Abstract
The amyloid β (Aβ) peptide associated with Alzheimer’s Disease results from processing of the amyloid precursor protein (APP) by secretases. Following cleavage of APP by β-secretase, a 99 amino acid C-terminal fragment of APP (C99) is produced consisting of a single transmembrane (TM) helix. Simulation studies of C99 congeners and structural studies of C99 in surfactant micelles and lipid vesicles have shown that a key peptide structural motif is a prominent “GG kink,” centered at two glycines and dividing the TM helix. The flexibility of the GG kink is important in the processing of C99 by γ-secretase. We performed multiscale simulations of C9915–55 in a DPC surfactant micelle and POPC lipid bilayer in order to elucidate the role of membrane surface curvature in modulating the peptide structure. C9915–55 in a DPC surfactant micelle possesses a “GG kink” in the TM domain near the dynamic hinge located at G37/G38, while such a kink is not observed in C9915–55 in a POPC lipid bilayer. Intramolecular interaction between the extracellular and TM domains of C9915–55 is enhanced in the micelle environment, influencing helical stability, TM helix extension, exposure to water, and depth of insertion in the lipophilic region. Our results point to critical differences in C9915–55 structures in micelle and bilayer environments, and show that the fluctuations of the structural ensemble of APP are strongly influenced by membrane surface curvature.
There has been a great deal of interest in understanding the structure and kinetics of aggregation of Amyloid β (Aβ) peptides associated with Alzheimer’s Disease (AD).1,2 In contrast, much less attention has been paid to the production of Aβ peptide, which occurs by proteolytic cleavage of the membrane associated APP-C99 (C99), a ninty-nine amino acid C-terminal fragment of the Amyloid Precursor Protein (APP), by γ-secretase. 3-5
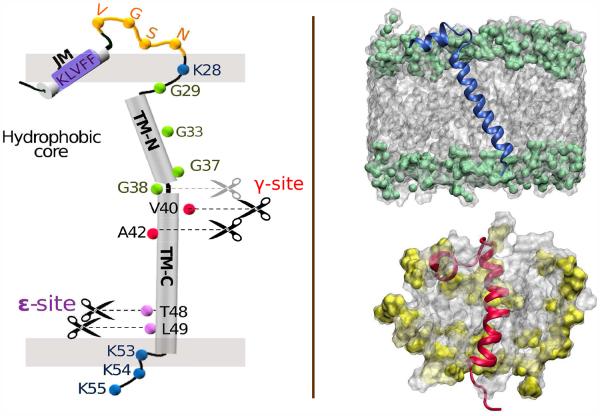
C99 consists (see Fig. 1) of a central TM helical domain (K28-K53/K699-K724 for C99/APP) flanked by an extracellular N-terminal region, including asparagine glycosylation sites and a juxtamembrane (JM) helix (Q15-V24/Q686-V695), and an intracellular domain, including a C-terminal helix (T90-N99/T761-N770).3-6 Cleavage by γ-secretase is initiated at the ∊-site (T49/L49) and precedes processively until termination. The point of termination varies leading to Aβ lengths of 38 to 43 amino acids. 7 Aβ40, the dominant isoform, 8,9 and Aβ42, considered the most amyloidogenic isoform, normally occur in a 10:1 ratio.10,11 How changes in C99 sequence and variations in membrane environment impact the distribution of Aβ isoforms is critical to our understanding of the etiology of AD.12,13
Figure 1.
(Left) Schematic of C99 showing key sequence information, likely secondary structure regions, ∊- and γ- cleavage sites, and approximate insertion within the membrane bilayer. The break in the TM helix at the “GG kink” between G37/G38 is indicated. (Right) Depiction of C9915–55 monomer in a POPC lipid bilayer (above) showing an average tilt angle of 22.5 (deg) with respect to the bilayer normal and (below) C9915–55 in a DPC micelle. The phosphocholine group is shaded green (POPC) or yellow (DPC).
