Abstract
Aminoacyl-tRNA synthetases remove (proofread) incorrect substrates and thereby prevent errors in protein synthesis. We report enzyme-catalyzed pre-transfer editing by pimeloyl-CoA ligase (BioW), a biotin synthetic enzyme that converts pimelate, a seven carbon dicarboxylic acid, to its CoA ester. The noncognate BioW substrate, glutaric acid, results in hydrolysis of ATP to AMP with formation of only trace amounts of glutaryl-CoA thereby mimicking pre-transfer editing of incorrect aminoacyl-adenylates by aminoacyl-tRNA synthetases.
A major determinant of the low error rate of protein synthesis is the specific aminoacyl-tRNA synthetase-catalyzed acylation of tRNAs by only the cognate amino acid. Aminoacyl-tRNA synthetases can ensure faithful protein synthesis by having high substrate selectivity. However, as first pointed out by Pauling (Pauling, 1958) small differences in binding energy between aliphatic amino acids cannot provide the discrimination necessary for faithful protein synthesis. For amino acids having closely related structures such as isoleucine and valine, discrimination against the smaller amino acid would be incomplete. Indeed, Baldwin and Berg (Baldwin and Berg, 1966) showed that isoleucine tRNA synthetase converted both isoleucine and valine to their adenylates. However, only isoleucine was transferred to tRNA because valyl-adenylate was hydrolyzed to AMP and valine. These acyl-adenylate editing reactions proceed either by eviction of the non-cognate acyl-adenylate into solution where it undergoes spontaneous hydrolysis or by catalyzed hydrolysis within the active site (or in a separate editing site). In some cases enzymatic hydrolysis is stimulated by the presence of an acceptor tRNA. These reactions are generally referred to as editing, but proofreading is more accurate because proofreading only eliminates errors whereas editing improves the product.
We report what we believe is the first case of proofreading by an acyl-CoA synthetase/ligase (synthetase and ligase are approved synonyms). The acyl-CoA ligase is BioW, an enzyme required for biotin synthesis in Bacillus subtilis and closely related bacteria (Bower, et al., 1996). The physiological reaction of BioW is the ATP-dependent conversion of pimelate, a seven carbon α, ω-dicarboxylic acid to its CoA monothioester (Fig. 1a). Although BioW contains none of the sequence motifs characteristic of acyl-CoA ligases, the reaction was demonstrated to proceed through the canonical acyl-adenylate intermediate. However when presented with glutaric acid, the C5 homologue of pimelic acid, only traces of adenylate and CoA thioester were formed and much of the ATP was converted to AMP. Conversion of ATP to AMP in the absence of final product synthesis is the hallmark of pre-transfer editing in protein synthesis (Yadavalli and Ibba, 2012). Our data argue that the glutaryl-adenylate intermediate is largely cleaved within the BioW active site. A similar, but less efficient, proofreading of the adenylate of a second incorrect substrate, the C8 α, ω-dicarboxylic acid, suberic acid, also occurred.
