Abstract
Drosophila has recently become a powerful model system to understand the mechanisms of temporal patterning of neural progenitors called neuroblasts (NBs). Two different temporal sequences of transcription factors (TFs) have been found to be sequentially expressed in NBs of two different systems: the Hunchback, Krüppel, Pdm1/Pdm2, Castor, and Grainyhead sequence in the Drosophila ventral nerve cord; and the Homothorax, Klumpfuss, Eyeless, Sloppy-paired, Dichaete, and Tailless sequence that patterns medulla NBs. In addition, the intermediate neural progenitors of type II NB lineages are patterned by a different sequence: Dichaete, Grainyhead, and Eyeless. These three examples suggest that temporal patterning of neural precursors by sequences of TFs is a common theme to generate neural diversity. Cross-regulations, including negative feedback regulation and positive feedforward regulation among the temporal factors, can facilitate the progression of the sequence. However, there are many remaining questions to understand the mechanism of temporal transitions. The temporal sequence progression is intimately linked to the progressive restriction of NB competence, and eventually determines the end of neurogenesis. Temporal identity has to be integrated with spatial identity information, as well as with the Notch-dependent binary fate choices, in order to generate specific neuron fates.
1. INTRODUCTION
One fundamental question in developmental neurobiology is to understand how to generate the remarkable diversity of neurons and glia present in adult brains, from a small number of seemingly homogenous neural stem cells in the embryo. Spatial patterning of neural stem cells that is achieved by various morphogens and their signaling cascades contributes to the generation of neural diversity (Bhat, 1999a; Dessaud, McMahon, & Briscoe, 2008). Furthermore, single neural stem cells can generate different neural types in a stereotyped order: This is achieved by temporal patterning of neural stem cells. Extensive studies in the vertebrate central nervous system (CNS), especially cerebral cortex and retina, have revealed that birth order correlates with distinct neuronal/glial identity (reviewed in Jacob, Maurange, & Gould, 2008; Livesey & Cepko, 2001; Molyneaux, Arlotta, Menezes, & Macklis, 2007; Okano & Temple, 2009; Pearson & Doe, 2004). Although both extrinsic and intrinsic factors are required for the correct specification of temporal identify, isolated neural stem cells cultured in vitro can recapitulate the sequential generation of different neuron types, thus underscoring a cell-intrinsic temporal control of stem cells (Gaspard et al., 2008; Naka, Nakamura, Shimazaki, & Okano, 2008; Shen et al., 2006).
Neural stem cells are called neuroblasts (NBs) in Drosophila. Drosophila NBs usually undergo several rounds of Notch-dependent asymmetric divisions to produce a self-renewed NB, and a smaller daughter cell called ganglion mother cell (GMC). GMCs generally divide asymmetrically once to produce two postmitotic progeny with different fates (reviewed in Doe, 2008; Karcavich, 2005; Knoblich, 2010; Lin & Lee, 2012). Several Drosophila NB lineages in the CNS have been partially or completely characterized. They generate multiple distinct progeny in a birth-order-dependent manner (Akiyama-Oda, Hosoya, & Hotta, 1999; Baek & Mann, 2009; Bossing, Udolph, Doe, & Technau, 1996; Isshiki, Pearson, Holbrook, & Doe, 2001; Jefferis, Marin, Stocker, & Luo, 2001; Karcavich & Doe, 2005; Lee, Lee, & Luo, 1999; Lundell & Hirsh, 1998; Pearson & Doe, 2003; Schmid, Chiba, & Doe, 1999; Schmidt et al., 1997; Skeath & Thor, 2003; Yu, Chen, Shi, Huang, & Lee, 2009; Yu et al., 2010). Drosophila has recently become a powerful model system to understand the mechanisms of temporal specification of neurons and there have been several extensive and excellent reviews on this topic (Brand & Livesey, 2011; Kao & Lee, 2010; Maurange, 2012; Pearson & Doe, 2004). This review will therefore focus on the most recent advances in temporal patterning of NBs in several systems of Drosophila that are now well characterized. Below is a brief introduction of each system to be described.
1.1. Embryonic and larval ventral nerve cord
Classic work on the temporal patterning of NBs in the Drosophila embryonic ventral nerve cord (VNC) has made it undoubtedly the best-characterized system. The VNC of Drosophila, spanning three thoracic and eight abdominal segments, is the counterpart of the vertebrate spinal cord. During early embryonic development, NBs delaminate from a neuroectoderm at stereotypical positions. Each hemi-segment contains approximately 30 NBs arranged in seven rows, and each NB is identifiable based on its position (e.g., NB1-1 is in row1, column1) and molecular markers (Bhat, 1999b; Doe, 1992; Skeath & Thor, 2003; Technau, Berger, & Urbach, 2006). The distinction between these NBs is achieved by spatial patterning that will not be discussed in detail here. The neuronal and glial progeny composition of each embryonic NB lineage have been determined by early Dil NB labeling experiments: A single NB can generate multiple neuron types and glia (Bossing et al., 1996; Schmid et al., 1999, 1997). At the embryo to larval transition, some of these NBs commit to apoptosis, while others enter into a quiescence phase. They are then reactivated during the larval stage and continue to proliferate during larval and pupal stages. These postembryonic NBs generate ~90% of neurons that constitute the adult CNS (Prokop & Technau, 1991; Truman & Bate, 1988). Relatively less is known about the postembryonic lineages as compared to embryonic lineages, but several recent studies have shed some light on the temporal control of larval lineages (see below).
1.2. Olfactory system
Another well-characterized system is the olfactory system, including the antennal lobe, which receives olfactory inputs from olfactory receptor neurons (ORNs), and the mushroom body, which receives inputs from projection neurons (PNs) in the antenna lobe. Systematic clonal analysis of mushroom body neurons using the mosaic analysis with a repressible cell marker (MARCM) technique has demonstrated that each of the four mushroom body NBs sequentially generates at least three types of neurons, γ neurons first, followed by α′β′, and then αβ neurons (Lee et al., 1999), suggesting birth-order-dependent neuronal specification. Similarly, the antennal lobe PNs that are derived from three NBs (an anterodorsal, a lateral, and a ventral NB) are specified by lineage and birth order to connect with specific classes of ORN axons ( Jefferis et al., 2001). These three lineages have been extensively characterized (Das, Reichert, & Rodrigues, 2010; Das et al., 2008; Lai, Awasaki, Ito, & Lee, 2008; Lin, Kao, Yu, Huang, & Lee, 2012; Lin et al., 2010; Yu et al., 2010). For example, the anterodorsal lineage produces 40 types of PNs in a stereotypic order, while the lateral lineage sequentially generates 48 pairs of local interneurons paired with distinct PNs.
1.3. Medulla in the optic lobes
The Drosophila optic lobes, composed of lamina, medulla, and lobula complex, are the processing centers of visual information in the brain. Among them, the medulla is the largest neuropil. It contains approximately 40,000 neurons, belonging to more than 70 different cell types (Fischbach & Dittrich, 1989; Morante & Desplan, 2008). These neurons are generated by NBs derived from a crescent-shaped single-layered neuroepithelium (NE) in the larval brain called the outer proliferation center (Green, Hartenstein, & Hartenstein, 1993; White & Kankel, 1978). During development, a wave of neurogenesis progresses from the edge of the crescent toward its center, and sequentially converts NE cells into medulla NBs. Therefore, NBs of different ages can be observed together in one snapshot (Egger, Boone, Stevens, Brand, & Doe, 2007; Yasugi, Umetsu, Murakami, Sato, & Tabata, 2008). These NBs generate GMCs, which then produce medulla neurons that form chains below each NB, with the first-born neurons located in the deepest position and latest-born neurons in the most superficial position, near the NB (Hasegawa et al., 2011; Morante, Erclik, & Desplan, 2011). Recently, the developing medulla has emerged as a new powerful system for studying temporal patterning of NBs (Li et al., 2013; Suzuki, Kaido, Takayama, & Sato, 2013).
1.4. Type II NBs in the central brain
In contrast to the regular type I NBs that divide asymmetrically to generate a series of GMCs that each divides once to produce two postmitotic cells, type II NBs divide asymmetrically to self-renew and to generate a series of transit amplifying GMCs called intermediate neural progenitors (INPs), with each INP dividing asymmetrically to generate several GMCs and eventually giving rise to 6–12 neurons and/or glia (Bayraktar, Boone, Drummond, & Doe, 2010; Boone & Doe, 2008; Viktorin, Riebli, Popkova, Giangrande, & Reichert, 2011; Weng, Golden, & Lee, 2010; Zhu, Barshow, Wildonger, Jan, & Jan, 2011). Type II lineages allow more neurons than type I lineages to be quickly generated.
