgkinia cicadicola Dsem | Proteobacteria | Bacteria |
| 682 | Ochrobactrum anthropi ATCC 49188 | Proteobacteria | Bacteria |
| 683 | Brucella microti CCM 4915 | Proteobacteria | Bacteria |
| 684 | Brucella canis ATCC 23365 | Proteobacteria | Bacteria |
| 685 | Brucella suis 1330 | Proteobacteria | Bacteria |
| 686 | Brucella melitensis bv. 1 16M | Proteobacteria | Bacteria |
| 687 | Brucella ovis ATCC 25840 | Proteobacteria | Bacteria |
| 688 | Brucella abortus bv. 1 9-941 | Proteobacteria | Bacteria |
| 689 | Rhizobium sp. NGR234 | Proteobacteria | Bacteria |
| 690 | Sinorhizobium medicae WSM419 | Proteobacteria | Bacteria |
| 691 | Sinorhizobium meliloti 1021 | Proteobacteria | Bacteria |
| 692 | Rhizobium etli CFN 42 | Proteobacteria | Bacteria |
| 693 | Rhizobium leguminosarum bv. viciae 3841 | Proteobacteria | Bacteria |
| 694 | Agrobacterium vitis S4 | Proteobacteria | Bacteria |
| 695 | Agrobacterium radiobacter K84 | Proteobacteria | Bacteria |
| 696 | Agrobacterium tumefaciens C58 | Proteobacteria | Bacteria |
| 697 | Candidatus Liberibacter asiaticus psy62 | Proteobacteria | Bacteria |
| 698 | Chelativorans sp. BNC1 | Proteobacteria | Bacteria |
| 699 | Parvibaculum lavamentivorans DS-1 | Proteobacteria | Bacteria |
| 700 | Mesorhizobium loti MAFF303099 | Proteobacteria | Bacteria |
| 701 | Methylocella silvestris BL2 | Proteobacteria | Bacteria |
| 702 | Beijerinckia indica ssp. indica ATCC 9039 | Proteobacteria | Bacteria |
| 703 | Oligotropha carboxidovorans OM5 | Proteobacteria | Bacteria |
| 704 | Rhodopseudomonas palustris CGA009 | Proteobacteria | Bacteria |
| 705 | Nitrobacter winogradskyi Nb-255 | Proteobacteria | Bacteria |
| 706 | Nitrobacter hamburgensis X14 | Proteobacteria | Bacteria |
| 707 | Bradyrhizobium sp. ORS278 | Proteobacteria | Bacteria |
| 708 | Bradyrhizobium japonicum USDA 110 | Proteobacteria | Bacteria |
| 709 | Bartonella tribocorum CIP 105476 | Proteobacteria | Bacteria |
| 710 | Bartonella henselae Houston-1 | Proteobacteria | Bacteria |
| 711 | Bartonella grahamii as4aup | Proteobacteria | Bacteria |
| 712 | Bartonella quintana Toulouse | Proteobacteria | Bacteria |
| 713 | Bartonella bacilliformis KC583 | Proteobacteria | Bacteria |
| 714 | Acidithiobacillus ferrooxidans ATCC 23270 | Proteobacteria | Bacteria |
| 715 | Mannheimia succiniciproducens MBEL55E | Proteobacteria | Bacteria |
| 716 | Aggregatibacter aphrophilus NJ8700 | Proteobacteria | Bacteria |
| 717 | Aggregatibacter actinomycetemcomitans D11S-1 | Proteobacteria | Bacteria |
| 718 | Haemophilus somnus 129PT | Proteobacteria | Bacteria |
| 719 | Pasteurella multocida ssp. multocida Pm70 | Proteobacteria | Bacteria |
| 720 | Haemophilus parasuis SH0165 | Proteobacteria | Bacteria |
| 721 | Haemophilus ducreyi 35000HP | Proteobacteria | Bacteria |
| 722 | Haemophilus influenzae Rd KW20 | Proteobacteria | Bacteria |
| 723 | Actinobacillus succinogenes 130Z | Proteobacteria | Bacteria |
| 724 | Actinobacillus pleuropneumoniae L20 | Proteobacteria | Bacteria |
| 725 | Tolumonas auensis DSM 9187 | Proteobacteria | Bacteria |
| 726 | Aeromonas salmonicida ssp. salmonicida A449 | Proteobacteria | Bacteria |
| 727 | Aeromonas hydrophila ssp. hydrophila ATCC 7966 | Proteobacteria | Bacteria |
| 728 | Aliivibrio salmonicida LFI1238 | Proteobacteria | Bacteria |
| 729 | Vibrio fischeri ES114 | Proteobacteria | Bacteria |
| 730 | Vibrio parahaemolyticus RIMD 2210633 | Proteobacteria | Bacteria |
| 731 | Vibrio harveyi ATCC BAA-1116 | Proteobacteria | Bacteria |
| 732 | Vibrio sp. Ex25 | Proteobacteria | Bacteria |
| 733 | Vibrio splendidus LGP32 | Proteobacteria | Bacteria |
| 734 | Vibrio vulnificus YJ016 | Proteobacteria | Bacteria |
