Abstract
This paper investigates a new test for normality that is easy for biomedical researchers to understand and easy to implement in all dimensions. In terms of power comparison against a broad range of alternatives, the new test outperforms the best known competitors in the literature as demonstrated by simulation results. In addition, the proposed test is illustrated using data from real biomedical studies.
Keywords: Goodness of fit, Normal distribution, Projection, Shapiro-Wilk test, Power
1. Introduction
Normal distribution is widely used in many applications. The problem of testing whether a sample of observations comes from a normal distribution has been studied extensively by many generations of statisticians, including [3, 6, 10, 13, 16, 17, 20, 23]. For instance, in a recent monograph devoted to the topic of testing for normality, Thode [23] reviewed more than thirty formal procedures proposed specifically for testing normality. Briey, in terms of power performance against a broad range of alternatives, the Shapiro-Wilk (SW) test [20] is the benchmark of omnibus tests for univariate data [3, 6, 23]. For testing multivariate normality, the Henze-Zirkler (HZ) test [13] is recommended by Thode [23, pp. 220]. In many practical applications, researchers often prefer to use tests that are both informative and easy to understand [4]. Although generally quite powerful as a multivariate test, the HZ test has the drawback of not being as easy to understand as the simple skewness or kurtosis based tests, or as the SW test which is known to many researchers and is generally powerful to detect outliers or influential observations as well as to skewed distributions. To the best of our knowledge, there is no known test that is both informative and has competitive power in all dimensions. In this paper, we introduce a simple informative test that is easy to understand and to implement in all dimensions. Simulations studies indicate that the new test has very competitive power compared to the HZ test and other best known tests in both univariate and multivariate cases.
The rest of this paper is organized as follows. Section 2 gives a brief review of some well-known tests for normality. Section 3 introduces our new test. Numerical studies on power comparison against a broad range of alternatives are reported in Section 4. Real data examples are given to illustrate the newly proposed test in Section 5. Some concluding remarks are in Section 6.
2. Some Well-known Tests for Normality
Consider independent observations X1,…,Xn from a p-variate random vector X, we want to test H0 : X has a p-variate normal density, against the general alternative H1 : X has a p-variate non-normal Lebesgue density. Throughout this paper, we consider the cases where n > p. Moreover, if we say that H0 is rejected when the test statistic is too extreme, we mean that, towards the direction of opposing normality, it exceeds the critical value, which can be accurately obtained by simple Monte Carlo simulations. Let X̄n and Sn be the sample mean and sample covariance, respectively. Denote the transpose of any vector x by xT, and its norm by ‖x‖ = (xTx)1/2. Given the existence of a p-variate Lebesgue density of X, Sn is nonsingular, i.e. exists, almost surely [8]. Thus, without loss of generality, we can reject the null hypothesis of normality when Sn is singular. In the rest of this paper we consider the case where Sn is nonsingular, use to denote the symmetric square root of , and consider the standardized data:
| (1) |
2.1 The Shapiro-Wilk test
The Shapiro-Wilk (SW) test [20] was originally designed for testing univariate normality. Given univariate data z1,…,zn, the SW statistic has the following form:
| (2) |
where z(1),…,z(n) are the order statistics of z1,…,zn, z̄n is the sample mean, and the constants an,i are (an,1,…,an,n) = (mTV−1V−1m)−1/2mTV−1, with m = (m1,…,mn)T and V the mean and covariance of the order statistics of a random standard normal sample of size n, respectively. The SW statistic can be easily evaluated using the free software R and many other statistical packages.
The SW test statistic can be regarded as the ratio of two variance estimators, the best linear unbiased estimator (BLUE) and the maximum likelihood estimator (MLE). Specifically, if X1,…,Xn are independent and identically distributed (i.i.d) as N(μ,σ2), then Xi = μ + σ Zi, i = 1,…,n, where Zi’s are i.i.d N(0, 1). The order statistics then have similar identities, namely X(i) = μ + σZ(i). Thus, X(i) = μ + σmi + εi, where mi = E(Z(i)) and εi = σ(Z(i) − mi). Under the normality assumption, the joint distribution of (ε1,…, εn)T has zero mean and covariance matrix σ2Vn, where Vn is the covariance matrix of the order statistics (Z(1),…,Z(n))T from N(0, 1). In addition, under the normality assumption, one can estimate the unknown parameter σ by using either the MLE s or the BLUE σ̂BL. Then Wn in (2) has an equivalent form as the variance ratio . Under the alternative model of non-normality, the BLUE σ̂BL tends to be smaller than the MLE s. Thus, the SW test rejects the hypothesis of normality for small values of Wn.
Moreover, from the derivation of Wn via linear regression of observed order statistics on the means of the standard normal order statistics, and the fact that the linear regression line is very sensitive to outliers or observations corresponding to extreme values in the x-direction, we can expect that the SW test is sensitive (or powerful) for detecting the non-normality due to outliers, or against a density with heavier tail than the normal density (e.g., the Cauchy distribution). These features of the SW test are quite easy to understand even for most biomedical researchers.
