Abstract
The brain of Drosophila is formed by approximately 100 lineages, each lineage being derived from a stem cell-like neuroblast that segregates from the procephalic neurectoderm of the early embryo. A neuroblast map has been established in great detail for the early embryo, and a suite of molecular markers has been defined for all neuroblasts included in this map (Urbach and Technau, 2003a). However, the expression of these markers was not followed into later embryonic or larval stages, mainly due to the fact that anatomical landmarks to which expression patterns could be related had not been defined. Such markers, in the form of stereotyped clusters of neurons whose axons project along cohesive bundles (“primary axon bundles” or “PABs”) are now available (Younossi-Hartenstein et al., 2006). In the present study we have mapped the expression of molecular markers in relationship to primary neuronal clusters and their PABs. The markers we analyzed include many of the genes involved in patterning of the brain along the anteroposterior axis (cephalic gap genes, segment polarity genes) and dorso-ventral axis (columnar patterning genes), as well as genes expressed in the dorsal protocerebrum and visual system (early eye genes). Our analysis represents an important step along the way to identify neuronal lineages of the mature brain with genes expressed in the early embryo in discrete neuroblasts. Furthermore, the analysis helped us to reconstruct the morphogenetic movements that transform the two-dimensional neuroblast layer of the early embryo into the three-dimensional larval brain and provides the basis for deeper understanding of how the embryonic brain develops.
Keywords: Drosophila, embryonic brain, brain development, Hox genes, pair rule genes, segment polarity genes, head gap genes, retinal patterning genes, columnar patterning genes
1. RESULTS AND DISCUSSION
The brain of Drosophila is formed by structurally complex compartments, including (among many others) the mushroom body (Strausfeld et al., 1998; Jefferis et al., 2002), the central complex (Strauss and Heisenberg, 1993; Renn et al., 1999), the optic ganglia (Meinertzhagen, 1993; Morante and Desplan, 2004) and the olfactory glomeruli of the antennal lobe (Stocker, 1994; Laissue et al., 1999). The neurons of the mushroom body and some of the antennal lobe projection neurons have been traced back to embryonic brain elements (Lee et al., 1999; Noveen et al., 2000; Kurusu et al., 2002; Marin et al., 2005; Ramaekers et al., 2005). However, for most of the mature brain compartments, the embryonic origin remains elusive, and as a result, information about the cellular and molecular mechanisms that control their development is lacking.
The compartments of the mature Drosophila brain are formed by a stereotyped set of approximately 100 lineages. Lineages are derived from neuroblasts that delaminate from the neurectoderm of the early embryo and undergo stereotyped series of divisions. The population of brain NBs has been investigated extensively, and characteristic patterns of gene expression have been defined for most neuroblasts (Younossi-Hartenstein et al., 1996; Urbach and Technau, 2003a, b, Urbach et al., 2003; reviewed in Urbach and Technau, 2004). In the late embryo, lineages comprise in the order of 10–20 primary neurons; during a second phase of neuroblast proliferation that takes place in the larva, this number is increased ten-fold. In the late larval brain (Pereanu and Hartenstein, 2006) cells of one lineage are grouped together and project axons that fasciculate into a cohesive bundle with characteristic trajectory in the neuropile. These features enabled us to generate a map of larval lineages. It is our goal to follow lineages forward, into the adult brain, to establish how lineages contribute to functionally or structurally defined compartments. Furthermore, we want to go backward in development to discover how lineages relate to the embryonic neuroblast map, and thereby to establish for each lineage the “history” of gene expression.
One step in following larval lineages backward in development was taken in a recent study that defined stereotyped axon bundles, called primary axon bundles (PABs), emanating from clusters of primary neurons, which most likely represent individual lineages (Younossi-Hartenstein et al., 2006). PABs form a pattern that foreshadows the pattern of lineages recognized in the larval brain. To clearly establish the relationship between late embryonic primary neuronal clusters and larval lineages we need to utilize molecular markers that are continuously expressed from embryonic to late larval stages. We searched for such markers among the large number of patterning genes defined for the embryonic neuroblast map (Urbach and Technau, 2003a), and we describe here 18 genes whose expression persists in the late embryonic brain. Since in the early embryo, most of these genes are expressed in subsets of neuroblasts localized at discrete position along the anteroposterior axis (e.g., head gap genes) and dorso-ventral axis (e.g., columnar genes, eye specification genes), the comparison of their early and late expression pattern allowed us to infer certain conclusions regarding the morphogenetic movements that shape the embryonic brain.