2. SEQUENTIALLY EXPRESSED TRANSCRIPTION FACTORS IN NBs CONTROL BIRTH-ORDER-DEPENDENT NEURON FATES
2.1. Temporal sequence of transcription factors in Drosophila embryonic VNC NBs
How does a NB generate a stereotypical order of different neuron types as it ages? Extensive studies in the Drosophila embryonic VNC have led to the identification of a molecular mechanism of temporal specification, where a series of transcription factors (TFs) are sequentially and transiently expressed in NBs as they age. Postmitotic progeny born during each time window maintains the expression of the TF. This temporal sequence of TFs includes Hunchback (Hb), Krüppel (Kr), Pdm1/Pdm2 (Pdm), Castor (Cas), and Grainyhead (Grh) (Baumgardt, Karlsson, Terriente, Diaz-Benjumea, & Thor, 2009; Brody & Odenwald, 2000; Cui & Doe, 1992; Grosskortenhaus, Pearson, Marusich, & Doe, 2005; Isshiki et al., 2001; Kambadur et al., 1998; Mellerick, Kassis, Zhang, & Odenwald, 1992; Romani et al., 1996; Yang, Yeo, Dick, & Chia, 1993). Although different NBs in the VNC delaminate and start differentiation at different times, most of them follow the same TF sequence. These temporally expressed TFs have been shown to be both required and sufficient for birth-order-dependent neuronal specification in several different NB lineages (Baumgardt et al., 2009; Benito-Sipos et al., 2010; Cleary & Doe, 2006; Grosskortenhaus, Robinson, & Doe, 2006; Isshiki et al., 2001; Kambadur et al., 1998; Novotny, Eiselt, & Urban, 2002; Pearson & Doe, 2003; Tran & Doe, 2008). For example, each of the first five GMCs in the NB7-1 lineage gives rise to a motor neuron (U1–U5) and a sibling cell. The unique identity of each of the U1–U5 motor neurons is specified by the sequentially expressed TFs: Hb is necessary and sufficient to specify U1 and U2 fates (high Hb: U1, low Hb: U2); Kr is necessary andsufficienttospecifytheU3fate;PdmalonespecifiesU4;whilePdmandCas together specify U5 (Cleary & Doe, 2006; Grosskortenhaus et al., 2005; Isshiki et al., 2001; Pearson & Doe, 2003; Fig. 3.1). In the NB7-3 lineage, there are only three GMCs generated: GMC-1 is Hb+,Kr+, and generates the EW1 interneuron and the GW motor neuron; GMC-2 is Kr+, and generates the EW2 interneuron and a sibling that undergoes apoptosis; GMC-3 is Pdm+, and differentiates directly into the EW3 interneuron (Isshiki et al., 2001; Karcavich & Doe, 2005; Lundell & Hirsh, 1998). In this case, Hb is necessary and sufficient for the first-born neuron fates (EW1/GW). Loss of hb results in a lack of the EW1/GW, while EW2 and EW3 are still present. Prolonged expression of Hb in the NB leads to additional neurons with EW1/GW fates attheexpenseofEW2andEW3(Isshikietal.,2001; Novotnyetal.,2002).Kris necessary and sufficient for the second-born neuron fate EW2 (Isshiki et al., 2001).
Figure 3.1.
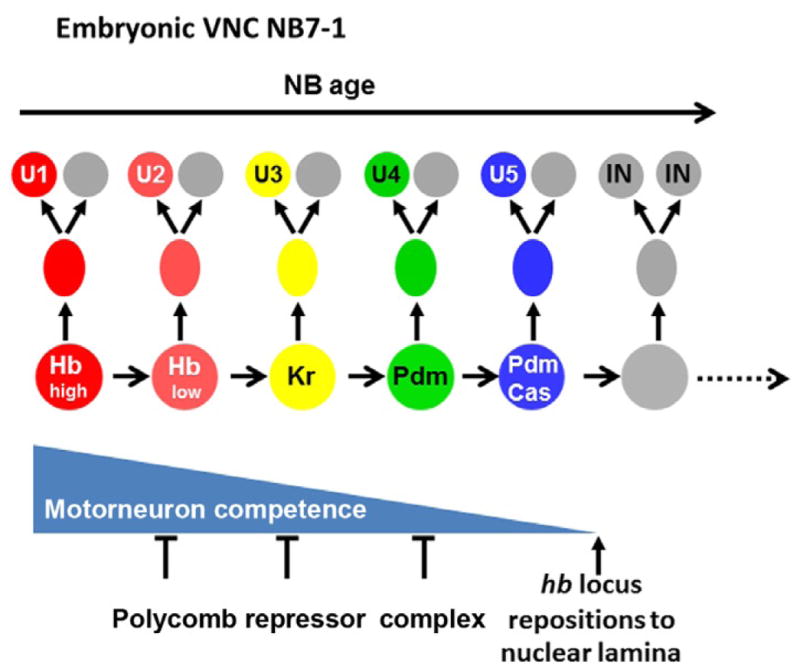
Schematic model showing the sequential expression of Hb, Kr, Pdm, and Cas in the embryonic VNC NB7-1. These TFs control the sequential generation of different U motor neurons. The Polycomb repressor complex is required for the progressive loss of NB competence to generate motor neurons. At the end of the first five divisions, the hb locus relocates to the nuclear periphery, and the NB completely loses its competence to generate motor neurons.
There are variations in the function of the temporal TFs: In the NB3-1 lineage, Pdm is not required for specification of the third temporal identity, but instead to close the preceding Kr+ temporal identity window (Tran & Doe, 2008). The first four GMCs of NB3-1 lineage produce HB9+, Islet+ RP motor neurons with a birth order of RP1→RP4→RP3→RP5 (and their non-RP siblings): Hb specifies RP1 and RP4 (high Hb: RP1; low Hb: RP4); Kr specifies RP3; while Pdm is not required for specifying RP5, because there is only a modest expansion of Kr expression in NBs in pdm mutants, with a few extra RP3 neurons produced, but RP5 neurons are still specified. Similarly, Cas is required for closing the third (RP5) temporal identity window: In cas mutants, there are ectopic RP5 neurons (Tran & Doe, 2008).
Thus, a series of TFs sequentially expressed in NBs control the sequential generation of different neural types in multiple NB lineages. Since different NBs generate different lineages, these TFs do not specify a certain neuron type, but control the birth-order-dependent neuronal identity. The birth-order-dependent temporal identity is integrated with the spatial identity of the NB within each segment or between different segments, and is translated into specific cell types. For example, hb controls the first-born cell fates in multiple lineages, which can be motor neurons, interneurons or glial cells, depending on the NB lineage (Isshiki et al., 2001).
2.2. Similar or different TF sequence in other systems?
Since birth-order-dependent neuronal specification has been widely observed in various systems, is the same or a similar temporal TF sequence utilized to pattern neural stem cells of other systems?
In the mushroom body, which is generated by four NBs, each NB sequentially generates at least three types of neurons. Thus far, no temporal sequence of TFs that controls the fate of these neurons has been identified in the NB. However, a novel bric-a-brac, tramtrack, broad (BTB)-zinc finger protein, named Chinmo (chronologically inappropriate morphogenesis), acts in GMCs or young neurons to control the temporal identity of mushroom body neurons (Zhu et al., 2006). Although Chinmo mRNA is equally expressed throughout the entire NB lineage, Chinmo protein shows a temporal gradient in the neuronal progeny. It is absent in NBs and is expressed at its highest levels in the early-born neurons, lower in the next-born neurons, and undetectable in the latest-born neurons. Reducing or increasing Chinmo levels causes transformation of neurons toward later or earlier fates, respectively (Zhu et al., 2006). Recently, microRNAs of the Let-7-complex, the heterochronic miRNAs originally identified in Caenorhabditis elegans, were found to target chinmo to regulate the temporal identity of Drosophila mushroom body neurons (Wu, Chen, Mercer, & Sokol, 2012). Since Chinmo protein or Let-7 miRNAs are not detected in NBs, how the Chinmo gradient in neurons is regulated as the NB ages is not understood.