2.2 The skewness and kurtosis tests
The skewness and kurtosis have long been suggested for detecting non-normality in the univariate setting [17]. For general multivariate data, Mardia [16] constructed two statistics for measuring multivariate skewness and kurtosis. Using the notation for standardized data in (1), the skewness statistic MS is:
| (3) |
The kurtosis statistic MK is:
| (4) |
The skewness test rejects the hypothesis of normality if MS is too large, and the test based on the centralized kurtosis statistic MK rejects the null hypothesis of normality if its absolute value |MK| is too large, that is, it exceeds the appropriate critical value. Both the skewness and kurtosis tests are simple and informative [4], and provide specific information about non-normality of the data. In the univariate case, D’Agostino and Pearson [5] proposed the K2 test, by combining the skewness and kurtosis measures. Bowman and Shenton [2] considered an alternative way of combining skewness and kurtosis equivalent to the following statistic
| (5) |
which has an asymptotic distribution in the univariate case. In the multivariate case, Doornik and Hansen [7] demonstrated some practical utility and good power performance of MSK, especially against generalized Burr-Pareto-logistic distribution with normal marginals [7, Table 4]. However, these tests are not consistent for testing general alternatives and can have very low power against many alternatives.
2.3 The Henze-Zirkler test
Extending the work of [1, 10], Henze and Zirkler [13] proposed a test for normality using
| (6) |
where β ∈ R, IE,IEc are indicator functions with E = {Sn is singular} and in terms of Yi in (1),
The Henze-Zirkler (HZ) test rejects normality if HZβ is too large. They also proposed an optimal choice of the parameter β in using HZβ in the p-variate case as
A drawback of the HZ test is that, when H0 is rejected, the possible violation of normality is generally not clear. Thus many biomedical researchers would prefer an more informative and equally or more powerful test than the HZ test.
3. The new test
The main goal here is to construct a simple test with a combination of easy-to-understand components. Starting from univariate data, there have been many known tests obtained via combinations of kurtosis with other tests, among which the most well-known one [5] combines the kurtosis statistic MK with the skewness statistic MS. However, MS is powerful only when the alternative is skewed, thus it generally lacks power to test against non-skewed alternatives. On the other hand, SW has excellent power to detect skewed alternatives. Moreover, SW is also consistent against general alternatives [14]. Thus, we propose to combine kurtosis with SW, instead of with MS.
For a p-dimensional random vector Y, it is well known that Y is normal if and only if θTY is univariate normal for all θ ∈ {θ ∈ Rp: ‖θ‖ = 1}. It can also be proved that, if Y is not normal, the event that θTY is univariate normal can only occur for θ in a zero measure subset of {θ ∈ Rp: ‖θ‖ = 1}, which is a null set if one picks a θ randomly as discussed in [19, Theorem 1]. Thus it seems reasonable to pick a few data-driven directions, say θ ∈ Θ, and use SW test to detect non-normality in univariate projections on those directions. More specifically, for multivariate data X1,…,Xn, using the standardization discussed in (1) to obtain {Yi}1≤i≤n, we can then consider the the following statistic based on projection of Yi’s in the direction θ:
| (7) |
where Wn is the Shapiro-Wilk function in (2). Fattorini [11] considered a test which rejects normality for small values of following statistic
| (8) |
The Fattorini (FA) test is recommended by [23, pp.220] for testing multivariate normality in addition to the HZ test. It is clear that the FA test is based on detecting non-normality of multivariate data in the most “extreme” directions corresponding to the smallest Gn values evaluated at random directions {‖Yj‖−1Yj}1≤j≤n.
More generally, it seems natural to detect non-normality of the p most “extreme” directions corresponding to p smallest Gn values evaluated at random directions {‖Yj‖−1Yj}1≤j≤n, denoted Θ1. It is also reasonable to consider the p marginal variates which often have specific physical meanings and are of interest to practitioners. As a result, our new test statistic for normality is:
| (9) |
where Gn is the function in (7), Θ1 consists of the p most “extreme” directions corresponding to p smallest Gn values evaluated at random directions {‖Yj‖−1Yj}1≤j≤n and Θ2 = {ej}1≤j≤p with ej = (0,…,0, 1, 0,…,0)T (the unit vector with its j-th opponent being 1 and all others being 0), and
| (10) |
with c1, c2 being certain percentiles of MK in (4) under H0, e.g., c1 and c2 are 1% and 99% quantiles of MK as used in Section 4 for power simulation.
Note that, large values of 1 − Wn, |MK| all indicate departure from normality, thus the new test will reject H0 when Tn in (9) is too large. The statistic Tn is easy to evaluate using functions in R for Wn. Simulation results reported in the next section show that the new test has competitive power in all dimensions compared to some best known tests.
4. Numerical Studies
For power comparison of our proposed test Tn in (9), competing tests considered here include the Shapiro-Wilk test Wn in (2) (univariate case), the MSK test in (5), the Fatorrini test FA in (8), and the Henze-Zirkler test HZβ in (6) with the optimal choice of β in [13]. A broad range of alternative models are considered in the simulation. In the univariate case, the alternatives include those from Table 3 of the classic goodness-of-fit paper by Stephens [22], and also the Pearson type II, Pearson type VII distributions, and some other spherically symmetric distributions as considered in [13]. In the multivariate cases, alternative distributions include those considered in Table 6.4 in [13] and other similar ones. Significance levels of all tests are set at α = 5%.
