Abstract
Early neurogenesis comprises the phase of nervous system development during which neural progenitor cells are born. In early development, the embryonic ectoderm is subdivided by a conserved signaling mechanism into two main domains, the epidermal ectoderm and the neurectoderm. Subsequently, cells of the neurectoderm are internalized and form a cell layer of proliferating neural progenitors. In vertebrates, the entire neurectoderm folds into the embryo to give rise to the neural tube. In Drosophila and many other invertebrates, a subset of neurectodermal cells, called neuroblasts (NBs), delaminates and forms the neural primordium inside the embryo where they divide in an asymmetric, stem cell-like mode. The remainder of the neuroectodermal cells that stay behind at the surface loose their neurogenic potential and later give rise to the ventral part of the epidermis. The genetic and molecular analysis of the mechanisms controlling specification and proliferation of NBs in the Drosophila embryo, which played a significant part in pioneering the field of modern developmental neurobiology, represents the topic of this review.
NBs with their stem cell-like proliferative characteristics were recognized by histological methods for a number of insect species more than 100 years ago1 (Fig. 1A). An important step forward was the reconstruction of the pattern of NBs in the grasshopper ventral nerve cord2 (Fig. 1A). This study showed that NBs are uniquely identifiable cells. In each half-segment (hemi-neuromere) of the grasshopper embryo, NBs form a regular array of four columns and seven rows. Subsequent studies in Drosophila3, 4 and other insects showed very similar patterns. With the advent of molecular probes and antibodies, it became clear that each NB expresses a unique combination of transcriptional regulators4, 5. The genetic analysis of early neurogenesis was initiated in the 1930s by Poulson who noted that mutations in the Notch (N) gene resulted in a massive overproduction of NBs6. More than 40 years later, systematic genetic screens for mutations7–10, combined with improved methods to study embryos11 and to analyze gene function molecularly, resulted in the definition of several groups of genes which (1) made clusters of neurectodermal cells competent to generate NBs (“proneural genes”, expressed in “proneural clusters”); (2) determined the spacing of NBs by lateral inhibition (“neurogenic genes”); (3) controlled the expression pattern of proneural genes and thereby of NBs (“prepatterning genes”); (4) controlled the asymmetric division of the delaminated NBs (Inscuteable complex, Par complex).
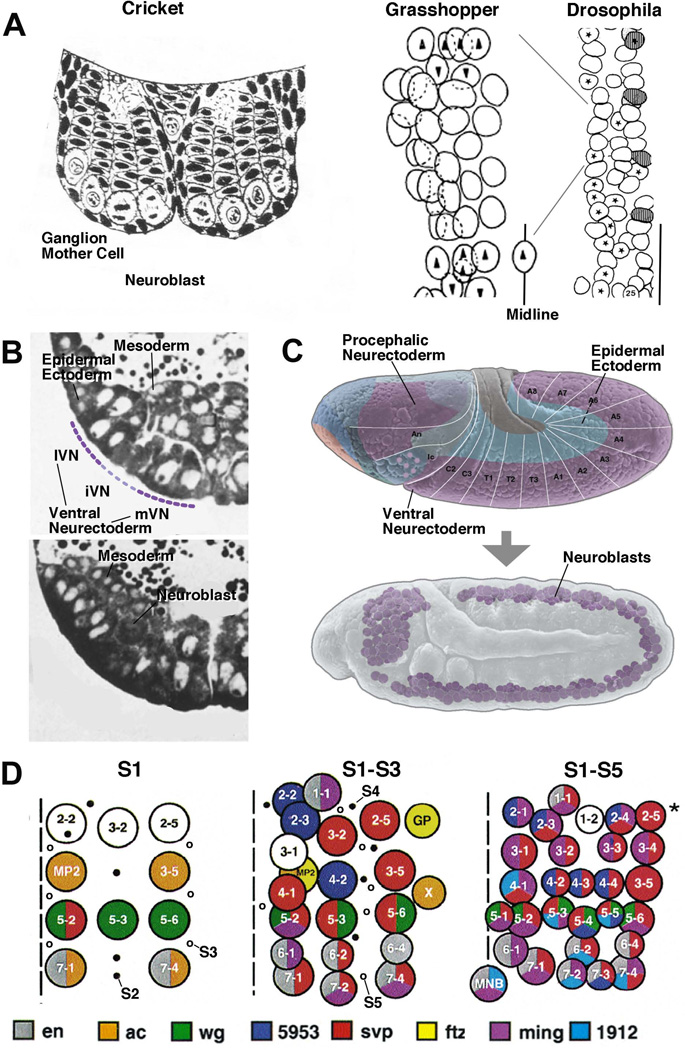
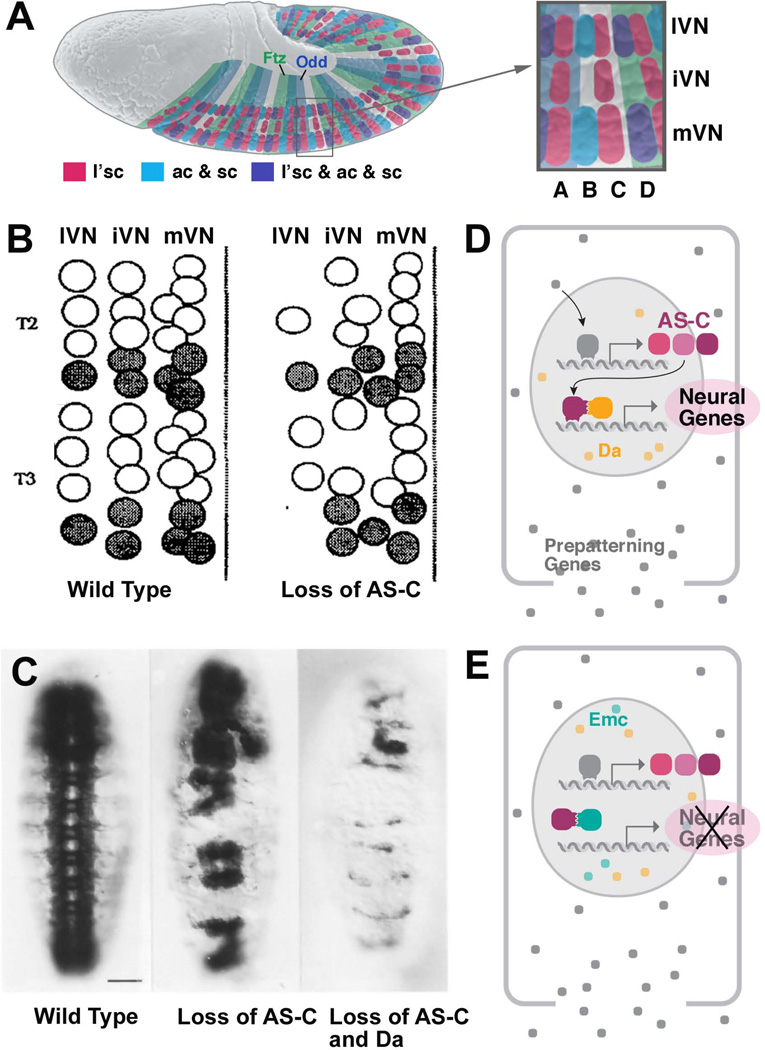
Figure 1. Development and pattern of neuroblasts.
(A) On the left is a schematic drawing of a cross section of the embryonic nervous system of the cricket. This is one of the first depictions of neural lineages, consisting of NBs and stacks of GMCs and neurons (from1). In the center is a drawing by M. Bate2, depicting the full set of NBs in one hemisegment of the grasshopper embryo. The drawing on the right (from3) shows the NB pattern of several hemisegments of the Drosophila embryo, drawn to the same scale. Only S1/S2 NBs, forming four rows and three columns, have formed at the stage depicted. (B) Histological cross sections of the Drosophila embryo prior to (upper panel) and after (lower panel) NB delamination. Only left ventral quadrant of the embryo is shown. The ventral neurectoderm can be distinguished from the dorsal ectoderm by its tall cylindrical cells. Prior to NB delamination, a division in medial column, intermediate column, and lateral column (mVN, iVN, lVN) is evident. (C) Lateral view of Drosophila embryo prior to (upper panel) and after (lower panel) NB delamination. In this and all other figures, anterior is to the left, dorsal is up. Neurectoderm and dorsal ectoderm are shaded in purple, and blue, respectively; white lines and letters indicate segments. (D) NB map of one abdominal hemisegment (from4). Left: S1 NBs; locations where S2 and S3 will appear are indicated by filled and open circles, respectively. Center: S1-S3 NBs; location where S4 and S5 NBs will appear indicated by filled and open circles, respectively. Right: All NBs have delaminated. Midline is represented by hatched line. NBs are individually identified by numbers and gene expression pattern (coloring).
Morphogenetic movements and genes that shape early neurogenesis: A synopsis
Following gastrulation, the ventral neurectoderm makes its appearance as a columnar epithelium of approximately 100 cells in length and 8–9 cells in width3, 12 (Fig. 1B, C). It gives rise to the 15 neuromeres of the ventral nerve cord. A smaller domain in the embryonic head (procephalic neurectoderm) gives rise to the brain hemispheres. Molecularly, the ventral neurectoderm is defined by the expression of the chordin homolog, short gastrulation (sog)13, 14. Sog is turned on by high activity levels of the ventral morphogen, Dorsal, and antagonizes the function of the dorsally expressed morphogen, Decapentaplegic (Dpp), thereby setting the stage for the expression of prepatterning genes and proneural genes discussed further below.
NBs delaminate from the ventral neurectoderm in five successive waves, S1–S5. S1 NBs are born within a medial and lateral columnar domain of the ventral neurectoderm (mVN and lVN, respectively)3, 4, 15, 16 (Fig. 1B, D). From each of these columns, four regularly spaced NBs delaminate per hemi-neuromere. Two additional S1 NBs derive from the neurectodermal domain enclosed by mVN and lVN, called the intermediate ventral neurectoderm (iVN). Thus, the early pattern of S1 NBs forms a regular, orthogonal grid of three columns (medial, intermediate, lateral NBs), and four rows (A, B, C, D) in each half-segment. Shortly after S1, five S2 NBs delaminate from the intermediate column, followed by six S3 NBs originating from the medial and lateral columns (Fig. 1D). S4 and S5 NBs are more numerous and, based on their position within the map, delaminate from the intermediate domain of the neurectoderm (Fig. 1D). Aside from these NBs, a double row of cells, called mesectoderm or “midline”, located in between the medial columns of either side, also gives rise to neural progenitors, the majority developing into neurons, and some into glia cells17. Note that the mechanism of NB specification, reviewed in this paper, has been worked out only for the first set (S1) of NBs, which are spaced apart and can be each assigned to its own proneural cluster.
Proneural genes and prepatterning genes are expressed in neurectodermal domains that match the orthogonal grid of S1 NBs, and are responsible for distinct subsets of S1 NBs. Note that the role of these genes for the later waves of S2-S4 NBs has not been studied in detail to date. Combinations of three proneural genes, achaete (ac), scute (sc) and lethal of scute (l’sc), are expressed prior to NB delamination in 10 groups of 6–8 cells each per hemisegment18–20 (Fig. 2A–D). These groups were defined as “proneural clusters”. Each proneural cluster gives rise to one S1 NB. Prepatterning genes, responsible for activating proneural genes and/or allowing them to function, are expressed in regular longitudinal and transverse stripes21, 22. Among the former are the homeobox genes ventral nerve cord defective (vnd; vertebrate Nkx 2.2)23, 24, intermediate neuroblasts defective (ind; vertebrate Gsh-1)25, and muscle-specific homeobox gene (msh; vertebrate Msx)26, whose expression domain coincides with the medial, intermediate, and lateral column of the neurectoderm, respectively. The transverse rows A–D coincide with the expression domains of the pair rule and segment polarity genes, and proneural gene expression in these rows is directly controlled by pair rule genes20, 27.