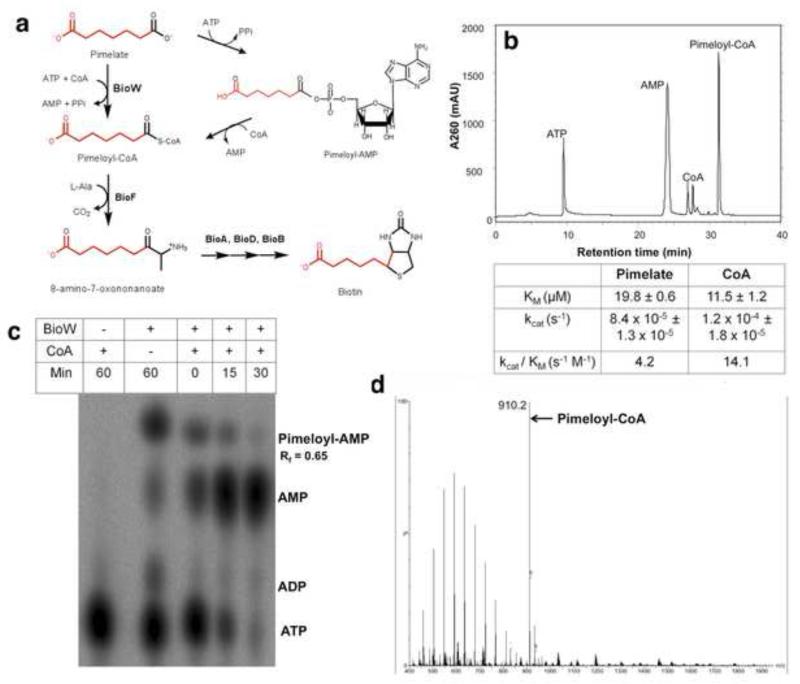
Figure 1. Schematic of B. subtilis biotin biosynthesis and the BioW reaction.
(a) The pathway begins with the conversion of pimelate to pimeloyl-CoA by an ATP-dependent reaction catalyzed by BioW with the adenylate as intermediate. The pimelate thioester provides most the carbon atoms of the biotin as indicated in red. The pimelate thioester reacts with L-alanine in a decarboxylative condensation catalyzed by BioF to form 8-amino-7-oxononanoate, the first intermediate in formation of the fused heterocyclic rings of biotin. Ring formation is completed by successive reactions catalyzed by BioA, BioD and BioB to generate biotin. (b) The BioW reaction with pimelate as substrate with separation of substrates and products by reverse phase HPLC. The absorbance peaks were identified using authentic standards of ATP, AMP and CoA. Below the chromatogram the steady state kinetic parameters for pimelate and CoA determined in sodium HEPES buffer pH 7 with 50 nM BioW are given. The parameters were determined by fitting the data from three independent experiments to Lineweaver-Burk plots and are given as the mean ± standard error. (c) Thin layer chromatography of α-32P-ATP labeled reactions in the absence and presence of CoA. A time course is shown for the CoA reactions. (d) Identification of pimeloyl-CoA (calculated mass 909.7) by electrospray ionization mass spectroscopy. The sharp peak eluting after 31 min in (b) was collected dried under nitrogen and submitted for mass spectroscopy. See Figs. S1, S2 and S3 for BioW purification and characterization.
Pimelic acid (heptanedioic acid) is a seven carbon α, ω-dicarboxylic acid that contributes most of the carbon atoms of biotin, the others coming from alanine and CO2 (Lin and Cronan, 2011) (Fig. 1a). To be used as a biotin precursor one of the pimelate carboxyl groups must be activated by conversion to a thioester (of either CoA or acyl carrier protein) (Fig. 1a). This thioester then reacts with alanine in the decarboxylative condensation reaction catalyzed by BioF to form 8-amino-7-oxononanoate, the first intermediate in formation of the fused heterocyclic rings of biotin (Fig. 1a).
Although in E. coli pimelate is formed by a modified fatty acid synthetic pathway (Lin, et al., 2010), the source of pimelate in B. subtilis remains unknown. However, the role of BioW is clear; strains lacking BioW are defective in biotin synthesis (unpublished data). Moreover, upon expression of BioW in E. coli, pimelate supplementation allows bypass of mutations in pimelate synthesis (Bower, et al., 1996; Lin, et al., 2010). BioW is an extremely unusual acyl-CoA ligase in that it lacks all of the well characterized motifs of this highly conserved enzyme family (Gulick, 2009) and is half the typical size. This remarkable divergence from the canonical enzymes led us to purify BioW and characterize the pimeloyl-CoA synthesis reaction in detail.