| 735 | Vibrio cholerae O1 biov. El Tor N16961 | Proteobacteria | Bacteria |
| 736 | Photobacterium profundum SS9 | Proteobacteria | Bacteria |
| 737 | Psychromonas ingrahamii 37 | Proteobacteria | Bacteria |
| 738 | Idiomarina loihiensis L2TR | Proteobacteria | Bacteria |
| 739 | Shewanella piezotolerans WP3 | Proteobacteria | Bacteria |
| 740 | Shewanella loihica PV-4 | Proteobacteria | Bacteria |
| 741 | Shewanella halifaxensis HAW-EB4 | Proteobacteria | Bacteria |
| 742 | Shewanella sediminis HAW-EB3 | Proteobacteria | Bacteria |
| 743 | Shewanella denitrificans OS217 | Proteobacteria | Bacteria |
| 744 | Shewanella pealeana ATCC 700345 | Proteobacteria | Bacteria |
| 745 | Shewanella oneidensis MR-1 | Proteobacteria | Bacteria |
| 746 | Shewanella baltica OS155 | Proteobacteria | Bacteria |
| 747 | Shewanella woodyi ATCC 51908 | Proteobacteria | Bacteria |
| 748 | Shewanella sp. MR-7 | Proteobacteria | Bacteria |
| 749 | Shewanella amazonensis SB2B | Proteobacteria | Bacteria |
| 750 | Shewanella violacea DSS12 | Proteobacteria | Bacteria |
| 751 | Shewanella frigidimarina NCIMB 400 | Proteobacteria | Bacteria |
| 752 | Shewanella putrefaciens CN-32 | Proteobacteria | Bacteria |
| 753 | Colwellia psychrerythraea 34H | Proteobacteria | Bacteria |
| 754 | Pseudoalteromonas atlantica T6c | Proteobacteria | Bacteria |
| 755 | Pseudoalteromonas haloplanktis TAC125 | Proteobacteria | Bacteria |
| 756 | Teredinibacter turnerae T7901 | Proteobacteria | Bacteria |
| 757 | Saccharophagus degradans 2-40 | Proteobacteria | Bacteria |
| 758 | Marinobacter aquaeolei VT8 | Proteobacteria | Bacteria |
| 759 | Alteromonas macleodii Deep ecotype | Proteobacteria | Bacteria |
| 760 | Hahella chejuensis KCTC 2396 | Proteobacteria | Bacteria |
| 761 | Kangiella koreensis DSM 16069 | Proteobacteria | Bacteria |
| 762 | Alcanivorax borkumensis SK2 | Proteobacteria | Bacteria |
| 763 | Marinomonas sp. MWYL1 | Proteobacteria | Bacteria |
| 764 | Chromohalobacter salexigens DSM 3043 | Proteobacteria | Bacteria |
| 765 | Methylococcus capsulatus Bath | Proteobacteria | Bacteria |
| 766 | Dichelobacter nodosus VCS1703A | Proteobacteria | Bacteria |
| 767 | Stenotrophomonas maltophilia R551-3 | Proteobacteria | Bacteria |
| 768 | Xylella fastidiosa 9a5c | Proteobacteria | Bacteria |
| 769 | Xanthomonas axonopodis pv. citri 306 | Proteobacteria | Bacteria |
| 770 | Xanthomonas albilineans | Proteobacteria | Bacteria |
| 771 | Xanthomonas oryzae pv. oryzae KACC10331 | Proteobacteria | Bacteria |
| 772 | Xanthomonas campestris pv. campestris ATCC 33913 | Proteobacteria | Bacteria |
| 773 | Halothiobacillus neapolitanus c2 | Proteobacteria | Bacteria |
| 774 | Alkalilimnicola ehrlichii MLHE-1 | Proteobacteria | Bacteria |
| 775 | Thioalkalivibrio sp. HL-EbGR7 | Proteobacteria | Bacteria |
| 776 | Halorhodospira halophila SL1 | Proteobacteria | Bacteria |
| 777 | Allochromatium vinosum DSM 180 | Proteobacteria | Bacteria |
| 778 | Nitrosococcus halophilus Nc4 | Proteobacteria | Bacteria |
| 779 | Nitrosococcus oceani ATCC 19707 | Proteobacteria | Bacteria |
| 780 | Coxiella burnetii RSA 493 | Proteobacteria | Bacteria |
| 781 | Legionella longbeachae NSW150 | Proteobacteria | Bacteria |
| 782 | Legionella pneumophila ssp. pneumophila Philadelphia 1 | Proteobacteria | Bacteria |
| 783 | Baumannia cicadellinicola Hc | Proteobacteria | Bacteria |
| 784 | Candidatus Carsonella ruddii PV | Proteobacteria | Bacteria |
| 785 | Candidatus Vesicomyosocius okutanii HA | Proteobacteria | Bacteria |
| 786 | Candidatus Ruthia magnifica Cm | Proteobacteria | Bacteria |
| 787 | Cronobacter turicensis z3032 | Proteobacteria | Bacteria |
| 788 | Cronobacter sakazakii ATCC BAA-894 | Proteobacteria | Bacteria |