1.1 Atlas model of primary neuronal clusters and PABs
The labeling of primary neuron clusters and their primary axon bundles by elav::Gal4 driven UAS::Synaptobrevin-GFP has produced a map of the primary cell clusters in the late embryonic brain (Younossi-Hartenstein et al., 2006). To confirm that the elav::Gal4 driver line is expressed in all neuronal clusters and PABs we compared the brain structures labeled by elav::Gal4 driven UAS::Syn-GFP with the pan-neuronal labeling of the anti-HRP (horseradish peroxidase) antibody that reveals all cell bodies of the cortex, as well as their neurites in the neuropile (Jan and Jan, 1982; Fig. 1). Anti-HRP staining visualizes the same pattern of primary axon bundles that is seen following labeling elav::Gal4 driven UAS::Synaptobrevin-GFP (arrow in Fig. 1). Based on confocal optical sections of elav::Gal4 driven UAS::Synaptobrevin-GFP labeled preparations, a digital 3D atlas model of all of the primary cell clusters observed at late embryonic stage 15 was generated (Younossi-Hartenstein et al., 2006). Aside from primary neuronal clusters, the array of Fasciclin II-positive pioneer neurons (that represent early differentiating primary neurons of strategically positioned lineages; Nassif et al., 1998) was also incorporated into the atlas model. Figures 2A and 2B show a dorsal and lateral view of the model, respectively (compare to Fig. 2E, F, which shows the orientation of panels 2A and 2B within the stage 15 embryo). Individual clusters are represented in a color coding scheme that will be used throughout the subsequent description (for nomenclature of cell clusters, see Material and Methods).
Figure 1.
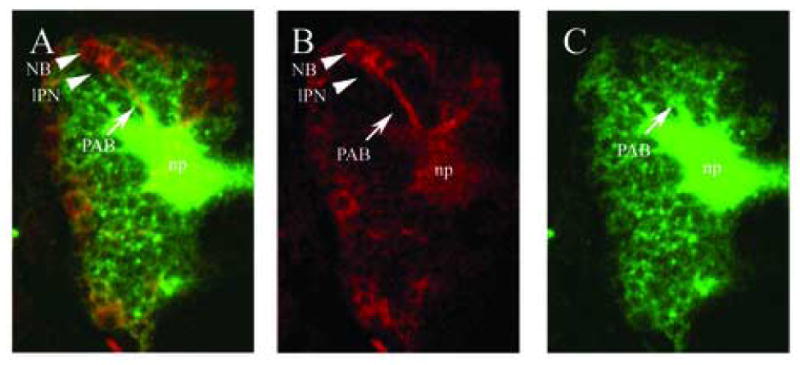
Comparison of labeling with a conventional anti-HRP antibody and with an elav::Gal4 driven UAS::syn-GFP. (A–C): dorsal views of brains of stage 15 embryos carrying the elav::Gal4 driven UAS::syn-GFP inserts, stained with anti-GFP (B; red) and anti-HRP (C; green); (A) is a merge of (B) and (C). Anti-GFP immunostaining (B) shows primary axon bundles (PAB; arrow) and late-born primary neurons (lPN) from which they emanate. Many clusters still include neuroblasts (NB). (C) Anti-HRP antibody immunoreactivity includes the PABs and the embryonic neuropile (np). Since all neuronal cell bodies show strong anti-HRP immunoreactivity, the primary axon bundles and their associated primary cell clusters can not be distinguished very clearly. In contrast, elav::Gal4 driven UAS::Synaptobrevin-GFP shows a marked temporal dynamics in that the more immature neurons of the primary cell clusters, which are located near the cortex surface, express the reporter gene at much higher levels than the more mature neurons and their neurites (Fig. 1B). This makes it possible to anatomically map the primary cell clusters and PABs individually.
Figure 2.