Attachment of hexahistidine tags to either end of BioW destroyed the ability of the protein to function in E. coli and thus the enzyme was purified by conventional column chromatography steps to obtain preparations that gave a single band in denaturing gel electrophoresis and a single peak on size exclusion chromatography that indicated BioW is a dimeric protein (Figs. S1 and S2). Pimeloyl-CoA synthesis was assayed by HPLC with detection by UV absorbance and required pimelate, ATP, Mg++ and CoA (Fig. 1b). Pimeloyl-CoA formation, verified by mass spectroscopy (Fig. 1d), was accompanied by production of AMP consistent with a pimeloyl-adenylate intermediate. Use of α-32P-ATP allowed direct demonstration of the pimeloyl-adenylate intermediate which accumulated in the absence of the CoA acceptor and was converted to AMP upon addition of CoA (Fig. 1c). The overall reaction had the stoichiometry expected of an acyl-CoA ligase; one pimeloyl-CoA molecule was formed per ATP hydrolyzed to AMP (Gulick, 2009). The reaction was most rapid at pH 7. ATP was the sole active nucleotide triphosphate and a metal ion (Mg++ > Mn++ > Co++) was required. (Fig. S3). Additionally, the kinetic parameters for pimelate and CoA were determined in sodium HEPES buffer at pH 7 using the α-32P labeled ATP assay (Fig. 1b). These data showed that BioW is a notably poor catalyst. This property is characteristic of biotin synthetic enzymes (Bar-Even, et al., 2011; Lin and Cronan, 2011; Lin, et al., 2010) consistent with the extremely low physiological demand for biotin.
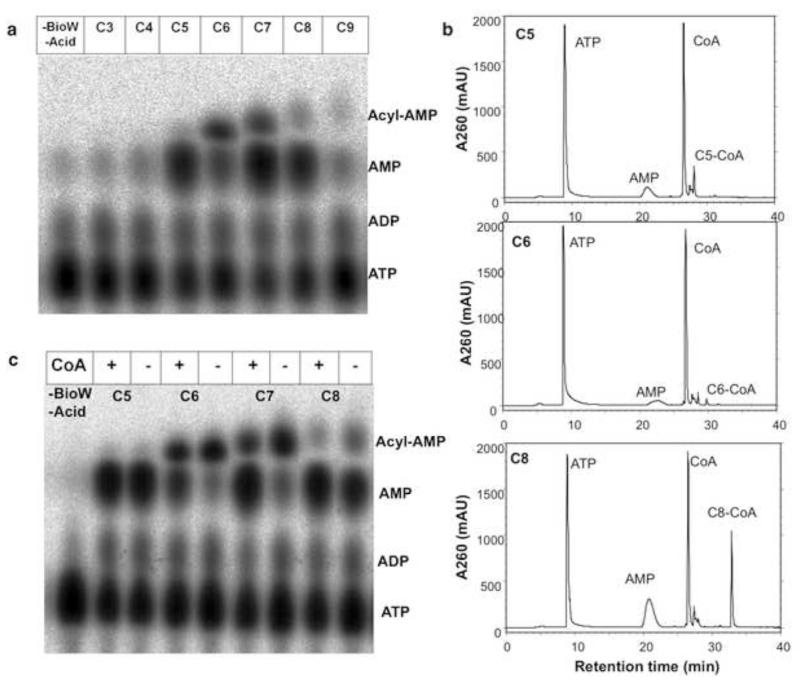
The BioW dicarboxylate substrate specificity was first determined in the overall reaction and as expected pimelate was the best substrate (Figs. 1a & 2b). No synthesis of the CoA ester was seen with the C6 homologue, adipic acid, whereas the C8 acid, suberic acid, was only weakly active and the C5 acid, glutaric acid, gave only traces of the CoA ester (Fig. 2b). To determine which partial reaction limited utilization of the noncognate substrates, α-32P-labeled ATP was used to assay conversion of the acids to their acyl-adenylates in the absence of CoA. Adipic (C6) and suberic (C8) acids synthesized detectable levels of their adenylates whereas the C5 homologue, glutaric acid, formed only trace amounts of glutaryl-adenylate (Fig. 2c). However, in the glutarate reactions AMP accumulated at the expense of ATP in a glutarate-dependent manner (Fig. 2c). AMP accumulation in reactions with a noncognate substrate is the hallmark of pre-transfer proofreading (editing) by aminoacyl-tRNA synthetases; the adenylate is synthesized and then cleaved to AMP (Martinis and Boniecki, 2010; Yadavalli and Ibba, 2012). For some aminoacyl-tRNA synthetases proofreading is largely or totally dependent on the presence of the tRNA cognate to the synthetase (Yadavalli and Ibba, 2012). However, the presence of CoA had no detectable effect on glutaryl-adenylate hydrolysis by BioW (Fig. 2c). Note that the low levels of glutaryl-CoA accumulation cannot be attributed to solvent hydrolysis of the thioester. Glutaryl-CoA is a well-studied enzyme substrate and is not an unusually labile acyl-CoA. Indeed, we monitored the thioester linkage by absorbance at 232 nm of a solution of glutaryl-CoA at 23°C for 3 h in assay buffer at pH 7.5 without detectable loss of absorbance.
