Summary
Advances in our understanding of the genetic determinants of leukemia have translated to better treatment options and improved survival of patients with acute myeloid and acute lymphoid leukemia. However, some leukemias, such as those bearing 11q23 (MLL) translocations, result in aggressive diseases with a relatively poor prognosis, despite improved treatments such as allogeneic hematopoietic stem cell transplantation. This article will briefly review the functions and regulation of wild-type MLL during normal hematopoiesis, while focusing on recent advances in our understanding of the molecular mechanisms governing MLL leukemias. The transcriptional targets, cooperating signaling pathways and molecular machinery involved in MLL-associated leukemias will be discussed, as well as how these may be harnessed for more personalized treatment of this disease.
MLL-associated leukemia
Since its discovery, the mixed-lineage leukemia (MLL) gene, located on chromosome 11q23, has garnered much attention both from a hematologic and a basic science standpoint because of its role in epigenetics and leukemia. A number of genetic aberrations including chromosomal translocations, internal tandem duplications, internal deletions and amplifications of the MLL gene have been observed in hematologic malignancies [1–6]. The most frequent abnormalities involving 11q23 are chromosomal translocations that constitute more than 70% of acute lymphoid leukemia in infants under the age of 1 year and between 35 and 50% of infant acute myeloid leukemia (AML) [7–9]. Greater than 70 recurrent translocations and more than 50 different partner genes have been discovered, leading to the expression of chimeric oncogenic fusion proteins [10,11]. Rearrangements of the MLL gene are also found in adult leukemias and therapy-related leukemias arising in patients after treatment with topoisomerase II inhibitors for other malignancies, and is seen in approximately 10% of human leukemias overall [12,13]. Generally, 11q23 abnormalities are considered a poor prognostic factor and patients are treated with high-risk protocols [14]. In a study of infant acute lymphoid leukemia, patients with germline MLL displayed a good response to treatment, with a 5-year event-free survival (EFS) of approximately 80% compared with an EFS of 15% for patients with rearranged MLL [15]. A slightly better 5-year EFS of approximately 44% is observed in AML patients with MLL rearrangements; however, survival rates vary largely within the 11q23 subgroup based on age, lineage, translocation partner and the presence of cooperating mutations [16]. For example, patients with a t(1;11)(q21;q23) coding for the MLL–AF1p fusion protein had a favorable outcome, with a 5-year EFS of 92% compared with 29% for patients with t(4;11)(q21;q23), which generates the MLL–AF4 fusion protein. These characteristics demonstrate the complexity of 11q23 leukemia, and underscore the importance of understanding the molecular mechanisms of these diseases.
Hematopoiesis & MLL
Cloning of the breakpoint region on chromosome 11q23 from leukemic cells led to the discovery of MLL, a gene orthologous to the Drosophila melanogaster trithorax [1–3]. Trithorax group proteins, including MLL, play fundamental roles in development, acting antagonistically to Polycomb group proteins through the epigenetic regulation of target genes such as the clustered Hox genes [17]. Hox proteins are spatio–temporally regulated transcription factors involved in a number of developmental processes including anterior–posterior patterning and hematopoietic differentiation (for review, see [18]). Deletion of Mll results in impaired expression of Hox genes, including Hoxc8 and Hoxa7, and embryonic lethality at E10.5 [19]. Within both the developing fetus and adult hematopoietic systems, Mll plays a central role in the survival and/or proliferation of the hematopoietic stem cell (HSC) and progenitor compartments through the proper regulation of Hox genes [20–22]. Defects to hematopoietic colony-forming potential are rescued by expression of Hox genes, confirming the importance of the MLL–HOX axis [23]. In the absence of Mll, Hox gene transcription is initiated normally but not properly maintained, suggesting a role for Mll in establishing epigenetic cellular memory [24].
The MLL protein is a 3969-amino-acid histone H3 lysine 4 (H3K4) methyl transferase transcription factor that is proteolytically cleaved by TASPASE1 into 320 kDa N-terminal (MLLN) and 180 kDa C-terminal (MLLC) fragments that self-associate in the nucleus [25]. MLL contains several highly conserved structural domains, including three AT-hooks and a CxxC domain that, in the latter case, binds specifically to nonmethylated DNA and protects from de novo methylation, and a C-terminal Su[var]3–9, enhancer of zeste, trithorax (SET) domain with intrinsic H3K4 methyltransferase activity [26–31]. A set of four plant homeo domain (PHD) fingers with an embedded atypical bromodomain located downstream of the CxxC domain play an important role in the localization and post-translation regulation of wild-type MLL [32]. The third PHD finger binds trimethylated H3K4 and contributes to MLL recruitment [33,34]. Recently, the second PHD finger of MLL and MLL's closest homolog, MLL4, were shown to possess intrinsic E3 ubiquitin ligase activity, leading to auto-degradation [35]. The bromodomain and PHD4 of MLL associate with the ECSASB2 E3 ubiquitin ligase complex, and together these activities play a role in auto-ubiquitination and proteasome-mediated degradation during hematopoietic differentiation [36]. MLL stability is also regulated through ATR-mediated phosphorylation events that control ubiquitination during cell-cycle progression by the SCFSkp2 and APCCdc20 E3 ligase complexes during S and M phase, respectively [37,38].