| 789 | Candidatus Riesia pediculicola USDA | Proteobacteria | Bacteria |
| 790 | Dickeya zeae Ech1591 | Proteobacteria | Bacteria |
| 791 | Dickeya dadantii Ech703 | Proteobacteria | Bacteria |
| 792 | Candidatus Hamiltonella defensa 5AT | Proteobacteria | Bacteria |
| 793 | Candidatus Blochmannia floridanus | Proteobacteria | Bacteria |
| 794 | Pectobacterium wasabiae WPP163 | Proteobacteria | Bacteria |
| 795 | Pectobacterium atrosepticum SCRI1043 | Proteobacteria | Bacteria |
| 796 | Pectobacterium carotovorum ssp. carotovorum PC1 | Proteobacteria | Bacteria |
| 797 | Sodalis glossinidius morsitans | Proteobacteria | Bacteria |
| 798 | Pantoea ananatis LMG 20103 | Proteobacteria | Bacteria |
| 799 | Wigglesworthia glossinidia | Proteobacteria | Bacteria |
| 800 | Buchnera aphidicola APS | Proteobacteria | Bacteria |
| 801 | Photorhabdus asymbiotica | Proteobacteria | Bacteria |
| 802 | Photorhabdus luminescens ssp. laumondii TTO1 | Proteobacteria | Bacteria |
| 803 | Edwardsiella ictaluri 93-146 | Proteobacteria | Bacteria |
| 804 | Edwardsiella tarda EIB202 | Proteobacteria | Bacteria |
| 805 | Yersinia pseudotuberculosis IP 32953 | Proteobacteria | Bacteria |
| 806 | Yersinia pestis CO92 | Proteobacteria | Bacteria |
| 807 | Yersinia enterocolitica ssp. enterocolitica 8081 | Proteobacteria | Bacteria |
| 808 | Xenorhabdus bovienii SS-2004 | Proteobacteria | Bacteria |
| 809 | Shigella sonnei Ss046 | Proteobacteria | Bacteria |
| 810 | Shigella flexneri 2a 2457T | Proteobacteria | Bacteria |
| 811 | Shigella dysenteriae Sd197 | Proteobacteria | Bacteria |
| 812 | Shigella boydii Sb227 | Proteobacteria | Bacteria |
| 813 | Serratia proteamaculans 568 | Proteobacteria | Bacteria |
| 814 | Salmonella enterica ssp. enterica ser. Typhimurium LT2 | Proteobacteria | Bacteria |
| 815 | Proteus mirabilis HI4320 | Proteobacteria | Bacteria |
| 816 | Klebsiella variicola At-22 | Proteobacteria | Bacteria |
| 817 | Klebsiella pneumoniae ssp. pneumoniae MGH 78578 | Proteobacteria | Bacteria |
| 818 | Escherichia fergusonii ATCC 35469 | Proteobacteria | Bacteria |
| 819 | Escherichia coli K-12 subMG1655 | Proteobacteria | Bacteria |
| 820 | Erwinia tasmaniensis Et1/99 | Proteobacteria | Bacteria |
| 821 | Erwinia pyrifoliae Ep1/96 | Proteobacteria | Bacteria |
| 822 | Erwinia amylovora ATCC 49946 | Proteobacteria | Bacteria |
| 823 | Enterobacter sp. 638 | Proteobacteria | Bacteria |
| 824 | Citrobacter rodentium ICC168 | Proteobacteria | Bacteria |
| 825 | Citrobacter koseri ATCC BAA-895 | Proteobacteria | Bacteria |
| 826 | Azotobacter vinelandii DJ | Proteobacteria | Bacteria |
| 827 | Pseudomonas entomophila L48 | Proteobacteria | Bacteria |
| 828 | Pseudomonas syringae pv. tomato DC3000 | Proteobacteria | Bacteria |
| 829 | Pseudomonas stutzeri A1501 | Proteobacteria | Bacteria |
| 830 | Pseudomonas putida KT2440 | Proteobacteria | Bacteria |
| 831 | Pseudomonas fluorescens Pf-5 | Proteobacteria | Bacteria |
| 832 | Pseudomonas mendocina ymp | Proteobacteria | Bacteria |
| 833 | Pseudomonas aeruginosa PAO1 | Proteobacteria | Bacteria |
| 834 | Cellvibrio japonicus Ueda107 | Proteobacteria | Bacteria |
| 835 | Psychrobacter sp. PRwf-1 | Proteobacteria | Bacteria |
| 836 | Psychrobacter arcticus 273-4 | Proteobacteria | Bacteria |
| 837 | Psychrobacter cryohalolentis K5 | Proteobacteria | Bacteria |
| 838 | Acinetobacter baumannii ATCC 17978 | Proteobacteria | Bacteria |
| 839 | Acinetobacter sp. ADP1 | Proteobacteria | Bacteria |
| 840 | Thiomicrospira crunogena XCL-2 | Proteobacteria | Bacteria |
| 841 | Francisella philomiragia ssp. philomiragia ATCC 25017 | Proteobacteria | Bacteria |