Figure 2. Dicarboxylate specificity of BioW.
(a) BioW reactions were performed with dicarboxylic acids from C3 to C9 to determine acyl chain substrate specificity. The reactions were carried out as described in Experimental Procedures and 1 μl of each reaction was spotted onto cellulose TLC plate. (b CoA thioester formation from the C5, C6 and C8 dicarboxylates after reaction with BioW at 37°C for 1 h. Panels from top to bottom show HPLC analyses of BioW reactions with the C5, C6 and C8 dicarboxylates, respectively. (c) Evidence of adenylate editing in absence of CoA. BioW reactions with the C5, C6, C7 and C8 dicarboxylates were carried out in presence or absence of CoA at 37°C for 1 h. Samples of 1 μl were spotted onto a TLC plate which was developed as described in Experimental Procedures. Note that the migration rates of the acyl-AMP intermediates vary with the dicarboxylate moiety. C8-CoA was identified by mass spectroscopy (Fig. S4) whereas the trace amounts of C5-CoA formed were insufficient for analysis.
The C8-adenylate also showed signs of hydrolysis to AMP whereas the C6-adenylate accumulated but showed no hydrolysis in the absence of CoA (Fig. 2c). Upon CoA addition AMP levels increased somewhat but no adipoyl-CoA was formed (Fig. 2b) suggesting that CoA increased solvent hydrolysis of the enzyme-bound C6-adenylate. The BioW reaction with adipic acid (C6) was not further studied because it is not a naturally occurring compound and thus physiologically irrelevant.
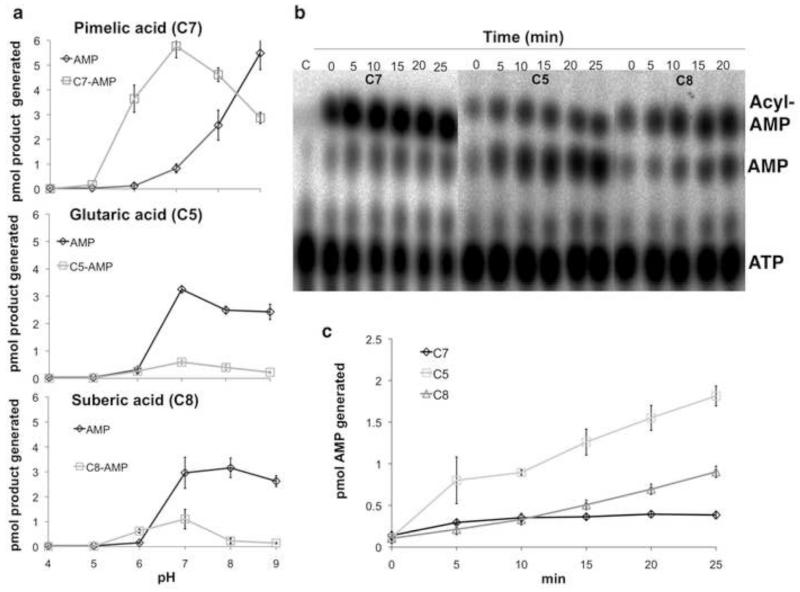
As reported for aminoacyl-tRNA synthetase pre-transfer editing reactions (Yadavalli and Ibba, 2012), adenylate hydrolysis could occur either by cleavage within the BioW active site or by release of the adenylate to solution where the mixed anhydride bond would be cleaved by hydroxide ion. The rate of acyl-adenylate hydrolysis increases as pH values rise above neutrality (Demoss, et al., 1956; Jencks, 1963; Schall, et al., 1998) and thus hydrolysis of adenylates released to solution should increase with pH. However, in the case of the C5- and C8-adenylates increased solvent pH had no effect on hydrolysis of the adenylate (Fig. 3a). The most straightforward interpretation of these data is that these adenylates are largely sequestered within the BioW active site. If so, then hydrolysis of the C5- and C8-adenylates must proceed within the enzyme active site. In contrast the level of the C7-adenylate declined at elevated pH values with a concomitant increase in AMP indicating that solvent could access this adenylate (Fig. 3a). A greater accessibility of C7-adenylate bound within the active site seems reasonable because the CoA thiol must enter the active site for pimeloyl-CoA production. However, a pH-induced conformational change that releases the C7-adenylate to solution could also occur. The BioW catalyzed hydrolysis of the C5- and C8-adenylates was linear with time whereas pimeloyl-adenylate filled the BioW active site (1 molecule of pimeloyl-AMP per BioW monomer) (Fig. 3c).