We had previously predicted 14 that the TM helix was rendered flexible through the presence of a dynamic “GG hinge” at G37/G38 using simulations of monomeric C991–55 in a model bilayer. The helix preceding G37 (referred to as Domain B) was found to be less helical than the helix following G38 (referred to as Domain C). We also proposed that the reduced helicity in Domain B facilitated close interpeptide backbone association and Cα hydrogen bonding stabilizing the homodimer. Subsequent experiments quantitatively confirmed these two key simulation predictions. 6
Based on structural studies of C99 in micelles 6,15 it has been conjectured that the TM helix flexibility, due to the presence of a dynamic GG hinge located at G37/38 near the center of the TM helix (see Fig. 1), may facilitate interaction of C99 with the active site of γ-secretase. 16
Although a structural kink, i.e a non dynamic bend in the structure, might be considered a constraint on the passage of C99 toward the active site, possibly signaling termination of cleavage, the presence of a dynamic hinge is expected to facilitate passage of the peptide during processive cleavage. This finding is in accord with our predictions, 14 although it is likely that the extent of the GG kink will depend on the membrane environment.
A recent study involving H/D exchange experiments on the C99 peptide complemented by molecular dynamics simulations of C9928–55 in a POPC bilayer, has provided further insights into the stability of helical regions of C99 including the TM helix. Considering the flexible GG hinge as dividing the TM helix, the N-terminal region of the TM domain (TM-N helix, Domain B) showed enhanced H/D exchange relative to the C-terminal portion (TM-C helix, Domain C). 17 The finding is in agreement with experimental studies of C99 monomer in LMPG micelles. 6
It should be noted that our simulations 14 and those of Pester and coworkers 17 were performed in bilayer environments, whereas the insightful NMR experiments used C99 in spatially constrained micelles. 6 The difference raises a crucial question, namely, how does the membrane, especially the surface curvature, effect the conformational fluctuations of C99? The flexibility of the GG kink likely determines the distribution of isoforms of Aβ peptide upon cleavage by γ-secretase. Thus, it is important to quantitatively determine the structural fluctuations of C99 in general and the GG hinge in particular as the membrane characteristics are varied.
To answer this open question quantitatively, we performed simulations of the structure and stability of monomeric C9915–55 in POPC lipid bilayer and DPC surfactant micelle environments. We show that although micelles are an important model system for probing the structure of C99, the extent of fluctuations in C99, proposed to be crucial in the Aβ product distribution upon cleavage by secretases, depends strongly on membrane surface curvature. Our results provide a detailed picture of the C9915–55 structural ensemble and the potential role for changes in structure to influence the function and processing of this critical amyloid precursor protein.
The structural ensemble of C9915–55 was characterized in POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) lipid bilayer and DPC (dodecylphosphocholine) surfactant micelle environments. To ensure that our conclusions are robust, a multiscale computational approach was employed combining μs time scale coarse-grained (CG) models of the protein, lipids, and solvent using the MARTINI force field 18,19 and 100 ns time scale all-atom CHARMM36 force field models for the protein, membrane, and solvent environments(see Supplemental Information for details). 20,21 The CG simulations were used to assess the long-time dynamics and the role of fluctuations in the protein and lipid conformational ensemble22-25. All atom simulations provided an atomically detailed picture of the protein structure, protein-membrane interactions, the lipid/water interface, and water dynamics allowing for a detailed comparison with experimental data for C99 in LMPG (lysomyristoylphosphatidylglycerol) micelles at pH 6.5 and 45C (see Fig. 1).