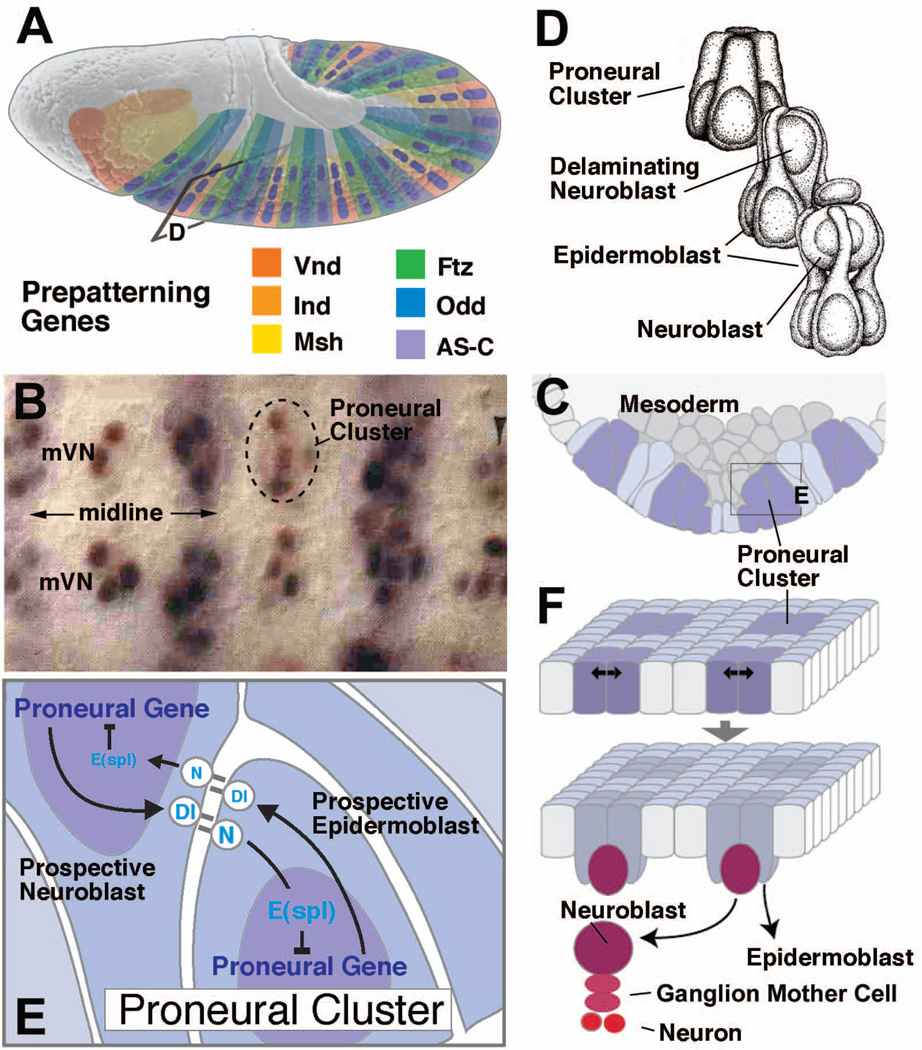
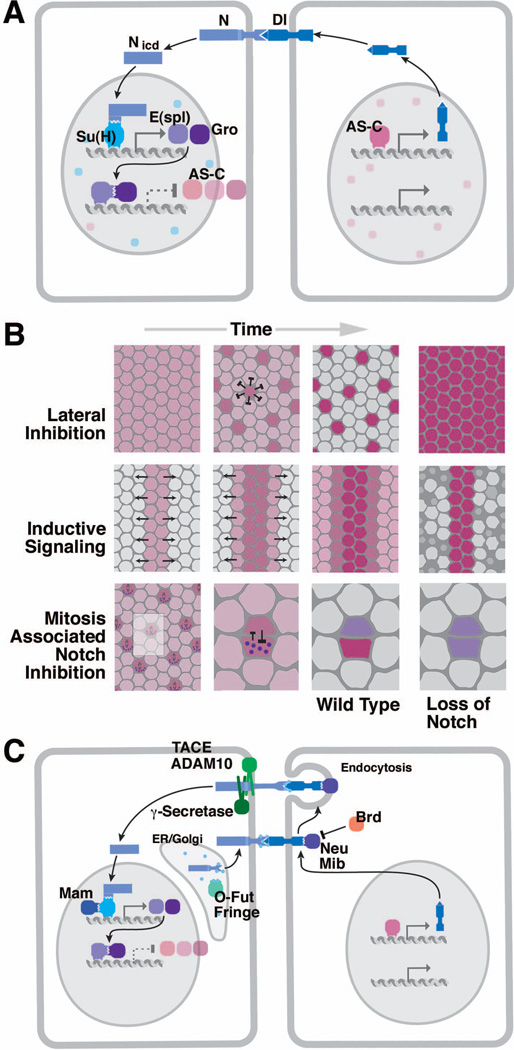
Figure 2. The proneural-neurogenic gene network controls NB patterning.
(A) Lateral view of Drosophila embryo prior to NB delamination. Expression of the prepatterning factors which trigger proneural gene expression is indicated by coloring. Along the antero-posterior axis, Vnd, Ind, and Msh are expressed in the medial, intermediate, and lateral column of the neurectoderm, respectively; segment polarity genes and pair rule genes (represented by Ftz and Odd) define transverse domains in each segment. (B) Photograph showing ventral view of medial neurectoderm (mVN: medial column); proneural clusters expressing ac are labeled brown; purple stripes indicate expression of segment polarity gene engrailed (from20). (C) Schematic cross section of neurectoderm, indicating pattern of proneural clusters (purple shading). Rectangle indicates frame shown in panel (E). (D) Artistic rendering of proneural cluster before (top), during (middle), and after NB delamination (bottom; from4). (E) Lateral inhibition within proneural cluster mediated by N, Dl and E(spl) genes. (F) Two-step model of NB specification. Expression of proneural genes defines proneural clusters (top); lateral inhibition in each cluster selects NB (bottom).
Inside the proneural clusters, the cellular interactions which result in the selection of one NB are accompanied by rapid changes in cell size, cell shape, and nuclear position. Delamination of NBs is strictly coordinated with mitotic division28. At the time when prepatterning of the neurectoderm takes place, all cells are in G2 of the 14th cell cycle (the first cell cycle after the blastoderm). The neighboring dorsal ectoderm and mesoderm have divided and are in cell cycle 15. S1 NBs start delaminating from the proneural clusters of the lVN and iVN when cells of these domains enter mitosis. Prior to mitosis, nuclei of all cells of a proneural cluster are located basally. Subsequently, nuclei of the non-neuronal cells move apically in preparation for mitosis, and only the nuclei of presumptive NBs remain basally and postpone mitosis. They maintain a slender process connected to the apical surface, and appear as the characteristic "bottle cells" (Fig. 7A). Within a few minutes S1 NBs segregate, lose connection to the apical surface, and enter mitosis with an orthogonally oriented spindle (Fig. 7B).
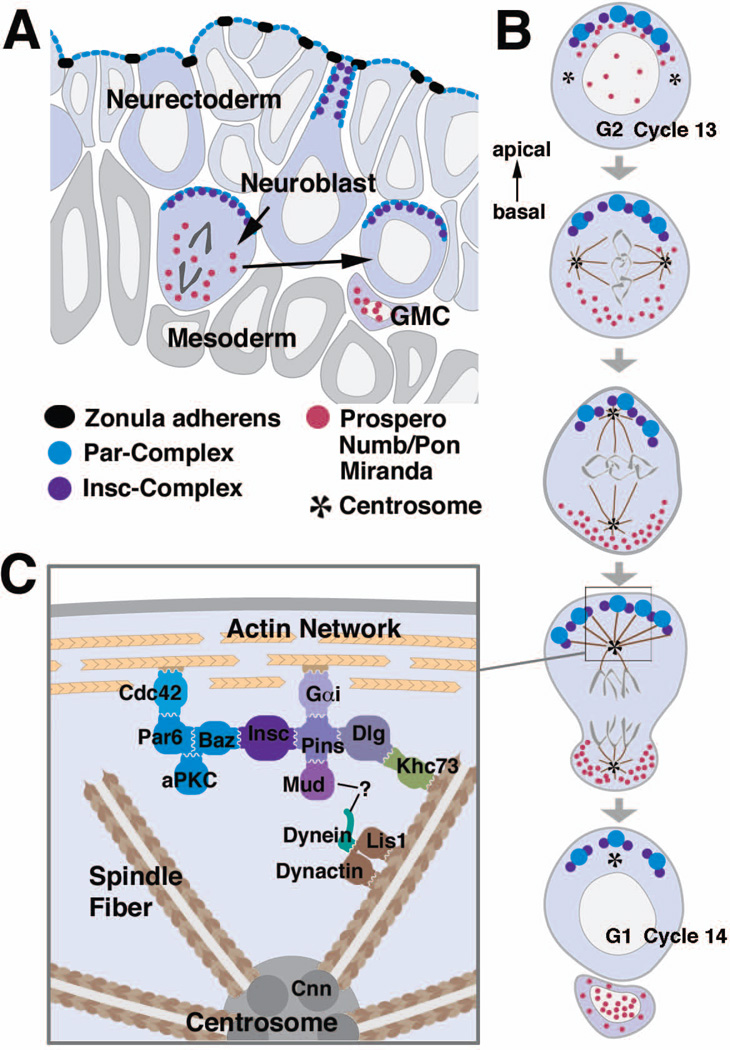
Figure 7. Asymmetric division of NBs.
(A) Schematic cross section of the neurectoderm showing NBs before, during and after delamination. (B) Time course of the first mitosis of a NB after delamination. Note that spindle rotation (compare second and third panel from the top) does only occur in the first mitosis after delamination, but not in subsequent mitoses. (C) Interactions between the cortex and astral microtubules. For details see text. Color coding of protein complexes is the same for all three panels.
The spatial separation of cells destined to become NBs and epidermoblasts is accompanied by rapid changes in the expression pattern of proneural genes, and is mediated by the function of neurogenic genes that encode members of the N signaling pathway. ac, sc and l’sc are transiently and rapidly upregulated in NBs, and downregulated in the surrounding epidermoblasts20, 29, 30. Subsequently they disappear from NBs; at the same time, another set of regulatory genes (“pan-neural genes”), including the basic helix-loop-helix (bHLH) genes asense (ase)31, 32 and snail (sna)33, 34 and the Hox gene prospero (pros)35, 36 are turned on in all NBs. The receptor N and its ligand, Delta (Dl), are expressed ubiquitously in the neurectoderm throughout early neurogenesis. Activation of N by Dl results in the expression of another class of bHLH transcription factors, encoded by the Enhancer of Split [E(spl)] gene complex37–41. E(spl) genes directly repress transcription of proneural genes (Fig. 2E). In addition to this negative interaction between the E(spl) and proneural genes there exists a finely tuned feedback loop between proneural genes and Dl at the transcriptional level that is instrumental in selection of one NB in each proneural cluster (Fig. 2F). The dynamic interactions between proneural genes, Dl, N activity and E(spl) genes constitute the core element of the mechanism regulating early neurogenesis, discussed in more detail in the following section. However, before proceeding directly there, we want to briefly summarize a second phase of early neurogenesis, namely the formation of sensory organ progenitors (SOPs), which in many ways resembles NB development. Historically, the molecular genetic-analysis of SOP and NB specification went hand in hand, and findings in each of these paradigms cross-fertilized each other.