We mention briefly in Section 2 that the null critical values can be simulated using empirical distribution via the Monte Carlo approach. This is due to the following two facts: First of all, under H0, as functions of the standardized data in (1), all the test statistics considered here have null distributions that do not depend on the location vector and the covariance matrix [13]. Thus, we can simulate their null distributions directly using the zero location vector and identity covariance matrix; Secondly, the empirical distribution function Fm(t) for the Monte Carlo sample (of size m) is uniformly (in the Kolmogorov-Smirnov distance) close to the true cumulative distribution function F(t). In fact, by the well-known Dvoretzky-Kiefer-Wolfowitz inequality [18, Lemma 5.1],
where C is a constant independent of F. Then, by the Borel-Cantelli lemma, almost surely, we have . Note that m is the number of replicates in Monte Carlo, which can easily be chosen as 5 million or more if needed. Therefore, the Monte Carlo approximated critical values, calculated as quantiles of Fm, can be as accurate as desired by choosing a sufficiently large m.
For HZn,β, Henze and Zirkler [13] developed an optimal choice β* for β, where β* = 2−1/2{[(2p + 1)n]/4}1/(p+4). Different β values in (6) yield tests that are sensitive to different types of alternatives. Simulations indicated that β* results in a test having good power against a broad range of alternatives, thus as suggested in [13], it is a preferred choice of β as an omnibus test for normality here. We use the 1% and 99% empirical quantiles of MK under H0, obtained through Monte Carlo simulation, for c1, c2 respectively. For example, when n = 50, p = 2, c1 = −1.455, c2 = 2.551 Note that in this way, for the level α = 5% test, the critical value of Tn is less than 1. Critical values of all test statistics are calculated using 100,000 simple random samples from the p-variate standard normal distribution. Those critical values are then used to calculate the empirical powers in Table A1-A4, and are given in Table 1. In addition, for practical convenience, the critical values of all test statistics considered corresponding to the level α = 5% test are presented in Table A5 for more sample size choices.
Table 1.
Empirical critical values and corresponding type I errors for the level α = 5% test of p dimensional normality when n = 50.
| Critical Value |
Type I Error |
|||||||
|---|---|---|---|---|---|---|---|---|
| Dimension | MSK | FA | HZ | Tn | MSK | FA | HZ | Tn |
| p = 1 | 5.329 | 0.046 | 0.729 | 0.048 | 0.0497 | 0.0498 | 0.0499 | 0.0488 |
| p = 2 | 10.712 | 0.068 | 0.873 | 0.054 | 0.0490 | 0.0493 | 0.0495 | 0.0488 |
| p = 5 | 49.509 | 0.106 | 0.963 | 0.053 | 0.0490 | 0.0496 | 0.0499 | 0.0489 |
| p = 10 | 244.326 | 0.187 | 0.993 | 0.063 | 0.0496 | 0.0496 | 0.0490 | 0.0485 |
Empirical powers are obtained based on the percentage of 5,000 Monte Carlo samples declared significant. Let θ be the true power, the standard error of the empirical power is then . Power comparison results corresponding to sample size n = 50 are reported. In addition, average ranks are appended at the end of each power table, where smaller values indicate better power performance. Average power across the alternatives are also included for comparison.
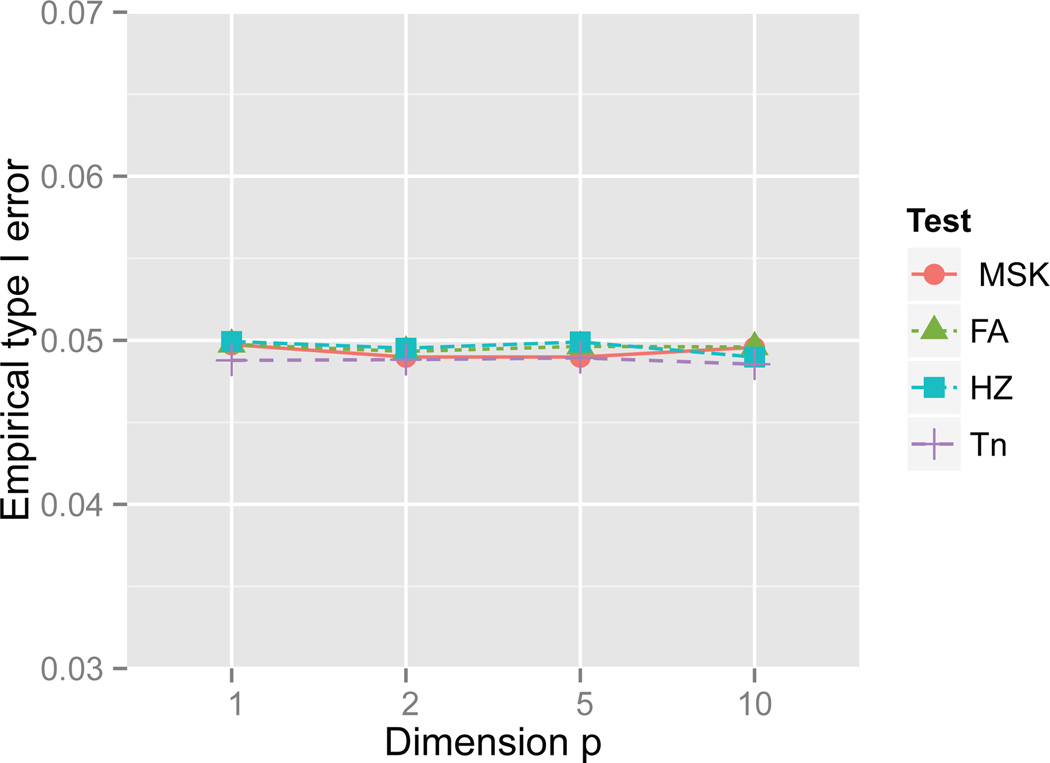
For all the critical values used in the empirical power calculation, the corresponding empirical type I errors, calculated as the proportion of rejection using 100,000 Monte Carlo normal samples, are also provided in Table 1. In the meanwhile, those type I errors are visualized in Figure 1. Power results of the univariate case are summarized in Table A1, where NormMix(p, μ2,σ2) stands for the univariate normal mixture distribution whose density is given by pϕ(x) + (1 − p)ϕ((x − μ2)/σ2), with ϕ(x) being the density of standard normal distribution. In this case, the FA test reduces to Wn since the Shapiro-Wilk statistic is location-scale invariant. From Table A1, even in the univariate case, the new test Tn is never seriously less powerful than Wn for all alternatives investigated. Moreover, Tn can be significantly more powerful than Wn for several alternatives considered here. For examples, Tn is 60% more powerful than Wn for Beta(2, 2) and PSII(1). Whether measuring by average rank or average power, Tn slightly outperforms the benchmark Shapiro-Wilk Wn test in the univariate case. Also, as demonstrated in Table A1, Tn is generally more powerful than the HZ test and MSK test.