| 842 | Francisella tularensis ssp. tularensis SCHU S4 | Proteobacteria | Bacteria |
| 843 | Brachyspira hyodysenteriae WA1 | Spirochaetes | Bacteria |
| 844 | Leptospira borgpetersenii ser. Hardjo-bovis L550 | Spirochaetes | Bacteria |
| 845 | Leptospira interrogans ser. Lai 56601 | Spirochaetes | Bacteria |
| 846 | Leptospira biflexa ser. Patoc Patoc 1 (Paris) | Spirochaetes | Bacteria |
| 847 | Treponema pallidum ssp. pallidum Nichols | Spirochaetes | Bacteria |
| 848 | Treponema denticola ATCC 35405 | Spirochaetes | Bacteria |
| 849 | Borrelia garinii PBi | Spirochaetes | Bacteria |
| 850 | Borrelia afzelii PKo | Spirochaetes | Bacteria |
| 851 | Borrelia burgdorferi B31 | Spirochaetes | Bacteria |
| 852 | Borrelia recurrentis A1 | Spirochaetes | Bacteria |
| 853 | Borrelia duttonii Ly | Spirochaetes | Bacteria |
| 854 | Borrelia turicatae 91E135 | Spirochaetes | Bacteria |
| 855 | Borrelia hermsii DAH | Spirochaetes | Bacteria |
| 856 | Aminobacterium colombiense DSM 12261 | Synergistetes | Bacteria |
| 857 | Thermanaerovibrio acidaminovorans DSM 6589 | Synergistetes | Bacteria |
| 858 | Candidatus Phytoplasma mali | Tenericutes | Bacteria |
| 859 | Aster yellows witches-broom phytoplasma AYWB | Tenericutes | Bacteria |
| 860 | Onion yellows phytoplasma OY-M | Tenericutes | Bacteria |
| 861 | Acholeplasma laidlawii PG-8A | Tenericutes | Bacteria |
| 862 | Mesoplasma florum L1 | Tenericutes | Bacteria |
| 863 | Ureaplasma parvum ser. 3 ATCC 700970 | Tenericutes | Bacteria |
| 864 | Ureaplasma urealyticum ser. 10 ATCC 33699 | Tenericutes | Bacteria |
| 865 | Mycoplasma mycoides ssp. mycoides SC PG1 | Tenericutes | Bacteria |
| 866 | Mycoplasma capricolum ssp. capricolum ATCC 27343 | Tenericutes | Bacteria |
| 867 | Mycoplasma crocodyli MP145 | Tenericutes | Bacteria |
| 868 | Mycoplasma conjunctivae HRC/581 | Tenericutes | Bacteria |
| 869 | Mycoplasma penetrans HF-2 | Tenericutes | Bacteria |
| 870 | Mycoplasma mobile 163K | Tenericutes | Bacteria |
| 871 | Mycoplasma arthritidis 158L3-1 | Tenericutes | Bacteria |
| 872 | Mycoplasma agalactiae PG2 | Tenericutes | Bacteria |
| 873 | Mycoplasma synoviae 53 | Tenericutes | Bacteria |
| 874 | Mycoplasma pulmonis UAB CTIP | Tenericutes | Bacteria |
| 875 | Mycoplasma pneumoniae M129 | Tenericutes | Bacteria |
| 876 | Mycoplasma hyopneumoniae 232 | Tenericutes | Bacteria |
| 877 | Mycoplasma hominis | Tenericutes | Bacteria |
| 878 | Mycoplasma genitalium G37 | Tenericutes | Bacteria |
| 879 | Mycoplasma gallisepticum R(low) | Tenericutes | Bacteria |
| 880 | Kosmotoga olearia TBF 19.5.1 | Thermotogae | Bacteria |
| 881 | Petrotoga mobilis SJ95 | Thermotogae | Bacteria |
| 882 | Fervidobacterium nodosum Rt17-B1 | Thermotogae | Bacteria |
| 883 | Thermosipho melanesiensis BI429 | Thermotogae | Bacteria |
| 884 | Thermosipho africanus TCF52B | Thermotogae | Bacteria |
| 885 | Thermotoga lettingae TMO | Thermotogae | Bacteria |
| 886 | Thermotoga sp. RQ2 | Thermotogae | Bacteria |
| 887 | Thermotoga naphthophila RKU-10 | Thermotogae | Bacteria |
| 888 | Thermotoga petrophila RKU-1 | Thermotogae | Bacteria |
| 889 | Thermotoga neapolitana DSM 4359 | Thermotogae | Bacteria |
| 890 | Thermotoga maritima MSB8 | Thermotogae | Bacteria |
| 891 | Coraliomargarita akajimensis DSM 45221 | Verrucomicrobia | Bacteria |
| 892 | Opitutus terrae PB90-1 | Verrucomicrobia | Bacteria |
| 893 | Methylacidiphilum infernorum V4 | Verrucomicrobia | Bacteria |
| 894 | Akkermansia muciniphila ATCC BAA-835 | Verrucomicrobia | Bacteria |
| 895 | Thermobaculum terrenum ATCC BAA-798 | Bacteria | |
| 896 | Hyperthermus butylicus DSM 5456 | Crenarchaeota | Archaea |
| 897 | Aeropyrum pernix K1 | Crenarchaeota | Archaea |