Development of sensilla in the peripheral nervous system
The insect sensory system is formed by widely distributed and diverse cell clusters (“sensilla”) built according to a similar scheme (reviewed in42, 43). For example, mechanosensitive sensilla, represented by the small and large bristles (microchaetes and macrochaetes) on the fly thorax, each contain a bipolar sensory neuron surrounded by a pair of inner and outer accessory cells. The inner accessory cells form a sheath around the dendrite (thecogen cell) and, variably, soma/proximal axon of the sensory neuron (glial cell). The outer accessory cells are arranged concentrically around the inner ones (Fig. 3A) and form processes that secrete cuticle in the shape of hairs or bristles. One of the outer accessory cells forms the shaft (shaft forming cell) and the other one forms a socket around the shaft base (socket forming cell). All cells of a sensillum originate from a single cell, the SOP, which divides in a stereotyped pattern. SOPs appear in the epidermal ectoderm of the embryo at a stage after all NBs of the CNS have formed44, 45. In the larva and early pupa, SOPs for the sensilla of the adult arise in the imaginal discs45–47. Unlike NBs, SOPs do not delaminate fully, remaining instead as bottle-shaped cells integrated in the epidermal ectoderm (Fig. 3B). Only following the final division of one of the SOP daughter cells, the sensory neuron and glia cells move underneath the epidermis. Similar to the NBs of the CNS, SOPs are selected from the ectoderm in a two-step mechanism controlled by the proneural and neurogenic genes45, 48. Prepatterning genes and proneural genes establish proneural clusters in which neurogenic genes set up the selection mechanism for individual SOPs. In some cases, such as the macrochaetes on the Drosophila notum, each sensillum is represented by a proneural cluster49 (Fig. 3C, D). More typically, arrays of closely spaced sensilla, such as the microchaetes on the notum, or the sensilla on the antenna, legs and wing margin, originate from stripe-like proneural domains. In these cases, evenly distributed cells which are slightly variable in number are selected as SOPs in each proneural domain50–53.
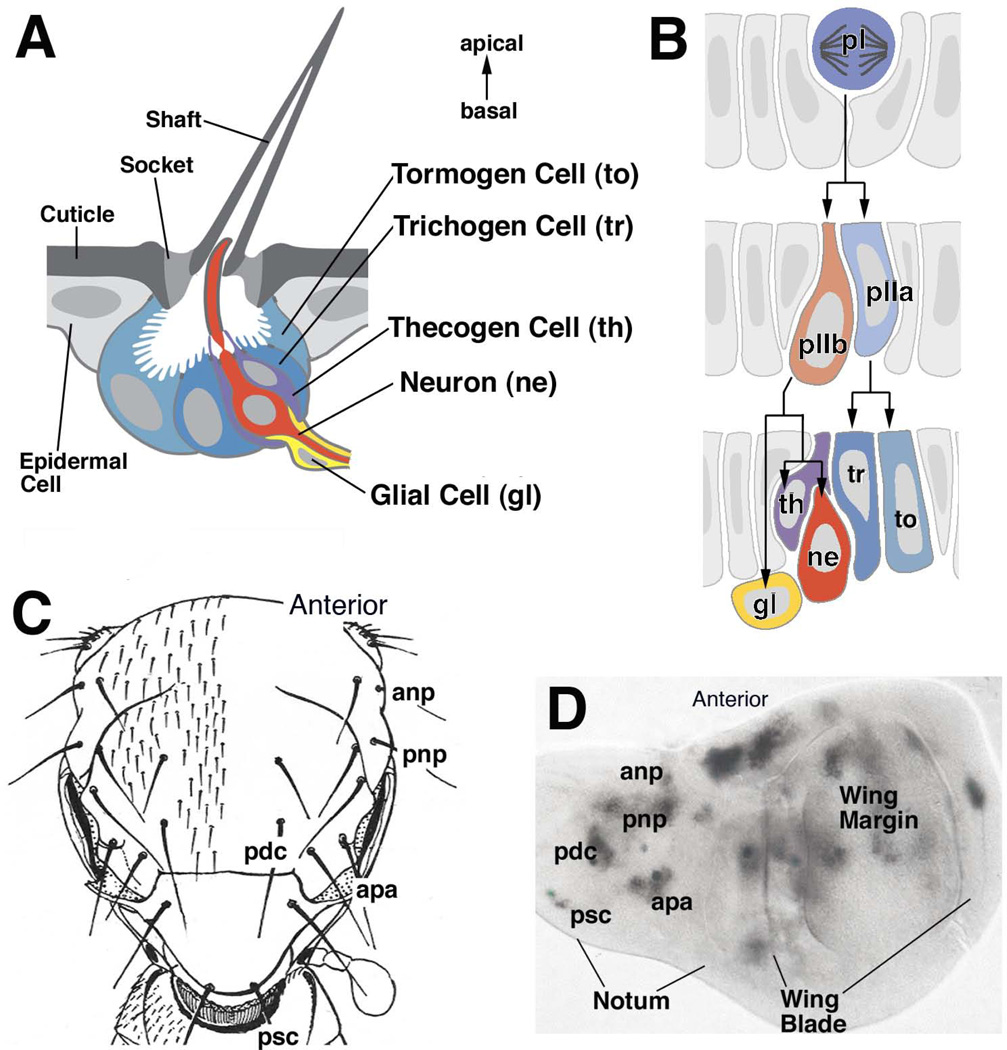
Figure 3. Development of a Drosophila mechanosensory bristle (sensillum).
(A) Schematic cross section of mature sensillum, showing cuticular apparatus (bristle shaft and socket) and underlying cells. (B) Schematic cross sections of developing sensillum. Sensillum progenitor (SOP; pI) divides into two daughters, pIIa and pIIb. pIIa divides into the support cells forming the sensillum shaft and socket (tr, to). pIIb gives rise to the neuron (ne), glial cell (gl), and inner sheath cell (th). (C) Drawing of the back of the Drosophila thorax (notum), showing pattern of mechanosensory bristles. Large bristles (machrochaetae) form an invariant pattern and are individually named (anp, anterior notopleural; apa, anterior postalar ; pdc, posterior dorsocentral; pnp, posterior notopleural; psc, posterior scutellar). (D) Larval imaginal wing disc giving rise to wing blade and notum. Proneural clusters are labeled by expression of proneural gene scute. Each one of the macrochaetae can be assigned to one proneural cluster.
Specifying the NB pattern: The proneural genes
Classical genetic studies carried out since the early decades of the 20th century (reviewed in54) led to identification of three genes, ac, sc, and l’sc, with distinct phenotypes affecting the pattern of sensory bristles. The three genes are located next to each other on the same chromosome and together with a few additional genes they are collectively referred to as the Achaete-Scute complex (AS-C, see below). The genes of the AS-C, in conjunction with several other genes that also affect specific subsets of neural progenitors, were later dubbed “proneural genes”45, 55. That embryos carrying a deficiency at the sc locus have severe defects in the central nervous system was first published in 195656, and was rediscovered twenty five years later in two independent studies7, 57. Similar to the AS-C, the N locus was shown to be involved in early neurogenesis during the early part of the 20th century6 and was redescribed, together with other mutations (“neurogenic mutations”) exhibiting a similar phenotype, several decades later58–60. With the introduction of technologies for positional cloning, the AS-C and several neurogenic genes [N, Dl, E(spl)-C] were among the first gene loci subjected to intensive molecular analysis. These studies were among the pioneering efforts yielding insight into the role of activating and inhibitory transcriptional regulators and signaling pathways during cell fate determination.
The AS-C locus comprises six transcripts (T1a-T6), of which four encode transcription factors of the bHLH family [T3: l’sc; T4: sc; T5: ac; T1a (also called T8): ase]31, 32, 54, 61, 62. T2 and T6 encode enzymes unrelated to proneural function. ac, sc and l’sc are expressed in proneural clusters preceding the first wave of NB delamination. One can distinguish 10 clusters per hemi-segment. 6 of these express l’sc, two express ac and sc, and 2 express all three genes (Fig. 4A)27. ase is expressed in all NBs at a stage when these cells become singled out from the proneural cluster. The expression pattern of proneural genes, placed in the context of previous and subsequent experimental studies, helped in formulating the two-step model of early neurogenesis still valid today (Fig. 2F): (1) expression of proneural genes defines equivalence groups of cells (the “proneural clusters”) which are all competent to generate NBs; (2) lateral inhibition within these clusters restricts the neural fate to just one cell of each cluster.
Figure 4. Proneural genes promote neural development.
(A) Lateral view of embryo prior to NB delamination. Expression pattern of the proneural genes ac, sc and l’sc in proneural clusters corresponding to S1 NBs. (B) Map of S1-S3 NBs of two hemisegments (T2, T3) in wild type embryo (left) and embryo deficient for the AS-C (right) (from65). (C) Photographs of ventral nerve cord labeled with anti- HRP in wild type (left), loss of AS-C (center), loss of AS-C and da (right). (D, E) Role of proneural gene products of AS-C and interacting factors (Da and Emc).
A number of important details which proved of general significance for our understanding of molecular mechanisms controlling cell fate also materialized with the continued analysis of proneural gene function. First, a number of other, more widely expressed transcriptional regulators bind to proteins encoded by the AS-C and act in concert with or antagonistically to them (Fig. 4D). Daughterless (Da) is a ubiquitously expressed bHLH protein that forms heterodimers with Ac, Sc and L’sc and, if deleted, also results in reduction of the number of NBs63–66. Da was one of the first developmental regulators recognized widely to be “promiscuous”, that is, involved in developmental steps that were completely unrelated to each other67: prior to appearing on the horizon of neurobiologists, Da had been known for a long time as a central part of the mechanism controlling germline development. Extramacrochaete (Emc) represents another class of HLH proteins that lacks the basic, DNA-binding domain68–70. By dimerizing with AS-C proteins, Emc inhibits these factors from binding to DNA and exerting their proneural function (Fig. 4E).
The second important lesson taught by careful analysis of the phenotypes resulting from the loss of proneural genes concerns redundancy and cell type specificity of gene function. Initial observations of specific bristle defects associated with mutations in AS-C genes, as well as the expression pattern of these genes in the embryonic neurectoderm (see above) suggested that a particular proneural gene is responsible for the development of a specific type of NB/sensillum precursor. In short: gene A, expressed in precursor A’, is required for the commitment of this cell; lack of A results in lack of A’. Subsequent studies, however, considerably weakened this view. Even after loss of the entire AS-C, only a relatively small fraction (20–25%) of NBs fail to appear65 (Fig. 4B, C). Which of the NBs are deleted is variable. Secondly, ectopic expression of a proneural gene normally not used for a certain type of neural progenitor (as shown for the SOPs of the thoracic sensilla) can induce this progenitor71. These findings indicate that multiple genes (e.g., proneural genes, da, genes upstream and downstream of proneural genes) act redundantly in promoting specific NB and SOP fates.