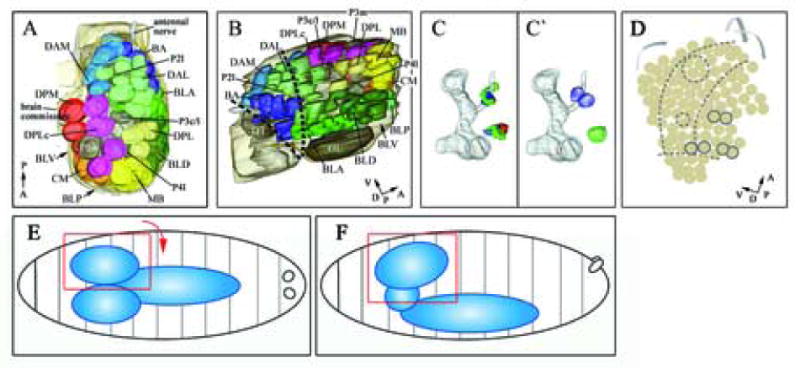
Digital 3D model of the stage 15 embryonic brain in dorsal view (A; anterior to the top) and lateral view (B, anterior to the left). Color code of primary cell clusters according to Table 1 (FasII expressing clusters, which are normally deeper in the brain are in dark grey; optic Lobe and the DT clusters are visible). (C) Dorsal view of the morphed en b1- and en b2-stripes of three independent brain models; red, blue and green show the en expression of the three individual brains (neuropile shown in grey). (C′) Generalized model of en-expressing cell clusters (neuropile shown in grey). (D) Morphogenetic deformation of the neuroblast map between early and late embryonic stages indicated by arrows (For major landmarks engrailed expression (grey circles) and mushroom bodies (large dashed circle) and FasII cluster (small dashed circle) are displayed; dashed lines indicate columnar domains of the embryonic neurectoderm). (B) Dark dashed lines indicated neuromeric boundaries between protocerebral (anterior), deuterocerebral (middle) and tritocerebral (posterior) domains. Perpendicular arrows show the anteroposterior and dorsoventral orientation of the brain according to neuraxis in A, B and D. Schematic overview of the CNS at stage 15 embryo in a dorsal view (E) and a lateral view (F). In the following (as shown in A, B) a close-up view of the hemi-brain will be shown as indicated by rectangles (red) in E (orientation for panel A rotated 90° clockwise) and F (orientation for panel B).
In order to relate the expression of molecular markers to primary neuronal clusters, volume renderings of gene expression pattern obtained from serial confocal sections were warped into the digital 3D model and subsequently annotated (see Material and Methods; see also Pereanu and Hartenstein, 2004). To verify that the process of warping results in reproducible mapping of expression domains onto primary cell cluster models at the same location, we warped the neuronal clusters expressing the engrailed (en) gene of three different embryos into the atlas model (Fig. 2C; see Material and Methods). Even though there are minor inter-individual differences in the location of the en expression domains we find that in each case they map to the same primary neuronal clusters (Fig. 2C, C′): the primary cell clusters BA2 and BA3 of the posterior deuterocerebrum, and the protocerebral primary cell cluster BLA6/7. We therefore assume that despite minor variability in gene expression patterns and in the morphology of embryonic brains (mostly due to slight differences in age), a reproducible mapping of gene expression onto the primary neuronal clusters of the atlas model is possible.
Most molecular markers are expressed in NBs and their progeny in a highly dynamic pattern. For instance engrailed, even though roughly expressed in all cells of the described projection clusters, shows a temporally dynamic expression pattern, in that it declines as neurons maturate. Other markers, such as orthodenticle (otd) or eyes absent (eya), remain expressed in all neurons of some clusters, whereas they are confined to only a few cells in other clusters. Only few, if any, of the molecular markers described in this study show a “simple” continuous expression pattern which includes all neurons of a fixed set of primary clusters. That being said, our analysis revealed that clusters that by late stage 11 [the stage represented in the neuroblast map of Urbach et al. (2003)] express a given gene, generally maintain some level of expression of that gene in the late embryonic brain. Likewise, only in rare cases does de novo expression of a gene in a cluster begin later than the end of stage 11 (V.H., unpublished observation). This allowed us to relate, even if at a relatively low level of resolution, the pattern of NBs expressing a gene with the pattern of clusters descended from the NBs (see below).
1.2 Expression of molecular markers in primary cell clusters
We have analyzed the expression patterns of 18 key developmental control genes in relation to about 72 identified primary cell clusters in the late embryonic brain cortex. The genes studied include members of the cephalic gap genes, segment polarity genes, pair rule genes, Hox and Pax genes, and columnar patterning genes. An overview of the expression patterns of all of the genes studied in relation to all of the primary cell clusters is given in Table 1. Detailed 3D models of the primary cell clusters that express a selected subset of these genes are presented in Figs 3, 4 and 5. The expression of the two genes en and ventral nervous system defective (vnd) in the late embryonic brain are shown at high magnification in individual confocal sections (Fig. 3).
Table 1.