Insertion depth
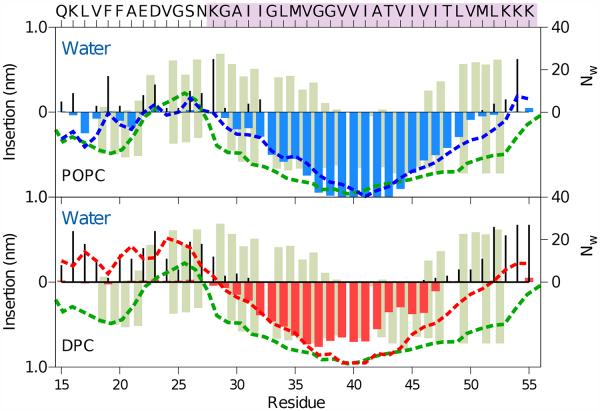
To characterize the relative degree of insertion of C9915–55 in the bilayer and micelle, the per residue accessibility of the peptides to water and lipids was computed in terms of the minimum distance of encounter between a given Cα atom to any POPC lipid or DPC surfactant at a given time. The average over time provides a measure of the depth of insertion of the Cα into the water or lipid region (see Fig. 2). Comparison of the simulation results with experiments of Sanders and coworkers, derived from NMR experiments with water-soluble and lipophilic paramagnetic probes,15 shows good agreement.
Figure 2.
Water and lipid accessibility of each residue of C9915–55 in POPC bilayer (top) and a DPC micelle (bottom) derived from all-atom simulations. The green bars depict the degree of contact with the water-soluble paramagnetic probe (Gd-DTPA, positive values) and the lipophilic probe (16-DSA, negative values) reported by Sanders and coworkers for C99 in LMPG micelles.15 The EPR power saturation data from experimental measurements6 is presented (green dashed line). Shown for comparison are simulation results for the depth of insertion in the lipid phase (colored bar), number of water molecules, Nw, (black lines) within 4 Å of each amino acid, and the insertion depth calculated by taking the location of the membrane width or the micelle size for POPC (blue dashed line) and DPC (red dashed line). The TM sequence is marked with orchid shading.
Our study was designed to compare the impact of geometry and membrane surface curvature on peptide structure, using a POPC lipid and DPC surfactant that share similar zwitterionic head groups and alkyl chain lengths. Because the experimental results were obtained for C99 in LMPG micelles, 6 which have an anionic (rather than zwitterionic) head group and longer (by 2 carbons) alkyl chain length, a direct comparison with our predictions cannot be made.
In particular, the simulation results for C9915–55 in a DPC micelle captures the essential features of the experimental measurements for full length C99 in an LMPG micelle (see Fig. 2). Differences primarily occur near the N- and C-terminal regions of C9915–55 (mid-sequence in full-length C99) and for residues localized near the head group region (chemically distinct in neural DPC and anionic LMPG). Comparison of the results between the bilayer and micelle (Fig. 2, top and bottom) shows the quantitative difference, which is the first indication that the surface curvature of the membrane affects the conformational ensemble of C9915–55. We observe remarkable agreement in JM helix insertion and peptide solvation between the all-atom and CG simulations in POPC bilayer (Fig. 2 and S2). Having established that our simulations represent the water and lipid accessibility in a micellar environment, we further examined the role of membrane surface curvature on other structural features. In simulations, residues at the interface in the DPC micelle present higher accessibility to water than the same residues in the POPC bilayer. Our simulations in a POPC bilayer show that the JM domain of C9915–55 (residues Q15-N27) is localized at the membrane interface, which allows the central hydrophobic residues V17FFA20 to insert into the head group region. In contrast, in the DPC micelle the residues of the JM domain are significantly more accessible to water. In both the bilayer and micelle environments, K28 is localized near the interface. However, in the DPC micelle the C-terminal amino acids L49VMLKKK55 are significantly more exposed to water than in the POPC bilayer.