| 898 | Ignicoccus hospitalis KIN4/I | Crenarchaeota | Archaea |
| 899 | Staphylothermus marinus F1 | Crenarchaeota | Archaea |
| 900 | Desulfurococcus kamchatkensis 1221n | Crenarchaeota | Archaea |
| 901 | Metallosphaera sedula DSM 5348 | Crenarchaeota | Archaea |
| 902 | Sulfolobus tokodaii 7 | Crenarchaeota | Archaea |
| 903 | Sulfolobus islandicus Y.N.15.51 | Crenarchaeota | Archaea |
| 904 | Sulfolobus solfataricus P2 | Crenarchaeota | Archaea |
| 905 | Sulfolobus acidocaldarius DSM 639 | Crenarchaeota | Archaea |
| 906 | Thermofilum pendens Hrk 5 | Crenarchaeota | Archaea |
| 907 | Caldivirga maquilingensis IC-167 | Crenarchaeota | Archaea |
| 908 | Pyrobaculum calidifontis JCM 11548 | Crenarchaeota | Archaea |
| 909 | Pyrobaculum arsenaticum DSM 13514 | Crenarchaeota | Archaea |
| 910 | Pyrobaculum aerophilum IM2 | Crenarchaeota | Archaea |
| 911 | Pyrobaculum islandicum DSM 4184 | Crenarchaeota | Archaea |
| 912 | Thermoproteus neutrophilus V24Sta | Crenarchaeota | Archaea |
| 913 | Methanocella paludicola SANAE | Euryarchaeota | Archaea |
| 914 | Methanosaeta thermophila PT | Euryarchaeota | Archaea |
| 915 | Methanococcoides burtonii DSM 6242 | Euryarchaeota | Archaea |
| 916 | Methanosarcina acetivorans C2A | Euryarchaeota | Archaea |
| 917 | Methanosarcina mazei Go1 | Euryarchaeota | Archaea |
| 918 | Methanosarcina barkeri Fusaro | Euryarchaeota | Archaea |
| 919 | Methanohalophilus mahii DSM 5219 | Euryarchaeota | Archaea |
| 920 | Methanosphaerula palustris E1-9c | Euryarchaeota | Archaea |
| 921 | Candidatus Methanoregula boonei 6A8 | Euryarchaeota | Archaea |
| 922 | Methanospirillum hungatei JF-1 | Euryarchaeota | Archaea |
| 923 | Methanocorpusculum labreanum Z | Euryarchaeota | Archaea |
| 924 | Methanoculleus marisnigri JR1 | Euryarchaeota | Archaea |
| 925 | Methanopyrus kandleri AV19 | Euryarchaeota | Archaea |
| 926 | Ferroglobus placidus DSM 10642 | Euryarchaeota | Archaea |
| 927 | Archaeoglobus profundus DSM 5631 | Euryarchaeota | Archaea |
| 928 | Archaeoglobus fulgidus DSM 4304 | Euryarchaeota | Archaea |
| 929 | Thermococcus onnurineus NA1 | Euryarchaeota | Archaea |
| 930 | Thermococcus kodakarensis KOD1 | Euryarchaeota | Archaea |
| 931 | Thermococcus gammatolerans EJ3 | Euryarchaeota | Archaea |
| 932 | Thermococcus sibiricus MM 739 | Euryarchaeota | Archaea |
| 933 | Pyrococcus horikoshii OT3 | Euryarchaeota | Archaea |
| 934 | Pyrococcus abyssi GE5 | Euryarchaeota | Archaea |
| 935 | Pyrococcus furiosus DSM 3638 | Euryarchaeota | Archaea |
| 936 | Thermoplasma volcanium GSS1 | Euryarchaeota | Archaea |
| 937 | Thermoplasma acidophilum DSM 1728 | Euryarchaeota | Archaea |
| 938 | Picrophilus torridus DSM 9790 | Euryarchaeota | Archaea |
| 939 | Haloquadratum walsbyi DSM 16790 | Euryarchaeota | Archaea |
| 940 | Halomicrobium mukohataei DSM 12286 | Euryarchaeota | Archaea |
| 941 | Halorhabdus utahensis DSM 12940 | Euryarchaeota | Archaea |
| 942 | Haloterrigena turkmenica DSM 5511 | Euryarchaeota | Archaea |
| 943 | Natronomonas pharaonis DSM 2160 | Euryarchaeota | Archaea |
| 944 | Natrialba magadii ATCC 43099 | Euryarchaeota | Archaea |
| 945 | Halorubrum lacusprofundi ATCC 49239 | Euryarchaeota | Archaea |
| 946 | Haloferax volcanii DS2 | Euryarchaeota | Archaea |
| 947 | Halobacterium salinarum R1 | Euryarchaeota | Archaea |
| 948 | Halobacterium sp. NRC-1 | Euryarchaeota | Archaea |
| 949 | Haloarcula marismortui ATCC 43049 | Euryarchaeota | Archaea |
| 950 | Methanocaldococcus sp. FS406-22 | Euryarchaeota | Archaea |
| 951 | Methanocaldococcus fervens AG86 | Euryarchaeota | Archaea |
| 952 | Methanocaldococcus vulcanius M7 | Euryarchaeota | Archaea |
| 953 | Methanocaldococcus jannaschii DSM 2661 | Euryarchaeota | Archaea |