The MLL protein functions within a large protein complex that makes several direct associations with MLL. A core complex of proteins including ASH2l, WDR5 and RbBP5 assemble around the SET domain of MLL and are required for full methyltransferase activity [39–41]. Several histone acetyltransferases, including the H4K16 acetyltransferase MOF and CBP/p300, also associate with MLL and contribute to full transcriptional activation of target genes such as Hoxa9 [42,43]. In addition to the functions listed above, PHD3 of MLL mediates binding of the nuclear peptidyl-prolyl isomerase Cyp33 that regulates the binding of a number of co-repressor proteins including CtBP, HPC2, BMI-1 and HDAC1 [32,44]. Importantly, the sequences mediating these interactions are deleted in MLL leukemia and do not contribute to MLL fusion protein function. However, a number of interactions are preserved in MLL fusion proteins and relate to the mechanisms of MLL leukemia.
MLL mechanisms of disease
Molecular machinery of MLL leukemia
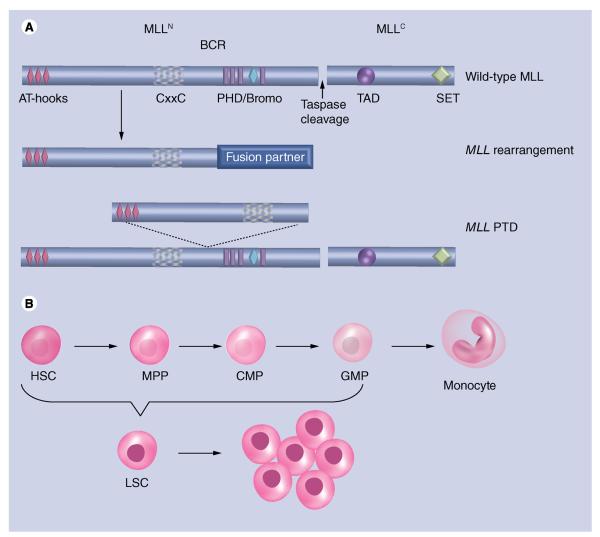
With the identification of more than 70 different MLL translocation partners, developing a unifying model for MLL-mediated transformation is challenging. Still, some recurring themes have emerged. In the event of an MLL rearrangement, amino terminal sequences of MLL up to and including the CxxC domain are retained, while the PHD fingers and beyond are deleted, resulting in the loss of both recruitment and transcriptional activation activities (Figure 1). Despite truncation of MLL and loss of H3K4 methyltransferase activity, the resultant chimeric MLL fusion protein leads to deregulated activation of leukemic target genes such as HOXA9 and MEIS1 that is observed in the vast majority of MLL-associated leukemia [45,46].
Figure 1. Normal and mutated forms of the mixed-lineage leukemia protein.
(A) The wild-type MLL protein is shown schematically with conserved domains indicated and labeled. MLL is cleaved into two fragments, MLLN and MLLC. The site of taspase-mediated protein cleavage is indicated. AT-hooks and the CxxC domain of MLLN bind to DNA, while the plant homeodomain/bromo region aids in localization and protein regulation. A SET and TAD domain contributes to histone methylation and recruitment of histone acetyltransferases. BCR is the site of fusion in the event of chromosomal translocations. MLL fusion proteins contain a sequence of a fusion partner protein fused in-frame to the MLLN fragment at the BCR. MLL-PTDs duplicate the MLL sequence from the AT-hooks through the CxxC domain that is inserted at the BCR. Proteins are not drawn to scale. (B) HSC differentiation to a monocyte is shown, including intermediate progenitor cells: MPPs, CMPs and GMPs. MLL fusion proteins are capable of transforming HSCs through GMPs into LSCs that gives rise to leukemia.
BCR: Breakpoint cluster region; CMP: Common myeloid progenitor; GMP: Granulocyte/monocyte progenitor; HSC: Hematopoietic stem cell; LSC: Leukemic stem cells; MLL: Mixed-lineage leukemia; MLL-PTD: MLL partial tandem duplications; MPP: Multipotent progenitor.
DNA-binding domains, such as the AT-hooks and CxxC domain, are retained in the MLL fusion protein and, in the case of the CxxC domain, are essential for MLL fusion protein transformation [47]. In the case of MLL partial tandem duplications, the N-terminal region of MLL containing these domains is duplicated at the breakpoint cluster region, leading to increased self-renewal and transformation of hematopoietic stem and progenitor cells (Figure 1) [48,49]. Other than preferential binding to nonmethylated CpG islands by the CxxC domain [26], these domains lack sequence specificity, necessitating other mechanisms for the recruitment of MLL fusion proteins to target loci. One mechanism that has attracted interest from a therapeutic standpoint occurs between the extreme N-terminus of MLL and the tumor suppressor protein Menin [50,51]. Menin is coded by the MEN1 gene and commonly mutated in multiple endocrine neoplasia type 1. Two Menin-binding motifs reside within the N-terminus of MLL and mediate the formation of a trimolecular complex consisting of MLL, Menin and the chromatin-associated LEDGF protein [52,53]. The proper formation of this complex is critical to the targeting of MLL fusion proteins to target loci, such as Hoxa9, and necessary for MLL fusion protein-mediated leukemogenesis. It has been demonstrated that MLL fusion proteins also rely on an interaction with the PAF complex (PAFc) for proper targeting to gene loci. The PAFc interaction domain of MLL flanks the CxxC domain of MLL and is invariably retained in all MLL fusion proteins [34,54]. The PAFc is a transcriptional activation complex that associates directly with RNA pol II and promotes elongation by facilitating the deposition of several epigenetic marks associated with active transcription, including H2BK120 ubiquitination, H3K4, H3K36 and H3K79 methylation [55,56]. Like the Menin–LEDGF interaction, MLL fusion proteins are dependent on the PAFc interaction for leukemogenesis.