Postembryonic neuroblasts (pNBs) in the VNC generate 90% of neurons that constitute the adult CNS. Although a complete temporal TF sequence has not been identified, recent studies identified two members of the post-embryonic TF sequence, Cas and Seven-up (Svp) (Maurange, Cheng, & Gould, 2008; Tsuji, Hasegawa, & Isshiki, 2008). They are required for a temporal switch of pNBs from generating small Chinmo+ neurons to instead producing large Br-C+ (Broad Complex) neurons in many different lineages. Since the switch from Chinmo+ to Br-C+ happens later than the transient expression of Cas or Svp, the switch must be directly controlled by an unknown member of the TF sequence whose expression is promoted by cas and svp by feedforward regulation (Maurange et al., 2008). Cas and Svp are also required for the NBs to eventually end neurogenesis. This is discussed in detail below.
In the anterodorsal lineage of the antennal lobe, Kr was shown to act in the NB to define one out of 40 temporal fates of PNs (Kao, Yu, He, Kao, & Lee, 2012). Loss of Kr from the NB causes a single PN fate to be skipped. However, loss of Hb, Pdm or Cas does not produce detectable phenotypes (Kao et al., 2012). It will be interesting to identify more TFs that are temporally expressed in antennal lobe NBs and control the temporal specification of a large number of distinct PNs and interneurons.
2.3. Different TF sequence in medulla NBs
In the developing medulla, NBs of different ages can be visualized in one snapshot, and thus the medulla provides another powerful system to study temporal patterning of NBs. Recent studies identified a different series of TFs expressed in medulla NBs: Homothorax (Hth), Klumpfuss (Klu), Eyeless (Ey), Sloppy paired (Slp), Dichaete (D), and Tailless (Tll) are sequentially expressed in NBs of increasing ages, with Hth expressed in newly differentiated NBs, Klu, Ey, Slp, and D expressed in increasingly older NBs, and Tll in the oldest NBs. Hth, Ey, and Slp were shown to control the generation of specific neuron types born during each time window (Li et al., 2013; Suzuki et al., 2013; Fig. 3.2). This identification of a second TF sequence that is different from the one in the embryonic VNC suggests that TF sequence-dependent temporal patterning of NBs is likely to be broadly utilized, and that different TF sequences can be recruited in different systems.
Figure 3.2.
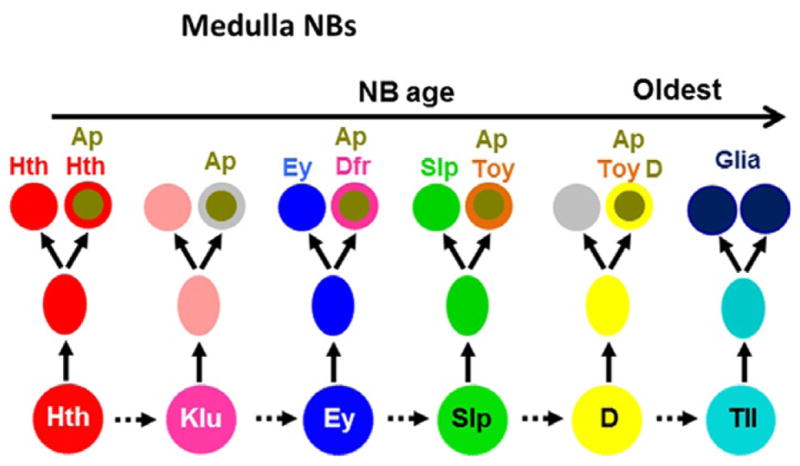
Schematic model showing the sequential expression of Hth, Klu, Ey, Slp, D, and Tll in medulla NBs. These TFs control the sequential generation of different neuronal types marked by specific combinations of neuronal markers. Ap is expressed in the N-on daughters of ganglion mother cells. Tll+ NBs generate glia.
2.4. Combinatorial temporal patterning of INPs and type II NBs
A recent elegant paper showed that, in type II NB lineages, both NBs and INPs are temporally patterned by different temporal series of TFs (Bayraktar & Doe, 2013). Type II NBs sequentially express D/Cas and Svp as well as other not yet identified members of a TF sequence. Along the second temporal axis, INPs sequentially express D, Grh, and Ey as they age: These TFs are required for the sequential production of distinct neural subtypes. Thus, TF sequences on the two temporal axes act combinatorially to generate larger neural diversity (Bayraktar & Doe, 2013; Fig. 3.3).
Figure 3.3.
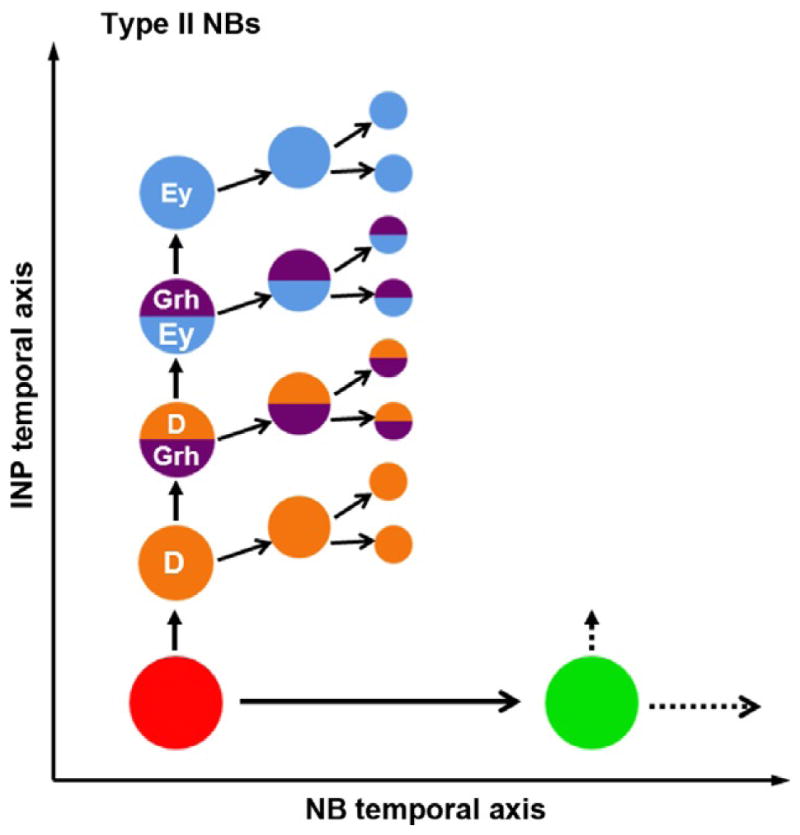
Schematic model showing combinatorial temporal patterning in type II NB lineages in the central brain. Both NBs and INPs are patterned by distinct temporal series of TFs. INPs sequentially express D, Grh, and Ey as they age. These TFs are required for the sequential production of distinct neuronal subtypes.
3. HOW ARE TEMPORAL TRANSITIONS IN NBs CONTROLLED?
3.1. Cross-regulations between the TFs play important roles in the transitions
In the embryonic VNC, the Hb→Kr→Pdm→Cas→Grh sequence can be recapitulated in isolated NBs cultured in vitro, suggesting a lineage-intrinsic mechanism for the TF transitions (Brody & Odenwald, 2000; Grosskortenhaus et al., 2005). Gain and loss of function studies suggest that multiple cross-regulations between these temporal TFs exist: one TF activates the next TF, while repressing the previous TF and the next-plus-one TF (Fig. 3.4). This, in theory, could allow the TF transitions (Baumgardt et al., 2009; Brody & Odenwald, 2000; Grosskortenhaus et al., 2006; Isshiki et al., 2001; Kambadur et al., 1998; Nakajima, Isshiki, Kaneko, & Ishihara, 2010; Tran & Doe, 2008). However, in most lineages, loss of Hb, Kr, or Pdm simply causes the corresponding temporal identity to be skipped, rather than a blockage of the temporal progression, suggesting that the cross-regulations are not essential and that other factors are involved in the switches. However, loss of Cas does lead to a block in the temporal progression and to persistent Pdm expression (Brody & Odenwald, 2000; Grosskortenhaus et al., 2006; Isshiki et al., 2001; Kambadur et al., 1998; Maurange et al., 2008; Tran & Doe, 2008).
Figure 3.4.