The neurogenic genes: the saga of N signaling in early neurogenesis
A pivotal role of lateral inhibition in NB specification emerged from a set of experimental studies done in grasshopper in the early 1980s where a laser microbeam was used to ablate individual NBs, which then were replaced by neurectodermal cells72. These experiments demonstrated that normally, NBs send out signals to the adjacent neurectoderm from where they delaminated earlier, and inhibit neurectoderm to produce more NBs. These findings complemented the picture emerging around the same time from developmental observations and genetic studies in Drosophila embryos. Here, it became clear that presumptive NBs and epidermal progenitors (“epidermoblasts”) were intermingled within the neurectoderm3, and that an entire group of genes when mutated increased the ratio of NBs at the expense of epidermoblasts (neurogenic genes58, 59). The rapidly ensuing molecular-genetic characterization of three neurogenic genes [N, Dl, E(spl)] set the stage for a detailed in-vivo model of cellcell interactions, possibly the first of its kind, where molecular sequence and expression data and classical genetic data were combined with experimental studies (transplantation of genetically different cells73; mosaic and clonal analysis74–76). N and Dl encode transmembrane proteins whose extracellular domain contains multiple epidermal growth factor (EGF)-like repeats required for binding of the two molecules60, 77–87. Another EGF-containing transmembrane protein, Serrate (Ser), turned out to be the second N ligand in Drosophila88, 89. The findings that part of the N protein is cleaved at the membrane and enters the nucleus90–92, and that Dl expression accumulates in proneural clusters93, 94, led to the idea that Dl represents a ligand, and N its receptor (Fig. 5A). Since both N and Dl are transmembrane proteins, it was predicted that cell-cell interactions mediated by N and Dl are local (between neighboring cells), and this was confirmed by clonal analysis76.
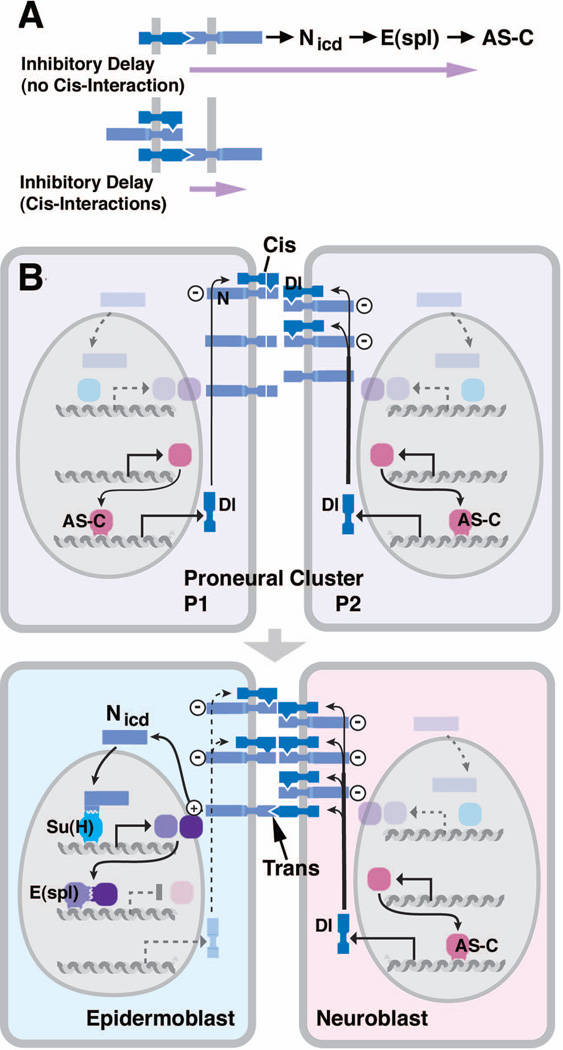
Figure 5. The N pathway and signaling during early neurogenesis.
(A) Core elements of the N pathway (AS-C, transcripts of AS-C; E(spl), transcripts of E(spl)-C; Nicd, cleaved intracellular domain of N). (B) Three types of Notch signaling (lateral inhibition, inductive signaling, mitosis-associated Notch inhibition). Rectangles represent neurectoderm; red shading indicates potential cell fate. In lateral inhibition, all cells start out with a neurogenic fate (left); Notch-mediated lateral inhibition restricts neural fate to NBs (center); loss of Notch results in neural hyperplasia (right). Inductive signaling presupposes a population of cells that are already committed to a specific fate (e.g., wing margin; photoreceptors; left panel). These cells send a N signal to their neighbors inducing in them the expression of other fate determinants (center). Loss of N results in the failure of induction, often accompanied by cell death (right). The signaling scenario at the bottom links N signaling to cell division: prior to mitosis, N pathway is active in all cells (e.g. NBs; SOPs; left); at the same time, the N inhibitor Numb (purple) is channeled into one daughter cell, which thereupon shuts off the N pathway and adopts a different fate (center); loss of N results in both cells retaining their original fate (right). (C) Modulators and signal transducers of the N pathway.
The E(spl) locus, previously known for (and named after) its agonistic interaction with N, resembles the AS-C. It contains multiple tandemly arranged genes encoding bHLH transcription factors (mδ, mγ, mβ, m3, m5, m7, m8), in addition to several other genes with unrelated sequences (mα, m1, m2, m4, m9/10)37–40, 95, 96. Of these, m9/10 (groucho, gro) plays an important role in early neurogenesis. The bHLH genes of the E(spl)-C, as well as gro, are expressed in the neurectoderm prior to NB appearance; as NBs delaminate, these genes become restricted to the surface ectoderm38, 97, 98. In other words, from an initial overlapping pattern of expression of the genes of the AS-C and E(spl)-C, there emerges a separation into complementary domains, with the AS-C genes (transiently) being upregulated in delaminating NBs, and E(spl) genes being expressed in epidermoblasts. Based on this complementary expression pattern, as well as genetic interaction studies, a model was proposed according to which segregation of NBs and epidermoblasts is controlled by reciprocal interactions between the products of the E(spl)-C and the AS-C99 (Fig. 5A).
It quickly became apparent that the reciprocal interaction between AS-C and E(spl)-C is mediated by a feedback loop involving the N-Dl signaling interaction. An important component of this loop connecting the N receptor/signal transducer to E(spl) is the Suppressor of Hairless [Su(H)] protein, discovered in genetic screens to interact with N and Dl during the specification of SOPs. As mentioned above, it was clear from earlier studies that the proneural and neurogenic genes play a role not only during early neurogenesis, but also during related developmental events occurring in the larva and pupa, notably the formation of sensory organs. Two genes, Hairless (H) and Su(H), act antagonistically during the specification of sensory bristles on the wing and notum100, 101. Su(H), homolog of the mammalian JκRBP protein, is directly bound by the Nintra domain once it has been cleaved following N-Dl interaction102 (Fig. 5A) and, as a result, activates the transcription of E(spl)-C genes103. H, a nuclear protein with no known homologs in vertebrates100, 104, represses the binding of Su(H) to its target genes in some developmental scenarios, including the formation of sensory bristles, but plays no role in early neurogenesis. The protein encoded by the “classical” neurogenic gene mastermind (mam) also binds to Su(H), but enhances its role as a transcriptional activator105, 106.
The molecular feedback loop underlying the distinction of NBs from epidermoblasts was further extended with the discovery that the bHLH proteins encoded by several E(spl) genes directly bind to and repress transcription of ASC genes107–109. Along with a number of additional Drosophila genes, notably hairy (h), the E(spl) genes became known as the HES-class of (inhibitory) bHLH genes, as opposed to the activating bHLH genes represented by the AS-C and several other genes discovered during the late 1980s (e.g., myoD110). Binding of E(spl) to its targets requires the co-repressor Gro, which is encoded by the m9/10 transcript of the E(spl)-C111.
N activation via Su(H) activates E(spl), which in turn inhibits AS-C, resulting in the abrogation of neural fate and the initiation of epidermal fate; but what maintains expression of the signal, Dl? This step, the final link in the feedback loop, occurs by transcriptional activation of Dl by the AS-C genes112, 113 (Fig. 5A). Together, these components represent the molecular underpinnings of a feed back loop, known as lateral inhibition (lateral signaling) that essentially consists of three steps114. (1) An equivalence group (the proneural cluster in case of early neurogenesis) expresses both signal and receptor in an overlapping pattern. All the cells have the equal potential to become neural or epidermal, further reflected in the overlapping expression of the corresponding “fate determinants” [AS-C and E(spl), respectively]. (2) One cell of the equivalence group acquires a bias to become either the signaling or receiving cell. The mechanism by which this happens, still unknown today, could involve stochastic fluctuations in the expression of ligand and receptor. (3) Any emerging small bias is immediately amplified by the inhibitory feedback loop, resulting in only one cell maintaining ligand expressing neural determinants (AS-C) while all others maintaining the activated N receptor expressing epidermal determinants [E(spl)].
Lateral inhibition mediated by N signaling is a widespread type of cell-cell interaction occurring in virtually all organ systems. Two variants of N-dependent interactions, inductive signaling, and asymmetric distribution of N inhibitors (Fig. 5B), have been defined and will only briefly be mentioned here. Inductive signaling starts out with a state that is the end-product of lateral signaling: two (or more) cell populations which are already set apart by the differential expression of fate determinants. Here, expression of ligand (Dl or Ser) in one type acts on its neighbors where it activates the N pathway and “induces” a different fate. This occurs, for instance, in the developing Drosophila wing margin115, 116 or eye117. In the latter, epidermal growth factor receptor (EGFR) signaling specifies certain types of photoreceptors which switch on Dl. Dl activates N in neighboring cells which do not express Dl. N activation causes cells to adopt the cone cell fate instead of becoming photoreceptors. The second N-dependent interaction, asymmetrically distributed N inhibitors, occurs at many occasions where N signaling is tied to cell division. The first (and best) known case is that of SOPs in the Drosophila wing. Here, receptor and ligand are expressed in the SOP, and after its division in both daughters. However, during the division, several cytoplasmic molecules (e.g., Numb) are segregated into only one daughter, in which they inhibit N from entering the nucleus118. This process, touched upon again in the last part of this review, results in two daughter cells expressing two different fates.
Active research since the mid 1990s has elucidated the role of numerous additional molecules, which form part of the N signaling pathway. We will only briefly mention these studies, because they mostly utilized paradigms other than early neurogenesis (such as the imaginal disc, or cell culture), even though the resulting mechanistic insights will most likely also apply to N signaling in the early embryo (for a comprehensive review that does justice to the known complexity of the N pathway, see119). First, the proteolytic enzymes responsible for the cleavage of activated N receptor became known [two metalloproteases, Kuzbanian/ADAM10120, 121 and tumor necrosis factor α-converting enzyme (TACE)/ADAM17122, and a γ-secretase, Presenilin123, 124; Fig. 5C]. Secondly, it became clear that glycosylating enzymes acting on the extracellular domains of N and its ligands modify the affinity of ligand/receptor binding (the glycosyl transferase Fringe125 and O-fucosyl transferase126). Finally, a role for endocytotic internalization of ligand in the signaling cells emerged as a crucial element of receptor activation127, and many of the genes known for a long time as members of the “neurogenic complex” turned out to be involved in ligand endocytosis. For example, the “classical” neurogenic gene neuralized (neu) encodes a ubiquitin ligase promoting internalization of Dl128, 129 (Fig. 5C). Bearded, among other factors, acts as an inhibitor of Neu130. Among the questions whose answer is still unclear is the mechanism by which Dl (or Ser) endocytosis influences N activation. One model discussed is that Dl endocytosis exerts a mechanical force on the extracellular domain of N while it is bound to Dl, and that this force is needed to allow for the proteolytic enzymes to cleave N119, 131.