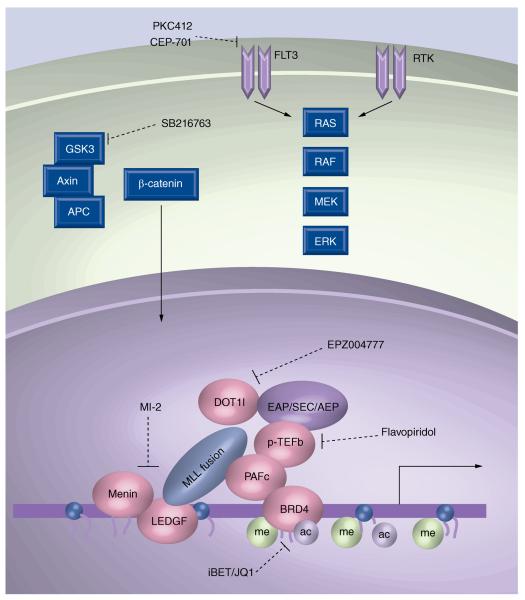
Truncation of MLL is not sufficient for transformation, indicating the presence of a translocation partner is required for leukemia [57]. While the above interactions likely aid in MLL fusion protein recruitment, the translocation partner plays a significant role in transcriptional activation. MLL translocation partners include both nuclear proteins implicated in transcriptional activation and cytoplasmic proteins. Despite the apparent complexity of revealing a mechanism for transformation, some themes have emerged. For example, many of the most common MLL translocation partners, including MLLT3 (AF9), MLLT1 (ENL), AFF4 (AF5q31) and AFF1 (AF4), function within a common transcriptional elongation complex that includes p-TEFb (Figure 2) [58]. The p-TEFb is composed of cyclin T1 and CDK9 and functions in transcriptional elongation by phosphorylating the C-terminal domain of RNA pol II. A variation of this complex has been reported to include the elongation factors ELL2, ELL3, EAF1 and EAF2 [59]. Interestingly, the Polycomb group protein CBX8, normally associated with transcriptional repression within the PRC1 complex, associates with this complex and contributes to transcriptional activation through recruitment of the HAT protein TIP60 [60]. Notably, HAT activity also contributes to MLL leukemias by direct fusion of CBP/p300 to MLL [61]. The AF9 and ENL translocation partners also reside within a protein complex termed DotCom that includes MLLT10 (AF10) and MLLT6 (AF17), two nuclear translocation partners of MLL, and the histone H3K79 methyltransferase DOT1l leading to transcriptional activation [62]. Furthermore, DOT1L and p-TEFb have been found together in transcription activation complexes including AF4, ENL, AF9 and AF5q31 [63–65]. Recruitment of DOT1l is required for transformation by some MLL fusion proteins by upregulating HOXA9 and MEIS1, as well as the WNT signaling pathway that has been shown to be essential for MLL-mediated leukemogenesis [66–69]. Indeed, MLL leukemic cells display robust H3K79 methylation patterns across the HoxA locus indicative of Dot1l recruitment. These studies have distilled a common mechanism for many MLL fusion proteins that implicates p-TEFb and DOT1l in the upregulation of HOX genes.
Figure 2. Targeting mixed-lineage leukemia fusion protein complexes and cooperating pathways.
A representative MLL fusion protein complex is shown that recruits a transcriptional activation complex. Varieties of this complex have been termed EAP [122], SEC [59] and AEP [58]. Included in the recruitment are p-TEFb and the histone H3K79 methyltransferase DOT1l. Menin and PAFc bind directly to MLL fusion proteins, aiding in target recognition. BRD4 associates with PAFc and is required for MLL fusion protein function. GSK3 is shown associated with Axin and APC, which are involved in several signaling pathways including the WNT/β-catenin pathway. FLT3 activates several signaling pathways, including the Ras pathway that cooperates with MLL fusion proteins leading to leukemia. Compounds known to inhibit some of these processes and protein interactions are shown.
ac: Acetylation; me: Methylation; MLL: Mixed-lineage leukemia; PAFc: PAF complex.
Cooperating events
Deep sequencing has revealed MLL leukemias show increased genetic stability compared with other leukemias [70], which may reflect the highly potent oncogenicity of MLL fusion proteins. Still, analyses of 11q23 leukemias have revealed strong correlations with activation of both the FLT3 and the Ras signaling pathway, suggesting cooperating events contribute to MLL leukemia [71]. Consistent with the two-hit leukemic model whereby activated kinase pathways cooperate with altered DNA-binding transcriptional machinery [72], MLL mutations found in utero require a latency period before giving rise to leukemia, likely reflecting the need for cooperating mutations [73]. FLT3 mutations are found in up to 30% of AML, leading to enhanced proliferation and survival [74]. Although activating mutations are less common in MLL leukemias [75], an independent analysis of FLT3 expression in cytogenetic AML subgroups showed the highest correlation with MLL leukemias [76]. Furthermore, recent data has demonstrated up to approximately 30% of pediatric MLL-rearranged AML also contains Ras mutations [16]. Experimentally, the disease latency in mouse models of MLL leukemia is significantly shortened with the introduction of constitutively active FLT–internal tandem duplications or k-RasG12V in support of a cooperative model of leukemogenesis [77–79]. Supporting the involvement of the Ras signaling pathway, the Ras-like proteins Rac1 and Rac2 GTPases were shown to be necessary for MLL–AF9 transformed cells through regulation of the antiapoptotic Bcl2 proteins [80]. A full characterization of the cooperating events in MLL rearranged cells will present a more comprehensive view of the mechanisms and potential therapeutic targets of these leukemias.