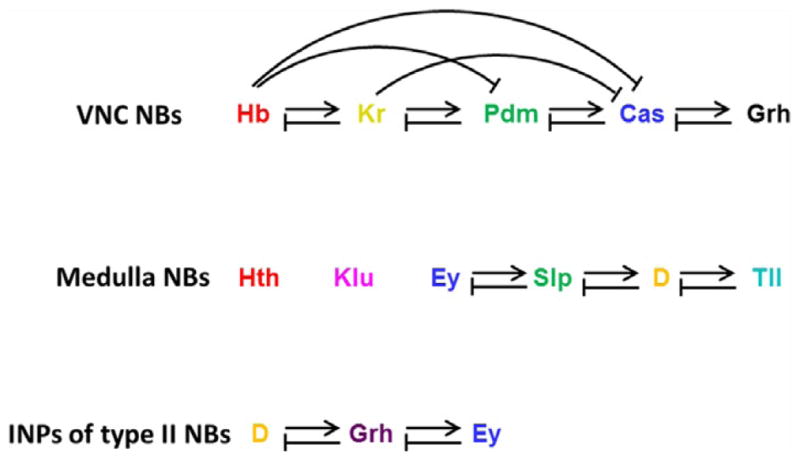
Cross-regulations between TFs in the three temporal sequences that pattern NBs or INPs. Each TF activates the next TF and represses the previous TF (with some exceptions).
In the Hth→Klu→Ey→Slp→D→Tll sequence that patterns medulla NBs, loss of Ey, Slp, or D does disrupt essential cross-regulations, and the NBs keep dividing but do not progress to the next TF stage. Slp and D are also required to repress the previous TF (Li et al., 2013; Suzuki et al., 2013). Thus, Ey, Slp, and D are required for the sequential TF switching, and, in this regard, they are similar to Cas in the embryonic sequence. However, there are a few exceptions to these modes of regulation: No cross-regulation is observed among Hth, Klu, and Ey; and Tll is not required, but it is sufficient to repress D (Fig. 3.4). This suggests that there are more TFs temporally expressed in medulla NBs that work together with the Hth→Klu→Ey→Slp→D→Tll sequence to ensure temporal transitions (Li et al., 2013; Suzuki et al., 2013).
In the D→Grh→Ey sequence that patterns INPs of type II NB lineages, D is necessary but not sufficient to activate the next TF, Grh. In turn, Grh is required to repress D and activate Ey. Ey is required and sufficient to repress Grh. Thus, a “feedforward activation/feedback repression” model for D→Grh→Ey cross-regulation appears to control the transitions of TFs (Bayraktar & Doe, 2013; Fig. 3.4).
Considering the three know sequences of TFs in the VNC, medulla and type II NBs, it is clear that a cross-regulatory network among the temporally expressed TFs does have important roles in the transition between TFs. However, more factors or timing mechanisms must be involved and may vary case by case. Redundant mechanisms might also be present to ensure the robustness of transitions.
3.2. Differential requirements for cell cycle and asymmetric division
Since the NB progresses through the TF sequence as it undergoes asymmetric divisions to produce neuronal progeny, cell cycle and cell division might play a role in these transitions, either by cell-cycle dependent signaling event, asymmetric localization of certain mRNA or protein, or by feedback signal from the progeny. Indeed, the first Hb to Kr transition requires cell cycle and cytokinesis, but it does not require asymmetric localization of hb mRNA or Hb protein. However, the Kr→Pdm→Cas transitions are regulated in a cell-cycle independent manner, and progress normally in cell-cycle arrested NBs (Grosskortenhaus et al., 2005).
3.3. Switching factors involved in TF transitions
As discussed above, Hb expression is downregulated at the transcription level in a cell-division dependent manner (Grosskortenhaus et al., 2005; Kanai, Okabe, & Hiromi, 2005; Kohwi, Hiebert, & Doe, 2011; Mettler, Vogler, & Urban, 2006). The orphan nuclear receptor TF Svp is required for the Hb to Kr switch in several NB lineages of the Drosophila embryonic CNS (Kanai et al., 2005). Loss of svp causes prolonged Hb expression in NBs as well as ectopic early-born neurons. The nuclear export of svp mRNA (and thus its efficient translation) is dependent on the cell cycle (Mettler et al., 2006), providing an explanation for the requirement of cell division for the Hb to Kr transition.
In addition to Svp, the nuclear proteins distal antenna and distal antenna-related proteins (Dan and Dan-r) function in parallel pathways with Svp to downregulate Hb expression, and ensure the timely transition of the NBs from making early-born neurons to generating late-born neurons (Kohwi et al., 2011).
In the medulla NB TF sequence, both Slp and D are required to turn off the preceding TF and turn on the next TF, therefore, acting as switching factors for themselves. Additional transition factors remain to be identified for other transitions (Li et al., 2013; Suzuki et al., 2013).
4. RELATIONSHIP BETWEEN TEMPORAL SEQUENCE AND NB COMPETENCE
4.1. Restriction of NB competence
Early studies in mammalian neurogenesis have demonstrated that neural stem cells undergo progressive restriction in their competence to generate different types of progeny in response to extrinsic signals (Desai & McConnell, 2000; Livesey & Cepko, 2001). For example, during development of the cerebral cortex, neural stem cells generate neurons in the six cortical layers in an “inside-out” defined order, in which layer 6 and 5 neurons are generated first, followed by layer 4, 3 and 2. The fate of the progeny also depends on environmental cues to which the cells respond prior to cell division. Transplantation experiments showed that mid-stage neural stem cells that are producing layer 4 neurons have the competence to generate layer 2/3 (later-born) neurons when transplanted into older brains. However, when these neural stem cells are transplanted into younger brains in which layer 6 neurons are being generated, they have lost the competence to generate layer 6 neurons. Instead, they produce layer 4 and 5 neurons. Thus, the competence of neural stem cells to generate a given cell type is progressively lost, although this loss of competence lags behinds the completion of generation of this neural type (Desai & McConnell, 2000).
4.2. TF sequence and regulation of NB competence
Loss of competence is also observed in Drosophila NBs, although, in this case, competence responds to intrinsic TFs rather than extrinsic cues. In the VNC, providing ectopic Hb to the NB after Hb has been downregulated is sufficient to generate additional early-born neurons. However, this competence to respond to Hb progressively decreases over time. In the NB7-1 lineage, a pulse of ectopic Hb at the time U1 and U2 are normally born leads to the generation of several ectopic U1 and U2 neurons. If the pulse of ectopic Hb is provided later, when U3 and U4 are normally born, only one ectopic U2 neuron can be generated; and if Hb is given after the fifth division when the NB switches to generate interneurons, the NB fails to generate ectopic U neurons (Pearson & Doe, 2003). Further studies showed that NB7-1 has a single early competence window spanning the first five divisions for responding to Hb, Kr, Pdm, or Cas to make the corresponding U motor neurons. Then, NBs completely lose the competence to produce U motor neurons in response to these TFs (Cleary & Doe, 2006; Fig. 3.1). Similarly in the NB3-1 lineage, when the NB switches to generate interneurons at the fifth division, the competence to generate RP3 motor neurons in response to Kr is lost (Tran & Doe, 2008). This suggests that NBs can go sequentially through several competence windows in which they respond differently to the same temporal TF, allowing the repeated use of the same TF within one lineage to specify different fates (Cleary & Doe, 2006; Pearson & Doe, 2003).
The NB competence window can be extended if ectopic Hb is continuously provided before endogenous Hb is downregulated. In this case, NBs generate many U1/U2 neurons. They also retain their competence to generate later-born U motor neurons, and the lineage is extended. Thus, downregulation of Hb expression is required for the gradual loss of NB competence to respond to Hb (Cleary & Doe, 2006; Grosskortenhaus et al., 2005; Isshiki et al., 2001). Repression of multiple target genes (including Pdm2) by sustained Hb is necessary and sufficient for the maintenance of NB competence. Indeed, sustained expression of a form of Hb that functions solely as a transcription activator cannot significantly extend the competence window (Tran, Miller, & Doe, 2010).
Therefore, loss of NB competence to generate certain neuron fates lags behind the progression of the TF sequence (e.g., for cell fate specified by Hb, it is 3 cell divisions later) (Kohwi, Lupton, Lai, Miller, & Doe, 2013; Pearson & Doe, 2003). However, progression of the TF sequence, that is, downregulation of Hb relieving its transcription repression on multiple target genes, including a later temporal TF (Pdm), is required for the NB to close the current competence window and transit to the next.
4.3. Epigenetic mechanism for loss of NB competence
Recent studies uncovered a close relationship between epigenetic mechanism and loss of NB competence. Mammalian cortical neural stem cells generate cortical neurons that populate the different layers, and then lose the competence to generate neurons and switch to producing glia. This is due to the Polycomb repressor complexes (PRCs) that are required to suppress transcription of the neural fate TF neurogenin 1 in late-stage progenitors (Hirabayashi et al., 2009). PRCs induce repressive chromatin marks at the neurogenin 1 locus that gradually accumulate over time, and thus may provide a timer for the loss of competence to make neurons (Hirabayashi et al., 2009).