Figure 3. Cleavage of cognate and noncognate adenylates.
(a) The pH profiles of BioW activity with pimelate (top), glutarate (middle) and suberate (bottom) in absence of CoA. Buffers of sodium acetate (pH 4 and pH 5), sodium 2-(N-morpholino)ethanesulfonic acid (pH 6), sodium HEPES (pH 7) and Tris-HCl (pH 8 and pH 9) were used to determine the pH profile of the BioW reaction for 1 h at 37°C. (b) BioW catalyzed glutaryl- and suberyl-adenylate hydrolysis was assayed by determining the rate of AMP production with time. Reactions with pimelate (C7) were included for comparison. At different time points 1 μl samples of BioW reactions performed at pH 7.0 and 37°C were spotted onto cellulose TLC plates followed by development and autoradiography. (c) Quantitation of the data points of panel b were used to show the rate of formation of AMP with increasing time with C7, C5 and C8 as substrates. The intensities of the spots were quantitated using the phosphor-imaging software of the Fujifilm FLA-3000 phosphoimager. The percent of photostimulated luminescence of AMP and acyl-AMP spots obtained from the reaction was compared to that of the ATP spot obtained from a control reaction lacking enzyme included on each cellulose TLC plate to derive the amounts of products generated. All reactions were repeated three times and the bars denote standard deviations.
BioW proofreading by cleavage of glutaryl-adenylate is reminiscent of valine activation by isoleucine-tRNA synthetase, a situation first addressed by Pauling (Pauling, 1958) who calculated that the small differences in binding energy among aliphatic amino acids could not provide the discrimination necessary for accurate protein synthesis. He estimated that incorporation of isoleucine would be favored over valine by a factor of only 100-200. Direct binding measurements to isoleucyl-tRNA synthetase confirmed Pauling’s prediction (Loftfield and Eigner, 1966). However, the error rate in insertion of valine for isoleucine is 1 in 3000 (Loftfield and Vanderjagt, 1972) due to proofreading (Fersht, 1999). Pauling’s argument also pertains to the BioW reaction with glutarate. Glutarate is smaller than pimelate precluding steric occlusion and the two additional methylene groups of pimelate would not provide sufficient binding energy to insure specificity. Moreover, glutaric acid occurs in nature and is particularly abundant in seeds and root vegetables. Thus, it seems likely that B. subtilis, a soil bacterium, could encounter high levels of this dicarboxylate and proofreading of the adenylate would act to prevent possible short circuiting of biotin synthesis by glutarate.