| 954 | Methanococcus aeolicus Nankai-3 | Euryarchaeota | Archaea |
| 955 | Methanococcus maripaludis S2 | Euryarchaeota | Archaea |
| 956 | Methanococcus vannielii SB | Euryarchaeota | Archaea |
| 957 | Methanothermobacter thermautotrophicus Delta H | Euryarchaeota | Archaea |
| 958 | Methanosphaera stadtmanae DSM 3091 | Euryarchaeota | Archaea |
| 959 | Methanobrevibacter ruminantium M1 | Euryarchaeota | Archaea |
| 960 | Methanobrevibacter smithii ATCC 35061 | Euryarchaeota | Archaea |
| 961 | uncultured methanogenic archaeon RC-I | Euryarchaeota | Archaea |
| 962 | Aciduliprofundum boonei T469 | Euryarchaeota | Archaea |
| 963 | Candidatus Korarchaeum cryptofilum OPF8 | Korarchaeota | Archaea |
| 964 | Nanoarchaeum equitans Kin4-M | Nanoarchaeota | Archaea |
| 965 | Nitrosopumilus maritimus SCM1 | Thaumarchaeota | Archaea |
References
- 1.Caetano-Anolles D., Kim K.M., Mittenthal J.E., Caetano-Anolles G. Proteome evolution and the metabolic origins of translation and cellular life. J. Mol. Evol. 2011;72:14–33. doi: 10.1007/s00239-010-9400-9. [DOI] [PubMed] [Google Scholar]
- 2.Lesk A.M. Introduction to Protein Architecture. Oxford University Press; New York, NY, USA: 2001. [Google Scholar]
- 3.Cordes M.H., Davidson A.R., Sauer R.T. Sequence space, folding and protein design. Curr. Opin. Struct. Biol. 1996;6:3–10. doi: 10.1016/s0959-440x(96)80088-1. [DOI] [PubMed] [Google Scholar]
- 4.Linderstrom-Lang K.U., Schellman J.A. The Enzymes. Academic Press; New York, NY, USA: 1959. pp. 443–510. [Google Scholar]
- 5.Wang M., Caetano-Anolles G. The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure. 2009;17:66–78. doi: 10.1016/j.str.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Vogel C., Bashton M., Kerrison N.D., Chothia C., Teichmann S.A. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Wang M., Yafremava L.S., Caetano-Anolles D., Mittenthal J.E., Caetano-Anolles G. Reductive evolution of architectural repertoires in proteomes and the birth of the tripartite world. Genome Res. 2007;17:1572–1585. doi: 10.1101/gr.6454307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerstein M., Hegyi H. Comparing genomes in terms of protein structure: Surveys of a finite parts list. FEMS Microbiol. Rev. 1998;22:277–304. doi: 10.1111/j.1574-6976.1998.tb00371.x. [DOI] [PubMed] [Google Scholar]
- 9.Chothia C., Gough J., Vogel C., Teichmann S.A. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 10.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. Scop: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 11.Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. Cath—A hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 12.Riley M., Labedan B. Protein evolution viewed through escherichia coli protein sequences: Introducing the notion of a structural segment of homology, the module. J. Mol. Biol. 1997;268:857–868. doi: 10.1006/jmbi.1997.1003. [DOI] [PubMed] [Google Scholar]
- 13.Ponting C.P., Russell R.R. The natural history of protein domains. Annu. Rev. Biophys. Biomol. Struct. 2002;31:45–71. doi: 10.1146/annurev.biophys.31.082901.134314. [DOI] [PubMed] [Google Scholar]
- 14.Andreeva A., Howorth D., Chandonia J.M., Brenner S.E., Hubbard T.J., Chothia C., Murzin A.G. Data growth and its impact on the scop database: New developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caetano-Anolles G., Wang M., Caetano-Anolles D., Mittenthal J.E. The origin, evolution and structure of the protein world. Biochem. J. 2009;417:621–637. doi: 10.1042/BJ20082063. [DOI] [PubMed] [Google Scholar]