The robustness of lateral inhibition and cis-interaction between N and its ligands
The prepatterning mechanism adjusts the size of the first proneural clusters emerging within the ventral neurectoderm to 6–8 cells. These small proneural clusters each produce one NB. Similarly, small proneural clusters in the ectoderm of the embryo or imaginal discs produce single SOPs giving rise to certain types of sensilla. The error rate of NB or SOP production, that is the frequency of cases where a proneural cluster gives rise to more than one, or to none, NB/SOP, is very low, less than 1%132. In other words, the mechanism of NB/SOP selection, despite its presumed stochastic nature, is very robust. Mathematical modeling of the lateral inhibition process, based on the known molecular circuits involved, predicts that the time delay in inhibitory activity must be short, and that the negative feedback loop involving transcriptional activation of E(spl) is too long for it to be the single factor responsible for the inhibition.
To clarify this postulate, let us assume that proneural cells upregulate production of ligand at similar time points. Ligands then activate N in neighboring cells, which as a result transcribe E(spl). One cell (P1) may be slightly ahead in producing the ligand which would then activate N and E(spl) in neighboring cells (P2) slightly ahead of time. When this inhibition is set in motion in P1, there will be a delay in inhibition, since it takes a certain amount of time for E(spl) to be transcribed and translated, and to reach a threshold that allows it to inhibit AS-C in P2. Just like the build up of ligand, that we assumed to be slightly faster in P1, there is a stochastic variation in the build-up of E(spl), that is, in the inhibitory delay. Let’s assume this delay is greater in P2 than in P1: in that case, P2 can “catch up” with P1 in terms of the amount of Dl ligand it produces. What has happened is that the initial slight advantage of P1 over P2 is lost; both cells may be able to maintain relatively high amounts of Dl which then inhibits other cells of the proneural cluster, with the result that the cluster gives rise to two NBs. To avoid these mistakes, one has to postulate that the inhibitory delay be kept short.
The delay in inhibition is dramatically shortened by a cis-inhibitory interaction between N and its ligands which was first postulated based on genetic studies133, 134, and has since then be substantiated by a number of direct experimental observations. The emerging model bypasses the long delay that would ensue from the build-up of inhibitory E(spl) (Fig. 6A) and postulates that, as a proneural cell accumulates ligand, it first binds to N receptor present in the same cell132. Such cis-interactions are inhibitory: N bound by ligand in cis cannot form Nintra, and cannot trigger E(spl) transcription (Fig. 6B, top). As a result, during the ligand build-up phase, no N-activation occurs in any of the cells of the proneural cluster. Now let us assume that in one cell, P2, the number of ligand molecules starts to exceed the number of N molecules in the same cell (Fig. 6B, bottom right). These excess ligand molecules will now interact in trans with N on neighboring cells (P1), and will cause an effective pulse of Nintra production in these cells. At the same time, P2 itself is refractory to any ligand that might arrive a few seconds later from a neighboring cell which has reached a level of ligand exceeding the level of N. P2 is refractory because all N molecules in this cell are bound in cis by ligand. Therefore, Nintra will never be formed in P2. By contrast, in all neighboring cells, the transient pulse of Nintra has occurred, and will trigger the build-up of E(spl). The cis-interactions between N and ligand as depicted here allow for a short delay in inhibitory signaling, which in turn explains the high accuracy in NB/SOP number.
Figure 6. The role of cis-interactions between N and Dl.
(A) Without cis-interactions the inhibitory delay (purple arrow) is long, including the time interval from N-Dl interaction to build-up of inhibitory E(spl); assuming cis-interaction, the delay is shortened to just N-Dl interaction and N cleavage. (B) Mechanism of cis-interaction between N and Dl. For detail, see text.
Generation of neurons and glia cells by asymmetric division of NBs
After the determination of NBs in the ventral neuroectoderm of the embryo by the mechanisms discussed above, the NBs enter mitosis and begin a series of asymmetric cell divisions. In each of these divisions, one larger daughter cell is produced, which maintains NB fate and the capacity to self renew, and one smaller daughter, the so-called ganglion mother cell (GMC). The GMC will divide only once more and gives rise to a pair of neurons or glia cells that become postmitotic and enter a program of terminal differentiation. In the next paragraphs, we will discuss the following aspects of NB division: 1) How is cortical polarity of the NB established? 2) How is the orientation of the mitotic spindle coordinated with the polarity axis of the NB? 3) How do GMCs, neurons and glia cells that are the progeny of a single NB obtain different fates and differentiation programs? 4) What are the factors that determine whether a NB becomes quiescent or undergoes apoptosis 5) How are NBs reactivated after a period of quiescence?
Establishment of cortical polarity in NBs
Drosophila NBs represent one of the best-studied model systems to investigate asymmetric cell division135. In asymmetric cell division, cell fate determinants are localized asymmetrically during mitosis and segregate into only one of the daughter cells upon cytokinesis. This asymmetric segregation of determinants is responsible for the different identities of the two daughter cells generated in an asymmetric cell division, e. g. a NB and a GMC. In NBs, three cell fate determinants have been identified so far: 1) the transcription factor Prospero (Pros), which activates genes that promote differentiation and suppresses genes that promote proliferation136–139, 2) the protein Numb, which suppresses signal transduction by the N receptor118, 138, 140, and 3) the Brain tumor (Brat) protein, which appears to function as a translational regulator141–143. These three cell fate determinants localize to the basal cortex of the NB in mitosis and segregate into the GMC upon cytokinesis (Fig. 7A, B). Their association with the cortex is indirect and requires the adapter proteins Miranda, which binds to Pros and Brat142–148, and Partner of Numb (Pon), which binds to Numb149. For the proper localization of cell fate determinants and their adapters to the basal NB cortex, a group of proteins is essential that not only controls the polarity of NBs but also of epithelia. These proteins are the components of the Bazooka (Baz)/Par3-Par6-atypical protein kinase C (aPKC) complex150–154 and the tumor suppressor proteins Lethal giant larvae (Lgl), Discs large (Dlg) and Scribble (Scrib)155–157. Proteins of the Baz-Par6-aPKC complex localize to the apical cortex of the NB opposite to the cell fate determinants and their adapters (Fig. 7A, B). Since the Baz-Par6-aPKC complex also localizes to the apical junctional region in the neuroectodermal epithelium from which the NBs delaminate, it appears that NB polarity is directly inherited from the polarity of the epithelium during delamination.
How do these polarity regulators control the basal localization of cell fate determinants? It turns out that this is mainly achieved via a cascade of phosphorylation events affecting the activity, binding affinity and subcellular localization of several components in this system. In brief, at the onset of mitosis the mitotic kinase Aurora A phosphorylates Par6, which reliefs the inhibition of the kinase activity of aPKC158. aPKC now phosphorylates Lgl, leading to its release from the complex with aPKC and Par6 and its exchange for Baz158, 159. In the Baz-Par6-aPKC complex, Baz binds directly to Numb and allows its phosphorylation by aPKC, which causes its dissociation from the apical NB cortex158, 160. Similarly, aPKC directly phosphorylates Miranda, causing its exclusion from the apical NB cortex161. Additional kinases contribute to the generation of cortical polarity, including Polo, which phosphorylates Pon162, and Par1, which phosphorylates Baz163. These kinases are antagonized by phosphatases, including PP2A163–166 and PP4167, which are also required for proper polarization of NBs.
Control of spindle orientation and unequal daughter cell size in NBs
In order to segregate cell fate determinants and apical polarity regulators properly during asymmetric cell division, the mitotic spindle has to align precisely with the polarity axis of the NB. In the first division after NB delamination, the spindle initially orients parallelly to the overlying neurectodermal epithelium and then rotates by 90° to obtain its final orientation perpendicular to the epithelium168, 169 (Fig. 7B). In all subsequent NB divisions, the spindle forms directly in the correct orientation without rotation169. In addition to its proper alignment with the polarity axis of the NB, the spindle has to be positioned asymmetrically to generate two differently sized daughter cells (Fig. 7B). A protein complex consisting of Partner of Inscuteable (Pins), Gαi, Canoe/Afadin (Cno) and Mushroom body defect (Mud) is required for proper spindle orientation in NBs170–177. Mutations in the respective genes cause randomization of spindle orientation. Like the components of the Baz-Par6-aPKC complex, Pins, Gαi, Cno and Mud all localize to the apical NB cortex (Fig. 7C). The colocalization between these two protein complexes depends on the protein Inscuteable (Insc), which binds to both Baz and Pins151, 152, 178 (Fig. 7C). By the interaction of Mud with astral microtubules, one pole of the mitotic spindle is pulled close to the center of the apical Mud crescent, ensuring proper alignment of the spindle with the axis of cortical polarity. However, the formation of a Pins, Gαi and Mud cortical crescent is not completely dependent on Insc. In insc mutants, astral microtubules together with the kinesin Khc-73 and the Discs large (Dlg) protein can induce the formation of Pins, Gαi and Mud cortical crescents179 (Fig. 7C).
In wild type embryos, the polarity axis of NBs is always perpendicular to the plane of the overlying neurectodermal epithelium, leading to the stratification of the developing CNS. Experiments with dissociated NBs in cell culture demonstrated that spindle orientation in NBs is affected by contact to adjacent cells, providing the first experimental evidence for an influence of the neurectodermal epithelium on NB spindle orientation180. This hypothesis has recently been corroborated by the finding that the G protein coupled receptor Tre1, together with the G protein oα subunit, recruits Pins and is required for proper spindle orientation in NBs181. However, the ligand that activates Tre1 and is presumably produced by the epithelium remains to be identified.
In consecutive NB divisions, the GMCs always bud off at a basal position very close to the budding site of the previous GMC, raising the question of how NB polarity is maintained over many cell division cycles. Recent work has beautifully demonstrated that one centrosome remains apically positioned throughout the cell cycle and is responsible for the de novo formation of an apical Par3-Par6-aPKC protein crescent prior to the onset of the next mitosis182. This function of the centrosome is microtubule dependent, demonstrating the importance of the interaction between astral microtubules and the cortex for cortical polarization. After centriole duplication in early interphase, the second centrosome fails to nucleate pericentriolar material (PCM), has no microtubule organizing activity and wanders around in the cytosol. Only at the beginning of mitosis, this centriole starts to assemble PCM, gains microtubule organizing activity and forms one pole of the spindle183, 184. Elegant in vivo studies demonstrated that the dominant apical centrosome which remains in the NB contains the daughter centriole, whereas the wandering centrosome which ends up in the GMC contains the mother centriole, contrary to other types of stem cells where the mother centriole is inherited by the stem cell185, 186. Centrosomes are not only responsible for the maintenance of spindle orientation in consecutive division cycles but also for the asymmetric shape of the mitotic spindle in NBs. Because the apical centrosome nucleates more and longer astral microtubules than the basal one, the spindle midzone is displaced basally, leading to the asymmetric positioning of the cleavage plane, which is the reason for the difference in cell size between the NB and the GMC. However, recent work has shown that there is a second, spindle-independent pathway for cleavage plane positioning, which depends on cortical polarity regulators including Pins187.