Transcriptional pathways
The clustered HOX genes are some of the best-understood targets of MLL and MLL fusion proteins. High-level expression of A cluster HOX genes, as well as the HOX protein cofactor, MEIS1, is highly characteristic of MLL rearranged leukemia. In fact, gene-expression profiling has demonstrated HOXA9 is the most highly correlated gene for poor prognosis in AML [81]. The importance of Hox proteins in MLL leukemia is highlighted by studies showing Hoxa9-deficient cells are not capable of transformation by MLL fusion proteins [82]. Furthermore, overexpression of Hoxa9 is sufficient to induce leukemia in mice; however, disease onset is accelerated substantially by co-expression of Meis1 [83]. It should be noted that some experimental systems involving MLL fusion proteins are capable of initiating leukemia independent of Hoxa9, which is likely due to compensation by other HoxA cluster genes [84,85]. Indeed, overexpression of almost all HOXA cluster genes leads to immortalization in serial replating assays with the exception of HOXA2 and HOXA5 [86]. HOX proteins contribute to leukemogenesis by blocking differentiation and promoting survival of transformed cells [18]. Hoxa9 was recently shown to associate with enhancer regions, in collaboration with lineage-specific transcription factors such as PU.1, controlling the expression of known proto-oncogenes, such as Flt3, Lmo2, Runx1 and Sox4 [87]. Interestingly, PU.1, which is required for hematopoietic differentiation, is regulated by RUNX1-mediated recruitment of MLL methyltransferase activity, suggesting a possible feedback mechanism involving HOX proteins [88]. In addition, following direct activation of the RUNX1 locus by MLL–AF4 fusion proteins, the RUNX1 protein associates with the reciprocal chromosomal fusion product AF4–MLL [89]. This association can contribute to both activation of RUNX1 target genes and the proleukemic capabilities demonstrated for the AF4–MLL reciprocal fusion protein [90].
Several additional targets of MLL fusion proteins have been identified that also contribute to leukemogenesis. For example, upregulation of the transcription regulatory factors Myb, Hmgb3 and Cbx5 by MLL fusion proteins establish an embryonic stem cell-like gene-expression program in hematopoietic progenitor cells, leading to transformation independent of Hoxa9/Meis1 [91,92]. MLL fusion proteins are also reported to regulate MEF2C and EVI-1, whose expression contributes to leukemic stem cell homing and tumor growth [93–95]. Interestingly, the MLL fusion proteins may recognize only a small subset of wild-type MLL target genes, and the localization of the fusion protein may be dependent on the fusion partner [66,96]. Some additional target genes that aid in MLL-mediated leukemogenesis include Myc and Epha7 [97–99]; however, with additional genome-wide localization studies this list is likely to expand.
Recent work has uncovered unique miRNA-expression patterns associated with leukemias bearing MLL rearrangements. miR-196b, which resides within the HOXA cluster locus, is directly regulated by MLL and MLL fusion proteins. Hoxa9/Meis1 and Fas are targeted by miR-196b, which likely plays a role in primitive hematopoietic cells [100]. The importance of miR-196b expression has been demonstrated in MLL leukemia (and others with high-level HOX expression), where mi-196b expression is functionally linked with upregulation of HOXA genes, thus contributing to MLL-mediated transformation [101,102]. The overexpression and inhibition of miR-196b can delay MLL leukemia, which may reflect a delicate balance of targets such as HOX and FAS in leukemic cells [100]. Likewise, the miRNA cluster (miR-17–92) is frequently overexpressed in various hematologic and solid tumors, including MLL rearranged leukemias, and is directly bound by MLL fusion proteins. This cluster appears to regulate p21 expression, leading to inhibition of both cell differentiation and apoptosis of the MLL transformed cells [103,104]. Important MLL collaborating proteins, such as FLT3 and MYB, are targeted by miR-150, which is negatively regulated by MLL fusion proteins through a MYC/LIN28 axis. miR-150, along with miR-495, both appear to be down-reulgated in MLL leukemias and contribute to disease progression [105].
Targeting MLL in the leukemic stem cell
Inhibition of MLL fusion protein complexes
Evolution of leukemic stem cells poses a serious challenge to effectively targeting MLL rearranged leukemia. This model suggests the self-renewing leukemic stem cell (LSC) gives rise to the leukemic cell burden in a patient that can further evolve both genetically and epigenetically, leading to various clonal subpopulations of different frequency. Selective pressures, such as chemotherapy, redefine the leukemic population in favor of previously existing subclones that can vary significantly from the disease characterized at the time of diagnosis [106,107]. However, since LSCs will likely remain dependent on initiating driving genetic events, MLL fusion proteins represent an excellent therapeutic target.