Molecular marker expression in primary projection clusters. Anatomical terminology according to Younossi-Hartenstein et al. (Younossi-Hartenstein et al., 2006). Clusters include DAM (dorsal anterior medial) and BA (basal anterior) of the deuterocerebrum; DAL (dorsal anterior lateral), DPM (dorsal posterior medial), DPL (dorsal posterior lateral), DPLc (dorsal posterior lateral central), CP (central posterior), CM (central medial), BLA (basal lateral anterior), BLD (basal lateral dorsal), BLP (basal lateral posterior), MB (mushroom body). The clusters D/T, P1, P2, P3, P4, P5 and the optic lobe primordium (grey) also express Fas II. Clusters which will split later (sub-clusters) are indicated by slashed numbers, for more information on nomenclature see material and methods.
| cluster nr | en | hh | wg | fkh | eve | odd | ems | otd | tll | Dll | vnd | msh | ey | toy | dac | eya | lab | exd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Optic lobe | ● | ● | ● | ● | ● | ● | ||||||||||||
| MB1 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| MB2 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| MB3 | ● | ● | ● | ● | ● | ● | ||||||||||||
| MB4 | ● | ● | ● | ● | ● | ● | ||||||||||||
| DPM1/2 | ● | ● | ● | |||||||||||||||
| DPM3 | ● | ● | ||||||||||||||||
| DPM4 | ● | ● | ||||||||||||||||
| DPLc1/2 | ● | ● | ● | ● | ● | |||||||||||||
| DPLc3 | ● | ● | ● | ● | ● | ● | ||||||||||||
| DPLc4/5 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| DPLc6/7 | ● | ● | ● | ● | ● | |||||||||||||
| DPL1/2 | ● | ● | ● | ● | ● | |||||||||||||
| DPL3 | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| DPL4/5 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| CPv1 | ● | ● | ● | ● | ● | ● | ||||||||||||
| CPv2 | ● | ● | ● | ● | ● | |||||||||||||
| CPd | ● | ● | ● | ● | ● | ● | ||||||||||||
| CPm | ● | ● | ● | ● | ||||||||||||||
| CM1/2 | ● | ● | ||||||||||||||||
| CM3/4/5 | ● | ● | ● | ● | ||||||||||||||
| P31 | ● | ● | ● | ● | ● | ● | ||||||||||||
| P31p | ● | ● | ● | ● | ● | |||||||||||||
| P41 | ● | ● | ● | ● | ● | |||||||||||||
| P51 | ● | ● | ● | ● | ||||||||||||||
| BLA1 | ● | ● | ● | ● | ● | |||||||||||||
| BLA2/3 | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| BLA4/5 | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| BLA6/7 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |||||||
| BLD1/2 | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| BLD3/4 | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| BLD5/6 | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| BLD7/8 | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| BLD9/10 | ● | ● | ● | ● | ||||||||||||||
| BLD11 | ● | ● | ● | ● | ● | |||||||||||||
| BLV1/2/3 | ● | ● | ||||||||||||||||
| BLV4/5/6 | ● | ● | ● | ● | ||||||||||||||
| BLP1/2 | ● | ● | ● | ● | ||||||||||||||
| BLP3/4 | ● | ● | ● | ● | ● | ● | ||||||||||||
| BLP5/6 | ● | ● | ● | ● | ● | |||||||||||||
| BLP7/8 | ● | ● | ● | ● | ● | ● | ||||||||||||
| DAL1 | ● | ● | ||||||||||||||||
| DAL2 | ● | ● | ● | |||||||||||||||
| DAL3 | ● | ● | ● | |||||||||||||||
| DAL4 | ● | ● | ● | ● | ||||||||||||||
| DAL5 | ● | ● | ● | ● | ● | |||||||||||||
| DAL6 | ● | ● | ● | ● | ● | |||||||||||||
| DAL7 | ● | ● | ● | |||||||||||||||
| DAL8 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| DAL9 | ● | ● | ● | ● | ● | ● | ||||||||||||
| DAL10 | ● | ● | ● | ● | ● | ● | ||||||||||||
| DAM1 | ● | ● | ● | |||||||||||||||
| DAM2 | ● | ● | ● | |||||||||||||||
| DAM3 | ● | ● | ● | ● | ● | |||||||||||||
| DAM4 | ● | ● | ● | |||||||||||||||
| HA1 | ● | ● | ● | ● | ||||||||||||||
| HA2 | ● | ● | ● | ● | ● | ● | ||||||||||||
| HA3 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| HA4 | ● | ● | ● | ● | ● | ● | ● | ● | ● | |||||||||
| HA5 | ● | ● | ● | ● | ||||||||||||||
| HA6 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| HA7 | ● | ● | ● | ● | ● | ● | ● | |||||||||||
| P4m | ● | ● | ● | |||||||||||||||
| P3m | ● | ● | ● | ● | ||||||||||||||
| P11v | ||||||||||||||||||
| P11a | ||||||||||||||||||
| P11p | ● | ● | ● | |||||||||||||||
| P21a | ● | ● | ● | ● | ||||||||||||||
| P21 | ● | ● | ● | ● | ||||||||||||||
| D/Tda | ● | ● | ● | ● | ● | ● | ● | ● | ||||||||||
| D/Ta | ● | ● | ● | ● | ● | ● | ||||||||||||
| D/T1 | ● | ● | ● | ● | ● | ● | ● |
Figure 3.