Figure 1.
Empirical type I error of tests for normality at significance level α = 5% when n = 50.
In the multivariate case, F1 ⊗ F2 denotes the distribution with independent marginal distributions F1 and F2, and the product of k independent copies of F1 will be denoted by . Table A2 displays the power performances in the bivariate case. MVNMIX(a. b. c. d) stands for the multivariate normal mixture distribution with density aN(0,Σ1) + (1 − a)N(b1,Σ2), where 1 is the column vector with all elements being 1, Σ1 = (1 − c)I+c11T and Σ2 = (1 − d)I + d11T. Table A3 and Table A4 show results of testing 5 and 10 dimensional normality. It is clear that the new test outperforms the MSK, FA, HZ tests for an overwhelming majority of the examined alternatives. The new test also has higher power than the HZ test in more than 90% of alternatives studied, and the power difference are often quite substantial. Overall, the new test Tn is superior to HZ in terms of both average power and average rank. For example, when p = 5, the new test Tn is the most powerful among the four competitors for testing 46 out of the 48 alternatives, and in the two alternatives the new test is second most powerful. Thus Tn by far has the best average rank. When the dimension p = 10, the average power of Tn among these 48 alternatives is 73.04% which is far above the 47.9% average power of the HZ test. As illustrated using these alternatives, the power advantage of Tn over the best known competitor HZ test is overwhelming. The overall power advantage of the new test over MSK and FA is even more striking.
5. Examples
5.1 The Ramus Bone Data
Elston and Grizzle [9] gave a very interesting data of ramus height (in millimeters) of 20 boys each measured at age 8, 8.5, 9 and 9.5 years old. The data is fairly well known because it has a few interesting features. The recorded heights of the ramus bone marginally appear to be normally distributed while not jointly normal [24, pp. 126-130]. Specifically, we consider testing for bivariate normality of the ramus height data corresponding to the measurements at ages 8 and 9. The p-values corresponding to MSK, FA, HZ, Tn are 0.146, 0.017, 0.002, 0.037 respectively. Thus, at level α = 5%, our test, FA test and HZ test reject the null hypothesis of bivariate normality, while MSK fails to reject H0. On the other hand, if we test the multivariate normality of ages 8, 8.5, 9, 9.5, the p-values, for MSK, FA, HZ, Tn are 0.002, 0.054, < 0.001, < 0.001 respectively. The potential reasons for the non-normality include the latent genetic heterogeneities which lead to different growth profiles. For example, Timm [24] pointed out that observation 9 appears to be an outlier, that is, the #9 boy’s growth profile appears to difier from many others.
5.2 Fisher’s Iris Data
Fisher’s Iris data set is a multivariate data set introduced by Fisher [12] to demonstrate the use of unclassified observations in estimating discriminant function. Looney [15] considered testing univariate normality of each of the four measurements (sepal length, sepal width, petal length, and petal width) corresponding to variety Iris setosa, and found that normality assumption is tenable for all the variables except the petal width. Consider the multivariate normality of the four measurements, When MSK, FA, HZ, Tn are applied to all the 150 observations, consisting of all three species: Iris setosa, Iris versicolor, Iris virginica, the multivariate normality of the four measurements is rejected by all tests (all p-values are less than 2%). If we only use the first 50 observations, corresponding to Iris setosa, to test the four dimensional normality, we obtain corresponding p-values of 0.085, 0.065, 0.049, 0.037 for MSK, FA,HZ, Tn respectively. Thus at level α = 5%, the HZ test and our test Tn are able to reject the normality assumption of the four measurements on variety Iris setosa even with only 1/3 of the data. Rejecting multivariate normality for this data is consistent with the findings in the literature [21].
6. Conclusion
This paper presents a new test for normality that is very competitive in terms of power in both univariate and multivariate cases. As demonstrated by the numerical study, the new test outperforms the best known competitors in the literature, and the power advantage of the new test over its best known competitors is overwhelming in high dimensions. In addition, the practical utility of the proposed test is also illustrated using two well-known real data examples. Moreover, our projection-based statistic is very easy to understand and to implement using existing freely available numerical tools such as the shapiro.test function in the R computing platform. The R program for computing the p-value of the proposed test will be freely available at authors’s websites upon publication of the manuscript.