Residue-dependent helix probability
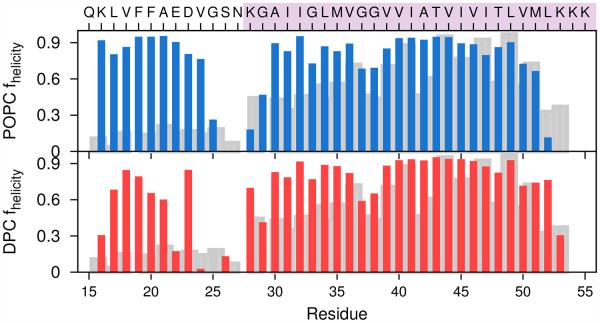
Fig. 3 relates fluctuations in the TM and JM helical regions for C9915–55 in the POPC bilayer (upper) and DPC micelle (lower) derived from simulation and compared with measures of helicity from experimental studies of C99 in an LMPG micelle at pH 6.5 and 45C. 15 In both the bilayer and the micelle environments, two helical regions are observed with the interhelical region located at V23GSN27. This region is often observed to form bends or turns in Aβ fibrils and monomers.26 However, there are also significant differences. In the POPC bilayer the integrity of the JM helix is somewhat greater than in the DPC micelle. Moreover, there is a critical break in the TM helix observed in both simulation and experiment in the micelle environment near G37/G38 that is largely absent for the peptide in the POPC bilayer. This break in the TM helix has previously been identified as a structural GG kink that might be critical in the processing of C99 by γ-secretase. 15 K28 in POPC bilayer and DPC micelle serves as an anchor residue interacting with the lipid headgroups and superficial waters. Simulations using implicit solvent models had suggested that K28 can be involved in critical salt bridges with E22 and D2314 whose stability might be dependent on the headgroup composition. 27,28
Figure 3.
α-helicity of each residue in the C9915–55 monomer simulation in DPC micelle (red) and POPC bilayer (blue) derived from all-atom simulations. The α-helical residues were assigned with DSSP. Shown in grey is the degree of helicity determined experimentally from Cα chemical shifts for the C99 peptide in LMPG micelles.15
Fluctuations in the GG hinge
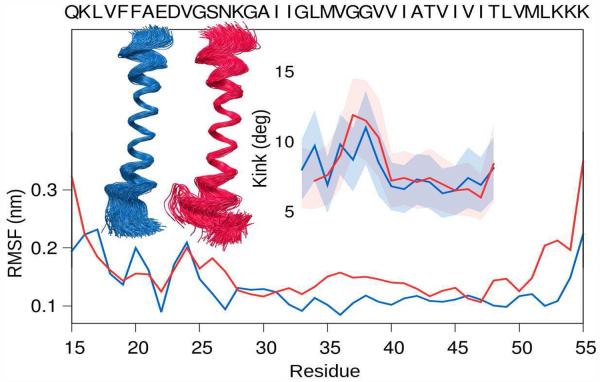
Our results (See Fig. 4) establish that a structural kink is prominent in the peptide in a micelle environment, but less pronounced in the bilayer, which may approximate the biological membrane more closely. Our results suggest that differences in the packing of lipid tail groups in the bilayer and micelle environments, along with substantial surface curvature of the surfactant/solvent interface of the micelle, introduces strain leading to a structural GG kink in the TM helix. In the bilayer there is greater integrity of the JM helix, less interhelical interaction, and a more extended TM helix. In contrast, in the micelle environment there are more substantial fluctuations in the V23GSN27 region of C9915–55 (see Fig. 4).
Figure 4.
Measurement of the (inset) structural kink derived from all-atom simulations in the peptide and (lower) root-mean-square fluctuations in Cα atoms as a function of residue for the C9915–55 monomer in POPC (blue) and DPC (red). Shaded regions indicate ± one standard deviation in uncertainty.
An experimental assessment of the distance between the end residues of the TM helix was performed using spin labels at G29 (G700) and L52 (L723) and a pulsed EPR double electron-electron resonance measurement.6 Average G29-L52 distances derived from simulation are 34.3 Å in the micelle and 34.5 Å in the bilayer (see Fig. 5). The experimentally determined average G29-L52 distances of 33.5 ± 1.0 Å for the WT peptide and 35.3 ± 0.5 Å for the G37L/G38L double mutant peptide (in which the structural kink is expected to be diminished) in 1:4 POPG:POPC lipid vesicles compare well with our simulated results for the POPC bilayer. This is particularly true as it is known that addition of POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol) to POPC increases the thickness of the lipid vesicle wall.