Several recent studies have reported either enzymatic inhibition or disruption of protein interactions within the MLL fusion protein complex with small-molecule inhibitors. The identification of a common biochemical complex shared by many of the most common MLL fusion proteins (discussed above) (Figure 2) raises the prospect of targeting MLL fusion protein complexes. One attractive target of this complex is the transcriptional elongation complex, p-TEFb. The flavonoid flavopiridol shows activity against this kinase and may be effective in treating MLL leukemia, but clinical trials have raised concerns about efficacy and toxicity [108]. The H3K79 methyltransferase DOT1l also associates with the AEP complex and has recently been targeted with the small molecule EPZ004777 (Figure 2). This compound binds as a SAM structural analog and inhibits DOT1l function and selectively slows the growth of human MLL rearranged leukemic cells in xenograft transplantation assays [109]. Another target, which is shared among all MLL fusion proteins, is the MLL–Menin interaction. Small molecules developed to bind Menin and inhibit the MLL–Menin interaction have shown remarkable specificity for inducing both the apoptosis and differentiation of MLL leukemic cells (Figure 2) [110]. The PAFc is predicted to associate with all MLL fusion proteins and poses another potential therapeutic target. A physical association between BRD4 and the PAFc, as well as p-TEFb, has demonstrated the usefulness of targeting the histone-acetyl-binding pocket of BRD4 [111]. Exposure of MLL leukemia cells to small molecules (I-BET151 and JQ1) designed to bind the acetyl-binding bromodomain of BRD4 led to cell cycle arrest and monocytic differentiation (Figure 2) [97,98]. These data confirm the importance of DOT1l, Menin and BRD4 in MLL leukemias as potential therapeutic targets, and suggests critical interacting proteins, such as CBX8 and the PAFc, may show similar promise.
Signaling pathways necessary for MLL leukemias
Therapeutic targeting of cooperating signaling pathways may work synergistically with pharmaco logic inhibition of the MLL fusion protein complex to help eradicate MLL LSCs. MLL fusion proteins are capable of transforming both HSCs and committed granulocyte/monocyte progenitors into leukemic stem cells, in part, through the reactivation of the WNT/β-catenin pathway [112–114]. The targeting of Wnt/β-catenin has attracted attention due to the fact that β-catenin is dispensible for normal HSC self-renewal, but required for MLL-transformed LSCs [114,115]. Indeed, pharmacologic inhibition of β-catenin by modulating prostaglandin signaling with the cyclooxygenase inhibitor indomethacin is effective in impairing MLL–AF9 LSCs, as well as BCR–ABL-positive myeloid leukemia stem cells [114,116]. Paradoxically, GSK-3, which functions in complex with Axin and APC to degrade β-catenin, is required for the survival of MLL leukemias by promoting CREB binding to the HOX co-factor protein MEIS1. As such, GSK-3 inhibition with SB216763 displayed a highly selective growth inhibition of leukemic cells dependent on high HOX expression (Figure 2) [117,118]. Recent work has identified a requirement by MLL leukemic cells for the H3K4 demethylase KDM1a (LSD1). LSD1 inhibition with tranylcypromine slowed the growth of MLL–AF9 cells in vivo, but spares the clonogenic potential for normal hematopoietic progenitor cells [119]. Other cooperating pathways, such as the FLT3 signaling pathway described above, have been targeted with the inhibitor molecules PKC412 and CEP-701 and shown effectiveness in differentially killing MLL transformed cells especially following chemotherapy treatment [78,120,121].
Conclusion & future perspective
Rapid advances in our understanding of the biochemical and biological processes involved in MLL leukemia have revealed molecular contacts necessary for transformation, and ushered in new prospective therapeutic options. Exploiting these molecular points of susceptibility with drugs designed to specifically inhibit these functions shows promise for more individualized treatment options. Future studies are likely to focus on the changes to the epigenome and epigenetic regulators that distinguish leukemic stem cells from their normal counterparts. The integration of both genetic and epigenetic models of leukemogenesis and the interplay between the two is likely to provide a more unified understanding of cancer progression, while also unlocking new therapies that may help to reverse the aggressiveness of MLL leukemia.
Practice Points
-
■
Mixed-lineage leukemia (MLL) is a histone H3K4 methyltransferase critical for normal hematopoietic development.
-
■
Rearrangements of the MLL locus (11q23) are common in acute myeloid and lymphoid leukemias, and are associated with a poor prognosis.
-
■
Leukemias harboring MLL fusion proteins display altered epigenetic profiles that contribute to disease.
-
■
MLL fusion protein-mediated activation of proleukemic target genes, such as HOXA9, MEIS1 and miRNAs, is critical for leukemogenesis.
-
■
MLL fusion proteins associate with a transcriptional elongation complex, containing DOT1L and p-TEFb, leading to deregulated transcription.
-
■
Targeted chemical disruption of the MLL fusion protein supercomplex effectively inhibits the growth of MLL-associated leukemic cells.
Acknowledgments
AG Muntean is supported by NIH grant R00 CA158136 and an American Society of Hematology Scholar Award.
No writing assistance was utilized in the production of this manuscript.