Suberic acid also occurs naturally and thus hydrolytic proofreading of its adenylate is appropriate. Superficially, isoleucine activation by valine-tRNA synthetase would seem an appropriate comparison, because the noncognate substrate has one additional methylene group. However, the chiral amino acid must bind the enzyme with strict stereochemistry whereas the nonchiral and flexible dicarboxylate is able to bind in a manner that allows formation of the adenylate and low levels of the CoA ester.
The broad question is whether or not BioW proofreading has an evolutionary relationship with aminoacyl-tRNA synthetase pre-transfer editing. This may be the case but the extant protein sequences provide no support for a relationship. BioW is a most unusual protein in that database searches by a large number of search algorithms including those designed to detect very remotely homologous sequences (e.g., HMMER http://hmmer.janelia.org/) give only BioW sequences. Moreover, as mentioned above, BioW has no recognized sequence motifs. This is not surprising for pimelate, an unusual ligand, but is surprising for CoA and especially so for ATP. There may be structural homologies that could speak for (or against) a relationship between BioW and aminoacyl-tRNA synthetases but this will require a BioW crystal structure.
Other examples of hydrolytic proofreading are known although they involve different chemistry. For example in polyketide synthesis a recent report demonstrated that a protein previously thought to be an acyltransferase was a thioesterase that specifically cleaved non-cognate acyl-thioesters (Jensen, et al., 2012). It seems reasonable that proofreading reactions may occur most often in the early steps in synthesis of complex molecules such as biotin or an intricate polyketide to prevent embarking on the expensive synthesis of inactive molecules.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
The bioW gene was PCR amplified from Bacillus subtilis subsp. subtilis str. 168 genomic DNA using primers pMM1 5′-GGA GAT ATA CCA TGA TGC AAG AAG AAA C-3′ and pMM2 5′-CTT CTT GCA TCA TGG TAT ATC TCC TTC-3′ and Pfu Turbo DNA polymerase from Stratagene. The amplified gene was digested with restriction enzymes, NcoI and XhoI, and inserted between the NcoI and XhoI sites of vector pET28b (Novagen). The sequence verified plasmid (pMM8) was transformed into E. coli BL21 Tuner Novagen) and the transformants were plated on LB medium containing 50 μg/ml of kanamycin sulfate.
Expression and Purification of BioW
A two liter LB culture of E. coli BL21 Tuner transformed with plasmid pMM8 was grown to an OD600 of 0.6 at 37°C. followed by induction with 1 mM isopropyl-β-D-thiogalactopyranoside for 5 h at 37°C. The cells were harvested by centrifugation and resuspended in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol and 5 mM tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and then lysed by passage through a French pressure cell. The supernatant obtained after centrifugation of cell lysate at 39,000×g for 30 min at 4°C, was applied to a 5 mL HiTrap Q FF ion exchange chromatography column (GE Healthcare Life Sciences) equilibrated with the buffer used to suspend the harvested cells. The column was washed with 50 mM Tris-HCl (pH 7.5), 1 M NaCl, 10% glycerol and 5 mM TCEP. The protein fraction eluted at about 250 mM NaCl and was analyzed by SDS/PAGE. Fractions containing the highly purified protein were pooled, concentrated and resuspended in 50 mM Tris (pH 7.5), 100 mM NaCl, 10% glycerol, 5 mM TCEP and 1 M ammonium sulfate and applied to a 5 mL HiTrap Phenyl HP hydrophobic interaction chromatography column (GE Healthcare Life Sciences) equilibrated with the same buffer. The column was washed with 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol and 5 mM TCEP and the protein was eluted with a linear gradient from 1 M to 0 ammonium sulfate. After analyzing samples by SDS/PAGE, the fractions having the highest degree of purity were pooled and applied to a HiLoad 26/60 Superdex 200 size exclusion chromatography column. The column was equilibrated with 50 mM Tris-HCl (pH 8.0) containing 150 mM NaCl and the protein was allowed to elute over one column volume. The protein fractions of greatest purity were pooled and dialyzed against a storage buffer of 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20% glycerol and 5 mM TCEP. Finally, the dialyzed protein was concentrated to 2.7 mg/mL, flash frozen and stored at −80°C.