A temporal cascade of transcription factor expression controls cell fate in NB lineages
A fundamental question in neurobiology is how neural stem cells give rise to the huge variety of neurons and glia cells that form the mature nervous system. In Drosophila, the question is how the different GMCs and their neural and glial progeny that are produced by a series of asymmetric divisions from a single NB obtain different fates and enter highly specific differentiation pathways. An answer to this question came from the observation that single identified NBs change the expression of a small group of transcription factors in a stereotypic temporal manner188–191. Typically, a freshly delaminated NB first expresses the transcription factor Hunchback (Hb). After one or two cell cycles it abandons Hb expression and starts expressing Krueppel (Kr). In one of the next cell cycles, Kr is shut off and another factor, Pdm, is switched on. The last factor in this cascade is Castor. Importantly, the GMC born at a time when a certain transcription factor is expressed in the NB will also express the same factor, thus distinguishing it molecularly from the GMCs that were born earlier and from those that will be born later. Thus, the temporal gene expression cascade in NBs allows the precise determination of each GMC derived from a specific NB. Together with the positional information provided by Hox genes, dorsal-ventral patterning genes and segmentation genes, this mechanism ensures that each GMC acquires its unique identity. A detailed discussion of this topic is beyond the scope of this article, so the reader is referred to an excellent recent review highlighting this issue192.
The transition from embryonic to larval neurogenesis
At the end of embryonic neurogenesis, NBs either undergo apoptosis, which is the case for the majority of NBs in the abdominal neuromeres, or they arrest in the G1 phase of the cell cycle and become quiescent. The decision between apoptosis and quiescence is regulated intrinsically by the combined action of Hox proteins and temporal identity factors193. The quiescent NBs get activated again during late first instar or early second instar larval stages and continue dividing to generate the adult nervous system194. Interestingly, the temporal transcription factor cascade is only paused upon entry into quiescence and continues during larval NB divisions193. The exit of NBs from quiescence depends on the nutritional state of the larva and is triggered by a signal of unknown nature from the fat body195. Recent work has shown that the fat body derived signal is relayed via glia cells that surround the NBs. The glia cells secrete insulin-like peptides that activate the insulin receptor/PI3-kinase pathway in NBs196, 197, demonstrating that larval NBs depend on niche-derived signals for exit from quiescence.
Conclusions
The determination of NBs in the Drosophila embryo was one of the first examples for a scientific problem in developmental neurobiology that was approached by a combination of careful observation and description of the process as such, systematic genetics, molecular analysis of the relevant genes and finally, biochemical elucidation of the underlying signaling mechanisms. This revolutionary approach has transformed the whole field of developmental biology and has united the formerly separate fields of developmental biology and cell biology. The Drosophila NB is not only an excellent system to study cell fate determination and lateral inhibition, but is also one of the best-studied systems to investigate the mechanisms that control asymmetric cell division135. Intriguingly, all the proteins that regulate cortical polarity and spindle orientation in NBs are highly conserved in evolution and appear to have similar functions in vertebrates. With the finding that defects in the asymmetric division of larval Drosophila NBs can cause brain tumors in the fly198, the mechanisms that ensure the balance between self renewal and differentiation in the developing fly brain have gained the attention of cancer biologists199. The highly sophisticated toolbox of Drosophila genetics, combined with state-of-the-art whole genome analysis tools now opens many opportunities to study the molecular differences between selfrenewing and differentiating cells136, 200, 201. Looking back at the past 30 years of studying neurogenesis in the fly, we expect that this model system will continue to reveal exciting insights in the fields of stem cells and cancer biology.
Acknowledgements
We thank M. Bate, C. Q. Doe and J. Skeath for permission to use published material for some of our figures. We also thank Manu Tiwari for comments on the manuscript and all members of the Hartenstein and Wodarz labs for discussion. The work of VH is supported by the National Institutes of Health (grant NIH/1 R01 NS054814). The work of AW is supported by the Deutsche Forschungsgemeinschaft [DFG Research Center Molecular Physiology of the Brain (CMPB), DFG Research Group 942 and DFG Research Group 1756].
References
- 1.Wheeler WM. Neuroblasts in the arthropod embryo. J Morph. 1891;4:337–343. [Google Scholar]
- 2.Bate CM. Embryogenesis of an insect nervous system. I. A map of the thoracic and abdominal neuroblasts in Locusta migratoria. J Embryol Exp Morphol. 1976;35:107–123. [PubMed] [Google Scholar]
- 3.Hartenstein V, Campos-Ortega JA. Early neurogenesis in wildtype Drosophila melanogaster. Roux’s Arch Dev Biol. 1984;193:308–325. doi: 10.1007/BF00848159. [DOI] [PubMed] [Google Scholar]
- 4.Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- 5.Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- 6.Poulson DF. Chromosomal Deficiencies and the Embryonic Development of Drosophila Melanogaster. Proc Natl Acad Sci U S A. 1937;23:133–137. doi: 10.1073/pnas.23.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jimenez F, Campos-Ortega JA. A region of the Drosophila genome necessary for CNS development. Nature. 1979;282:310–312. doi: 10.1038/282310a0. [DOI] [PubMed] [Google Scholar]
- 8.Jürgens G, Wieschaus E, Nüsslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle of Drosophila melanogaster. II. Zygotic loci on the third chromosome. Roux’s Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- 9.Nüsslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Roux’s Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 10.Wieschaus E, Nüsslein-Volhard C, Jürgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. III. Zygotic loci on the X chromosome and fourth chromosome. Roux's Arch Dev Biol. 1984;193:296–307. doi: 10.1007/BF00848158. [DOI] [PubMed] [Google Scholar]
- 11.Zalokar M, Erk I. Phase-partition fixation and staining of Drosophila eggs. Stain Technol. 1977;52:89–95. doi: 10.3109/10520297709116753. [DOI] [PubMed] [Google Scholar]
- 12.Hartenstein V, Campos-Ortega JA. Fate mapping in wildtype Drosophila melanogaster. I. The pattern of embryonic cell division. Roux’s Arch Dev Biol. 1985;194:181–195. [Google Scholar]
- 13.Francois V, Solloway M, O'Neill JW, Emery J, Bier E. Dorsal-ventral patterning of the Drosophila embryo depends on a putative negative growth factor encoded by the short gastrulation gene. Genes Dev. 1994;8:2602–2616. doi: 10.1101/gad.8.21.2602. [DOI] [PubMed] [Google Scholar]
- 14.Rusch J, Levine M. Threshold responses to the dorsal regulatory gradient and the subdivision of primary tissue territories in the Drosophila embryo. Curr Opin Genet Dev. 1996;6:416–423. doi: 10.1016/s0959-437x(96)80062-1. [DOI] [PubMed] [Google Scholar]
- 15.Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189:186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- 17.Bossing T, Technau GM. The fate of the CNS midline progenitors in Drosophila as revealed by a new method for single cell labelling. Development. 1994;120:1895–1906. doi: 10.1242/dev.120.7.1895. [DOI] [PubMed] [Google Scholar]
- 18.Cabrera CV, Martinez-Arias A, Bate M. The expression of three members of the achaete-scute gene complex correlates with neuroblast segregation in Drosophila. Cell. 1987;50:425–433. doi: 10.1016/0092-8674(87)90496-x. [DOI] [PubMed] [Google Scholar]
- 19.Romani S, Campuzano S, Modolell J. The achaete-scute complex is expressed in neurogenic regions of Drosophila embryos. Embo J. 1987;6:2085–2092. doi: 10.1002/j.1460-2075.1987.tb02474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skeath JB, Carroll SB. Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo. Development. 1992;114:939–946. doi: 10.1242/dev.114.4.939. [DOI] [PubMed] [Google Scholar]
- 21.Von Ohlen T, Doe CQ. Convergence of Dorsal, Dpp, Egfr Signaling Pathways Subdivides the Drosophila Neuroectoderm into Three Dorsal-Ventral Columns. Dev Biol. 2000;224:362–372. doi: 10.1006/dbio.2000.9789. [DOI] [PubMed] [Google Scholar]
- 22.Zhao G, Wheeler SR, Skeath JB. Genetic control of dorsoventral patterning and neuroblast specification in the Drosophila Central Nervous System. Int J Dev Biol. 2007;51:107–115. doi: 10.1387/ijdb.062188gz. [DOI] [PubMed] [Google Scholar]
- 23.Chu H, Parras C, White K, Jimenez F. Formation and specification of ventral neuroblasts is controlled by vnd in Drosophila neurogenesis. Genes Dev. 1998;12:3613–3624. doi: 10.1101/gad.12.22.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald JA, Holbrook S, Isshiki T, Weiss J, Doe CQ, Mellerick DM. Dorsoventral patterning in the Drosophila central nervous system: the vnd homeobox gene specifies ventral column identity. Genes Dev. 1998;12:3603–3612. doi: 10.1101/gad.12.22.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss JB, Von Ohlen T, Mellerick DM, Dressler G, Doe CQ, Scott MP. Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 1998;12:3591–3602. doi: 10.1101/gad.12.22.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isshiki T, Takeichi M, Nose A. The role of the msh homeobox gene during Drosophila neurogenesis: implication for the dorsoventral specification of the neuroectoderm. Development. 1997;124:3099–3109. doi: 10.1242/dev.124.16.3099. [DOI] [PubMed] [Google Scholar]
- 27.Skeath JB, Panganiban G, Selegue J, Carroll SB. Gene regulation in two dimensions: the proneural achaete and scute genes are controlled by combinations of axis-patterning genes through a common intergenic control region. Genes Dev. 1992;6:2606–2619. doi: 10.1101/gad.6.12b.2606. [DOI] [PubMed] [Google Scholar]
- 28.Hartenstein V, Younossi-Hartenstein A, Lekven A. Delamination and division in the Drosophila neurectoderm: spatiotemporal pattern, cytoskeletal dynamics, and common control by neurogenic and segment polarity genes. Dev Biol. 1994;165:480–499. doi: 10.1006/dbio.1994.1269. [DOI] [PubMed] [Google Scholar]
- 29.Cabrera CV. Lateral inhibition and cell fate during neurogenesis in Drosophila: the interactions between scute, Notch and Delta. Development. 1990;110:733–742. [PubMed] [Google Scholar]
- 30.Martin-Bermudo MD, Martinez C, Rodriguez A, Jimenez F. Distribution and function of the lethal of scute gene product during early neurogenesis in Drosophila. Development. 1991;113:445–454. doi: 10.1242/dev.113.2.445. [DOI] [PubMed] [Google Scholar]
- 31.Alonso MC, Cabrera CV. The achaete-scute gene complex of Drosophila melanogaster comprises four homologous genes. Embo J. 1988;7:2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez F, Romani S, Cubas P, Modolell J, Campuzano S. Molecular analysis of the asense gene, a member of the achaete-scute complex of Drosophila melanogaster, and its novel role in optic lobe development. Embo J. 1989;8:3553–3562. doi: 10.1002/j.1460-2075.1989.tb08527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 34.Ip YT, Levine M, Bier E. Neurogenic expression of snail is controlled by separable CNS and PNS promoter elements. Development. 1994;120:199–207. doi: 10.1242/dev.120.1.199. [DOI] [PubMed] [Google Scholar]
- 35.Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- 36.Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 37.Knust E, Bremer KA, Vassin H, Ziemer A, Tepass U, Campos-Ortega JA. The enhancer of split locus and neurogenesis in Drosophila melanogaster. Dev Biol. 1987;122:262–273. doi: 10.1016/0012-1606(87)90351-4. [DOI] [PubMed] [Google Scholar]