Appendix A. Power Tables
Table A1.
Empirical power of tests for univariate normality at significance level α = 5% when n = 50.
| Alternatives | MSK | Wn | HZ | Tn |
|---|---|---|---|---|
| N(0,1) | 5.0 | 4.8 | 5.1 | 4.7 |
| Exp(1) | 96 | 100 | 99 | 100 |
| Lognormal(0,1) | 100 | 100 | 100 | 100 |
| Lognormal(0,0.52) | 83 | 93 | 84 | 91 |
| Gamma(5,1) | 46 | 59 | 45 | 55 |
| χ2(1) | 100 | 100 | 100 | 100 |
| χ2(2) | 97 | 100 | 99 | 100 |
| χ2(5) | 72 | 89 | 78 | 88 |
| χ2(10) | 47 | 59 | 46 | 57 |
| Cauchy(0,1) | 100 | 100 | 100 | 100 |
| t(2) | 88 | 86 | 84 | 86 |
| t(5) | 44 | 35 | 26 | 36 |
| Beta(1,1) | 0 | 76 | 62 | 84 |
| Beta(1,2) | 14 | 83 | 70 | 80 |
| Beta(2,2) | 0 | 15 | 17 | 25 |
| Logistic(0,1) | 26 | 19 | 14 | 19 |
| Halfnormal | 59 | 94 | 80 | 92 |
| Weibull(0.8) | 100 | 100 | 100 | 100 |
| Weibull(1) | 97 | 100 | 99 | 100 |
| Weibull(1.5) | 63 | 88 | 73 | 86 |
| NormMix(.5,4,1) | 0 | 90 | 96 | 94 |
| NormMix(.5,3,1) | 0 | 38 | 51 | 49 |
| NormMix(.5,5,1) | 2 | 100 | 100 | 100 |
| NormMix(.5,6,3) | 35 | 99 | 98 | 98 |
| NormMix(.9,6,3) | 98 | 98 | 96 | 98 |
| NormMix(.5,2,1/3) | 36 | 99 | 98 | 98 |
| NormMix(.9,2,1/3) | 1 | 8 | 10 | 9 |
| PSII(0) | 0 | 76 | 62 | 85 |
| PSII(1) | 0 | 14 | 16 | 24 |
| PSVII(4) | 31 | 23 | 15 | 23 |
| PSVII(5) | 24 | 17 | 11 | 17 |
| SPH(Gamma(5,1)) | 0 | 94 | 98 | 94 |
| SPH(Beta(1,1)) | 0 | 75 | 62 | 84 |
| SPH(Beta(1,2)) | 0 | 5 | 6 | 7 |
| SPH(Beta(2,2)) | 1 | 99 | 99 | 100 |
| Average Power | 42.92 | 71.47 | 67.5 | 72.85 |
| Average Rank | 3.059 | 1.676 | 2.529 | 1.529 |
Table A2.
Empirical power of tests for bivariate normality at significance level α = 5% when n = 50.
| Alternatives | MSK | FA | HZ | Tn |
|---|---|---|---|---|
| N(0,1)2 | 5.2 | 5.3 | 5.2 | 4.8 |
| Exp(1)2 | 100 | 100 | 100 | 100 |
| Lognormal(0,1)2 | 100 | 100 | 100 | 100 |
| Lognormal(0, .52)2 | 96 | 97 | 94 | 99 |
| Gamma(.5,1)2 | 100 | 100 | 100 | 100 |
| Gamma(5,1)2 | 60 | 64 | 52 | 71 |
| χ2(1)2 | 100 | 100 | 100 | 100 |
| χ2(2)2 | 100 | 100 | 100 | 100 |
| χ2(5)2 | 88 | 93 | 86 | 96 |
| χ2(10)2 | 60 | 63 | 51 | 70 |
| χ2(15)2 | 44 | 45 | 35 | 51 |
| Cauchy(0,1)2 | 100 | 100 | 100 | 100 |
| t(2)2 | 96 | 95 | 95 | 97 |
| t(5)2 | 55 | 46 | 33 | 52 |
| Logistic(0,1)2 | 30 | 23 | 14 | 26 |
| Beta(1,1)2 | 0 | 53 | 67 | 90 |
| Beta(1,2)2 | 17 | 76 | 80 | 91 |
| Beta(2,2)2 | 0 | 6 | 19 | 33 |
| Halfnormal2 | 76 | 93 | 87 | 97 |
| Weibull(.8)2 | 100 | 100 | 100 | 100 |
| Weibull(1)2 | 100 | 100 | 100 | 100 |
| Weibull(1.5)2 | 79 | 89 | 83 | 95 |
| N(0,1)⊗Exp(1) | 89 | 99 | 93 | 99 |
| N(0,1)⊗χ2(5) | 58 | 75 | 52 | 77 |
| N(0,1)⊗t(5) | 31 | 27 | 17 | 29 |
| N(0,1)⊗Beta(1,1) | 1 | 33 | 31 | 43 |
| N(0,1)⊗Beta(1,2) | 7 | 51 | 41 | 54 |
| MVNMIX(.5,2,0,0) | 2 | 10 | 20 | 12 |
| MVNMIX(.5,4,0,0) | 1 | 100 | 100 | 98 |
| MVNMIX(.5,2,.9,0) | 73 | 67 | 81 | 62 |
| MVNMIX(.5,.5,.9,0) | 34 | 39 | 34 | 28 |
| MVNMIX(.5,.5,.9,−.9) | 72 | 90 | 93 | 74 |
| MVNMIX(.7,2,.9,.3) | 76 | 73 | 66 | 75 |
| MVNMIX(.3,1,.9,−.9) | 97 | 100 | 99 | 99 |
| PSII(0) | 0 | 27 | 71 | 96 |
| PSII(1) | 0 | 6 | 24 | 52 |
| PSVII(2) | 98 | 97 | 97 | 98 |
| PSVII(3) | 75 | 67 | 59 | 71 |
| PSVII(5) | 41 | 32 | 19 | 34 |
| SPH(Exp(1)) | 98 | 98 | 100 | 99 |
| SPH(Gamma(5,1)) | 4 | 8 | 18 | 13 |
| SPH(Beta(1,1)) | 0 | 3 | 15 | 4 |
| SPH(Beta(1,2)) | 24 | 29 | 66 | 31 |
| SPH(Beta(2,2)) | 0 | 3 | 10 | 24 |
| Average Power | 55.35 | 64.59 | 65.12 | 70.63 |
| Average Rank | 2.651 | 2.186 | 2.349 | 1.512 |
Table A3.