- 16.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 17.Wilson D., Madera M., Vogel C., Chothia C., Gough J. The superfamily database in 2007: Families and functions. Nucleic Acids Res. 2007;35:D308–D313. doi: 10.1093/nar/gkl910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karplus K. Sam-t08, hmm-based protein structure prediction. Nucleic Acids Res. 2009;37:W492–W497. doi: 10.1093/nar/gkp403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim K.M., Caetano-Anolles G. The proteomic complexity and rise of the primordial ancestor of diversified life. BMC Evol. Biol. 2011;11:140:1–140:24. doi: 10.1186/1471-2148-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogel C., Berzuini C., Bashton M., Gough J., Teichmann S.A. Supra-domains: Evolutionary units larger than single protein domains. J. Mol. Biol. 2004;336:809–823. doi: 10.1016/j.jmb.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 21.Vogel C., Teichmann S.A., Pereira-Leal J. The relationship between domain duplication and recombination. J. Mol. Biol. 2005;346:355–365. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 22.Vogel C., Chothia C. Protein family expansions and biological complexity. PLoS Comput. Biol. 2006;2:e48:0370–e48:0382. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel C. Function annotation of SCOP domain superfamilies 1.73. Superfamily-HMM library and genome assignments server. Available online: http://supfam.cs.bris.ac.uk/SUPERFAMILY/function.html (accessed on 28 October 2011)
- 24.Moreira D., Lopez-Garcia P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 25.Wang M., Kurland C.G., Caetano-Anolles G. Reductive evolution of proteomes and protein structures. Proc. Natl. Acad. Sci. USA. 2011;108:11954–11958. doi: 10.1073/pnas.1017361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin E.V., Wolf Y.I., Nagasaki K., Dolja V.V. The big bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 2008;6:925–939. doi: 10.1038/nrmicro2030. [DOI] [PubMed] [Google Scholar]
- 27.Das S., Paul S., Bag S.K., Dutta C. Analysis of nanoarchaeum equitans genome and proteome composition: Indications for hyperthermophilic and parasitic adaptation. BMC Genomics. 2006;7:186:1–186:16. doi: 10.1186/1471-2164-7-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huber H., Hohn M.J., Rachel R., Fuchs T., Wimmer V.C., Stetter K.O. A new phylum of archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 29.Waters E., Hohn M.J., Ahel I., Graham D.E., Adams M.D., Barnstead M., Beeson K.Y., Bibbs L., Bolanos R., Keller M., Kretz K., Lin X., Mathur E., Ni J., Podar M., Richardson T., Sutton G.G., Simon M., Soll D., Stetter K.O., Short J.M., Noordewier M. The genome of Nanoarchaeum equitans: Insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randau L., Munch R., Hohn M.J., Jahn D., Soll D. Nanoarchaeum equitans creates functional trnas from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 31.Randau L., Schroder I., Soll D. Life without rnase p. Nature. 2008;453:120–123. doi: 10.1038/nature06833. [DOI] [PubMed] [Google Scholar]
- 32.Di Giulio M. Nanoarchaeum equitans is a living fossil. J. Theor. Biol. 2006;242:257–260. doi: 10.1016/j.jtbi.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 33.Di Giulio M. The tree of life might be rooted in the branch leading to nanoarchaeota. Gene. 2007;401:108–113. doi: 10.1016/j.gene.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Kim K.M., Caetano-Anolles G. The evolutionary history of protein fold families and proteomes confirms Archaea is the most ancient superkingdom. Ms. submitted. doi: 10.1186/1471-2148-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese C.R., Maniloff J., Zablen L.B. Phylogenetic analysis of the mycoplasmas. Proc. Natl. Acad. Sci. USA. 1980;77:494–498. doi: 10.1073/pnas.77.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambaud I., Heilig R., Ferris S., Barbe V., Samson D., Galisson F., Moszer I., Dybvig K., Wróblewski H., Viari A., Rocha E.P., Blanchard A. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 2001;29:2145–2153. doi: 10.1093/nar/29.10.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gibson D.G., Smith H.O., Hutchison C.A., III, Venter J.C., Merryman C. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods. 2010;7:901–903. doi: 10.1038/nmeth.1515. [DOI] [PubMed] [Google Scholar]