Figure 5.
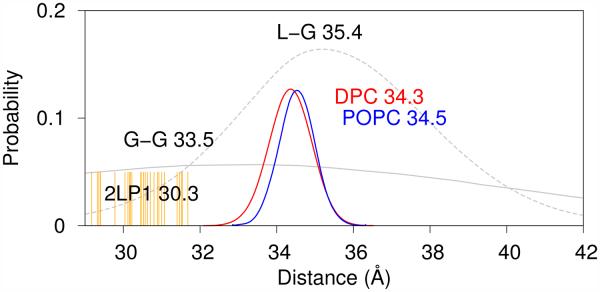
Distribution of distances between the backbone N atoms of residues G29 and L52 derived from all-atom simulations of C9915–55 in a DPC micelle and POPC bilayer, compared with experimental results derived from EPR studies (dotted line) of spin-labeled C99 in 1:4 POPG:POPC lipid vesicles.6 The G29-L52 distance derived from the deposited PDB structures6 (orange lines) are found to be significantly shorter than the average distance derived from the EPR data and simulations of C9915–55 in DPC micelle and POPC bilayer.
The degree of GG hinge motion observed in the simulation, while not consistent with the presence of a structural kink in the TM helix, accords well with experimental observations. Based on our simulation results, G29-L52 distances in the range of 33-34 Å are inconsistent with the presence of a structural kink in the TM helix near G37/G38. Consistent with this view, the G29-L52 distances derived from the deposited PDB structures (orange lines in Fig. 5) lead to a significantly shorter average distance. Interestingly, the distances reported for the WT peptide in 1:4 POPG:POPC lipid vesicles 6 are consistent with a more modest bend near a hinge located at G37/G3814 and in agreement with our simulations results for the POPC bilayer. Due to the substantial size of the lipid vesicles, the surface curvature is reduced relative to micelles, allowing for a more direct comparison between the vesicle and bilayer results. The uncertainty in the experimental model leads to a broad distribution of distances. However, the center of the distribution is in agreement with simulation results for the POPC bilayer. Results from simulations of C9915–55 in POPC bilayers and DPC micelles without the CMAP correction (see Fig. S1) are consistent with these results.
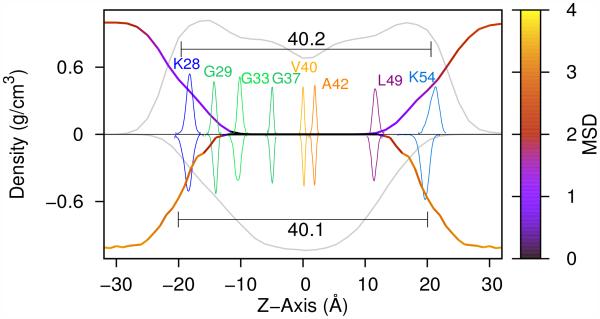
Profiles of the lipid density in the POPC bilayer and DPC micelle are presented in Fig. 6 alongside the related distributions of key residues in the C9915–55 peptide. In our simulations, the average lipid phase in the POPC bilayer is 40.2 Å and the widest micelle diameter in the DPC micelle is 40.1 Å (see Methods in Supplemental Information). While the DPC micelle and POPC bilayer have a small difference in the hydrophobic width, the most significant difference comes from the curvature of the interface shape, which is greater for the micelle. The substantial concavity of the interface facilitates interaction between the JM domain and the TM helix (see Fig. S3, Supplemental Information). Correspondingly, measurable differences are observed in the depths of insertion of particular residues of C9915–55 relative to the solvent interface in the bilayer and micelle environments. As shown in Fig. 6 (MSD), waters closer to the POPC bilayer interface show smaller MSD values compared to those close to the DPC micelle interface. Given these combined results, it appears that restrained waters at the POPC interface (1) reduce the lipid/solvent interfacial fluctuations and (2) localize and stabilize the C9915–55 JM helix relative to the more dynamic surfactant/solvent interface of the DPC micelle.