In Drosophila, PRCs are also involved in progressively restricting competence for generating motor neurons in NB7-1 and NB3-1: PRC loss of function extends the competence window to generate the corresponding motor neuron fate in response to Kr, while PRC gain of function precociously restricts this competence. In contrast, PRC activity does not affect the production of interneurons in multiple lineages (Touma, Weckerle, & Cleary, 2012), suggesting that it is involved specifically in the repression of multiple target genes involved in motor neuron specification (Fig. 3.1).
Chromosomal architecture has also recently been shown to be involved in the loss of competence of NB7-1 to respond to Hb, three NB divisions after hb transcription have stopped. At this stage, the hb gene locus relocates to the nuclear periphery of the NB, a repressive subnuclear compartment, preventing further activation of the hb gene by ectopic Hb, which is a requirement for the specification of early-born neurons (Kohwi et al., 2013; Fig. 3.1). The timing of the relocation correlates with downregulation of the Pipsqueak domain nuclear protein, Dan. Prolonging the expression of Dan can extend the NB competence by preventing relocalization of the hb locus at the periphery. This study proposed that Dan might competitively inhibit other Pipsqueak-domain factors, for example, Pipsqueak, a GAGA-binding factor essential for sequence-specific targeting of PRCs (Huang, Chang, Yang, Pan, & King, 2002), from binding to and recruiting hb and other loci to the nuclear lamina (Kohwi et al., 2013). This is consistent with the model that PRCs are involved in restricting NB competence (Touma et al., 2012).
In the medulla temporal sequence, NB competence has not yet been well characterized. Although mis-expressing the first NB TF Hth in all NBs leads to ectopic neurons with early-born cell fate, the phenotype becomes less obvious in later part of the lineage, suggesting that the competence of NBs to generate early-born cell fate in response to the first NB TF decreases with time. Whether the progressive loss of NB competence involves similar epigenetic mechanism remains to be studied. Interestingly, similar to the mammalian cortical neural stem cells, the medulla NBs switch from neurogenic to gliogenic at their final temporal stage when they express Tll (Li et al., 2013). Whether and how they lose the competence to generate neurons at this stage awaits further study.
5. FEEDFORWARD LOOPS DOWNSTREAM OF THE TEMPORAL SEQUENCE CONTROL NEURON FATES
Some temporal genes are expressed in a broad temporal window, during which multiple cell types are specified. How does a given temporal gene control multiple cell fates? An elegant timing mechanism subdivides the broad castor window in the NB5-6 lineage (Baumgardt et al., 2009). This NB lineage (producing in total ~20 progeny cells) ends with a broad castor window, during which ~10 cells are born, including the four last-born neurons that express the LIM-homeodomain TF Apterous (Ap), designated the Ap cluster (Ap1-4). The broad Cas window actually consists of two phases of Cas expression with a brief interruption of Cas expression when the Ap1 neuron is born. Ap1 and Ap4 are peptidergic neurons expressing the Nplp1 and FMRFamide neuropeptides, respectively. Ap2 and Ap3 are interneurons (Baumgardt, Miguel-Aliaga, Karlsson, Ekman, & Thor, 2007; Benveniste, Thor, Thomas, & Taghert, 1998; Park, Han, Kim, Han, & Taghert, 2004). The Ap cluster neurons are each born directly from a NB without a GMC intermediate (Baumgardt et al., 2009). Cell fate diversification of the four Ap neurons is achieved through three regulatory events triggered by Cas: (1) Cas activates the COE class TF Collier/Knot (Col) in the NB throughout the time Ap cluster neurons are born. It is initially inherited in all four Ap neurons, but it is only maintained in Ap1 where it acts in a feedforward loop involving sequential activation of Ap/Eyes absent, Dimmed, and Nplp1 to specify this cell fate (Baumgardt et al., 2007). (2) Cas also activates the zinc-finger protein squeeze (Sqz) in NBs, which is maintained in all four Ap neurons. After Sqz had time to accumulate in slightly later NBs, it acts with Cas to activate Nab, which is thus only inherited in Ap2-4. Sqz and Nab together rapidly downregulate Col in Ap2-4 neurons, and allow them to adopt fates different from Ap1 (Baumgardt et al., 2009). (3) Cas activates Grh expression, leading to gradual increase of Grh levels. Grh is required for the specification of Ap4 (Baumgardt et al., 2009). Thus, the broad Cas window is subdivided by these opposing feedforward loops that ensure the generation of neurons with different identities (Fig. 3.5).
Figure 3.5.
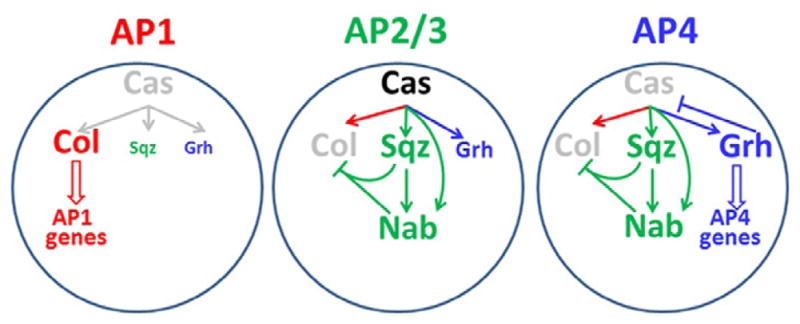
Model showing how three opposing feedforward loops activated by Cas subdivide the broad TF window. Ap1 loop (red): Transient Cas expression activates Col, which is initially expressed in all four Ap neurons, but is only maintained in Ap1, where it activates a feedforward loop that determines Ap1 fate. AP2-4 loop (green): Cas expression activates Sqz, and together they activate Nab in Ap2-4 neurons. Sqz and Nab together inactivate Col. AP4 loop (blue): Sustained Cas expression activates Grh, which is required for Ap4 neuron specification. Then, Grh represses Cas expression.
Similar regulatory logic might be widely used in other NB lineages to increase neural diversity. In the NB3-3 lineage, Nab and Sqz also function to specify temporal identities in the Cas window (Tsuji et al., 2008). In the medulla, NBs are estimated to generate 2–6 GMCs at each temporal TF stage, and there is evidence that the progeny from each GMC generated sequentially at the same TF stage express different TF combinations, and thus adopt different fates (Li et al., 2013). The temporal genes might act through multiple feedforward loops similar to the one described for the Ap cluster to subdivide each broad temporal window.
6. PROGRESSION OF THE TF SEQUENCE REQUIRED FOR THE END OF NEUROGENESIS
In order to generate the right number of neurons and avoid over-proliferation, NBs must end neurogenesis after completion of their lineages. Studies in pNBs in the VNC and central brain show that the temporal progression of TF sequence is also required for a timely ending of neurogenesis.
6.1. Apoptosis of abdominal pNBs
In the VNC, pNBs have lineages of different lengths: Abdominal pNBs have much smaller lineages (4–12 neurons) than thoracic and central brain pNBs (~100 progeny each) (Bello, Hirth, & Gould, 2003; Truman & Bate, 1988). This is due to Reaper, Grim, and Head involution defective (hid)-dependent apoptosis of abdominal pNBs at larval stages, which is induced by a burst of expression of the Hox protein Abdominal-A (Bello et al., 2003; Peterson, Carney, Taylor, & White, 2002). This apoptosis also requires inputs from the temporal sequence. At late stages of embryogenesis, Cas turns on the expression of Grh, and turns off the expression of D in NBs. Thus, Grh is expressed in NBs that enter quiescence and is subsequently maintained in pNBs through larval stages. The state of Grh+ D− is required for the abdominal pNBs to undergo apoptosis in response to the burst of Abdominal-A (Almeida & Bray, 2005; Cenci & Gould, 2005; Maurange et al., 2008). Furthermore, completion of the larval temporal sequence is required for the pNBs to undergo apoptosis: Either loss of postembryonic Svp or persistent expression of Cas can block the larval temporal sequence progression, and these NBs do not die in spite of normal Abdominal-A expression (Maurange et al., 2008; Fig. 3.6).
Figure 3.6.