Empirical power of tests for 5 dimensional normality at level α = 5% when n = 50.
| Alternatives | MSK | FA | HZ | Tn |
|---|---|---|---|---|
| N(0,1)5 | 5.0 | 5.1 | 5.1 | 5.0 |
| Exp(1)5 | 100 | 100 | 100 | 100 |
| Lognormal(0,1)5 | 100 | 100 | 100 | 100 |
| Lognormal(0,.52)5 | 99 | 96 | 96 | 100 |
| Gamma(.5,1)5 | 100 | 100 | 100 | 100 |
| Gamma(5,1)5 | 69 | 50 | 48 | 86 |
| χ2(1)5 | 100 | 100 | 100 | 100 |
| χ2(2)5 | 100 | 100 | 100 | 100 |
| χ2(5)5 | 96 | 82 | 88 | 100 |
| χ2(10)5 | 69 | 50 | 48 | 88 |
| χ2(15)5 | 48 | 35 | 28 | 66 |
| Cauchy(0,1)5 | 100 | 100 | 100 | 100 |
| t(2)5 | 100 | 99 | 99 | 100 |
| t(5)5 | 68 | 59 | 32 | 74 |
| Logistic(0,1)5 | 38 | 29 | 14 | 40 |
| Beta(1,1)5 | 0 | 0 | 52 | 86 |
| Beta(1,2)5 | 6 | 7 | 68 | 87 |
| Beta(2,2)5 | 0 | 0 | 14 | 35 |
| Halfnormal5 | 82 | 58 | 84 | 100 |
| Weibull(.8)5 | 100 | 100 | 100 | 100 |
| Weibull(1)5 | 100 | 100 | 100 | 100 |
| Weibull(1.5)5 | 88 | 67 | 80 | 99 |
| N(0,1)⊗Exp(1)4 | 100 | 99 | 100 | 100 |
| N(0,1)⊗χ2(5)4 | 88 | 74 | 74 | 98 |
| N(0,1)⊗t(5)4 | 57 | 51 | 24 | 64 |
| N(0,1)⊗Beta(1,1)4 | 0 | 1 | 39 | 58 |
| N(0,1)⊗Beta(1,2)4 | 5 | 6 | 54 | 67 |
| N(0,1)3⊗Exp(1)2 | 90 | 89 | 81 | 99 |
| N(0,1)3⊗χ2(5)2 | 53 | 50 | 35 | 76 |
| N(0,1)3⊗t(5)2 | 33 | 32 | 13 | 37 |
| N(0,1)3⊗Beta(1,1)2 | 1 | 3 | 17 | 16 |
| N(0,1)3⊗Beta(1,2)2 | 5 | 6 | 25 | 23 |
| MVNMIX(.5,2,0,0) | 3 | 4 | 22 | 8 |
| MVNMIX(.5,4,0,0) | 2 | 10 | 70 | 11 |
| MVNMIX(.5,2,.9,0) | 100 | 89 | 100 | 100 |
| MVNMIX(.5,.5,.9,0) | 92 | 76 | 99 | 94 |
| MVNMIX(.5,.5,.9,−.1) | 94 | 83 | 99 | 96 |
| MVNMIX(.7,2,.9,.3) | 100 | 96 | 99 | 100 |
| MVNMIX(.3,1,.9,−.1) | 86 | 76 | 91 | 89 |
| PSII(0) | 0 | 0 | 65 | 100 |
| PSII(1) | 0 | 0 | 30 | 88 |
| PSVII(4) | 99 | 97 | 98 | 99 |
| PSVII(5) | 92 | 82 | 71 | 92 |
| PSVII(10) | 38 | 29 | 13 | 33 |
| SPH(Exp(1)) | 100 | 100 | 100 | 100 |
| SPH(Gamma(5,1)) | 70 | 47 | 44 | 68 |
| SPH(Beta(1,1)) | 83 | 45 | 100 | 92 |
| SPH(Beta(1,2)) | 100 | 95 | 100 | 100 |
| SPH(Beta(2,2)) | 37 | 13 | 71 | 42 |
| Average Power | 64.38 | 57.97 | 68.41 | 79.4 |
| Average Rank | 2.125 | 2.896 | 2.208 | 1.229 |
Table A4.
