Abstract
Mesenchymal stem/stromal cells (MSCs) and osteoblasts are important niche cells for hematopoietic stem cells (HSCs) in bone marrow osteoblastic niche. Here, we aim to partially reconstitute the bone marrow HSC niche in vitro using collagen microencapsulation for investigation of the interactions between HSCs and MSCs. Mouse MSCs (mMSCs) microencapsulated in collagen were osteogenically differentiated to derive a bone-like matrix consisting of osteocalcin, osteopontin, and calcium deposits and secreted bone morphogenic protein 2 (BMP2). Decellularized bone-like matrix was seeded with fluorescence-labeled human MSCs and HSCs. Comparing with pure collagen scaffold, significantly more HSCs and HSC–MSC pairs per unit area were found in the decellularized bone-like matrix. Moreover, incubation with excess neutralizing antibody of BMP2 resulted in a significantly higher number of HSC per unit area than that without in the decellularized matrix. This work suggests that the osteogenic differentiated MSC–collagen microsphere is a valuable three-dimensional in vitro model to elucidate cell–cell and cell–matrix interactions in HSC niche.
Keywords: Osteogenic differentiation, hematopoietic stem cells, niche, mesenchymal stem/stromal cell, decellularization, in vitro model
Introduction
Hematopoietic stem cells (HSCs) are a subset of bone marrow cells that are capable of self-renewal and differentiation into all blood-cell lineages.1 HSCs reside in a specific microenvironment known as stem cell niche, composed of soluble signals, neighbor cells, and extracellular matrix.2 Two different HSC niches, namely, osteoblastic niche and vascular niche, are found in the bone marrow. In the osteoblastic niche, HSCs are associated with a subset of osteoblasts that line the endosteal surface of trabecular bone. In the vascular niche, HSCs are in close proximity to the sinusoidal endothelium of bone marrow. An orchestra of signals mediated by soluble factors and cell–cell interaction regulates the balance of quiescence,3–5 self-renewal,6,7 and differentiation of HSCs.8 Readers are directed to some excellent reviews on HSC niche.2,9–18
Osteoblastic cells are critical components in the osteoblastic niche in bone marrow. Calvi et al.6 found that activation of parathyroid hormone–related protein receptor, which is expressed in osteoblasts, associates with an increased number of HSCs and high levels of Notch ligand Jagged 1. Similarly, conditional ablation of bone morphogenetic protein receptor type IA (BMPRIA) augmented the number of osteoblasts with a simultaneous increase in HSC number.7 In contrast, a loss of HSC number was reported upon osteoblastic destruction in a genetically modified mouse model.19 These studies indicated the important role of osteoblasts in controlling HSC expansion.
Mesenchymal stem/stromal cells (MSCs), characterized by their multidifferentiation property and ability to act as feeder layer for HSC culture, are another essential HSC niche component. Recently, it was found that immature osteoblasts including MSCs and preosteoblasts or osteoprogenitors, rather than mature osteoblasts, actively participate as HSC niche cells.2,14,20 In a multicellular spheroid model, undifferentiated and osteogenically differentiated MSCs have been shown to form an informative niche for HSC lodgment and proliferation.21 Moreover, HSCs are closely associated with nestin-expressing (nestin+) MSCs.22 Depletion of nestin+ MSCs caused a reduction in bone marrow homing of hematopoietic progenitors. Furthermore, MSCs expressed factors such as stem cell factor (SCF), vascular cell-adhesion molecule 1 (VCAM1), and interleukin (IL)-7 that regulated HSC proliferation, adhesion, and homing.22,23 Specifically, engraftment of HSCs is promoted upon co-transplantation of HSC and MSC.23 Furthermore, conditioned medium of MSC supports HSC expansion,24 while osteogenically differentiating MSC acts as feeder layer of HSC via secretion of a chemoattractant CXCL12.25
Apart from niche cells, extracellular matrix niche factors present in the osteoblastic niche also involved in HSC regulation. Osteopontin (OPN), an extracellular matrix glycoprotein expressed by osteoblastic cells, has a negative effect on HSC proliferation as demonstrated by an increase in HSC number in OPN-deficient mice,3,4 suggesting its role in maintenance of HSC quiescence. Another extracellular matrix component, calcium also plays important role in the osteoblastic HSC niche. Calcium ions are important matrix niche for HSC because a high concentration of calcium ions in the bone marrow endosteum attracted HSCs through the calcium-sensing receptor expressed on cell surface, facilitating the lodgment and retention of HSC within the niche.26 Moreover, Ca2+ concentration also affects morphology and Ang 1 expression of osteoblast in the niche.27
Despite increasing knowledge on the HSC niche, the roles of cellular and extracellular components as well as their interactions with HSCs have not been fully elucidated. Much of the current understanding regarding HSC niche is characterized by using complex and time-consuming animal models. Furthermore, cells interact with their extracellular matrix differently when they are in three-dimensional (3D) configuration as compared with those in two-dimensional (2D) monolayer cultures.28 These suggest that in vitro model that engineers and recapitulates certain components in the HSC niche in 3D configuration is important29 and will significantly enhance our understanding of multicellular and cell–matrix interactions within the niche.
We previously developed a novel collagen microencapsulation technology,30 which entraps bone marrow–derived MSCs in a self-assembled biomimetic collagen fiber meshwork. This collagen meshwork provides a physiologically relevant microenvironment for MSCs to survive, proliferate, migrate, and differentiate.31 Upon induction of differentiation, committed MSCs are able to remodel the template collagen meshwork by depositing new extracellular matrices specific to the lineages that they are differentiating toward. For example, chondrogenically differentiating MSCs derived a type II collagen, glycosaminoglycan (GAG), and aggrecan-rich extracellular matrix,32 while osteogenically differentiating MSCs remodeled the collagen meshwork with osteogenic markers including alkaline phosphates (ALPs) and calcium phosphate nodules, producing bone-like matrices.33 In this study, we hypothesize that the bone-like matrices derived from osteogenically differentiating MSCs partially reconstitute important matrix niche for HSCs and represent an in vitro model for HSC niche studies. First, we aim to characterize the bone-like matrix reconstituted by osteogenically differentiating MSCs in collagen constructs. Second, decellularized bone-like matrix will be compared to pure collagen scaffold in supporting MSC–HSC interactions. Third, antibody neutralization will be used to study the importance of BMP2 during HSC–MSC interactions.
Materials and methods
Overall experimental design
To produce the in vitro model for HSC niche study, a bone-like matrix was reconstituted by osteogenically differentiating MSCs microencapsulated in collagen constructs. To study whether the bone-like matrix supports HSC and MSC interactions, the matrix was decellularized before seeding DiI fluorescence–labeled human MSCs and then green florescence protein (GFP)-labeled HSCs for co-cultures. Pure collagen scaffold was used as the control matrix. The number of HSCs bound to the scaffold and the number of MSC–HSC pairs with intimate proximity were evaluated using image analysis. To study the significance of BMP2 in the bone-like matrix, neutralizing antibody of BMP2 was supplemented to the culture medium preceding and during co-cultures before counting the number of HSC and MSC–HSC pairs.