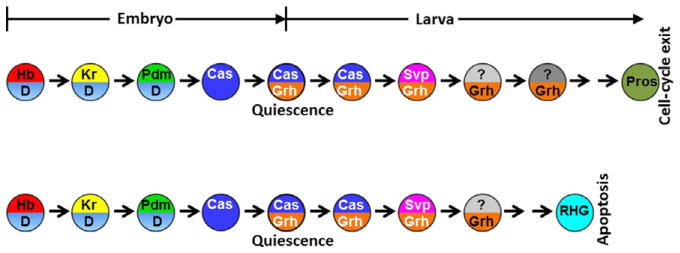
The temporal sequence in embryonic and larval VNC NBs is required for ending neurogenesis. At late stages of embryogenesis, Cas turns on Grh and turns off D in NBs. Grh, as well as two members of the larval temporal sequence, Cas and Svp, are required for the NBs to end neurogenesis at the correct time. Thoracic NB lineages are longer, and undergo pros-dependent cell-cycle exit. Abdominal NB lineages are shorter, and undergo apoptosis that depends on Reaper, Grim, and Hid.
6.2. Pros-dependent cell-cycle exit of thoracic and central brain pNBs
Most thoracic and central brain NBs (with the exception of mushroom body NBs) stop dividing at 120 h after larva hatching (~1 day into pupation). In contrast to the abdominal pNBs that undergo apoptosis, thoracic and central brain NBs end neurogenesis through cell-cycle exit that is dependent on a nuclear burst of expression of the homeodomain protein Prospero (Pros) (Maurange et al., 2008). Pros is tethered to cell membrane in NBs, and is asymmetrically localized to GMCs during asymmetric cell division. In GMCs, Pros is localized to nuclei where it promotes cell-cycle exit after one cell division (reviewed in Yu, Kuo, & Jan, 2006). Thoracic and central brain NBs exhibit a burst of expression of nuclear Pros at 120 h. pros mutant clones contain multiple NB-like cells that do not die at 120 h, and even continue dividing in the adult brain (Maurange et al., 2008).
Pros-dependent cell-cycle exit of thoracic and central brain NBs also requires inputs from both embryonic and larval temporal sequences: Sustained Grh expression in pNBs induced by embryonic Cas is required to prevent premature cell-cycle exit of NBs; while completion of the larval temporal sequence is required for the NBs to undergo Pros-dependent cell-cycle exit at 120 h. Remarkably, stalled temporal sequence caused by either loss of postembryonic Svp expression or persistent Cas expression prevents cell-cycle exit and NBs continue to divide even in 7-day-old adults (Maurange et al., 2008; Fig. 3.6).
6.3. Studies in other systems about the end of neurogenesis
In contrast to VNC and the majority of central brain NBs, mushroom body NBs that are born at embryonic stages do not have a quiescent stage and continue proliferating until the end of pupal stage. Tll is required for the uninterrupted and prolonged proliferation of mushroom body NBs: Loss of tll causes premature loss of mushroom body NBs in early pupal stage (Kurusu et al., 2009). Surprisingly, in the medulla NB temporal sequence, Tll appears to have the opposite function: Tll is the last TF that is expressed in the oldest NBs. These Tll+ NBs show nuclear localization of Pros, indicating that they undergo Pros-dependent cell-cycle exit at the end of their life, similar to the thoracic and central brain NBs (Li et al., 2013). Whether Tll is required or sufficient for ending medulla neurogenesis is currently not known. If so, it will be interesting to understand how Tll plays completely opposite roles in the mushroom body and in medulla NBs.
7. INTEGRATION OF TEMPORAL AND SPATIAL INFORMATION DETERMINES LINEAGES
Although almost all NBs in the embryonic VNC follow the same temporal sequence, they generate different lineages depending on their spatial identity. There are approximately 30 NBs in each hemisegment, and each NB has an individual fate based on its position and the expression of specific molecular markers. Intrasegmental specification is achieved by superimposed activities of segment polarity and dorso-ventral patterning genes (Technau et al., 2006). In addition to the intrasegmental spatial patterning, homologous NBs in different segments along the antero-posterior axis have slightly different lineages, although they also share significant similarities in their lineages and gene expression patterns. Hox genes are critical players in determining these differences. For example, in Section 6 we discussed how the abdA Hox gene, together with temporal factors, determines the length of lineages and the end of neurogenesis. Hox genes also work with temporal genes to control the generation of specific cell types only found in certain segments. For example, NB5-6 generates the Ap cluster neurons only in thoracic segments. Late temporal genes cas and grh, and the thoracic Hox gene Antennapedia (Antp), are required to specify the Ap cluster neurons (Karlsson, Baumgardt, & Thor, 2010). One of the target genes for integration of temporal and spatial information is Collier, which plays important roles in the feedforward loops that specify the Ap cluster neurons as discussed in Section 5. It will be interesting to further characterize the exact molecular mechanism of the integration (Karlsson et al., 2010).
Spatial identity also modulates the progression of the temporal sequence of TFs in NBs. For example, young NB3-3 never expresses Hb, but sequentially expresses Kr, Pdm, Cas, Cas/Grh, and Svp/Grh (Tsuji et al., 2008). The absence of Hb in the sequence is common to both thoracic and abdominal NB3-3, suggesting that this is modulated by the intrasegmental spatial patterning genes common to NB3-3 in each segment. Although the temporal sequence is the same in both thoracic and abdominal NB3-3, the speed of transitions between TFs is faster in abdominal NB3-3s than in thoracic NB3-3s. Particularly, the switch from Cas to Svp occurs in late embryo in abdominal NB3-3, while it occurs in the larval stage for thoracic NB3-3. This spatial difference in the timing of switching to Svp is also true for other NB lineages and is regulated by Hox genes. Loss of Antp or mis-expression of abd-A in thoracic NB3-3 causes precocious Svp expression during embryogenesis (Tsuji et al., 2008). How the Hox genes modulate the speed of the temporal progression and how the spatial–temporal information is integrated to regulate the NB lineage will be interesting questions for the future.
8. INTEGRATION OF TEMPORAL IDENTITY WITH BINARY FATE CHOICE
In embryonic NB lineages, GMCs usually divide asymmetrically to give rise to two progeny with different fates (either two different neuron types, one neuron and one glia, or one neuron and the other undergoing programmed cell death, etc.), and this depends on Notch signaling between the two GMC daughter cells (Buescher et al., 1998; Karcavich & Doe, 2005; Lundell, Lee, Perez, & Chadwell, 2003; Novotny et al., 2002; Schuldt & Brand, 1999; Skeath & Doe, 1998; Spana & Doe, 1996; Udolph, Rath, & Chia, 2001). During the asymmetric division of GMCs, Numb, a repressor of Notch signaling, is asymmetrically localized to one daughter cell, and this cell adopts a Notch-off (N-off ) fate, while the other cell adopts a Notch-on (N-on) fate (Buescher et al., 1998; Spana & Doe, 1996). All the N-on (or N-off ) cells within a lineage are collectively called a hemilineage. In postembryonic NB lineages, Notch/Numb also function in binary-fate choices of GMC progeny (Kumar, Bello, & Reichert, 2009; Li et al., 2013; Lin et al., 2010; Truman, Moats, Altman, Marin, & Williams, 2010).
Notch signaling only differentiates between two alternative fates, but the actual fates depend on the spatial and temporal identity of the NB. For example, Notch has distinct lineage-specific effects in the three antennal lobe NB lineages: antero-dorsal lineage (N-off: PN fates, N-on: apoptosis); ventral lineage (N-off: apoptosis, N-on: PN fates); and lateral lineage (N-off: PN fates, N-on: local interneuron fates) (Lin et al., 2010).
How does the Notch-dependent binary fate choice integrate with the temporal identity of NBs? In the developing medulla, the Notch pathway regulates the maintenance of the temporal TFs in postmitotic neurons and the expression of Ap. In the Ey and Slp NB stages, Ey or Slp is only maintained in the N-off daughter, while the N-on daughter turns on Ap. At the D NB stage, D is only maintained in the N-on daughter together with Ap. Although most if not all N-on daughters of medulla GMCs express Ap, they express different combinations of other TFs and adopt different fates depending on which NB stage they are born from. For example, the N-on progeny of Ey+ GMCs express Ap and Drifter (Dfr), and this is dependent on both Notch signaling and the expression of Ey in NBs. The N-on progeny of Slp+ GMCs express Ap and twin of eyeless (Toy), and similarly, this depends on both Notch signaling and Slp expression in NBs. Thus, the temporal TFs and the Notch pathway together control the expression of downstream TFs like Drifter and Toy to control neuron fates (Li et al., 2013; Fig. 3.2).
Studies in the antennal lobe lateral lineage illustrate another interesting question. In this lineage, PNs and local interneurons are produced as siblings of each GMC division (N-off: PN, N-on: local interneuron). However, as there is more diversity of PNs than of local interneurons, the same local interneuron can be the sibling of different PNs. Thus, it appears that the tempo of birth-order-dependent fate changes is different between the two N-off and N-on hemilineages (Lin et al., 2012). How the same temporal identity information regulates the independent pace of temporal switching of neuronal fates in the two hemilineages is an interesting question for the future.