Figure 6.
The mass density distribution (grey lines) of the lipid phases for the POPC bilayer (top) and DPC micelle (below) derived from all-atom simulations. The water density distribution is colored by the water mean square fluctuation (MSD) as a function of the bilayer normal for POPC and as a function of the distance to the center of the DPC micelle. Superimposed are distributions of Cα positions of key residues along the z-axis for C9915–55 in a POPC bilayer and DPC micelle.
γ-secretase initiates cleavage near the ∊-site (T48/L49) and typically terminates at the γ-site (V40/I41) to create Aβ40. However, changes in sequence and membrane composition can lead to variations in the termination point of cleavage leading to changes in the lengths of Aβ peptides liberated. That said, the mechanisms by which these changes occur, and, in particular, how changes in sequence and lipid environment inform structural changes in the TM helical region of C99, are less well understood.
Experimental and computational investigations of the structure of the TM helix in monomeric C99 congeners have provided insight into the structure of the peptide in a micelle environment, including its interactions with lipids, cholesterol, and water. 6,15,17 Those studies suggest the existence of a hinge at G37/G38 of the TM helix near the γ-site, observed in simulation studies. It has been proposed that the presence of a flexible GG hinge in the helix plays a significant role in the processive processing of C99 by γ-secretase. 14
We have discovered, using simulations of C9915–55 monomer in a DPC micelle and POPC bilayer, that the dynamic GG hinge leads to a structural kink in the TM helix. Fluctuations of the TM helix are observed to be significantly greater in the spatially constrained DPC micelle than in the POPC bilayer, facilitating enhanced interactions between the JM helical region and the TM helix. This in turn influences helical stability, TM helix extension, exposure to water, and depth of insertion in the lipophilic region. Our results underscore potential differences between the DPC micelle and the POPC bilayer, with the latter more accurately representing a biological membrane. Our simulations suggest that interfacial constraints of the micelle environment place strain on the TM helix and allow for its full extension only with some cost in free energy. The finding that the TM helix under strain forms the structural GG kink at the position of the dynamic hinge near G37/G38 confirms that the extent of fluctuations in the GG kink in the TM helix is controlled by the membrane curvature. 14,17
The inherent flexibility in the TM domain may ease its homodimerization and along with the position of a charged residue at K53 may facilitate the positioning of the peptide’s ∊-site near the active site of presinillin, the aspartyl protease that forms the active site of γ-secretase, during the initiation of processive cleavage. Flexibility of the TM domain may also facilitate translocation of the peptide during processive cleavage. Moreover, it is likely that termination of cleavage by γ-secretase is determined by the location of charged amino acids flanking the TM-N domain, particularly K28 in WT C99.7
Currently, relatively little is known regarding the sequence dependence of initiation and termination of processive cleavage by proteases such as γ-secretase. 29 It remains a challenge to define how peptide sequence and dynamics determine the termination of cleavage and the resultant isoform distribution of Aβ, so critical to the onset and evolution of AD. Our work forms the basis for understanding the link between structural fluctuations of C99 in a membrane and the resultant Aβ product distribution following cleavage by secretases. Future study in such areas could have an impact on our fundamental understanding of how these process affect the evolution of AD.
Supplementary Material
Acknowledgement
The authors gratefully acknowledge the support of the National Science Foundation (CHE-1114676 and CHE-0910433) and the National Institutes of Health (RO1 GM076688). JES and LD thank the Schlumberger Foundation “Faculty for the Future Program” and CONACYT for the generous support of our research. We are also thankful for the resources of the Center for Computational Science at Boston University. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number OCI-1053575.