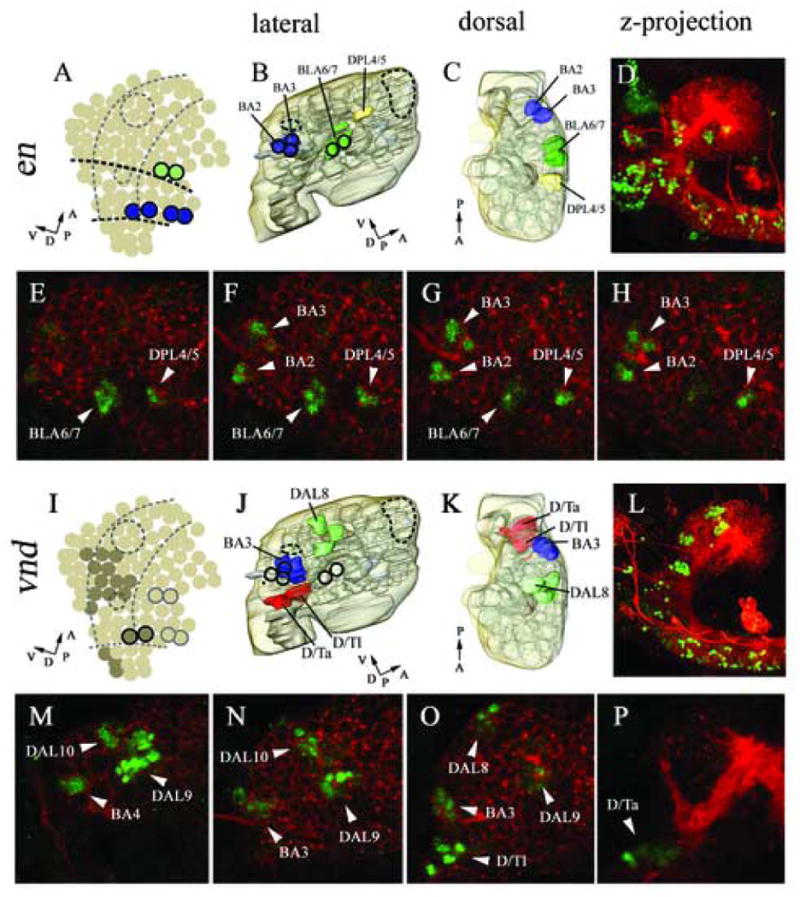
Expression in cortical cell clusters of engrailed (B–H), and ventral nervous system defective (J–P). Marker gene expression in neuroblasts according to Urbach et al., 2003 (A, I). 3D reconstructed models of molecular marker gene expression showing a lateral (B, J), and dorsal (C, K) view. The model includes the neuropile (light grey) and brain surface (transparent light brown). Lateral view of double immunostaining with anti-HRP (red) and anti-EN (green, D–H), anti-VND (green, L–P); confocal microscopic reconstructions of optical sections spanning from the midline to lateral most parts of the brain, lateral view (D, L). High magnification single confocal sections within the brain cortex from distal most (E, M) to medial most (H, P); intermediate sections (F, G, N, O). en expression is found at neuromeric boundary regions in specific cortical clusters (B, C, D). Anti-EN staining is found in the BLA6/7, BA3, BA4 and DPL4/5 clusters (E–H). vnd is expressed in specific clusters in the ventral part of the cortex (according to neuraxis) (J, K). Anti-VND staining is found in the DAL8, DAL9, DAL10, BA2, BA1, D/Ta and D/Tl clusters (M–P). For major landmarks engrailed expression (dark circles) and mushroom bodies (large dashed circle) and FasII cluster (small dashed circle) are displayed (A, B, I, J). (A, I) Dashed lines indicate columnar domains of the embryonic neurectoderm. (A) Dark dashed lines indicated neuromeric boundaries between protocerebral (anterior), deuterocerebral (middle) and tritocerebral (posterior) domains. (A, B, C, I, J, K) Perpendicular arrows show the anteroposterior and dorsoventral orientation of the brain according to neuraxis. For orientation specific clusters are labelled by name, for nomenclature see material and methods.