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Cimino G, Moir DT, Canaani O, et al. Cloning of ALL-1, the locus involved in leukemias with the t(4;11)(q21;q23), t(9;11) (p22;q23), and t(11;19)(q23;p13) chromosome translocations. Cancer Res. 1991;51(24):6712–6714. [PubMed] [Google Scholar]
- 2.Ziemin-van der Poel S, McCabe NR, Gill HJ, et al. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl Acad. Sci. USA. 1991;88(23):10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tkachuk DC, Kohler S, Cleary ML. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell. 1992;71(4):691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 4.Streubel B, Valent P, Jager U, et al. Amplification of the MLL gene on double minutes, a homogeneously staining region, and ring chromosomes in five patients with acute myeloid leukemia or myelodysplastic syndrome. Genes Chromosomes Cancer. 2000;27(4):380–386. [PubMed] [Google Scholar]
- 5.Caligiuri MA, Schichman SA, Strout MP, et al. Molecular rearrangement of the ALL-1 gene in acute myeloid leukemia without cytogenetic evidence of 11q23 chromosomal translocations. Cancer Res. 1994;54(2):370–373. [PubMed] [Google Scholar]
- 6.Raimondi SC, Frestedt JL, Pui CH, et al. Acute lymphoblastic leukemias with deletion of 11q23 or a novel inversion (11)(p13q23) lack MLL gene rearrangements and have favorable clinical features. Blood. 1995;86(5):1881–1886. [PubMed] [Google Scholar]
- 7.Rubnitz JE, Link MP, Shuster JJ, et al. Frequency and prognostic significance of HRX rearrangements in infant acute lymphoblastic leukemia: a pediatric oncology group study. Blood. 1994;84(2):570–573. [PubMed] [Google Scholar]
- 8.Chessells JM, Harrison CJ, Kempski H, et al. Clinical features, cytogenetics and outcome in acute lymphoblastic and myeloid leukaemia of infancy: report from the MRC Childhood Leukaemia working party. Leukemia. 2002;16(5):776–784. doi: 10.1038/sj.leu.2402468. [DOI] [PubMed] [Google Scholar]
- 9.Tien HF, Hsiao CH, Tang JL, et al. Characterization of acute myeloid leukemia with MLL rearrangements – no increase in the incidence of coexpression of lymphoid-associated antigens on leukemic blasts. Leukemia. 2000;14(6):1025–1030. doi: 10.1038/sj.leu.2401791. [DOI] [PubMed] [Google Scholar]
- 10.Huret JL, Dessen P, Le Minor S, Bernheim A. The “Atlas of genetics and cytogenetics in oncology and haematology” on the internet and a review on infant leukemias. Cancer Genet. Cytogenet. 2000;120(2):155–159. doi: 10.1016/s0165-4608(99)00250-2. [DOI] [PubMed] [Google Scholar]
- 11.Huret JL, Minor SL, Dorkeld F, Dessen P, Bernheim A. Atlas of genetics and cytogenetics in oncology and haematology, an interactive database. Nucleic Acids Res. 2000;28(1):349–351. doi: 10.1093/nar/28.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Super HJ, McCabe NR, Thirman MJ, et al. Rearrangements of the MLL gene in therapy-related acute myeloid leukemia in patients previously treated with agents targeting DNA-topoisomerase II. Blood. 1993;82(12):3705–3711. [PubMed] [Google Scholar]
- 13.Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T. AML with 11q23/MLL abnormalities as defined by the WHO classification: incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood. 2003;102(7):2395–2402. doi: 10.1182/blood-2003-02-0434. [DOI] [PubMed] [Google Scholar]
- 14.Pui CH, Behm FG, Downing JR, et al. 11q23/MLL rearrangement confers a poor prognosis in infants with acute lymphoblastic leukemia. J. Clin. Oncol. 1994;12(5):909–915. doi: 10.1200/JCO.1994.12.5.909. [DOI] [PubMed] [Google Scholar]
- 15.Chen CS, Sorensen PH, Domer PH, et al. Molecular rearrangements on chromosome 11q23 predominate in infant acute lymphoblastic leukemia and are associated with specific biologic variables and poor outcome. Blood. 1993;81(9):2386–2393. [PubMed] [Google Scholar]
- 16.Balgobind BV, Raimondi SC, Harbott J, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114(12):2489–2496. doi: 10.1182/blood-2009-04-215152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 18.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26(47):6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 19.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378(6556):505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]; First to describe homeotic transformations and hematopoietic defects in mice following genetic knockout of the Mll gene and linking Mll to Hox expression.
- 20.Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90(5):1799–1806. [PubMed] [Google Scholar]
- 21.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1(3):324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahon KA, Hiew SY, Hadjur S, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1(3):338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 23.Ernst P, Mabon M, Davidson AJ, Zon LI, Korsmeyer SJ. An Mll-dependent Hox program drives hematopoietic progenitor expansion. Curr. Biol. 2004;14(22):2063–2069. doi: 10.1016/j.cub.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl Acad. Sci. USA. 1998;95(18):10632–10636. doi: 10.1073/pnas.95.18.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh JJ, Cheng EH, Korsmeyer SJ. Taspase1: a threonine aspartase required for cleavage of MLL and proper HOX gene expression. Cell. 2003;115(3):293–303. doi: 10.1016/s0092-8674(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 26.Allen MD, Grummitt CG, Hilcenko C, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25(19):4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 2002;30(4):958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cierpicki T, Risner LE, Grembecka J, et al. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL–AF9 leukemia. Nat. Struct. Mol. Biol. 2010;17(1):62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erfurth FE, Popovic R, Grembecka J, et al. MLL protects CpG clusters from methylation within the Hoxa9 gene, maintaining transcript expression. Proc. Natl Acad. Sci. USA. 2008;105(21):7517–7522. doi: 10.1073/pnas.0800090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell. 2002;10(5):1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]; ■ Along with [31], the authors were the first to demonstrate intrinsic H3K4 methyltransferase activity of the MLL SET domain.