Fabrication and decellularization of bone-like matrix derived from osteogenically differentiating MSCs
All protocols involving animals were approved by the institutional ethical committee. Mouse MSCs (mMSCs) were isolated and cultured as previously described32 before encapsulating them in collagen constructs for subsequent osteogenic differentiation. In brief, bone marrow was flushed out from the femurs and tibias of imprinting control region (ICR) mice using a 30-gauge needle and then passed through a 70-µm nylon mesh cell strainer (BD Biosciences, Franklin Lakes, NJ, USA). Cells collected after centrifugation at 400g for 10 min were cultured using Dulbecco’s modified Eagle’s medium–low glucose (DMEM-LG) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM l-glutamine. Growth medium was changed after 3 days of culture and replenished every 3–4 days thereafter. Cells in passage 1 were used for microencapsulation as previously described.30 In brief, MSC suspension at a final density of 5 × 105 cells/mL was mixed with neutralized rat tail type I collagen solution (BD Biosciences, Bedford, MA, USA) at a final concentration of 2 mg/mL in an ice bath. An aliquot of 100 µL of the collagen–MSC mixture solution was dispensed into a custom-made cylindrical container with nonadhesive surface made by covering ultraviolet (UV)-irradiated parafilm. The collagen–MSC droplets were incubated at 37°C in a humidified atmosphere with 5% CO2 for 1 h to allow gelation before supplementing osteogenic differentiation medium for subsequent differentiation experiment. In order to derive bone-like matrices from mMSC, the cell-encapsulated constructs were cultured in osteogenic differentiation induction medium as described previously.33 DMEM-LG supplemented with 10% FBS, 100 nM dexamethasone, 50 µM l-ascorbic acid 2-phosphate, and 10 mM β-glycerophosphate was supplemented to culture wells containing the constructs for 3 weeks with regular replenishment every 3 days before subsequent decellularization. Decellularization was then conducted to remove the mMSCs from the bone-like matrix for subsequent experiments. Sodium deoxycholate was an effective ionic detergent for solubilizing cytoplasmic and nuclear membranes.34 Osteogenically differentiated mMSC–collagen constructs were decellularized with 4%, 5%, or 6% sodium (Na) deoxycholate for 1 h with constant agitation. The constructs were rinsed with phosphate-buffered saline (PBS) three times to remove excess detergent before subsequent evaluation and cell seeding. Decellularization was optimized based on calcium concentration, which represents important matrix component in the HSC niche26,27 and DNA content. Calcium deposits in the decellularized constructs were extracted with 1% trichloroacetic acid for 24 h before quantification by calcium assay kit (BioAssay Systems, Hayward, CA, USA). In brief, equal volumes of Reagent A and Reagent B were mixed and equilibrated to room temperature before use. An aliquot of 5 µL Ca2+ standard solutions or samples was transferred into wells of a clear-bottom 96-well plate, and 200 µL of working reagent was added. The mixture was incubated at room temperature for 3 min before measuring the absorbance at 612 nm. The amounts of Ca2+ present in the samples were determined by calibrating against the linear region of the standard curve, and maximal retention of calcium was used as the criteria for optimization. Significant removal of cellular components was also important for decellularization. DNA content in the scaffolds was measured with a fluorometric assay.35 Briefly, the constructs were washed with PBS for three times to remove excess culturing medium. Papain digestion solution consisting of papain (300 μg/mL), l-cysteine (5 mM), and ethylenediaminetetraacetic acid (EDTA) (5 mM) in phosphate buffer at pH 6.5 was used to digest the constructs at 60°C overnight, until the constructs were completely dissolved. The sample solutions were allowed to cool to room temperature. Standard DNA solutions of 0.25, 0.5, 1, 2, 4, 8, and 16 µg/mL were prepared by serial dilution with Tris EDTA buffer (pH 7.4). Hoechst 33258 dye (Invitrogen Catalog number: H21491) was diluted to 8 µg/mL with Tris NaCl EDTA buffer (pH 7.4). Aliquots of 100 µL of standards and samples were added to the wells of a black 96-well plate, followed by 100 µL of the Hoechst dye. Fluorescence signals were measured at 346 nm for excitation and 460 nm for emission using a microplate reader (Safire2; Tecan, Mannedorf, Switzerland). The amounts of DNA in the samples were then computed by calibration against the linear region of the standard curve, and significant reduction of DNA was used as criteria for decellularization optimization.
Characterization of the bone-like matrix
Part of the osteogenically differentiated mMSC–collagen constructs was cryosectioned into sections of 15 µm thick for histological and immunohistochemical evaluation on successful osteogenesis. Routine hematoxylin and eosin (H&E) and von Kossa staining were used to reveal the morphology and calcium deposition of the differentiated cells, respectively. In brief, slides were rehydrated by xylene and ethanol gradient to water, followed by 1% silver nitrate incubation under UV light for 1 h; 3% sodium thiosulfate was added afterward for 5 min and then rinsed with water. Nuclear fast red was used as counter stain. Immunohistochemistry against osterix, osteocalcin, and OPN were used to reveal intracellular and extracellular osteogenic markers. Sections were incubated with 10 mM sodium citrate buffer (pH 6) at 95°C for 20 min for antigen retrieval. They were then incubated with rabbit polyclonal antibodies against osterix (Abcam, Cambridge, MA, USA), osteocalcin (Millipore, Billerica, MA, USA), and OPN (Millipore, Billerica, MA, USA) at dilutions of 1:100, 1:200, and 1:1000, respectively. After overnight incubation at 4°C, sections were incubated with secondary antibody, biotinylated goat anti-rabbit IgG (BA-1000; Vector Laboratories, Burlingame, CA, USA), at a dilution of 1:400 for osterix and 1:500 for osteocalcin and OPN for 30 min at room temperature. For BMP2 immunohistochemistry, sections were incubated with 0.2% hyaluronidase in PBS (pH 7.4) at 37°C for 15 min for antigen retrieval. After overnight incubation at 4°C with rabbit polyclonal antibody against BMP2 (Abcam) at a dilution of 1:1000, sections were incubated with the anti-rabbit secondary antibody at a dilution of 1:800 for 30 min at room temperature. The VECTASTAIN ABC kit (Vector Laboratories) and the 3,3′-diaminobenzidine (DAB) substrate system (Daoko, Glostrup Denmark) were used for color development. Our previous study showed osteoinductive activity in osteogenically differentiated mMSC–collagen microspheres.33 Secretion of BMP2 by osteogenically differentiating MSCs was also measured. During osteogenic differentiation of mMSC in collagen constructs, conditioned medium from each construct cultured on 24-well plate was collected during medium replenishment. BMP2 enzyme-linked immunosorbent assay (ELISA) immunoassay (Quantikine; R&D Systems, Minneapolis, MN, USA) was conducted after debris removal by centrifugation according to the manufacturer’s instructions. In brief, BMP2 presented in conditioned medium bound to monoclonal antibody pre-coated on the dish. A second monoclonal antibody, which is enzyme-linked, against BMP2 was added afterward to bind the bound BMP2. Substrate solution was added to react with the linked enzyme for color development. Color intensity, which was proportional to BMP2 concentration, was measured by microplate reader (UVM 340; ASYS, Cambridge, UK). BMP2 secretion from the constructs from day 0 to , where C is the concentration of BMP2 in conditioned medium and V is the volume of conditioned medium (fixed). In order to evaluate the ultrastructure of the decellularized matrix, the constructs were fixed in 2.5% glutaraldehyde for 2 h at room temperature, dehydrated through a graded series of ethanol, followed by critical point drying. The constructs were fractured to expose their cross sections for mounting and gold sputtering before examination with scanning electron microscope (SEM) (LEO 1530; LEO Electron Microscopy, Cambridge, UK).
Fabrication of pure collagen scaffold
Pure porous collagen scaffold was fabricated as previously described36 and was used as the control group to study MSC–HSC interactions. Acid soluble rat tail type I collagen (Becton Dickinson) was diluted to 2 mg/mL with 0.2 M acetic acid to a final volume of 100 µL per construct in a 96-well culture plate. Collagen gel was allowed to undergo gelation in an ammonium chamber for 30 min. After brief rinse and overnight incubation in sterile distilled water, the collagen gel was then freeze-dried at −40°C for 24 h to form porous collagen scaffold. The freeze-dried constructs were sterilized with 70% ethanol for 15 min before equilibrating with 1× PBS prior to subsequent cell seeding.