Figure 4.
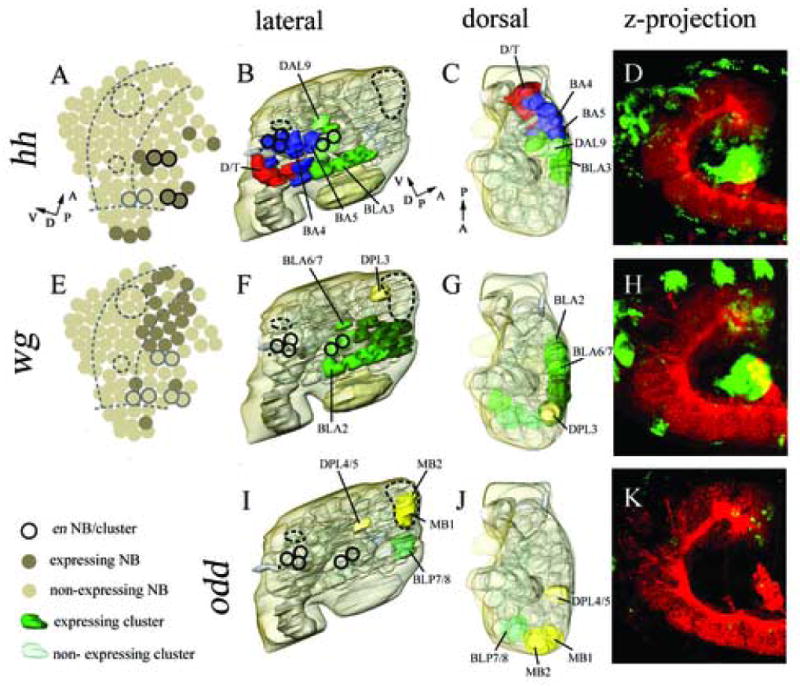
Expression in cortical cell clusters of, hedgehog (B–D), wingless (F–H), and odd skipped (I–K). Marker gene expression in neuroblasts according to Urbach et al., 2003 (A, E; note: no neuroblast expression data available on odd). 3D reconstructed models of molecular marker gene expression showing a lateral (B, F, I), and dorsal (C, G, J) view. The model includes the neuropile (light grey) and brain surface (transparent light brown). hh expression locates more posterior in a dorsoventral intermediate domain (B, C), whereas wg expression is restricted an antero-dorsal part of the protocerebrum (F, G) (according to neuraxis). odd is expressed in only 4 neuronal clusters, including two MB clusters and the DPL 4/5, BLP 7/8 clusters (I, J). Double immunostaining with anti-HRP (red) and anti-βGal (D), anti-βGal (H), anti-ODD (K). Confocal microscopic reconstructions of optical sections spanning from the midline to lateral most parts of the brain, lateral view. For major landmarks engrailed expression (dark circles) and mushroom bodies (large dashed circle) and FasII cluster (small dashed circle) are displayed (A, B, E, F, I). (A, I) Dashed lines indicate columnar domains of the embryonic neurectoderm. (A, B, C) Perpendicular arrows show the anteroposterior and dorsoventral orientation of the brain according to neuraxis. For orientation specific clusters are labelled by name, for nomenclature see material and methods.
Figure 5.