Empirical power of tests for 10 dimensional normality at level α = 5% when n = 50.
| Alternatives | MSK | FA | HZ | Tn |
|---|---|---|---|---|
| Normal10 | 5.1 | 4.8 | 4.9 | 5.1 |
| Exp(1)10 | 100 | 96 | 100 | 100 |
| Cauchy(0,1)10 | 100 | 100 | 100 | 100 |
| Gamma(.5,1)10 | 100 | 100 | 100 | 100 |
| Gamma(5,1)10 | 64 | 36 | 27 | 94 |
| Lognormal(0,.52)10 | 100 | 88 | 87 | 100 |
| Lognormal(0,1)10 | 100 | 100 | 100 | 100 |
| χ2(2)10 | 100 | 97 | 100 | 100 |
| χ2(5)10 | 94 | 63 | 65 | 100 |
| χ2(10)10 | 62 | 35 | 26 | 94 |
| χ2(15)10 | 43 | 25 | 16 | 76 |
| t(2)10 | 100 | 100 | 99 | 100 |
| t(5)10 | 77 | 62 | 20 | 89 |
| Logistic(0,1)10 | 39 | 25 | 8 | 50 |
| Beta(1,1)10 | 0 | 0 | 27 | 72 |
| Beta(1,2)10 | 2 | 2 | 35 | 83 |
| Beta(2,2)10 | 0 | 1 | 11 | 26 |
| Halfnormal10 | 69 | 29 | 52 | 100 |
| Weibull(.8)10 | 100 | 100 | 100 | 100 |
| Weibull(1)10 | 100 | 96 | 100 | 100 |
| Weibull(1.5)10 | 82 | 42 | 51 | 100 |
| N(0,1)9⊗Exp(1) | 27 | 29 | 11 | 49 |
| N(0,1)9⊗χ2(5) | 13 | 13 | 7 | 23 |
| N(0,1)9⊗t(5) | 12 | 14 | 6 | 15 |
| N(0,1)9⊗Beta(1,1) | 3 | 4 | 6 | 6 |
| N(0,1)9⊗Beta(1,2) | 5 | 5 | 7 | 6 |
| N(0,1)9⊗Beta(2,2) | 4 | 4 | 5 | 5 |
| N(0,1)5⊗Exp(1)5 | 96 | 79 | 70 | 100 |
| N(0,1)5⊗χ2(5)5 | 60 | 40 | 24 | 95 |
| N(0,1)5⊗t(5)5 | 43 | 39 | 9 | 55 |
| N(0,1)5⊗Beta(1,1)5 | 0 | 2 | 12 | 16 |
| N(0,1)5⊗Beta(1,2)5 | 3 | 4 | 15 | 22 |
| N(0,1)5⊗Beta(2,2)5 | 1 | 3 | 7 | 8 |
| N(0,1)2⊗Exp(1)8 | 100 | 92 | 96 | 100 |
| N(0,1)2⊗χ2(5)8 | 87 | 56 | 46 | 100 |
| N(0,1)2⊗t(5)8 | 65 | 53 | 15 | 79 |
| N(0,1)2⊗Beta(1,1)8 | 0 | 1 | 20 | 45 |
| N(0,1)2⊗Beta(1,2)8 | 2 | 2 | 26 | 57 |
| N(0,1)2⊗Beta(2,2)8 | 0 | 1 | 9 | 15 |
| PSII(0) | 0 | 0 | 48 | 100 |
| PSII(1) | 0 | 0 | 29 | 95 |
| PSII(4) | 0 | 0 | 11 | 48 |
| PSVII(6) | 100 | 100 | 100 | 100 |
| PSVII(8) | 98 | 89 | 71 | 98 |
| PSVII(10) | 86 | 62 | 29 | 84 |
| SPH(Gamma(5,1)) | 100 | 88 | 96 | 100 |
| SPH(Beta(1,1)) | 100 | 86 | 100 | 100 |
| SPH(Beta(1,2)) | 100 | 100 | 100 | 100 |
| SPH(Beta(2,2)) | 99 | 60 | 100 | 100 |
| Average Power | 54.91 | 46.28 | 47.9 | 73.04 |
| Average Rank | 2.188 | 2.854 | 2.521 | 1.042 |
Table A5.