Fluorescence labeling and seeding of human MSC
A human bone marrow mesenchymal stromal cell line (HS-5; American Type Culture Collection, Rockville, MD, USA) was used as the niche cell for HSC in this study. Our separate study showed that this cell line expresses osterix (Supplementary Information 1), an important transcription factor promoting osteogenic potential of MSCs.37 They were cultured in growth medium containing DMEM–high glucose (HG), 10% FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. Growth medium was replaced every 2–3 days. Cells were harvested before confluence and subcultured up to passage 23. Human MSCs (hMSCs) were labeled with fluorescent dye before seeding to either pure collagen scaffold or decellularized bone-like matrix. Prior to cell seeding, hMSCs were labeled with Vybrant® CM-DiI (a lipophilic vital dye) cell labeling solution (Molecular Probes; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s procedure. In brief, hMSCs were incubated with 1 µM labeling solution for 5 min at 37°C in a humidified atmosphere with 5% CO2 and a further 15 min at 4°C. Cells were rinsed with PBS twice, detached by 0.25% trypsin-EDTA and resuspended in growth medium for subsequent seeding experiment. Both freeze-dried pure collagen scaffold and decellularized constructs were washed with PBS and equilibrated with growth medium for 1 h prior to cell seeding. After removal of the medium from the scaffolds, 5 µL of labeled hMSCs suspension at a concentration of 2.4 × 107 cells/mL was directly dropped onto each construct. hMSCs were allowed to adhere to the construct for 45 min before carefully supplementing growth medium for culture of 3 days before seeding HSCs for co-culture.
Isolation, culture, and fluorescent labeling of human HSCs
Cord blood samples were collected from healthy donors. All procedures were approved by the Combined Clinical Ethics Committee of the University of Hong Kong and Hong Kong West Cluster Hospitals of Hospital Authority. Human HSCs (hHSCs) (CD34+ cells) were isolated by magnetic cell sorting using magnetic beads conjugated with anti-CD34 antibodies (MiltenyiBiotec, Auburn, CA, USA). hHSCs were cultured in StemSpan® H3000 medium (STEMCELL Technologies, Vancouver, BC, Canada) supplemented with StemSpan® H300 and StemSpan® CC110 (STEMCELL Technologies). hHSCs were then labeled by electroporation using a human CD34+ cell nucleofactor kit (VPA-1003; Lonza, Cologne, Germany). A GFP-containing plasmid was electroporated into the cells using a cocktail with a preset program (U-008) on the electroporation machine (Nucleofector™ 2b device; Lonza). The transfection efficiency was around 80%.
Co-culture of hHSCs with hMSCs
After culturing the DiI-labeled MSCs on either the pure collagen scaffold or the decellularized bone-like scaffold for 3 days, hHSCs were seeded on the constructs for co-culture. After removing all MSC culture medium, an aliquot of 5 µL of GFP-labeled hHSC suspension at a concentration of 9.6 × 106 cells/mL was dropped onto each construct, giving a final hMSC to hHSC ratio of around 5:1. hHSCs were allowed to adhere to the scaffolds for 45 min before carefully supplementing hHSC growth medium for further co-culture of 1 and 3 days.
Tracking fluorescence-labeled hMSC in decellularized matrix
In order to track the location of the seeded hMSCs in the decellularized scaffold, DiI membrane fluorescence–labeled hMSCs were seeded in decellularized scaffolds and cultured for 1 and 3 days as previously described. To distinguish newly seeded hMSCs from any incompletely discarded osteogenically differentiating mMSCs, which also express the intracellular osteogenic marker osterix, co-localization of DiI fluorescence and osterix was monitored. In brief, the constructs were cultured in hMSC growth medium for either 1 or 3 days and then fixed in 4% PBS-buffered paraformaldehyde for cryosections. Fluorescence-labeled hMSCs were imaged under confocal fluorescence microscope before processing for immunohistochemistry of osteogenic markers including osterix, OPN, and osteocalcin as described. Fluorescent images of labeled hMSCs and immunohistochemistry images of osterix positivity in the same sections were processed using Adobe Photoshop. By using the “Magic Wand Tool” with a tolerance value of 20, background of the fluorescence image was removed. The patterns of the fluorescent spots corresponding to labeled cells were then mapped to the immunopositive signals of osterix of the same sections in merged image by using the “Free Transform” function, during which the fluorescence image was resized and rotated without constraining the aspect ratio to match the respective signals.
Evaluation of the interactions between hHSCs and hMSCs
The distribution of hMSCs and hHSCs was visualized under a confocal microscopy (LSM 710; Carl Zeiss, Oberkochen, Germany). On days 1 and 3 of co-culture, confocal imaging was used to study the interactions between the hHSCs and hMSCs. Two constructs from each time point (days 1 and 3) were used for cell counting. A range of 3–8 views were randomly selected from each construct at different Z level under confocal microscope under the same magnifications. Then, the number of green HSCs per view (and hence per unit area) was counted. For quantification of cell number on the scaffolds, stack images of optical sections were obtained at 20× magnification, with objective Fluar 20×/0.75 M27. hHSCs and hMSCs with interactions were identified as pairs and the images were selected, zoomed, and exported as image file. The hHSC–hMSC pair was defined by either co-localization of or a less than one-cell-diameter (10 µm) distance between the green hHSC and the red hMSC,7,38–42 and the number of such pairs per view (and hence per unit area) was also counted. For imaging the hHSC and hMSC “pairs,” optical sections were obtained at 40× magnification, with an oil lens (EC Plan-Neofluar 40×/1.3 oil DIC M27). All GFP signals from hHSCs were excited by Argon 488 nm laser with bandpass slider ranging from 493 to 546 nm. All DiI signals from labeled hMSCs were excited by HeNe 543 nm laser with bandpass slider ranging from 548 to 673 nm. Raw files of images were exported for subsequent image analysis by Imaris software (Bitplane Scientific, Zurich, Switzerland), in which 3D view of stack images was reconstructed and presented as videos.
Effects of neutralizing antibody against BMP2
In order to study the importance of BMP2 in MSC–HSC interactions, a neutralization antibody experiment was conducted. In brief, decellularization should kill all osteogenically differentiating BMP2-releasing mMSCs. In order to assure that all BMP2 was removed during MSC and HSC binding and co-culture, excess neutralizing anti-human BMP-2/4 antibody at 2 µg/mL (R&D Systems) was supplemented to the HS-5 culture medium throughout 3 days of MSC culture period. Same amount of the antibody was then supplemented to the HSC culture medium throughout 3 days of MSC–HSC binding and co-culture period. Two constructs from each time point (days 1 and 3) and each group (with and without BMP2 antibody) were used for cell counting. A range of 6–9 views were randomly selected from each construct at different Z level under confocal microscope at the same magnification. Then, the number of HSCs and MSC–HSC pairs per view (and hence per unit area) was counted, using Imaris, in mMSC-derived bone-like matrices with and without BMP2 neutralizing antibody.
Data presentation and statistical analysis
Quantitative results such as quantity of Ca2+, DNA content, BMP2 secretion, and number of HSCs or HSC–MSC pairs were reported as mean ± 2 standard error (SE) of mean. Normality assumption was verified before using one-way or two-way analysis of variance (ANOVA) to reveal the differences among different treatment groups. A significant level was set as 0.05, and SPSS 19.0 was used for the analysis.