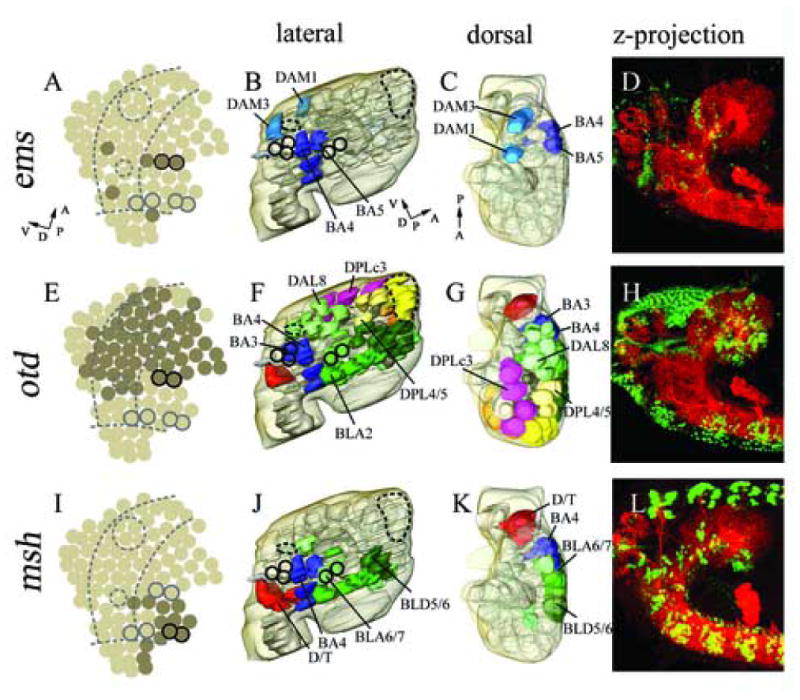
Expression in cortical cell clusters of ems (B–D), otd (F–H) and msh (J–L). Marker gene expression in neuroblasts according to Urbach et al., 2003 (A, E, I). 3D reconstructed models of molecular marker gene expression showing a lateral (B, F, J), and dorsal (C, G, K) view. The model includes the neuropile (light grey) and brain surface (transparent light brown). ems expression is restricted to a small number of cell clusters in an AP intermediate part (B, C), whereas otd expression covers large parts of the brain cortex (F, G). msh expression locates more in dorsal parts of the embryonic brain cortex (according to neuraxis) (J, K). Double immunostaining with anti-HRP (red) and anti-EMS (green, D), anti-OTD (H), anti-MSH (L); confocal microscopic reconstructions of optical sections spanning from the midline to lateral most parts of the brain, lateral view. For major landmarks engrailed expression (dark circles) and mushroom bodies (large dashed circle) and FasII cluster (small dashed circle) are displayed (A, B, E, F, I, J). (A, I) Dashed lines indicate columnar domains of the embryonic neurectoderm. (A, B, C) Perpendicular arrows show the anteroposterior and dorsoventral orientation of the brain according to neuraxis. For orientation specific clusters are labelled by name, for nomenclature see material and methods.
1.2.1 Segment polarity genes
In the neurectoderm and NBs of the ventral nerve cord, segment polarity gene expression forms stereotypical arranged segmental stripes which subdivide each neuromere along the anteroposterior axis (Bhat, 1996; Broadus et al., 1995; Skeath et al., 1995; Zhang et al., 1994). We have analyzed the expression of the segment polarity genes en, wingless (wg) and hedgehog (hh) in the cortical clusters of the late embryonic brain. The en gene is expressed in two rows of NBs demarcating the posterior boundary of the tritocerebrum (not shown) and deuterocerebrum (Fig. 3), respectively; further anterior, en appears in two NBs that form the “head spot” (Fig. 3). The head spot indicates the posterior boundary of the protocerebral neuromere. In the late embryonic brain, clusters descended from each of these segmentally organized groups of neuroblasts can be recognized (Hirth et al., 1995; Hirth et al., 2003). We find that the deuterocerebral en-positive clusters correspond to the BA2 and BA3 cell clusters (Fig. 3 B–D and F–H). The protocerebral head spot gives rise to two neuronal clusters. The more massive one of these coincides with the BLA6/7 cell cluster (Table 1; Fig. 3 B–D and E–G). The second cluster (“secondary head spot”; Hirth et al., 1995) is located further dorsally and coincides with the DPL4/5 cluster (Fig. 3 B–D and E–H).
For the analysis of wingless (wg) and hedgehog (hh) expression, we used wg-LacZ and hh-LacZ lines, which previously have been shown to mimic the endogenous expression of wg and hh (Mohler et al., 1995; Broadus et al., 1995; Page, 2002; Urbach and Technau, 2003b). hh expression is detected in the embryonic procephalic neurectoderm in a pattern that largely overlaps with that of en (Urbach et al., 2003b). Correspondingly, the pattern of stage 11 neuroblasts expressing hh roughly coincides with that of en-positive neuroblasts (Urbach and Technau, 2003b). During later stages hh expression appears to expand to neighboring regions that include a substantial number of deuterocerebral and posterior protocerebral clusters (Page, 2002). We also find hh expressed predominantly in the deuterocerebrum and in the posterior protocerebral BLA cell clusters (Table 1; Fig. 4B–D). hh expression is also detected in the tritocerebral D/T clusters and the optic lobe primordium (data not shown).
wg expression is detected in stripes or clusters of neurectodermal cells that are anteriorly adjacent to the en/hh stripes (Urbach and Technau, 2003b). In the deuterocerebrum, wg appears in only two neuroblasts; in the protocerebrum, a large group of posterior protocerebral neuroblasts express this gene (Fig. 4E). In the late embryonic brain, wg expression was reported to be only maintained in the protocerebral neuromere (Urbach and Technau, 2003b; Richter et al., 1998), and accordingly we find that wg expression covers many of the BLA and BLD cell clusters that represent part of the posterior protocerebrum (Table 1; Fig. 4 F–H).