High Performance Liquid Chromatography (HPLC) enzyme assays
Pimeloyl-CoA ligase activity was assayed by detection of pimeloyl-CoA formation. The reactions contained 100 mM sodium 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7), 10 mM MgCl2, 0.1M NaCl, 0.2 mM TCEP, 5 mM pimelate, 1 mM CoA and 1 mM ATP and were initiated by adding enzyme to 5 μM (all enzyme concentrations are given as monomeric protein). After incubation at 37°C for 1 h, the reaction was stopped by adding 10% trichloroacetic acid to precipitate the protein. The precipitated protein sample was removed by centrifugation and filtration through Amicon Ultra 3K devices (Millipore). The filtrate was loaded onto a μBondapak C18 cartridge (Waters) previously equilibrated with 50 mM ammonium acetate (pH 5). Solvent A was 50 mM ammonium acetate (pH 5) and solvent B was acetonitrile. The sample was eluted with 0% B for 10 min, 30 min of a gradient from 0%B to 50% B followed by 5 min gradient from 50% B to 80% B which was maintained for 10 min (Iram and Cronan, 2006). Eluent corresponding to the pimeloyl-CoA peak was dried under nitrogen and the mass was analyzed at the University of Illinois School of Chemical Sciences Mass Spectrometry Laboratory by electrospray ionization mass spectrometry performed on a Micromass Q-Tof Ultima instrument run in the positive mode.
Thin layer chromatographic enzyme assays
In general, the reactions contained 100 mM sodium HEPES (pH 7), 5-10 mM MgCl2, 0.1-0.2M NaCl, 0.2 mM TCEP, 1-10 mM pimelate, 0.25 mM CoA, 25-100 μM ATP wit [α-32P]ATP 600 to 1200-fold lower in concentration than the nonradioactive ATP and 5 μM BioW except where indicated. The specificity of BioW was tested with malonic acid (C3), succinic acid (C4), glutaric acid (C5), adipic acid (C6), pimelic acid (C7), suberic acid (C8), azelaic acid (C9). To assay non-cognate substrate editing by BioW, the rate the AMP formation in the absence of CoA was monitored in a time dependent manner. At each time point 1 μl of the reaction was spotted onto a thin layer plate. Kinetic parameters for pimelate and CoA were determined by varying the concentration (5-100 μM) of the respective substrate in a reaction containing 50 nM BioW in order to achieve a linear progression of enzymatic activity. Thin layer chromatography on cellulose plates were developed in isobutyric acid: ammonium hydroxide: water (66:1:33) (Christensen and Cronan, 2009). After drying the thin layer chromatograms were exposed to a phosphor imaging plate for 1-17 h and the results visualized using a Fujifilm FLA-3000 instrument. The chromatogram spots were quantitated by comparing them with a standard curve generated by serial dilution o known [α-32P]ATP concentration for each plate. All reactions were repeated three times and the kinetic parameters were determined by fitting the data points to Lineweaver-Burk plots.
Statistical Analysis
The error bars denote standard deviations.
Supplementary Material
Acyl-CoA ligase proofreading
Error correction
Analogy to tRNA synthetases
Acyl-adenylate hydrolysis
SIGNIFICANCE.