Results
Osteogenically differentiating mMSC remodeled the template collagen matrix
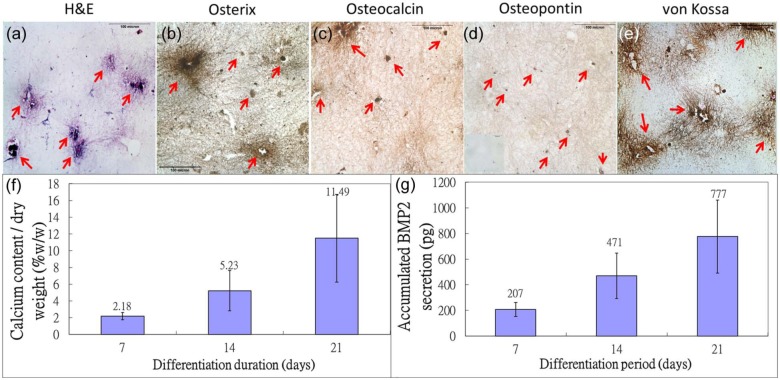
Figure 1 showed the histological, histochemical, and immunohistochemical staining of osteogenic markers in mMSC–collagen constructs exposed to osteogenic differentiation medium for 21 days. Differentiating mMSCs expressed osteogenic marker osterix (Figure 1(b)) and remodeled the template matrix by depositing new extracellular matrix components found in bones, including osteocalcin (Figure 1(c)), both expressed intracellularly and extracellularly, OPN (Figure 1(d)), which expressed extracellularly, calcium deposits (Figure 1(e)), and nodules (Figure 1(a)). Quantitative measurement of calcium deposits extracted from the constructs showed that the calcium content per unit dry weight increased up to ~12% (w/w) after 21 days of differentiation (Figure 1(f)). Moreover, Figure 1(g) showed that as the encapsulated mMSC committed toward osteogenic lineages, a major osteoinductive agent BMP2 was continuously released to the culture medium, echoing with our previous finding on the osteoinductivity of mMSC–collagen microspheres.32
Figure 1.
Osteogenic differentiation of mMSC in collagen constructs. Histological and immunohistochemical staining of osteogenic differentiated mMSC in collagen construct after 21 days of culture: (a) H&E staining; (b) osterix immunohistochemistry; (c) osteocalcin immunohistochemistry; (d) osteopontin immunohistochemistry (inset: negative control); and (e) von Kossa staining for calcium deposition (red arrows: cells and regions with positive staining). (f) Bar chart showing calcium content, normalized by dry weight, in constructs at 7, 14, and 21 days post osteogenic differentiation and (g) bar chart showing BMP2 secretion by constructs at 7, 14, and 21 days post osteogenic differentiation (n = 3, each with duplicates).
H&E: hematoxylin and eosin; BMP2: bone morphogenic protein 2; mMSC: mouse MSC.
Decellularization protocol with maximal retention of calcium and partial removal of cell remnants
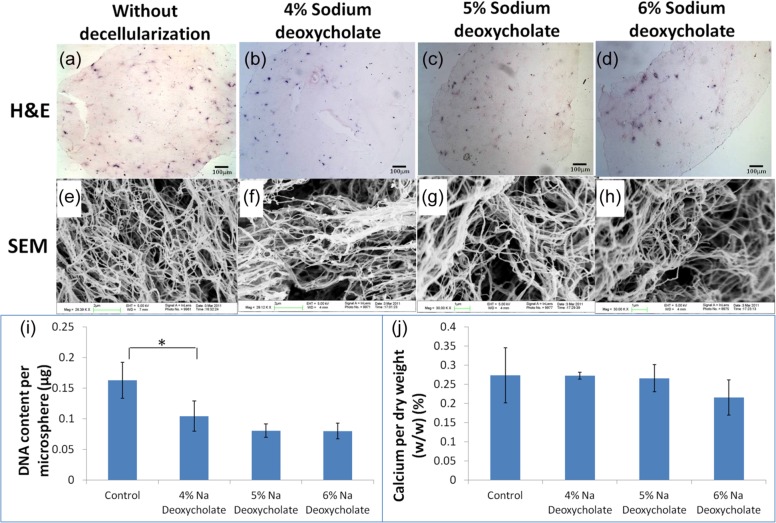
Different concentrations of sodium deoxycholate were used to decellularize the constructs, and the efficiency of decellularization was studied by SEM, calcium assay, and DNA assay. The decellularization protocol is said to be optimized if the physical properties of the scaffold and the critical extracellular matrices such as calcium, which has been shown to be important for HSC lodgment,26 are retained and cells are dead and largely removed. Figure 2(a)–(d) showed that the cellularity of osteogenic differentiating mMSC–collagen constructs was reduced after decellularization (Figure 2(b)–(d)). Figure 2(e)–(h) showed the SEM images while tiny calcium nodules were identified from the control (Figure 2(e)) and the lowest concentration of decellularization agent (Figure 2(f)), but hardly found in higher concentrations (Figure 2(g) and (h)). Fibers in 6% sodium deoxycholate group were thinner (Figure 2(h)), suggesting that the collagen meshwork might be destructed. Figure 2(i) showed that comparing with the control, a significant (p = 0.05) decrease in the amount of DNA was found in 4% sodium deoxycholate group, while further decrease in DNA was not significant at higher concentrations. Figure 2(j) showed the calcium content of the decellularized scaffolds. Calcium contents from 4% and 5% sodium deoxycholate groups were similar to the control group, while obvious decrease was noted in 6% group. However, no statistical significance (p > 0.05) was found among all groups. Decellularization with 4% sodium deoxycholate was therefore chosen as the optimal concentration since the collagen meshwork and calcium content were best preserved while removal of DNA was significant.
Figure 2.
Decellularization of osteogenic differentiated MSC–collagen constructs with detergent at different dosages. (a–d) Routine H&E staining (scale bar: 100 µm); (e–h) SEM images; (a and e) without decellularization; (b and f) decellularized with 4% sodium deoxycholate; (c and g) decellularized with 5% sodium deoxycholate; (d and h) decellularized with 6% deoxycholate; (i) DNA content after decellularization (*statistical significant difference: p = 0.05); (j) calcium content per dry weight after decellularization (n = 3, each with duplicates).
MSC: mesenchymal stem/stromal cell; H&E: hematoxylin and eosin; SEM: scanning electron microscope.
Repopulation of decellularized bone-like matrix with newly seeded hMSCs
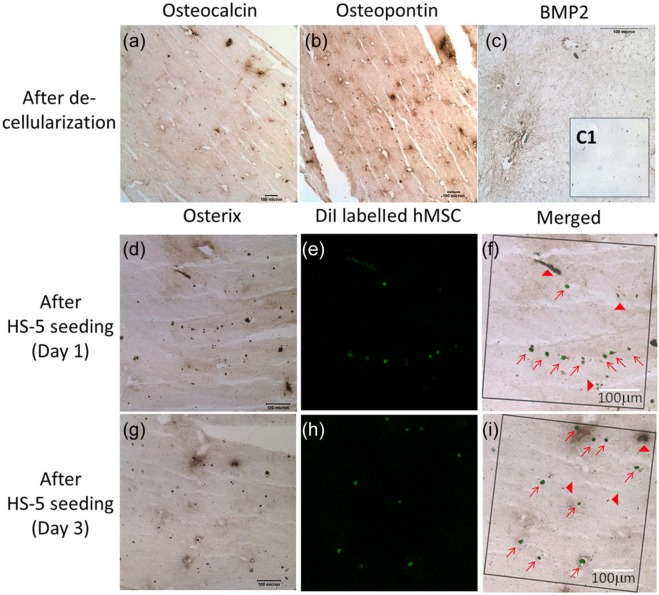
Figure 3(a)–(c) showed that the extracellular matrix osteocalcin (Figure 3(a)) and OPN (Figure 3(b)) were still retained in the decellularized matrix of osteogenic differentiating mMSC–collagen constructs. Moreover, the major osteoinductive agent BMP2 was also found immunopositive after decellularization (Figure 3(c)), contrasting to the negative control in the inset (Figure 3(c1)). These results suggest that the decellularized matrix still retains the bone-like microenvironment. Co-localization of the DiI-label (pseudo color: green) of hMSCs (Figure 3(e) and (h)) and the immunopositivity of osterix (Figure 3(d) and (g)) were found on both days 1 and 3 after seeding hMSCs to the decellularized matrix. Merged images (Figure 3(f) and (i)) showed that most cells found in the decellularized matrix are both DiI-positive and osterix-positive, suggesting that the newly seeded hMSCs were repopulating in the matrix. Occasionally, cells or remnants with osterix positivity but not DiI-label were identified, suggesting that further optimization of the decellularization protocol is necessary.