9. CONCLUSIONS AND FUTURE QUESTIONS
Currently, two different temporal sequences have been identified in NBs of two different systems: the Hb→Kr→Pdm→Cas→Grh sequence in the Drosophila VNC, and the Hth→Klu→Ey→Slp→D→Tll sequence that patterns medulla NBs. In addition, the INPs of type II NB lineages are patterned by a TF sequence D→Grh→Ey. These three examples suggest that TF-sequence-dependent temporal patterning of neural precursors is a common theme to generate neural diversity. This suggests that more temporal sequences will be identified in other systems. For example, in the antennal lobe antero-dorsal lineage, Kr specifies one out of 40 temporal identities in the NB. Further identification of a complete temporal sequence will rely on candidate gene approaches, and/or screening based on either gene expression or mutant phenotypes. Does TF-sequence-dependent temporal patterning of neural precursors also function in vertebrate systems? There is some evidence suggesting that this might be the case. In the vertebrate retina, one ortholog of hb, Ikaros, specifies early-born cell fates (Elliott, Jolicoeur, Ramamurthy, & Cayouette, 2008). In mammalian cortical neurogenesis, Foxg1, an ortholog of Slp, functions in cortical progenitors to suppress early-born cortical cell fates (Hanashima, Li, Shen, Lai, & Fishell, 2004). Although the vertebrate systems are much more complex than Drosophila, studies in flies have provided important concepts that might be applicable to vertebrates.
Cross-regulations between temporal TFs are important for temporal transitions: Feedback negative regulation and feedforward positive regulation among the temporal TFs can facilitate the progression of the sequence. There are many remaining questions to understand the mechanism of temporal transitions. Even in the best-known systems, there can be missing factors or timing mechanisms in addition to the identified TF sequence. When all factors involved and their regulatory relationships are characterized by genetic analysis, theoretical modeling will provide insights into how the genetic network precisely times the NB temporal progression.
Loss of NB competence is related to epigenetic changes, such as chromatin modifications and chromosome architecture. More studies are needed to examine how the expression dynamics of the temporal genes leads to epigenetic changes and thus regulates NB competence, and eventually determines the end of neurogenesis. Such studies are relevant for the development of cell replacement therapies using stem cells to treat various diseases.
Another big challenge is to elucidate how the temporal and spatial identity of NBs, as well as the Notch-dependent binary fate choices, are integrated to generate specific neuron fates.
Finally, how the temporal sequence evolved is a great question for evolutionary developmental neurobiologists. An interesting observation is that the Hb→Kr→Pdm→Cas temporal sequence in VNC NBs mimics the anterior to posterior spatial distribution of Hb→Kr→Pdm→Cas expression domains at cellular blastoderm in the embryo (Isshiki et al., 2001). Whether they use the same regulatory logic is not known. The Hth→ Klu→Ey→Slp→D→Tll or D→Grh→Ey regulatory cascades have not been described in other contexts. Evolutionary studies might provide clues as to how this powerful mechanism has evolved to pattern neural precursors to generate neural diversity.
References
- Akiyama-Oda Y, Hosoya T, Hotta Y. Asymmetric cell division of thoracic neuroblast 6-4 to bifurcate glial and neuronal lineage in Drosophila. Development. 1999;126:1967–1974. doi: 10.1242/dev.126.9.1967. [DOI] [PubMed] [Google Scholar]
- Almeida MS, Bray SJ. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mechanisms of Development. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Baek M, Mann RS. Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. The Journal of Neuroscience. 2009;29:6904–6916. doi: 10.1523/JNEUROSCI.1585-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Baumgardt M, Miguel-Aliaga I, Karlsson D, Ekman H, Thor S. Specification of neuronal identities by feedforward combinatorial coding. PLoS Biology. 2007;5:e37. doi: 10.1371/journal.pbio.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Development. 2010;5:26. doi: 10.1186/1749-8104-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Doe CQ. Temporal patterning in intermediate progenitors increases neural diversity. Nature. 2013;498:449–455. doi: 10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Benito-Sipos J, Estacio-Gomez A, Moris-Sanz M, Baumgardt M, Thor S, Diaz-Benjumea FJ. A genetic cascade involving klumpfuss, nab and castor specifies the abdominal leucokinergic neurons in the Drosophila CNS. Development. 2010;137:3327–3336. doi: 10.1242/dev.052233. [DOI] [PubMed] [Google Scholar]
- Benveniste RJ, Thor S, Thomas JB, Taghert PH. Cell type-specific regulation of the Drosophila FMRF-NH2 neuropeptide gene by Apterous, a LIM homeodomain transcription factor. Development. 1998;125:4757–4765. doi: 10.1242/dev.125.23.4757. [DOI] [PubMed] [Google Scholar]
- Bhat KM. Segment polarity genes in neuroblast formation and identity specification during Drosophila neurogenesis. Bioessays. 1999a;21:472–485. doi: 10.1002/(SICI)1521-1878(199906)21:6<472::AID-BIES4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Bhat KM. Segment polarity genes in neuroblast formation and identity specification during Drosophila neurogenesis. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 1999b;21:472–485. doi: 10.1002/(SICI)1521-1878(199906)21:6<472::AID-BIES4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Developmental Neurobiology. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Developmental Biology. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brand AH, Livesey FJ. Neural stem cell biology in vertebrates and invertebrates: More alike than different? Neuron. 2011;70:719–729. doi: 10.1016/j.neuron.2011.05.016. [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Developmental Biology. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Buescher M, Yeo SL, Udolph G, Zavortink M, Yang X, Tear G, et al. Binary sibling neuronal cell fate decisions in the Drosophila embryonic central nervous system are nonstochastic and require inscuteable-mediated asymmetry of ganglion mother cells. Genes & Development. 1998;12:1858–1870. doi: 10.1101/gad.12.12.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenci C, Gould AP. Drosophila Grainyhead specifies late programmes of neural proliferation by regulating the mitotic activity and Hox-dependent apoptosis of neuroblasts. Development. 2005;132:3835–3845. doi: 10.1242/dev.01932. [DOI] [PubMed] [Google Scholar]
- Cleary MD, Doe CQ. Regulation of neuroblast competence: Multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes & Development. 2006;20:429–434. doi: 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Doe CQ. ming is expressed in neuroblast sublineages and regulates gene expression in the Drosophila central nervous system. Development. 1992;116:943–952. doi: 10.1242/dev.116.4.943. [DOI] [PubMed] [Google Scholar]
- Das A, Reichert H, Rodrigues V. Notch regulates the generation of diverse cell types from the lateral lineage of Drosophila antennal lobe. Journal of Neurogenetics. 2010;24:42–53. doi: 10.3109/01677060903582202. [DOI] [PubMed] [Google Scholar]
- Das A, Sen S, Lichtneckert R, Okada R, Ito K, Rodrigues V, et al. Drosophila olfactory local interneurons and projection neurons derive from a common neuroblast lineage specified by the empty spiracles gene. Neural Development. 2008;3:33. doi: 10.1186/1749-8104-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai AR, McConnell SK. Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development. 2000;127:2863–2872. doi: 10.1242/dev.127.13.2863. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: A sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: Balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Development. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Fischbach KF, Dittrich APM. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell and Tissue Research. 1989;258:441–475. [Google Scholar]
- Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- Green P, Hartenstein AY, Hartenstein V. The embryonic development of the Drosophila visual system. Cell and Tissue Research. 1993;273:583–598. doi: 10.1007/BF00333712. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Developmental Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Grosskortenhaus R, Robinson KJ, Doe CQ. Pdm and Castor specify late-born motor neuron identity in the NB7-1 lineage. Genes & Development. 2006;20:2618–2627. doi: 10.1101/gad.1445306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Li SC, Shen L, Lai E, Fishell G. Foxg1 suppresses early cortical cell fate. Science. 2004;303:56–59. doi: 10.1126/science.1090674. [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Kitada Y, Kaido M, Takayama R, Awasaki T, Tabata T, et al. Concentric zones, cell migration and neuronal circuits in the Drosophila visual center. Development. 2011;138:983–993. doi: 10.1242/dev.058370. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Huang DH, Chang YL, Yang CC, Pan IC, King B. Pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Molecular Cell Biology. 2002;22:6261–6271. doi: 10.1128/MCB.22.17.6261-6271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jacob J, Maurange C, Gould AP. Temporal control of neuronal diversity: Common regulatory principles in insects and vertebrates? Development. 2008;135:3481–3489. doi: 10.1242/dev.016931. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes & Development. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai MI, Okabe M, Hiromi Y. Seven-up controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Developmental Cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Kao CF, Lee T. Birth time/order-dependent neuron type specification. Current Opinion in Neurobiology. 2010;20:14–21. doi: 10.1016/j.conb.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Yu HH, He Y, Kao JC, Lee T. Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron. 2012;73:677–684. doi: 10.1016/j.neuron.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcavich RE. Generating neuronal diversity in the Drosophila central nervous system: A view from the ganglion mother cells. Developmental Dynamics. 2005;232:609–616. doi: 10.1002/dvdy.20273. [DOI] [PubMed] [Google Scholar]