- 38.Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M. The 160-kilobase genome of the bacterial endosymbiont carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 39.Forterre P., Gribaldo S. Bacteria with a eukaryotic touch: A glimpse of ancient evolution? Proc. Natl. Acad. Sci. USA. 2010;107:12739–12740. doi: 10.1073/pnas.1007720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santarella-Mellwig R., Franke J., Jaedicke A., Gorjanacz M., Bauer U., Budd A., Mattaj I.W., Devos D.P. The compartmentalized bacteria of the planctomycetes-verrucomicrobia-chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 2010;8:e1000281:1–e1000281:11. doi: 10.1371/journal.pbio.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamneva O.K., Liberles D.A., Ward N.L. Genome-wide influence of indel substitutions on evolution of bacteria of the PVC superphylum, revealed using a novel computational method. Genome Biol. Evol. 2010;2:870–886. doi: 10.1093/gbe/evq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devos D.P., Reynaud E.G. Evolution. Intermediate steps. Science. 2010;330:1187–1188. doi: 10.1126/science.1196720. [DOI] [PubMed] [Google Scholar]
- 43.Katinka M.D., Duprat S., Cornillot E., Méténier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., Delbac F., El Alaoui H., Peyret P., Saurin W., Gouy M., Weissenbach J., Vivares C. P, Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 44.Corradi N., Pombert J.F., Farinelli L., Didier E.S., Keeling P.J. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun. 2010;1:77. doi: 10.1038/ncomms1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Douglas S., Zauner S., Fraunholz M., Beaton M., Penny S., Deng L.T., Wu X., Reith M., Cavalier-Smith T., Maier U.G. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- 46.Peyretaillade E., Biderre C., Peyret P., Duffieux F., Metenier G., Gouy M., Michot B., Vivares C.P. Microsporidian encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a lsu rrna reduced to the universal core. Nucleic Acids Res. 1998;26:3513–3520. doi: 10.1093/nar/26.15.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin W., Herrmann R.G. Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 1998;118:9–17. doi: 10.1104/pp.118.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keeling P.J., Slamovits C.H. Causes and effects of nuclear genome reduction. Curr. Opin. Genet. Dev. 2005;15:601–608. doi: 10.1016/j.gde.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Welch B.L. The significance of the difference between two means when the population variances are unequal. Biometrika. 1938;29:350–362. [Google Scholar]
- 50.Caetano-Anolles G., Kim H.S., Mittenthal J.E. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc. Natl. Acad. Sci. USA. 2007;104:9358–9363. doi: 10.1073/pnas.0701214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ingham P.W., Nokano Y., Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 52.Bürglin T.R. Evolution of hedgehog and hedgehog-related genes, their origin from Hog proteins in ancestral eukaryotes and discovery of a novel Hint motif. BMC Genomics. 2008;9:127:1–127:28. doi: 10.1186/1471-2164-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]