Figure 3.
Repopulation of decellularized bone-like matrix with newly seeded hMSCs. Immunohistochemistry of osteogenic matrix and osteoinductive markers in decellularized matrix derived from osteogenic differentiating mMSC–collagen constructs: (a) osteocalcin; (b) osteopontin; and (c) BMP2 (C1: negative control). Newly seeded hMSCs with co-localization of DiI-label and intracellular osterix after seeding for (d–f) 1 day and (g–i) 3 days; (d and g) osterix immunohistochemistry; (e and h) fluorescence staining of DiI-labeled hMSCs (pseudo color: green); (f and i) merged osterix immunohistochemistry and DiI-labeled hMSCs (squared frames: views being analyzed for co-localization; red arrows: cells and regions with double positive staining; red arrow heads: cell remnants without DiI from decellularization).
hMSCs: human mesenchymal stem/stromal cells; BMP2: bone morphogenic protein 2.
Decellularized bone-like matrix supports MSC–HSC interactions
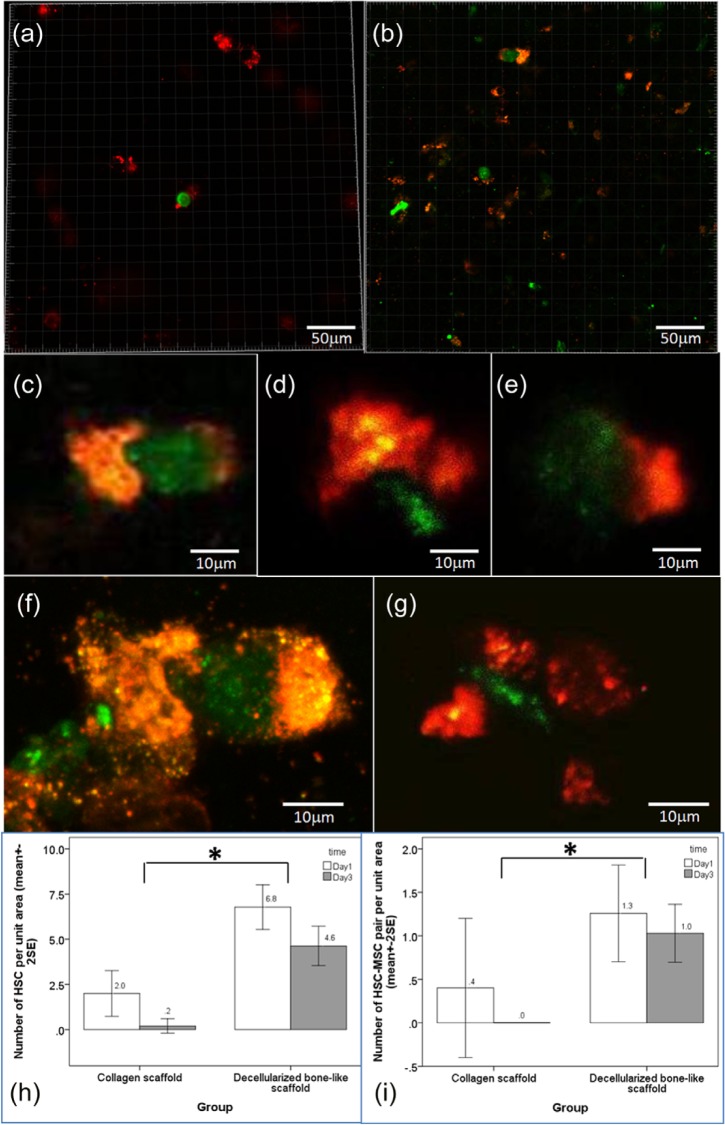
Figure 4 showed the distribution of DiI-labeled hMSCs (pseudo color: red fluorescence) and GFP-transfected hHSCs in pure collagen scaffold and decellularized bone-like matrix derived from osteogenically differentiating mMSC. There was less hMSCs (red) and hardly any HSC (green) found in pure collagen scaffolds (Figure 4(a)). However, in the bone-like matrix, more MSCs and MSC–HSC pairs were identified (Figure 4(b)). Figure 4(c)–(g) showed that the MSC–HSC pairs were in intimate proximity within one-cell diameter, suggesting that they were closely interacting with each other. In some pairs, orange color, which refers to co-localization of the green HSCs and the red MSCs, was found (Figure 4(c) and (f)). Supplementary Information 2 showed the video of the 3D reconstructed image of HSC–MSC pair shown in Figure 4(f). Figure 4(h) showed the mean number of HSCs and interacting HSC–MSC pairs per view found in each type of scaffold. In pure collagen scaffold, on average, there were only 2 ± 1.4 HSCs per unit area identified on day 1 post seeding where 0.4 ± 0.9 per unit area were paired up with MSC. On day 3 post seeding, there was hardly any HSC or HSC–MSC pair identified. In the decellularized bone-like matrices, significantly more HSCs, on average, 6.8 ± 3.4 per unit area, were identified on day 1 post seeding, out of which, 1.3 ± 1.5 per unit area were HSC–MSC interacting pairs. On day 3 post seeding, 4.6 ± 3.2 HSCs per unit area were identified, out of which, 1.0 ± 1.0 HSC–MSC pairs per unit area were identified. Two-way ANOVA showed that the number of HSC lodged in the decellularized bone-like scaffold was significantly different from that in collagen scaffold (p < 0.001) while the time factor was not significant (p = 0.07). Similarly, two-way ANOVA showed that the number of HSC–MSC pairs identified in the decellularized bone-like scaffold was also significantly different from that in pure collagen scaffold (p = 0.026) while the time factor was not significant (p = 0.452). These results suggest that the decellularized bone-like matrices supported HSCs’ attachment by increasing number of HSCs adhered and facilitated HSC–MSC interactions by increasing the number of HSC–MSC pairs while pure collagen scaffolds did not.
Figure 4.
MSC–HSC interactions on pure collagen scaffold and bone-like matrices derived from osteogenically differentiating MSCs. Dual-channel confocal microscopic images were captured at different magnifications for image analysis. (a) hMSCs and hHSCs co-cultured on pure collagen scaffolds and (b) hMSCs and hHSCs co-cultured on decellularized osteogenic differentiated mMSC–collagen constructs (red: hMSC; green: hHSC). Representative images showing hMSC–hHSC pairs on (c–e) day 1 (scale bar: 50 µm) and (f–g) day 3 (scale bar: 10 µm). Bar chart showing the mean number of (h) hHSCs and (i) hHSC–hMSC pairs per unit area in pure collagen scaffold and decellularized bone-like constructs (n = 2, each with 3–8 views, *statistical significant difference, p < 0.001 for (h) and p = 0.026 for (i)).
MSC: mesenchymal stem/stromal cell; HSC: hematopoietic stem cell; hMSCs: human mesenchymal stem/stromal cells; hHSC: human hematopoietic stem cells.