- 31.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell. 2002;10(5):1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Song J, Milne TA, et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141(7):1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang PY, Hom RA, Musselman CA, et al. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J. Mol. Biol. 2010;400(2):137–144. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne TA, Kim J, Wang GG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol. Cell. 2010;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Muntean AG, Wu L, Hess JL. A subset of mixed lineage leukemia proteins has plant homeodomain (PHD)-mediated E3 ligase activity. J. Biol. Chem. 2012;287(52):43410–43416. doi: 10.1074/jbc.M112.423855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Muntean AG, Hess JL. ECSASB2 mediates MLL degradation during hematopoietic differentiation. Blood. 2012;119(5):1151–1161. doi: 10.1182/blood-2011-06-362079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Takeda S, Kumar R, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467(7313):343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21(19):2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13(9):852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 40.Wysocka J, Swigut T, Milne TA, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 41.Dou Y, Milne TA, Ruthenburg AJ, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 42.Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121(6):873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol. Cell Biol. 2001;21(7):2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the Polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc. Natl Acad. Sci. USA. 2003;100(14):8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat. Genet. 2002;30(1):41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 46.Ferrando AA, Armstrong SA, Neuberg DS, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102(1):262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 47.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol. Cell Biol. 2004;24(23):10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Yan X, Sashida G, et al. Stress hematopoiesis reveals abnormal control of self-renewal, lineage bias, and myeloid differentiation in Mll partial tandem duplication (Mll-PTD) hematopoietic stem/progenitor cells. Blood. 2012;120(5):1118–1129. doi: 10.1182/blood-2012-02-412379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zorko NA, Bernot KM, Whitman SP, et al. Mll partial tandem duplication and Flt3 internal tandem duplication in a double knock-in mouse recapitulates features of counterpart human acute myeloid leukemias. Blood. 2012;120(5):1130–1136. doi: 10.1182/blood-2012-03-415067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 51.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67(15):7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grembecka J, He S, Shi A, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 2012;8(3):277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muntean AG, Tan J, Sitwala K, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17(6):609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799(5–6):379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomson BN, Arndt KM. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta. 2013;1829(1):116–126. doi: 10.1016/j.bbagrm.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobson CL, Warren AJ, Pannell R, Forster A, Rabbitts TH. Tumorigenesis in mice with a fusion of the leukaemia oncogene Mll and the bacterial lacZ gene. EMBO J. 2000;19(5):843–851. doi: 10.1093/emboj/19.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17(2):198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Along with [59,63,64,65], collectively found that several of the most common MLL translocation partner proteins exist within a transcriptional elongation complex, suggesting a common mechanism for MLL leukemogenesis.
- 59.Lin C, Smith ER, Takahashi H, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010;37(3):429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan J, Jones M, Koseki H, et al. CBX8, a Polycomb group protein, is essential for MLL–AF9-induced leukemogenesis. Cancer Cell. 2011;20(5):563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sobulo OM, Borrow J, Tomek R, et al. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc. Natl Acad. Sci. USA. 1997;94(16):8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohan M, Herz HM, Takahashi YH, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24(6):574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum. Mol. Genet. 2007;16(1):92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 64.Monroe SC, Jo SY, Sanders DS, et al. MLL–AF9 and MLL–ENL alter the dynamic association of transcriptional regulators with genes critical for leukemia. Exp. Hematol. 2011;39(1):77–86. e1–5. doi: 10.1016/j.exphem.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mueller D, Bach C, Zeisig D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110(13):4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernt KM, Zhu N, Sinha AU, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL–AF4 leukemias. Cancer Cell. 2008;14(5):355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■ Demonstrates altered epigenetic landscapes in leukemia cells bearing MLL translocations consistent with the deregulated recruitment of the DOT1l histone methyltransferase.
- 68.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang MJ, Wu H, Achille NJ, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70(24):10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide ana lysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 71.Liedtke M, Cleary ML. Therapeutic targeting of MLL. Blood. 2009;113(24):6061–6068. doi: 10.1182/blood-2008-12-197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilliland DG. Hematologic malignancies. Curr. Opin. Hematol. 2001;8(4):189–191. doi: 10.1097/00062752-200107000-00001. [DOI] [PubMed] [Google Scholar]; ■ Proposes the `two-hit' hypothesis for leukemogenesis where mutations that promote survival/proliferation collaborate with mutations that block differentation, resulting in acute myeloid leukemia.