Critical values for the level α = 5% test of p dimensional normality
| Sample Size | p = 1 | p = 2 | p = 5 | p = 10 |
|---|---|---|---|---|
| MSK | ||||
| n = 20 | 4.291 | 9.190 | 42.886 | 211.679 |
| n = 25 | 4.682 | 9.661 | 45.106 | 222.915 |
| n = 30 | 4.856 | 10.055 | 46.679 | 230.264 |
| n = 35 | 5.017 | 10.322 | 47.661 | 235.287 |
| n = 40 | 5.187 | 10.487 | 48.504 | 239.648 |
| n = 45 | 5.310 | 10.683 | 48.955 | 242.658 |
| n = 50 | 5.329 | 10.712 | 49.509 | 244.326 |
| FA | ||||
| n = 20 | 0.096 | 0.141 | 0.244 | 0.463 |
| n = 25 | 0.080 | 0.119 | 0.199 | 0.372 |
| n = 30 | 0.070 | 0.103 | 0.168 | 0.309 |
| n = 35 | 0.061 | 0.091 | 0.146 | 0.266 |
| n = 40 | 0.055 | 0.081 | 0.129 | 0.234 |
| n = 45 | 0.05 | 0.074 | 0.116 | 0.208 |
| n = 50 | 0.046 | 0.068 | 0.106 | 0.187 |
| HZ | ||||
| n = 20 | 0.546 | 0.728 | 0.910 | 0.985 |
| n = 25 | 0.591 | 0.766 | 0.924 | 0.987 |
| n = 30 | 0.624 | 0.798 | 0.936 | 0.989 |
| n = 35 | 0.658 | 0.820 | 0.944 | 0.991 |
| n = 40 | 0.685 | 0.840 | 0.952 | 0.992 |
| n = 45 | 0.717 | 0.861 | 0.957 | 0.992 |
| n = 50 | 0.729 | 0.873 | 0.963 | 0.993 |
| Tn | ||||
| n = 20 | 0.1001 | 0.1092 | 0.1018 | 0.1490 |
| n = 25 | 0.0842 | 0.0927 | 0.0864 | 0.1185 |
| n = 30 | 0.0729 | 0.0811 | 0.0759 | 0.0998 |
| n = 35 | 0.0644 | 0.0719 | 0.0681 | 0.0865 |
| n = 40 | 0.0581 | 0.0644 | 0.0618 | 0.0769 |
| n = 45 | 0.0526 | 0.0589 | 0.0567 | 0.0694 |
| n = 50 | 0.0483 | 0.0539 | 0.0525 | 0.0632 |
References
- 1.Baringhaus L, Henze N. A consistent test for multivariate normality based on the empirical characteristic function. Metrika. 1988;35:339–348. [Google Scholar]
- 2.Bowman KO, Shenton LR. Omnibus test contours for departures from normality based on √b1 and b2. Biometrika. 1975;62:243–250. [Google Scholar]
- 3.Coin D. A goodness-of-fit test for normality based on polynomial regression. Comput. Statist. Data Anal. 2008;52:2185–2198. [Google Scholar]
- 4.D’Agostino RB, Belanger A, D’agostino RB., Jr A suggestion for using powerful and informative tests of normality. The American Statistician. 1990;44:316–321. [Google Scholar]
- 5.D’Agostino RB, Pearson ES. Tests for departure from normality. emprical results for the distribution of b2 and √b1. Biometrika. 1973;62:613–622. [Google Scholar]
- 6.D’Agostino RB, Stephens MA. Goodness-of-Fit Techniques. New York: Marcel Dekker, Inc.; 1986. [Google Scholar]
- 7.Doornik JA, Hansen H. An omnibus test for univariate and multivariate normality. Oxford Bulletin of Economics and Statistics. 2008;70:927–939. [Google Scholar]
- 8.Eaton ML, Perlman MD. The non-singularity of generalized sample covariance matrices. Ann. Statist. 1973;1:710–717. [Google Scholar]
- 9.Elston RC, Grizzle JE. Estimation of time-response curves and their confidence bands. Biometrics. 1962;18:148–159. [Google Scholar]
- 10.Epps TW, Pulley LB. A test for normality based on the empirical characteristic function. Biometrika. 1983;70:723–726. [Google Scholar]
- 11.Fattorini L. Remarks on the use of the shapiro-wilk statistic for testing multivariate normality. Statistica. 1986;46:209–217. [Google Scholar]
- 12.Fisher RA. The use of multiple measurements in taxonomic problems. Annals of Eugenics. 1936;7:179–188. [Google Scholar]
- 13.Henze N, Zirkler B. A class of invariant consistent tests for multivariate normality. Comm. Statist. Theory Methods. 1990;19:3595–3617. [Google Scholar]
- 14.Leslie J, Stephens MA, Fotopoulos S. Asymptotic distribution of the shapiro-wilk w for testing for normality. Ann. Statist. 1986;14:1497–1506. [Google Scholar]
- 15.Looney SW. How to use tests for univariate normality to assess multivariate normality. The American Statistician. 1995;49:64–70. [Google Scholar]
- 16.Mardia KV. Measures of multivariate skewness and kurtosis with applications. Biometrika. 1970;57:519–530. [Google Scholar]
- 17.Pearson ES. A further development of tests for normality. Biometrika. 1930;22:239–249. [Google Scholar]
- 18.Shao J. Mathematical Statistics. 2nd ed. Springer-Verlag New York Inc; 2003. [Google Scholar]
- 19.Shao Y, Zhou M. A characterization of multivariate normality through univariate projections. Journal of Multivariate Analysis. 2010;101:2637–2640. doi: 10.1016/j.jmva.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- 21.Small N. Marginal skewness and kurtosis in testing multivariate normality. Applied Statistics. 1980;29:85–87. [Google Scholar]
- 22.Stephens MA. EDF statistics for goodness of fit and some comparisons. Journal of the American Statistical Association. 1974;69:730–737. [Google Scholar]
- 23.Thode HC., Jr . Testing for Normality. New York: Marcel Dekker, Inc.; 2002. [Google Scholar]
- 24.Timm NH. Applied multivariate analysis. New York: Springer; 2002. [Google Scholar]