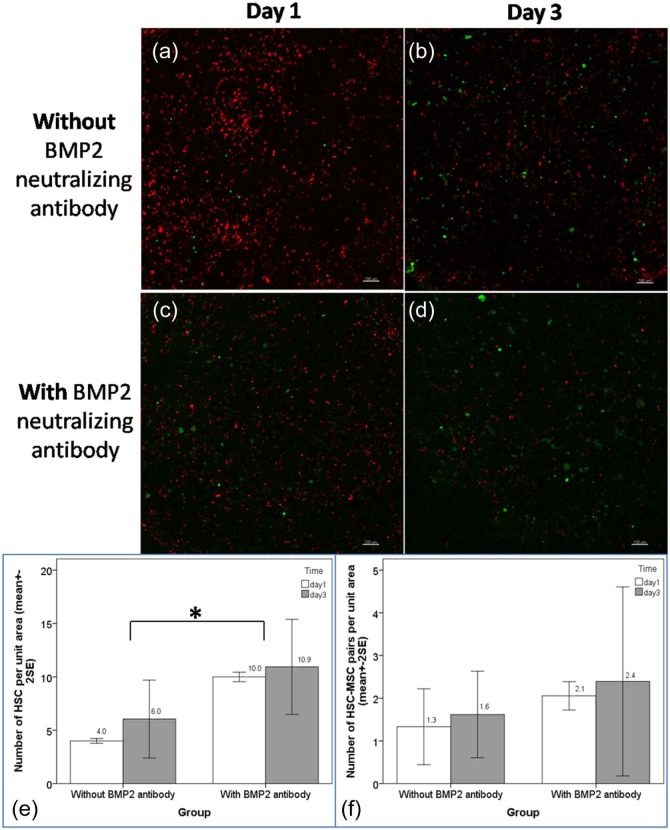
Neutralizing BMP2 antibody increases the number of HSCs
Figure 5 showed the qualitative and quantitative evaluation of HSCs and MSC–HSC pairs bound to the decellularized bone-like scaffold. Figure 5(a) and (b) showed that the number of green HSCs appeared increased over time when visualized. Figure 5(c) and (d) showed only a slight increase in the number of green HSCs over time. Figure 5(e) and (f) showed the bar charts of the number of HSCs and the percent of MSC–HSC pairs per unit area. Without BMP2 antibody, 4.0 ± 0.28 (mean + 2 SE) HSCs per unit area were found in the scaffold on day 1 and the number increased to 6.0 ± 3.6 per unit area on day 3. Out of these HSCs, 1.3 ± 0.85 and 1.6 ± 0.99 HSC–MSC pairs per unit area were found on days 1 and 3, respectively. With BMP2 antibody, there were significantly more HSCs, 10 ± 0.42 per unit area identified in the decellularized bone-like scaffolds on day 1 and the number slightly increased to 10.9 ± 4.3 per unit area on day 3. Among these HSCs, around 2.1 ± 0.28 and 2.4 ± 2.2 per unit area were paired up with MSCs on days 1 and 3, respectively. Two-way ANOVA showed that the number of HSC per unit area lodged in the decellularized bone-like matrix treated with BMP2 antibody was significantly different from that without (p = 0.02) while the time factor was not significant (p = 0.359). For the number of HSC–MSC pairs per unit area identified in the decellularized bone-like scaffold, both the BMP2 antibody group factor (p = 0.316) and the time factor (p = 0.659) were not significant. These data suggest that the presence of BMP2 in the osteogenic microenvironment is important in maintenance of HSC quiescence because removing the BMP2 signal through antibody neutralization resulted in significantly increased number of HSCs per unit area.
Figure 5.
Effects of neutralizing antibody of BMP2 on MSC–HSC interactions in bone-like matrices derived from osteogenically differentiating MSCs. Dual-channel confocal microscopy images of MSC–HSC interactions with and without neutralizing antibody of BMP2 (red: MSCs; green: HSCs). Without antibody on (a) day 1 and (b) day 3 post co-culture and with antibody on (c) day 1 and (d) day 3 post co-culture. Bar charts showing the mean number of (e) HSCs (*statistical significant difference: p = 0.02) and (f) MSC–HSC pairs per unit area with and without neutralizing antibody of BMP2 (n = 2, each with 6–9 views).
BMP2: bone morphogenic protein 2; MSC: mesenchymal stem/stromal cell; HSC: hematopoietic stem cell.
Discussion
Bone-like matrix partially represents important matrix components in the osteoblastic niche
A recent study reports that seeding decellularized bone with MSCs, the number of CD34+ HSCs increased,43 suggesting that the biological composition of the complex niche present in the decellularized bone is important for MSC–HSC interactions. This study reports in vitro reconstitution of a bone-like matrix by osteogenically differentiating MSC in collagen constructs and demonstrates that this bone-like matrix represents partially the osteoblastic niche as it is able to support HSC adhesion and MSC–HSC interactions. This bone-like matrix consists of calcium phosphate deposits, type I collagen, OPN, and osteocalcin. It may also immobilize soluble bioactive factors secreted by osteogenically differentiating MSCs such as BMP2, which is an osteoinductive agent and may also play a role in HSC niche. Importance of calcium ions for cell–matrix and cell–cell interactions between HSCs and their niche has been suggested by previous studies. First, calcium-sensing receptor promotes the adhesion and lodgment of HSCs to their niche.26 Another study27 showed that calcium plays a significant role in regulating the cell morphology of osteoblasts through cell–cell and cell–matrix interactions by expressing Angiopoietin 1 (Ang 1) and 2 (Ang 2). Since Ang 1 expressed by osteoblasts activates the Tie-2 receptors on HSCs, adhesion of HSCs to their niche is enhanced, supporting the quiescence and survival of HSCs. Similarly, it has been suggested that the calcium ions in the HSC niche might dictate the HSC location through regulation of HSC adhesion to collagen I.44 Corroborating with these studies, our results showed that the bone-like matrix derived from osteogenic differentiation of the MSCs encapsulated in collagen has significantly increased the number of HSCs bound to the matrix niche and the number of closely interacting MSC–HSC pairs when compared to that in the pure collagen scaffold. This suggests that calcium and other matrices deposited from previous osteogenic differentiation may promote the attachment and adhesion of HSCs onto the bone-like matrix and their binding and interactions with MSCs. Moreover, the importance of another bone extracellular matrix OPN in maintaining HSC quiescence has also been suggested as shown by increased HSC proliferation in OPN-deficient mice.3,4 Further studies should be carried out to investigate the contributions of individual matrix components in regulating HSC lodgment, expansion, and interactions with niche cells by approaches such as decalcification, competitive binding, or neutralization.
Importance of BMP2 signaling
BMPs are multifunctional proteins that are capable of regulating fate of many cells including MSC and HSC.45–47 BMP2 is an osteoinductive agent. Biologically active BMP2 is produced by osteogenically differentiating MSCs as shown in our previous study32 and the current report. BMP2 interferes with MSC function by inducing intracellular signaling via BMP-activated receptors48 and hence plays a role in promoting the osteogenic differentiation of MSC as well as the differentiation of osteoprogenitor cells.47,49,50 These MSC-derived osteoblastic cells are proven to be a critical component in HSC niche43,44,51 able to retain HSCs in the osteoblastic niche.6,7 Particularly, immature osteogenic lineages have been shown to play more important roles in maintaining HSCs in the osteoblastic niche.2,14,20 Specifically, osteoblastic cells anchor HSCs to the scaffold and maintain the quiescence signal of HSCs and hence regulating hematopoiesis.43,44,51 In this study, neutralization of BMP2 resulted in significantly higher number of HSCs retained in the scaffold, echoing with a previous study on conditional ablation of BMPRIA,7 which downregulates BMP signaling in the HSC niche and augments the number of osteoblasts with a simultaneous increase in HSC number.7 Therefore, our results also suggest the importance of BMP signaling on HSC maintenance and hence expansion. A second possible explanation is that blocking BMP2 may reduce the extent of seeded MSC in further committing to mature osteoblastic lineages, hence maintaining the immature or early osteoblastic niche cells, which have been shown to play an important role in supporting HSC lodgment. Further studies are warranted to delineate the role of BMP signaling in maintaining HSC niche. Moreover, apart from BMP2, other soluble factors secreted by MSCs such as chemokines attracting HSCs to bind and interact with the MSCs deserve investigation.