Error correction plays a major role in the fidelity of biological systems. It is difficult to build absolute specificity into some biochemical reactions because closely related, but smaller molecules can enter enzyme catalytic sites. In protein synthesis these errors are eliminated by going back and checking the properties of an intermediate in the reaction and, if incorrect, hydrolyzing the intermediate. We report that an enzyme unrelated to protein synthesis has this same property. This enzyme, called BioW, is required for synthesis of the vitamin, biotin, and cleaves incorrect intermediates that are too long or too short. This cleavage avoids making inactive and potentially toxic biotin analogues. BioW catalyzes a canonical acyl-CoA synthetase/ligase reaction with the cognate seven-carbon substrate whereas reactions with the homologous C5 and C8 substrates result in hydrolysis of the acyl-adenylate intermediate with little or no formation of the acyl-CoA product. Although BioW is an acyl-CoA synthetase/ligase, it lacks all of the motifs of this well understood enzyme family and is only half the size of the typical family member.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant AI15650 from the National Institute of Allergy and Infectious Diseases. We thank Dr. Steven Lin and Vandana Chakravartty for advice and support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Baldwin AN, Berg P. Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J Biol Chem. 1966;241:839–845. [PubMed] [Google Scholar]
- Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. The moderately efficient enzyme: evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50:4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- Bower S, Perkins JB, Yocum RR, Howitt CL, Rahaim P, Pero J. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J Bacteriol. 1996;178:4122–4130. doi: 10.1128/jb.178.14.4122-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen QH, Cronan JE. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J Biol Chem. 2009;284:21317–21326. doi: 10.1074/jbc.M109.015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoss JA, Genuth SM, Novelli GD. The enzymatic activation of amino acids via their acyl-adenylate derivatives. Proc Natl Acad Sci U S A. 1956;42:325–332. doi: 10.1073/pnas.42.6.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. W. H. Freeman and Company; New York, NY: 1999. [Google Scholar]
- Gulick AM. Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iram SH, Cronan JE. The ß-oxidation systems of Escherichia coli and Salmonella enterica are not functionally equivalent. J Bacteriol. 2006;188:599–608. doi: 10.1128/JB.188.2.599-608.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks W. Preparation and properties of acyl adenylates. Meth Enzymol. 1963;6:762–766. [Google Scholar]
- Jensen K, Niederkruger H, Zimmermann K, Vagstad AL, Moldenhauer J, Brendel N, Frank S, Poplau P, Kohlhaas C, Townsend CA, et al. Polyketide proofreading by an acyltransferase-like enzyme. Chem Biol. 2012;19:329–339. doi: 10.1016/j.chembiol.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Cronan JE. Closing in on complete pathways of biotin biosynthesis. Molec Biosyst. 2011;7:1811–1821. doi: 10.1039/c1mb05022b. [DOI] [PubMed] [Google Scholar]
- Lin S, Hanson RE, Cronan JE. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat Chem Biol. 2010;6:682–688. doi: 10.1038/nchembio.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftfield RB, Eigner EA. The specificity of enzymic reactions. Aminoacyl-soluble RNA ligases. Biochim Biophys Acta. 1966;130:426–448. doi: 10.1016/0304-4165(66)90239-x. [DOI] [PubMed] [Google Scholar]
- Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128:1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis SA, Boniecki MT. The balance between pre- and post-transfer editing in tRNA synthetases. FEBS Lett. 2010;584:455–459. doi: 10.1016/j.febslet.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L. Festschrift für Prof Dr. Arthur Stoll. Birkhauser Verlag; Basel, Switzerland: 1958. [Google Scholar]
- Schall OF, Suzuki I, Murray CL, Gordon JI, Gokel GW. Characterization of acyl adenyl anhydrides: Differences in the hydrolytic rates of fatty acyl-AMP and aminoacyl-AMP derivatives. J Org Chem. 1998;63:8661–8667. [Google Scholar]
- Yadavalli SS, Ibba M. Quality control in aminoacyl-tRNA synthesis: its role in translational fidelity. Adv Protein Chem Struct Biol. 2012;86:1–43. doi: 10.1016/B978-0-12-386497-0.00001-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.