- 73.Ford AM, Ridge SA, Cabrera ME, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363(6427):358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 74.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100(5):1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 75.Balgobind BV, Zwaan CM, Pieters R, Van Den Heuvel-Eibrink MM. The heterogeneity of pediatric MLL-rearranged acute myeloid leukemia. Leukemia. 2011;25(8):1239–1248. doi: 10.1038/leu.2011.90. [DOI] [PubMed] [Google Scholar]
- 76.Kuchenbauer F, Kern W, Schoch C, et al. Detailed ana lysis of FLT3 expression levels in acute myeloid leukemia. Haematologica. 2005;90(12):1617–1625. [PubMed] [Google Scholar]
- 77.Ono R, Nakajima H, Ozaki K, et al. Dimerization of MLL fusion proteins and FLT3 activation synergize to induce multiple-lineage leukemogenesis. J. Clin. Investig. 2005;115(4):919–929. doi: 10.1172/JCI22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stubbs MC, Kim YM, Krivtsov AV, et al. MLL–AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 2008;22(1):66–77. doi: 10.1038/sj.leu.2404951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamai H, Miyake K, Takatori M, et al. Activated K-Ras protein accelerates human MLL/AF4-induced leukemol-ymphomogenicity in a transgenic mouse model. Leukemia. 2011;25(5):888–891. doi: 10.1038/leu.2011.15. [DOI] [PubMed] [Google Scholar]
- 80.Mizukawa B, Wei J, Shrestha M, et al. Inhibition of Rac GTPase signaling and downstream prosurvival Bcl-2 proteins as combination targeted therapy in MLL–AF9 leukemia. Blood. 2011;118(19):5235–5245. doi: 10.1182/blood-2011-04-351817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 82.Ayton PM, Cleary ML. Transformation of myeloid progenitors by MLL oncoproteins is dependent on Hoxa7 and Hoxa9. Genes Dev. 2003;17(18):2298–2307. doi: 10.1101/gad.1111603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. Hoxa9 influences the phenotype but not the incidence of Mll–AF9 fusion gene leukemia. Blood. 2004;103(5):1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 85.So CW, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL–GAS7 in the absence of Hoxa7 or Hoxa9. Blood. 2004;103(8):3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 86.Bach C, Buhl S, Mueller D, Garcia-Cuellar MP, Maethner E, Slany RK. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115(14):2910–2918. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- 87.Huang Y, Sitwala K, Bronstein J, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119(2):388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang G, Zhao X, Wang L, et al. The ability of MLL to bind RUNX1 and methylate H3K4 at PU.1 regulatory regions is impaired by MDS/AML-associated RUNX1/AML1 mutations. Blood. 2011;118(25):6544–6552. doi: 10.1182/blood-2010-11-317909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilkinson AC, Ballabio E, Geng H, et al. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4–MLL complex interaction. Cell Rep. 2013;3(1):116–127. doi: 10.1016/j.celrep.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bursen A, Schwabe K, Ruster B, et al. The AF4. MLL fusion protein is capable of inducing ALL in mice without requirement of MLL.AF4. Blood. 2010;115(17):3570–3579. doi: 10.1182/blood-2009-06-229542. [DOI] [PubMed] [Google Scholar]
- 91.Hess JL, Bittner CB, Zeisig DT, et al. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108(1):297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Somervaille TC, Matheny CJ, Spencer GJ, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schwieger M, Schuler A, Forster M, et al. Homing and invasiveness of MLL/ENL leukemic cells is regulated by MEF2C. Blood. 2009;114(12):2476–2488. doi: 10.1182/blood-2008-05-158196. [DOI] [PubMed] [Google Scholar]
- 94.Faber J, Krivtsov AV, Stubbs MC, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bindels EM, Havermans M, Lugthart S, et al. EVI1 is critical for the pathogenesis of a subset of MLL–AF9-rearranged AMLs. Blood. 2012;119(24):5838–5849. doi: 10.1182/blood-2011-11-393827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang QF, Wu G, Mi S, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117(25):6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]; ■■ Along with [98,109], demonstrates that small-molecule-mediated inhibition of proteins within the MLL fusion protein complex inhibits the growth of MLL leukemia cells.
- 98.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakanishi H, Nakamura T, Canaani E, Croce CM. ALL1 fusion proteins induce deregulation of EphA7 and ERK phosphorylation in human acute leukemias. Proc. Natl Acad. Sci. USA. 2007;104(36):14442–14447. doi: 10.1073/pnas.0703211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li Z, Huang H, Chen P, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Popovic R, Riesbeck LE, Velu CS, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113(14):3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schotte D, Lange-Turenhout EA, Stumpel DJ, et al. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010;95(10):1675–1682. doi: 10.3324/haematol.2010.023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mi S, Li Z, Chen P, et al. Aberrant overexpression and function of the miR-17–92 cluster in MLL-rearranged acute leukemia. Proc. Natl Acad. Sci. USA. 2010;107(8):3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wong P, Iwasaki M, Somervaille TC, et al. The miR-17–92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70(9):3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang X, Huang H, Li Z, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22(4):524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Anderson K, Lutz C, van Delft FW, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]; ■■ Along with [107], experimentally reveals genetic heterogeneity within subpopulations of leukemia stem cells, and has important implications for treating leukemia.
- 107.Notta F, Mullighan CG, Wang JC, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469(7330):362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 108.Wang LM, Ren DM. Flavopiridol, the first cyclin-dependent kinase inhibitor: recent advances in combination chemotherapy. Mini Rev. Med. Chem. 2010;10(11):1058–1070. doi: 10.2174/1389557511009011058. [DOI] [PubMed] [Google Scholar]
- 109.Daigle SR, Olhava EJ, Therkelsen CA, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grembecka J, He S, Shi A, et al. Menin–MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 2012;8(3):277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Krivtsov AV, Twomey D, Feng Z, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL–AF9. Nature. 2006;442(7104):818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 113.Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev. 2003;17(24):3029–3035. doi: 10.1101/gad.1143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yeung J, Esposito MT, Gandillet A, et al. β-catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 2010;18(6):606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 116.Heidel FH, Bullinger L, Feng Z, et al. Genetic and pharmacologic inhibition of β-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 2012;10(4):412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Z, Iwasaki M, Ficara F, et al. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell. 2010;17(6):597–608. doi: 10.1016/j.ccr.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455(7217):1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harris WJ, Huang X, Lynch JT, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL–AF9 leukemia stem cells. Cancer Cell. 2012;21(4):473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 120.Armstrong SA, Kung AL, Mabon ME, et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2003;3(2):173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- 121.Brown P, Levis M, McIntyre E, Griesemer M, Small D. Combinations of the FLT3 inhibitor CEP-701 and chemotherapy synergistically kill infant and childhood MLL-rearranged ALL cells in a sequence-dependent manner. Leukemia. 2006;20(8):1368–1376. doi: 10.1038/sj.leu.2404277. [DOI] [PubMed] [Google Scholar]
- 122.Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7(11):e1000249. doi: 10.1371/journal.pbio.1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]