- 38.Knust E, Tietze K, Campos-Ortega JA. Molecular analysis of the neurogenic locus Enhancer of split of Drosophila melanogaster. Embo J. 1987;6:4113–4123. doi: 10.1002/j.1460-2075.1987.tb02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klambt C, Knust E, Tietze K, Campos-Ortega JA. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. Embo J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delidakis C, Artavanis-Tsakonas S. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci U S A. 1992;89:8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrons H, Knust E, Campos-Ortega JA. The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics. 1992;132:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai EC, Orgogozo V. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev Biol. 2004;269:1–17. doi: 10.1016/j.ydbio.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 43.Hartenstein V. Development of Sensilla. In: Gilbert L, Gill S, Kostas I, editors. Comprehensive Insect Physiology, Biochemistry, Pharmacology and Molecular Biology. 2005. [Google Scholar]
- 44.Younossi-Hartenstein A, Hartenstein V. Pattern, time of birth and morphogenesis of sensillum precursors in Drosophila. Micr Res Tech. 1997;39:479–491. doi: 10.1002/(SICI)1097-0029(19971215)39:6<479::AID-JEMT3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 45.Ghysen A, Dambly-Chaudiere C. Genesis of the Drosophila peripheral nervous system. Trends Genet. 1989;5:251–255. doi: 10.1016/0168-9525(89)90097-8. [DOI] [PubMed] [Google Scholar]
- 46.Hartenstein V, Posakony JW. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- 47.Le Borgne R, Bellaiche Y, Schweisguth F. Drosophila E-cadherin regulates the orientation of asymmetric cell division in the sensory organ lineage. Curr Biol. 2002;12:95–104. doi: 10.1016/s0960-9822(01)00648-0. [DOI] [PubMed] [Google Scholar]
- 48.Modolell J. Patterning of the adult peripheral nervous system of Drosophila. Perspect Dev Neurobiol. 1997;4:285–296. [PubMed] [Google Scholar]
- 49.Cubas P, de Celis JF, Campuzano S, Modolell J. Proneural clusters of achaetescute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- 50.Hartenstein V, Posakony JW. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol. 1990;142:13–30. doi: 10.1016/0012-1606(90)90147-b. [DOI] [PubMed] [Google Scholar]
- 51.Skaer N, Pistillo D, Simpson P. Transcriptional heterochrony of scute and changes in bristle pattern between two closely related species of blowfly. Dev Biol. 2002;252:31–45. doi: 10.1006/dbio.2002.0841. [DOI] [PubMed] [Google Scholar]
- 52.Joshi M, Buchanan KT, Shroff S, Orenic TV. Delta and Hairy establish a periodic prepattern that positions sensory bristles in Drosophila legs. Dev Biol. 2006;293:64–76. doi: 10.1016/j.ydbio.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Cohen M, Georgiou M, Stevenson NL, Miodownik M, Baum B. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19:78–89. doi: 10.1016/j.devcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 54.Campos-Ortega JA. The genetics of the Drosophila achaete-scute gene complex: a historical appraisal. Int J Dev Biol. 1998;42:291–297. [PubMed] [Google Scholar]
- 55.Romani S, Campuzano S, Macagno ER, Modolell J. Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes Dev. 1989;3:997–1007. doi: 10.1101/gad.3.7.997. [DOI] [PubMed] [Google Scholar]
- 56.Ede DA. Studies on the effects of some genetic lethal factors on the embryonic development of Drosophila melanogaster. Roux’s Arch Dev Biol. 1956;149:101–114. doi: 10.1007/BF01382509. [DOI] [PubMed] [Google Scholar]
- 57.White K. Defective neural development in Drosophila melanogaster embryos deficient for the tip of the X chromosome. Dev Biol. 1980;80:332–344. doi: 10.1016/0012-1606(80)90409-1. [DOI] [PubMed] [Google Scholar]
- 58.Lehmann R, Dietrich U, Jimenez F, Campos-Ortega JA. Mutations of early neurogenesis in Drosophila. Roux’s Arch Dev Biol. 1981;190:226–229. doi: 10.1007/BF00848307. [DOI] [PubMed] [Google Scholar]
- 59.Lehmann R, Jimenez F, Dietrich U, Campos-Ortega JA. On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster. Roux’s Arch Dev Biol. 1983;192:62–74. doi: 10.1007/BF00848482. [DOI] [PubMed] [Google Scholar]
- 60.Artavanis-Tsakonas S, Muskavitch MA, Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983;80:1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campuzano S, Carramolino L, Cabrera CV, Ruiz-Gomez M, Villares R, Boronat A, Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985;40:327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- 62.Villares R, Cabrera CV. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- 63.Caudy M, Grell EH, Dambly-Chaudiere C, Ghysen A, Jan LY, Jan YN. The maternal sex determination gene daughterless has zygotic activity necessary for the formation of peripheral neurons in Drosophila. Genes Dev. 1988;2:843–852. doi: 10.1101/gad.2.7.843. [DOI] [PubMed] [Google Scholar]
- 64.Caudy M, Vassin H, Brand M, Tuma R, Jan LY, Jan YN. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988;55:1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- 65.Jimenez F, Campos-Ortega JA. Defective neuroblast commitment in mutants of the achaete-scute complex and adjacent genes of D. melanogaster. Neuron. 1990;5:81–89. doi: 10.1016/0896-6273(90)90036-f. [DOI] [PubMed] [Google Scholar]
- 66.Cabrera CV, Alonso MC. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. Embo J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cline TW. The affairs of daughterless and the promiscuity of developmental regulators. Cell. 1989;59:231–234. doi: 10.1016/0092-8674(89)90280-8. [DOI] [PubMed] [Google Scholar]
- 68.Ellis HM, Spann DR, Posakony JW. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- 69.Garrell J, Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- 70.Van Doren M, Ellis HM, Posakony JW. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- 71.Hinz U, Giebel B, Campos-Ortega JA. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 72.Taghert PH, Doe CQ, Goodman CS. Cell determination and regulation during development of neuroblasts and neurones in grasshopper embryo. Nature. 1984;307:163–165. doi: 10.1038/307163a0. [DOI] [PubMed] [Google Scholar]
- 73.Technau GM, Campos-Ortega JA. Cell autonomy of expression of neurogenic genes of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1987;84:4500–4504. doi: 10.1073/pnas.84.13.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoppe PE, Greenspan RJ. Local function of the Notch gene for embryonic ectodermal pathway choice in Drosophila. Cell. 1986;46:773–783. doi: 10.1016/0092-8674(86)90353-3. [DOI] [PubMed] [Google Scholar]
- 75.Hoppe PE, Greenspan RJ. The Notch locus of Drosophila is required in epidermal cells for epidermal development. Development. 1990;109:875–885. doi: 10.1242/dev.109.4.875. [DOI] [PubMed] [Google Scholar]
- 76.Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- 77.Kidd S, Lockett TJ, Young MW. The Notch locus of Drosophila melanogaster. Cell. 1983;34:421–433. doi: 10.1016/0092-8674(83)90376-8. [DOI] [PubMed] [Google Scholar]
- 78.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 79.Knust E, Dietrich U, Tepass U, Bremer KA, Weigel D, Vassin H, Campos-Ortega JA. EGF homologous sequences encoded in the genome of Drosophila melanogaster, and their relation to neurogenic genes. Embo J. 1987;6:761–766. doi: 10.1002/j.1460-2075.1987.tb04818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hartley DA, Xu TA, Artavanis-Tsakonas S. The embryonic expression of the Notch locus of Drosophila melanogaster and the implications of point mutations in the extracellular EGF-like domain of the predicted protein. Embo J. 1987;6:3407–3417. doi: 10.1002/j.1460-2075.1987.tb02664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelley MR, Kidd S, Deutsch WA, Young MW. Mutations altering the structure of epidermal growth factor-like coding sequences at the Drosophila Notch locus. Cell. 1987;51:539–548. doi: 10.1016/0092-8674(87)90123-1. [DOI] [PubMed] [Google Scholar]
- 82.Vassin H, Bremer KA, Knust E, Campos-Ortega JA. The neurogenic gene Delta of Drosophila melanogaster is expressed in neurogenic territories and encodes a putative transmembrane protein with EGF-like repeats. Embo J. 1987;6:3431–3440. doi: 10.1002/j.1460-2075.1987.tb02666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kopczynski CC, Alton AK, Fechtel K, Kooh PJ, Muskavitch MA. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 1988;2:1723–1735. doi: 10.1101/gad.2.12b.1723. [DOI] [PubMed] [Google Scholar]
- 84.Kidd S, Baylies MK, Gasic GP, Young MW. Structure and distribution of the Notch protein in developing Drosophila. Genes Dev. 1989;3:1113–1129. doi: 10.1101/gad.3.8.1113. [DOI] [PubMed] [Google Scholar]
- 85.Johansen KM, Fehon RG, Artavanis-Tsakonas S. The notch gene product is a glycoprotein expressed on the cell surface of both epidermal and neuronal precursor cells during Drosophila development. J Cell Biol. 1989;109:2427–2440. doi: 10.1083/jcb.109.5.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-l. [DOI] [PubMed] [Google Scholar]
- 87.Lieber T, Wesley CS, Alcamo E, Hassel B, Krane JF, Campos-Ortega JA, Young MW. Single amino acid substitutions in EGF-like elements of Notch and Delta modify Drosophila development and affect cell adhesion in vitro. Neuron. 1992;9:847–859. doi: 10.1016/0896-6273(92)90238-9. [DOI] [PubMed] [Google Scholar]
- 88.Fleming RJ, Scottgale TN, Diederich RJ, Artavanis-Tsakonas S. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 1990;4:2188–2201. doi: 10.1101/gad.4.12a.2188. [DOI] [PubMed] [Google Scholar]
- 89.Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development. 1991;111:749–761. doi: 10.1242/dev.111.3.749. [DOI] [PubMed] [Google Scholar]
- 90.Lieber T, Kidd S, Alcamo E, Corbin V, Young MW. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 91.Rebay I, Fehon RG, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 92.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 93.Kopczynski CC, Muskavitch MA. Complex spatio-temporal accumulation of alternative transcripts from the neurogenic gene Delta during Drosophila embryogenesis. Development. 1989;107:623–636. doi: 10.1242/dev.107.3.623. [DOI] [PubMed] [Google Scholar]
- 94.Haenlin M, Kramatschek B, Campos-Ortega JA. The pattern of transcription of the neurogenic gene Delta of Drosophila melanogaster. Development. 1990;110:905–914. doi: 10.1242/dev.110.3.905. [DOI] [PubMed] [Google Scholar]
- 95.Hartley DA, Preiss A, Artavanis-Tsakonas S. A deduced gene product from the Drosophila neurogenic locus, enhancer of split, shows homology to mammalian G-protein beta subunit. Cell. 1988;55:785–795. doi: 10.1016/0092-8674(88)90134-1. [DOI] [PubMed] [Google Scholar]
- 96.Preiss A, Hartley DA, Artavanis-Tsakonas S. The molecular genetics of Enhancer of split, a gene required for embryonic neural development in Drosophila. Embo J. 1988;7:3917–3927. doi: 10.1002/j.1460-2075.1988.tb03278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Knust E, Schrons H, Grawe F, Campos-Ortega JA. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kramatschek B, Campos-Ortega JA. Neuroectodermal transcription of the Drosophila neurogenic genes E(spl) and HLH-m5 is regulated by proneural genes. Development. 1994;120:815–826. doi: 10.1242/dev.120.4.815. [DOI] [PubMed] [Google Scholar]
- 99.Campos-Ortega JA. Cellular interactions in the developing nervous system of Drosophila. Cell. 1994;77:969–975. doi: 10.1016/0092-8674(94)90437-5. [DOI] [PubMed] [Google Scholar]
- 100.Bang AG, Posakony JW. The Drosophila gene Hairless encodes a novel basic protein that controls alternative cell fates in adult sensory organ development. Genes Dev. 1992;6:1752–1769. doi: 10.1101/gad.6.9.1752. [DOI] [PubMed] [Google Scholar]
- 101.Schweisguth F, Posakony JW. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69:1199–1212. doi: 10.1016/0092-8674(92)90641-o. [DOI] [PubMed] [Google Scholar]
- 102.Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 103.Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 104.Maier D, Stumm G, Kuhn K, Preiss A. Hairless, a Drosophila gene involved in neural development, encodes a novel, serine rich protein. Mech Dev. 1992;38:143–156. doi: 10.1016/0925-4773(92)90006-6. [DOI] [PubMed] [Google Scholar]
- 105.Smoller D, Friedel C, Schmid A, Bettler D, Lam L, Yedvobnick B. The Drosophila neurogenic locus mastermind encodes a nuclear protein unusually rich in amino acid homopolymers. Genes Dev. 1990;4:1688–1700. doi: 10.1101/gad.4.10.1688. [DOI] [PubMed] [Google Scholar]
- 106.Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD. MAML1, a human homologue of Drosophila mastermind, is a transcriptional coactivator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 107.Oellers N, Dehio M, Knust E. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol Gen Genet. 1994;244:465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- 108.Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- 109.Gigliani F, Longo F, Gaddini L, Battaglia PA. Interactions among the bHLH domains of the proteins encoded by the Enhancer of split and achaete-scute gene complexes of Drosophila. Mol Gen Genet. 1996;251:628–634. doi: 10.1007/BF02174111. [DOI] [PubMed] [Google Scholar]