References
- 1.Selkoe DJ. J. Neuropath. 1991;53:438–447. doi: 10.1097/00005072-199409000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Kang J, Lemaire H, Unterbeck A, Salbaum J, Masters C, Grzeschik K, Multhaup G, Beyreuther K, Muller-Hill B. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 4.Nunan J, Small DH. FEBS Lett. 2000;483:6–10. doi: 10.1016/s0014-5793(00)02076-7. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe DJ. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 6.Barrett PJ, Song Y, van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czirr E, Cottrell BA, Leuchtenberger S, Kukar T, TB TBL, Esselmann H, Paul S, Schubenel R, Torpey JW, Pietrzik CU, Golde TE, Wiltfang J, Baumann K, Koo EH, Weggen S. J. Biol. Chem. 2008;283:17049–17054. doi: 10.1074/jbc.M802912200. [DOI] [PubMed] [Google Scholar]
- 8.Haass C, Schlossmacher M, Hung A, Vigo-Pelfrey C, Mellon A, Ostaszewski B, Lieberburg I, Koo E, Schenk D, Teplow D. Nature. 1992;359:322–325. doi: 10.1038/359322a0. [DOI] [PubMed] [Google Scholar]
- 9.Seubert P, Vigo-Pelfrey C, Esch F, Lee M, Dovey H, Davis D, Sinha S, Schlossmacher M, Whaley J, Swindlehurst C. Nature. 1992;359:325–357. doi: 10.1038/359325a0. [DOI] [PubMed] [Google Scholar]
- 10.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Proc. Natl. Acad. Sci. USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara Y. Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 12.Das C, Berezovska O, Diehl TS, Genet C, Buldyrev I, Tsai J-Y, Hyman BT, Wolfe M. J. Am. Chem. Soc. 2003;125:11794–11795. doi: 10.1021/ja037131v. [DOI] [PubMed] [Google Scholar]
- 13.Wolfe M, Guénette SY. J. Cell. Sci. 2007;120:3157–3161. doi: 10.1242/jcs.03481. [DOI] [PubMed] [Google Scholar]
- 14.Miyashita N, Straub JE, Thirumalai D. J. Am. Chem. Soc. 2009;131:17843–17852. doi: 10.1021/ja905457d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beel AJ, Mobley CK, Kim HJ, Tian F, Hadziselimovic A, Jap B, Prestegard JH, Sanders CR. Biochem. 2008;47:9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyashita N, Straub JE, Thirumalai D, Sugita Y. J. Am. Chem. Soc. 2009;131:3438–3439. doi: 10.1021/ja809227c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pester O, Barrett PJ, Hornburg D, Hornburg P, Pröobstle R, Widmaier S, Kutzner C, Dürrbaum M, Kapuirniotu A, Sanders CR, Scharnagl C, Langosch D. J. Am. Chem. Soc. 2013;135:1317–11329. doi: 10.1021/ja3112093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 19.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S-J. J. Chem. Theor. Compt. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 20.Klauda JB, Venable RM, Freites JA, O’Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Jr, Pastor RW. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J, MacKerell AD. Journal of Computational Chemistry. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayton GS, Voth GA. Curr. Opin. Struct. Biol. 2009;19:138–144. doi: 10.1016/j.sbi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyubartsev AP, Rabinovich AL. Soft Matter. 2011;7:25–39. [Google Scholar]
- 24.Marrink SJ, de Vries AH, Tieleman DP. Biochim, Biophys. Acta - Biomembranes. 2009;1788:149–168. doi: 10.1016/j.bbamem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Scott HL. Curr. Opin. Struct. Biol. 2002;12:495–502. doi: 10.1016/s0959-440x(02)00353-6. [DOI] [PubMed] [Google Scholar]
- 26.Straub JE, Thirumalai D. Ann. Rev. Phys. Chem. 2011;62:437–463. doi: 10.1146/annurev-physchem-032210-103526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tofoleanu F, Buchete N-V. Journal of Molecular Biology. 2012;421:572–586. doi: 10.1016/j.jmb.2011.12.063. [DOI] [PubMed] [Google Scholar]
- 28.Poojari C, Kukol A, Strodel B. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2013;1828:327–339. doi: 10.1016/j.bbamem.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Licht S, Lee I. Biochem. 2008;47:3595–3605. doi: 10.1021/bi800025g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.