Osteogenic differentiating MSC-derived bone-like matrix is a suitable 3D in vitro model to study HSC niche
Knowing that the real HSC niche in vivo is much more complex than what is reconstituted in the current bone-like matrix, this study reports a 3D model to investigate certain aspects of the HSC niche such as the niche cell and the extracellular matrix components of the niche. A few in vitro models have previously been used to study MSC–HSC interactions. First, an agarose gel allows HSC to migrate in and out, while MSC spheroid, osteogenic differentiated for 1 week, co-cultured with HSC has shown increased lodgment at vicinity to osteogenic-induced MSC and increased number of HSC lodged. Another report used PuraMatrix, a synthetic hydrogel made of self-assembled peptides, which allows quiescence maintenance and hypoxia gradient in HSCs.52 This work reports a bone-like microenvironment made by osteogenically differentiating MSCs in a type I collagen template, which acts as a suitable 3D in vitro model to study cellular and molecular events in HSC niche such as binding and interaction between HSCs and niche factors including the cell niche such as MSCs and osteoblastic cells derived from MSCs, the soluble niche such as BMP2, and matrix niche such as calcium deposits. The major advantage of the current in vitro model for HSC niche study is that the robust model allows studying of different niche factors including niche cells, matrix factors, soluble factors, and even mechanical factors as this model also fits to a high-throughput mechanoregulation loading system we previously developed.53,54
Limitations and future directions
A number of limitations in this study require future investigation. First, incomplete decellularization of the osteogenic differentiated mMSC, as shown by the remnant DNA content in the decellularized constructs, is a limitation of this study. mMSC was used to derive the bone-like matrix because they are more readily differentiating toward osteogenic lineages and remodeling the collagen scaffold with mineralized matrix than human MSCs as shown in our previous work.33 However, incomplete decellularization increases the difficulty to track the fate of newly seeded hMSCs. That is the reason why co-localization of the fluorescence label and the intracellular osterix was used to track the newly seeded hMSCs. Further evaluations on the expression of human genes related to osteogenic differentiation markers including osterix, OPN, and osteocalcin via real-time polymerase chain reaction (PCR) are needed to confirm the differentiation status of the seeded hMSCs. Second, the composition of the bone-like matrix reconstituted is complex. Apart from type I collagen, calcium, OPN, and remnant BMP2, other unknown components such as the mechanical stiffness of the scaffold might also contribute to the osteogenic differentiation of human MSCs seeded and the subsequent MSC–HSC interactions. Therefore, proteomic analysis on the detailed composition of the matrix and characterization of the mechanical stiffness in the in vitro model warrant further investigations. Third, BMP2 neutralization may not completely block the BMP signaling while other methods such as small interfering RNA (siRNA) may be more effective. Fourth, only short-term interacting events between MSCs and HSCs such as survival and intimate binding were investigated in this study, partly owing to the fact that the fluorescent dye DiI labeling the human MSCs would be photobleached upon long-term monitoring. Genetic labeling of MSCs for longer term co-culture between MSCs and HSCs in this model is necessary to investigate their functional interactions such as colony formation assay and secretion of soluble niche factors such as CXCL12. Last but not the least, an initial ratio of MSC:HSC of 5:1 was used in this study owing to the limited number of HSCs obtained. Different titrating ratios of MSCs to HSCs should be investigated in the future to find out the optimal ratio of the niche cell to the stem cell facilitating MSC–HSC interactions and HSC activities such as lodgment and quiescence.
Conclusion
This study reports an in vitro model to study HSC niche. First, by osteogenically differentiating MSCs encapsulated in collagen constructs, a bone-like matrix partially reconstituting the cellular and matrix components of the osteoblastic niche of HSCs was constructed. The differentiating MSCs express osteogenic markers including osterix, release osteoinductive agent BMP2, and remodel the scaffold to a complex matrix consisting of calcium deposits, OPN, and osteocalcin. Second, decellularized bone-like matrix was better than pure collagen scaffold in supporting HSCs’ adhesion and intimate binding with MSCs, as shown by a significantly higher number of HSC and MSC–HSC pairs per unit area, respectively. Third, BMP2 is an important factor for HSC lodgment and quiescence because neutralizing BMP2 signal by antibody resulted in a significant increase in the number of HSCs per unit area.
Acknowledgments
The authors would like to thank Ms Minting Yuan for assistance with the DNA assay and Annie Cheng for assistance with the calcium assay.
Footnotes
Supplementary Information 1: Immunocytochemistry of osterix in hMSCs (HS-5): (A) negative control without primary antibody and (B) immunopositivity of osterix with primary antibody at a dilution of 1:100 (scale bars: 50 µm).
Supplementary Information 2: Video showing the 3D reconstructed image of one hMSC–hHSC pair in the decellularized bone-like matrix on day 3 post seeding.
Declaration of conflicting interests: The sponsors have no involvement in the study design, collection, analysis and interpretation of data, writing the article, and the decision to submit the article for publication.
Funding: This work was supported by University Development Fund, Strategic Research Themes on Biomedical Engineering, The University of Hong Kong (HKU).
References
- 1. Domen J, Weissman IL. Self-renewal, differentiation or death: regulation and manipulation of hematopoietic stem cell fate. Mol Med Today 1999; 5: 201–208 [DOI] [PubMed] [Google Scholar]
- 2. Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol 2006; 6: 93–106 [DOI] [PubMed] [Google Scholar]
- 3. Stier S, Ko Y, Forkert R, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med 2005; 201: 1781–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nilsson SK, Johnston HM, Whitty GA, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood 2005; 106: 1232–1239 [DOI] [PubMed] [Google Scholar]
- 5. Arai F, Hirao A, Ohmura M, et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 2004; 118: 149–161 [DOI] [PubMed] [Google Scholar]
- 6. Calvi LM, Adams GB, Weibrecht KW, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 2003; 425: 841–846 [DOI] [PubMed] [Google Scholar]
- 7. Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 2003; 425: 836–841 [DOI] [PubMed] [Google Scholar]
- 8. Kopp HG, Avecilla ST, Hooper AT, et al. The bone marrow vascular niche: home of HSC differentiation and mobilization. Physiology (Bethesda) 2005; 20: 349–356 [DOI] [PubMed] [Google Scholar]
- 9. Taichman RS, Emerson SG. The role of osteoblasts in the hematopoietic microenvironment. Stem Cells 1998; 16: 7–15 [DOI] [PubMed] [Google Scholar]
- 10. Moore KA. Recent advances in defining the hematopoietic stem cell niche. Curr Opin Hematol 2004; 11: 107–111 [DOI] [PubMed] [Google Scholar]
- 11. Moore KA, Lemischka IR. Stem cells and their niches. Science 2006; 311: 1880–1885 [DOI] [PubMed] [Google Scholar]
- 12. Wilson A, Oser GM, Jaworski M, et al. Dormant and self-renewing hematopoietic stem cells and their niches. Ann N Y Acad Sci 2007; 1106: 64–75 [DOI] [PubMed] [Google Scholar]
- 13. Garrett RW, Emerson SG. Bone and blood vessels: the hard and the soft of hematopoietic stem cell niches. Cell Stem Cell 2009; 4: 503–506 [DOI] [PubMed] [Google Scholar]