- Karcavich R, Doe CQ. Drosophila neuroblast 7-3 cell lineage: A model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. The Journal of Comparative Neurology. 2005;481:240–251. doi: 10.1002/cne.20371. [DOI] [PubMed] [Google Scholar]
- Karlsson D, Baumgardt M, Thor S. Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues. PLoS Biology. 2010;8:e1000368. doi: 10.1371/journal.pbio.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Asymmetric cell division: Recent developments and their implications for tumour biology. Nature Reviews Molecular Cell Biology. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Hiebert LS, Doe CQ. The pipsqueak-domain proteins Distal antenna and Distal antenna-related restrict Hunchback neuroblast expression and early-born neuronal identity. Development. 2011;138:1727–1735. doi: 10.1242/dev.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ. Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell. 2013;152:97–108. doi: 10.1016/j.cell.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bello B, Reichert H. Lineage-specific cell death in postembryonic brain development of Drosophila. Development. 2009;136:3433–3442. doi: 10.1242/dev.037226. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Maruyama Y, Adachi Y, Okabe M, Suzuki E, Furukubo-Tokunaga K. A conserved nuclear receptor, Tailless, is required for efficient proliferation and prolonged maintenance of mushroom body progenitors in the Drosophila brain. Developmental Biology. 2009;326:224–236. doi: 10.1016/j.ydbio.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: Diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: Sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, et al. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013;498:456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Kao CF, Yu HH, Huang Y, Lee T. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS Biology. 2012;10:e1001425. doi: 10.1371/journal.pbio.1001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lai SL, Yu HH, Chihara T, Luo L, Lee T. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2010;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lee T. Generating neuronal diversity in the Drosophila central nervous system. Developmental Dynamics. 2012;241:57–68. doi: 10.1002/dvdy.22739. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: Lessons from the retina. Nature Reviews Neuroscience. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J. Eagle is required for the specification of serotonin neurons and other neuroblast 7-3 progeny in the Drosophila CNS. Development. 1998;125:463–472. doi: 10.1242/dev.125.3.463. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Lee HK, Perez E, Chadwell L. The regulation of apoptosis by Numb/Notch signaling in the serotonin lineage of Drosophila. Development. 2003;130:4109–4121. doi: 10.1242/dev.00593. [DOI] [PubMed] [Google Scholar]
- Maurange C. Temporal specification of neural stem cells: Insights from Drosophila neuroblasts. Current Topics in Developmental Biology. 2012;98:199–228. doi: 10.1016/B978-0-12-386499-4.00008-2. [DOI] [PubMed] [Google Scholar]
- Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Mellerick DM, Kassis JA, Zhang SD, Odenwald WF. Castor encodes a novel zinc finger protein required for the development of a subset of CNS neurons in Drosophila. Neuron. 1992;9:789–803. doi: 10.1016/0896-6273(92)90234-5. [DOI] [PubMed] [Google Scholar]
- Mettler U, Vogler G, Urban J. Timing of identity: Spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development. 2006;133:429–437. doi: 10.1242/dev.02229. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature Reviews Neuroscience. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Current Biology. 2008;18:553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morante J, Erclik T, Desplan C. Cell migration in Drosophila optic lobe neurons is controlled by eyeless/Pax6. Development. 2011;138:687–693. doi: 10.1242/dev.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nature Neuroscience. 2008;11:1014–1023. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- Nakajima A, Isshiki T, Kaneko K, Ishihara S. Robustness under functional constraint: The genetic network for temporal expression in Drosophila neurogenesis. PLoS Computational Biology. 2010;6:e1000760. doi: 10.1371/journal.pcbi.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- Okano H, Temple S. Cell types to order: Temporal specification of CNS stem cells. Current Opinion in Neurobiology. 2009;19:112–119. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Park D, Han M, Kim YC, Han KA, Taghert PH. Ap-let neurons –A peptidergic circuit potentially controlling ecdysial behavior in Drosophila. Developmental Biology. 2004;269:95–108. doi: 10.1016/j.ydbio.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annual Review of Cell and Developmental Biology. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Peterson C, Carney GE, Taylor BJ, White K. Reaper is required for neuroblast apoptosis during Drosophila development. Development. 2002;129:1467–1476. doi: 10.1242/dev.129.6.1467. [DOI] [PubMed] [Google Scholar]
- Prokop A, Technau GM. The origin of postembryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster. Development. 1991;111:79–88. doi: 10.1242/dev.111.1.79. [DOI] [PubMed] [Google Scholar]
- Romani S, Jimenez F, Hoch M, Patel NH, Taubert H, Jackle H. Kruppel, a Drosophila segmentation gene, participates in the specification of neurons and glial cells. Mechanisms of Development. 1996;60:95–107. doi: 10.1016/s0925-4773(96)00603-x. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: Neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Developmental Biology. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Schuldt AJ, Brand AH. Mastermind acts downstream of notch to specify neuronal cell fates in the Drosophila central nervous system. Developmental Biology. 1999;205:287–295. doi: 10.1006/dbio.1998.9014. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, et al. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nature Neuroscience. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Doe CQ. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S. Genetic control of Drosophila nerve cord development. Current Opinion in Neurobiology. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kaido M, Takayama R, Sato M. A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Developmental Biology. 2013;380:12–24. doi: 10.1016/j.ydbio.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Technau GM, Berger C, Urbach R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Developmental Dynamics. 2006;235:861–869. doi: 10.1002/dvdy.20566. [DOI] [PubMed] [Google Scholar]
- Touma JJ, Weckerle FF, Cleary MD. Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons. Development. 2012;139:657–666. doi: 10.1242/dev.071589. [DOI] [PubMed] [Google Scholar]
- Tran KD, Doe CQ. Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development. 2008;135:3491–3499. doi: 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KD, Miller MR, Doe CQ. Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development. 2010;137:1421–1430. doi: 10.1242/dev.048678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Developmental Biology. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Hasegawa E, Isshiki T. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development. 2008;135:3859–3869. doi: 10.1242/dev.025189. [DOI] [PubMed] [Google Scholar]
- Udolph G, Rath P, Chia W. A requirement for Notch in the genesis of a subset of glial cells in the Drosophila embryonic central nervous system which arise through asymmetric divisions. Development. 2001;128:1457–1466. doi: 10.1242/dev.128.8.1457. [DOI] [PubMed] [Google Scholar]
- Viktorin G, Riebli N, Popkova A, Giangrande A, Reichert H. Multipotent neural stem cells generate glial cells of the central complex through transit amplifying intermediate progenitors in Drosophila brain development. Developmental Biology. 2011;356:553–565. doi: 10.1016/j.ydbio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Weng M, Golden KL, Lee CY. dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Developmental Cell. 2010;18:126–135. doi: 10.1016/j.devcel.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Kankel DR. Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. Developmental Biology. 1978;65:296–321. doi: 10.1016/0012-1606(78)90029-5. [DOI] [PubMed] [Google Scholar]
- Wu YC, Chen CH, Mercer A, Sokol NS. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Developmental Cell. 2012;23:202–209. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yeo S, Dick T, Chia W. The role of a Drosophila POU homeo domain gene in the specification of neural precursor cell identity in the developing embryonic central nervous system. Genes & Development. 1993;7:504–516. doi: 10.1101/gad.7.3.504. [DOI] [PubMed] [Google Scholar]
- Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–1480. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nature Neuroscience. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Kao CF, He Y, Ding P, Kao JC, Lee T. A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS Biology. 2010;8:e1000461. doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Kuo CT, Jan YN. Drosophila neuroblast asymmetric cell division: Recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Zhu S, Barshow S, Wildonger J, Jan LY, Jan YN. Ets transcription factor Pointed promotes the generation of intermediate neural progenitors in Drosophila larval brains. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20615–20620. doi: 10.1073/pnas.1118595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]