- 110.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 111.Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 112.Haenlin M, Kunisch M, Kramatschek B, Campos-Ortega JA. Genomic regions regulating early embryonic expression of the Drosophila neurogenic gene Delta. Mech Dev. 1994;47:99–110. doi: 10.1016/0925-4773(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 113.Kunisch M, Haenlin M, Campos-Ortega JA. Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc Natl Acad Sci U S A. 1994;91:10139–10143. doi: 10.1073/pnas.91.21.10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kimble J, Simpson P. The LIN-12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol. 1997;13:333–361. doi: 10.1146/annurev.cellbio.13.1.333. [DOI] [PubMed] [Google Scholar]
- 115.Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- 116.Majumdar A, Nagaraj R, Banerjee U. strawberry notch encodes a conserved nuclear protein that functions downstream of Notch and regulates gene expression along the developing wing margin of Drosophila. Genes Dev. 1997;11:1341–1353. doi: 10.1101/gad.11.10.1341. [DOI] [PubMed] [Google Scholar]
- 117.Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 118.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 119.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 120.Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 121.Sotillos S, Roch F, Campuzano S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development. 1997;124:4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- 122.Jarriault S, Greenwald I. Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev Biol. 2005;287:1–10. doi: 10.1016/j.ydbio.2005.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 124.Ye Y, Lukinova N, Fortini ME. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–529. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 125.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notchligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 126.Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 127.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 128.Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- 129.Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. neuralized encodes a peripheral membrane protein involved in Delta signaling and endocytosis. Dev Cell. 2001;1:807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- 130.Bardin AJ, Schweisguth F. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev Cell. 2006;10:245–255. doi: 10.1016/j.devcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 131.Krahn MP, Wodarz A. Notch signaling: linking delta endocytosis and cell polarity. Dev Cell. 2009;17:153–154. doi: 10.1016/j.devcel.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 132.Barad O, Rosin D, Hornstein E, Barkai N. Error minimization in lateral inhibition circuits. Sci Signal. 2010;3:ra51. doi: 10.1126/scisignal.2000857. [DOI] [PubMed] [Google Scholar]
- 133.Jacobsen TL, Brennan K, Martinez-Arias A, Muskavitch MA. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development. 1998;125:4531–4540. doi: 10.1242/dev.125.22.4531. [DOI] [PubMed] [Google Scholar]
- 134.Li Y, Baker NE. The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev Biol. 2004;4:5. doi: 10.1186/1471-213X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 137.Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- 138.Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 139.Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- 140.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 141.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 142.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 143.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 144.Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- 145.Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 1998;125:4089–4098. doi: 10.1242/dev.125.20.4089. [DOI] [PubMed] [Google Scholar]
- 146.Schuldt AJ, Adams JH, Davidson CM, Micklem DR, Haseloff J, St Johnston D, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 1998;12:1847–1857. doi: 10.1101/gad.12.12.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 1997;90:449–458. doi: 10.1016/s0092-8674(00)80505-x. [DOI] [PubMed] [Google Scholar]
- 148.Shen CP, Knoblich JA, Chan YM, Jiang MM, Jan LY, Jan YN. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 1998;12:1837–1846. doi: 10.1101/gad.12.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lu B, Rothenberg M, Jan LY, Jan YN. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- 150.Kuchinke U, Grawe F, Knust E. Control of spindle orientation in Drosophila by the Par-3-related PDZ- domain protein Bazooka. Curr Biol. 1998;8:1357–1365. doi: 10.1016/s0960-9822(98)00016-5. [DOI] [PubMed] [Google Scholar]
- 151.Schober M, Schaefer M, Knoblich JA. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 1999;402:548–551. doi: 10.1038/990135. [DOI] [PubMed] [Google Scholar]
- 152.Wodarz A, Ramrath A, Kuchinke U, Knust E. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 1999;402:544–547. doi: 10.1038/990128. [DOI] [PubMed] [Google Scholar]
- 153.Wodarz A, Ramrath A, Grimm A, Knust E. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol. 2000;150:1361–1374. doi: 10.1083/jcb.150.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Petronczki M, Knoblich JA. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol. 2001;3:43–49. doi: 10.1038/35050550. [DOI] [PubMed] [Google Scholar]
- 155.Peng CY, Manning L, Albertson R, Doe CQ. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- 156.Ohshiro T, Yagami T, Zhang C, Matsuzaki F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 2000;408:593–596. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- 157.Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5:166–170. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- 158.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Betschinger J, Mechtler K, Knoblich JA. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 2003;422:326–330. doi: 10.1038/nature01486. [DOI] [PubMed] [Google Scholar]
- 160.Smith CA, Lau KM, Rahmani Z, Dho SE, Brothers G, She YM, Berry DM, Bonneil E, Thibault P, Schweisguth F, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. Embo J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Atwood SX, Prehoda KE. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr Biol. 2009;19:723–729. doi: 10.1016/j.cub.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature. 2007;449:96–100. doi: 10.1038/nature06056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Krahn MP, Egger-Adam D, Wodarz A. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev Cell. 2009;16:901–908. doi: 10.1016/j.devcel.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 164.Ogawa H, Ohta N, Moon W, Matsuzaki F. Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par-6 in Drosophila neuroblast asymmetric divisions. J Cell Sci. 2009;122:3242–3249. doi: 10.1242/jcs.050955. [DOI] [PubMed] [Google Scholar]
- 165.Wang C, Chang KC, Somers G, Virshup D, Ang BT, Tang C, Yu F, Wang H. Protein phosphatase 2A regulates self-renewal of Drosophila neural stem cells. Development. 2009;136:2287–2296. doi: 10.1242/dev.035758. [DOI] [PubMed] [Google Scholar]
- 166.Chabu C, Doe CQ. Twins/PP2A regulates aPKC to control neuroblast cell polarity and self-renewal. Dev Biol. 2009;330:399–405. doi: 10.1016/j.ydbio.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sousa-Nunes R, Chia W, Somers WG. Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions. Genes Dev. 2009;23:359–372. doi: 10.1101/gad.1723609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kaltschmidt JA, Davidson CM, Brown NH, Brand AH. Rotation and asymmetry of the mitotic spindle direct asymmetric cell division in the developing central nervous system. Nat Cell Biol. 2000;2:7–12. doi: 10.1038/71323. [DOI] [PubMed] [Google Scholar]
- 169.Rebollo E, Roldan M, Gonzalez C. Spindle alignment is achieved without rotation after the first cell cycle in Drosophila embryonic neuroblasts. Development. 2009;136:3393–3397. doi: 10.1242/dev.041822. [DOI] [PubMed] [Google Scholar]
- 170.Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- 171.Schaefer M, Shevchenko A, Knoblich JA. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol. 2000;10:353–362. doi: 10.1016/s0960-9822(00)00401-2. [DOI] [PubMed] [Google Scholar]
- 172.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 173.Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 174.Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- 175.Yu F, Cai Y, Kaushik R, Yang X, Chia W. Distinct roles of Galphai and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J Cell Biol. 2003;162:623–633. doi: 10.1083/jcb.200303174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Speicher S, Fischer A, Knoblich J, Carmena A. The PDZ protein Canoe regulates the asymmetric division of Drosophila neuroblasts and muscle progenitors. Curr Biol. 2008;18:831–837. doi: 10.1016/j.cub.2008.04.072. [DOI] [PubMed] [Google Scholar]
- 177.Carmena A, Makarova A, Speicher S. The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J Cell Biol. 2011;195:553–562. doi: 10.1083/jcb.201108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yu F, Morin X, Cai Y, Yang X, Chia W. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 2000;100:399–409. doi: 10.1016/s0092-8674(00)80676-5. [DOI] [PubMed] [Google Scholar]
- 179.Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 180.Siegrist SE, Doe CQ. Extrinsic cues orient the cell division axis in Drosophila embryonic neuroblasts. Development. 2006;133:529–536. doi: 10.1242/dev.02211. [DOI] [PubMed] [Google Scholar]
- 181.Yoshiura S, Ohta N, Matsuzaki F. Tre1 GPCR signaling orients stem cell divisions in the Drosophila central nervous system. Dev Cell. 2012;22:79–91. doi: 10.1016/j.devcel.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 182.Januschke J, Gonzalez C. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J Cell Biol. 2010;188:693–706. doi: 10.1083/jcb.200905024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 184.Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Conduit PT, Raff JW. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr Biol. 2010;20:2187–2192. doi: 10.1016/j.cub.2010.11.055. [DOI] [PubMed] [Google Scholar]
- 187.Cabernard C, Prehoda KE, Doe CQ. A spindle-independent cleavage furrow positioning pathway. Nature. 2010;467:91–94. doi: 10.1038/nature09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila Neuroblasts Sequentially Express Transcription Factors which Specify the Temporal Identity of Their Neuronal Progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 190.Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 191.Kanai MI, Okabe M, Hiromi Y. seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Dev Cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 192.Maurange C. Temporal specification of neural stem cells: insights from Drosophila neuroblasts. Curr Top Dev Biol. 2012;98:199–228. doi: 10.1016/B978-0-12-386499-4.00008-2. [DOI] [PubMed] [Google Scholar]
- 193.Tsuji T, Hasegawa E, Isshiki T. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development. 2008;135:3859–3869. doi: 10.1242/dev.025189. [DOI] [PubMed] [Google Scholar]
- 194.Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- 195.Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- 196.Sousa-Nunes R, Yee LL, Gould AP. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chell JM, Brand AH. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 199.Wodarz A, Nathke I. Cell polarity in development and cancer. Nat Cell Biol. 2007;9:1016–1024. doi: 10.1038/ncb433. [DOI] [PubMed] [Google Scholar]
- 200.Berger C, Harzer H, Burkard TR, Steinmann J, van der Horst S, Laurenson AS, Novatchkova M, Reichert H, Knoblich JA. FACS Purification and Transcriptome Analysis of Drosophila Neural Stem Cells Reveals a Role for Klumpfuss in Self-Renewal. Cell Rep. 2012;2:407–418. doi: 10.1016/j.celrep.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Southall TD, Brand AH. Neural stem cell transcriptional networks highlight genes essential for nervous system development. Embo J. 2009;28:3799–3807. doi: 10.1038/emboj.2009.309. [DOI] [PMC free article] [PubMed] [Google Scholar]