- 14. Askmyr M, Sims NA, Martin TJ, et al. What is the true nature of the osteoblastic hematopoietic stem cell niche? Trends Endocrinol Metab 2009; 20: 303–309, http://www.ncbi.nlm.nih.gov/pubmed?term=Purton%20LE%5BAuthor%5D&;cauthor=true&cauthor_uid=19595609 [DOI] [PubMed] [Google Scholar]
- 15. Lévesque JP, Helwani FM, Winkler IG. The endosteal “osteoblastic” niche and its role in hematopoietic stem cell homing and mobilization. Leukemia 2010; 24: 1979–1992 [DOI] [PubMed] [Google Scholar]
- 16. Oh IH, Kwon KR. Concise review: multiple niches for hematopoietic stem cell regulations. Stem Cells 2010; 28: 1243–1249 [DOI] [PubMed] [Google Scholar]
- 17. Bianco P. Bone and the hematopoietic niche: a tale of two stem cells. Blood 2011; 117: 5281–5288 [DOI] [PubMed] [Google Scholar]
- 18. Park D, Sykes DB, Scadden DT. The hematopoietic stem cell niche. Front Biosci 2012; 17: 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Visnjic D, Kalajzic Z, Rowe DW, et al. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood 2004; 103: 3258–3264 [DOI] [PubMed] [Google Scholar]
- 20. Raaijmakers MH, Mukherjee S, Guo S, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature 2010; 464: 852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Barros AP, Takiya CM, Garzoni LR, et al. Osteoblasts and bone marrow mesenchymal stromal cells control hematopoietic stem cell migration and proliferation in 3D in vitro model. PLoS One 2010; 5: e9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Méndez-Ferrer S, Michurina TV, Ferraro F, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010; 466: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li T, Wu Y. Paracrine molecules of mesenchymal stem cells for hematopoietic stem cell niche. Bone Marrow Res 2011; 2011: 353878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Torok-Storb B, Iwata M, Graf L, et al. Dissecting the marrow microenvironment. Ann N Y Acad Sci 1999; 872: 164–170 [DOI] [PubMed] [Google Scholar]
- 25. Mishima S, Nagai A, Abdullah S, et al. Effective ex vivo expansion of hematopoietic stem cells using osteoblast-differentiated mesenchymal stem cells is CXCL12 dependent. Eur J Haematol 2010; 84: 538–546 [DOI] [PubMed] [Google Scholar]
- 26. Adams GB, Chabner KT, Alley IR, et al. Stem cell engraftment at the endosteal niche is specified by the calcium-sensing receptor. Nature 2006; 439: 599–603 [DOI] [PubMed] [Google Scholar]
- 27. Nakamura S, Matsumoto T, Sasaki J, et al. Effect of calcium ion concentrations on osteogenic differentiation and hematopoietic stem cell niche-related protein expression in osteoblasts. Tissue Eng Part A 2010; 16: 2467–2673 [DOI] [PubMed] [Google Scholar]
- 28. Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J Cell Sci 2001; 114: 3583–3590 [DOI] [PubMed] [Google Scholar]
- 29. Peerani R, Zandstra PW. Enabling stem cell therapies through synthetic stem cell-niche engineering. J Clin Invest 2010; 120: 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan BP, Chan GCF, Wong HL, et al. Cell-matrix microsphere, associated products, methods for preparation and applications. Regular Patent application no. 11/750,863, USA, 7 February 2008
- 31. Chan BP, Hui TY, Yeung CW, et al. Self-assembled collagen-human mesenchymal stem cell microspheres for regenerative medicine. Biomaterials 2007; 28: 4652–4666 [DOI] [PubMed] [Google Scholar]
- 32. Hui TY, Cheung KM, Cheung WL, et al. In vitro chondrogenic differentiation of human mesenchymal stem cells in collagen microspheres: influence of cell seeding density and collagen concentration. Biomaterials 2008; 29: 3201–3212 [DOI] [PubMed] [Google Scholar]
- 33. Chan BP, Hui TY, Wong MY, et al. Mesenchymal stem cell-encapsulated collagen microspheres for bone tissue engineering. Tissue Eng Part C Methods 2010; 16: 225–235 [DOI] [PubMed] [Google Scholar]
- 34. Cheng HW, Tsui YK, Cheung KM, et al. Decellularization of chondrocyte-encapsulated collagen microspheres: a three-dimensional model to study the effects of acellular matrix on stem cell fate. Tissue Eng Part C Methods 2009; 15: 697–706 [DOI] [PubMed] [Google Scholar]
- 35. Kim YJ, Sah RL, Doong JY, et al. Fluorometric assay of DNA in cartilage explants using Hoechst 33258. Anal Biochem 1988; 174: 168–176 [DOI] [PubMed] [Google Scholar]
- 36. Chan BP, So KF. Photochemical crosslinking improves the physicochemical properties of collagen scaffolds. J Biomed Mater Res A 2005; 75: 689–701 [DOI] [PubMed] [Google Scholar]
- 37. Tu Q, Valverde P, Chen J. Osterix enhances proliferation and osteogenic potential of bone marrow stromal cells. Biochem Biophys Res Commun 2006; 341(4): 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sugiyama T, Kohara H, Noda M, et al. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 2006; 25: 977–988 [DOI] [PubMed] [Google Scholar]
- 39. Lo Celso C, Fleming HE, Wu JW, et al. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 2009; 457: 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xie Y, Yin T, Wiegraebe W, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature 2009; 457: 97–101 [DOI] [PubMed] [Google Scholar]
- 41. Köhler A, Schmithorst V, Filippi MD, et al. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood 2009; 114: 290–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jiang Y, Bonig H, Ulyanova T, et al. On the adaptation of endosteal stem cell niche function in response to stress. Blood 2009; 114: 3773–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tan J, Liu T, Hou L, et al. Maintenance and expansion of hematopoietic stem/progenitor cells in biomimetic osteoblast niche. Cytotechnology 2010; 62: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Z, Li L. Understanding hematopoietic stem-cell microenvironments. Trends Biochem Sci 2006; 31: 589–595 [DOI] [PubMed] [Google Scholar]
- 45. ten Dijke P, Korchynskyi O, Valdimarsdottir G, et al. Controlling cell fate by bone morphogenetic protein receptors. Mol Cell Endocrinol 2003; 211: 105–113 [DOI] [PubMed] [Google Scholar]
- 46. Calori GM, Donati D, Di Bella C, et al. Bone morphogenetic proteins and tissue engineering: future directions. Injury 2009; 40(Suppl. 3): S67–S76 [DOI] [PubMed] [Google Scholar]
- 47. Gautschi OP, Frey SP, Zellweger R. Bone morphogenetic proteins in clinical applications. ANZ J Surg 2007; 77: 626–631 [DOI] [PubMed] [Google Scholar]
- 48. Seib FP, Franke M, Jing D, et al. Endogenous bone morphogenetic proteins in human bone marrow-derived multipotent mesenchymal stromal cells. Eur J Cell Biol 2009; 88: 257–271 [DOI] [PubMed] [Google Scholar]
- 49. McKay WF, Peckham SM, Badura JM. A comprehensive clinical review of recombinant human bone morphogenetic protein-2 (INFUSE Bone Graft). Int Orthop 2007; 31: 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors 2004; 22: 233–241 [DOI] [PubMed] [Google Scholar]
- 51. Singbrant S, Askmyr M, Purton LE, et al. Defining the hematopoietic stem cell niche: the chicken and the egg conundrum. J Cell Biochem 2011; 112: 1486–1490 [DOI] [PubMed] [Google Scholar]
- 52. Sharma MB, Limaye LS, Kale VP. Mimicking the functional hematopoietic stem cell niche in vitro: recapitulation of marrow physiology by hydrogel-based three-dimensional cultures of mesenchymal stromal cells. Haematologica 2012; 97: 651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chan BP, Li CH, Au-Yeung KL, et al. A microplate compression method for elastic modulus measurement of soft and viscoelastic collagen microspheres. Ann Biomed Eng 2008; 36: 1254–1267 [DOI] [PubMed] [Google Scholar]
- 54. Kwok CB, Ho FC, Li CW, et al. Compression-induced alignment and elongation of human mesenchymal stem cell (hMSC) in 3D collagen constructs is collagen concentration dependent. J Biomed Mater Res A 2013; 101: 1716–1725 [DOI] [PubMed] [Google Scholar]