1.2.2 Pair rule genes
The pair-rule genes odd skipped (odd) and even skipped (eve) have been characterized in the developing ventral nerve cord (VNC). Both are expressed in segmentally reiterated, transient patterns in the VNC (Broadus et al., 1995; Skeath et al., 1995). In contrast, in the late embryonic brain cortex, odd and eve expression appears to be non-segmental. Odd is restricted to a few protocerebral clusters, including BPL7/8 and DPL4/5, as well as two of the four mushroom body (MB) clusters, namely MB1 and MB2 (Table 1; Fig. 4 I–K). eve is expressed at the trito-deuterocerebral boundary in the D/Tda cluster of Fas-II positive neurons (Table 1).
1.2.3 Cephalic gap genes
The cephalic gap genes empty spiracles (ems), orthodenticle (otd) and tailless (tll) are expressed in large domains of the procephalic region and play a crucial role in the patterning of the neurectoderm and the embryonic brain (Walldorf and Gehring, 1992; Hirth et al., 1995; Younossi-Hartenstein et al., 1997; Hartmann et al., 2000; Younossi-Hartenstein et al., 1996; Urbach et al., 2003). ems expression is present in a small number of neuroblasts that are closely associated with the deuterocerebral and protocerebral en-stripe (Hirth et al., 1995). Correspondingly, we find that in the late embryonic brain, ems appears in two DAM clusters (DAM1, DAM3) and two BA clusters (BA4, BA5) which are located in between the en-positive deuterocerebral and protocerebral clusters (BA2/3 and BLA6/7, respectively); in addition, ems is present in the clusters BA6 and BA7 which are located posterior to the deuterocerebral en-domain (Table 1; Fig. 5 B–D). Based on this topology, we assign all of the ems-positive clusters to the deuterocerebrum.
otd expression in the early embryo forms a large domain covering most of the protocerebral and part of the deuterocerebral neurectoderm, as well as all NBs that delaminate from this domain (Hirth et al., 1995; Younossi-Hartenstein et al., 1997; Urbach et al., 2003). During later embryonic stages otd expression has been described in large parts of the protocerebral and deuterocerebral neuromere (Hirth et al., 1995; Hirth et al., 2003). We find that otd is expressed in the majority of protocerebral clusters, as well as deuterocerebral clusters adjacent to the protocerebrum. Significantly, DAM clusters and DPM clusters are not otd-positive, suggesting that these clusters are derived from the group of anterior deuterocerebral and protocerebral neuroblasts that were also negative for otd (Table 1; Fig. 5 E–H). This notion is supported by analyzing the pattern of clusters expressing tll. This gene was mapped to the entirety of protocerebral neuroblasts, including those of the anterior protocerebrum (Rudolph et al., 1997; Younossi-Hartenstein et al., 1997; Urbach et al, 2003). Correspondingly, at embryonic stage 15 we find tll expression in a majority of the clusters in the protocerebral cortex, including the DPM group that is lacking from among the otd-positive neuroblasts (Table 1).
1.2.4 Columnar patterning genes
In the ventral neurectoderm of the trunk that gives rise to the neuroblasts of the VNC, the gene ventral nervous system defective (vnd) is expressed in a ventral columnar domain, whereas muscle specific homeobox (msh) expression is detected in a dorsal columnar domain (for review see Cornell and Von Ohlen, 2000; Mc Donald et al., 1998; Isshiki et al., 1997). In the neurectoderm and NBs of the head, vnd and msh are also expressed in columnar domains that represent the forward continuation of the columns of the trunk. Moreover, their anterior expression borders appear to coincide with neuromeric boundary regions, whereby msh is confined to the tritocerebrum and deuterocerebrum, and vnd expression extends up to the protocerebrum (Urbach and Technau, 2003; Fig. 3I). In the late embryonic brain, vnd is expressed in the BA3 and BA4 clusters of the posterior deuterocerebrum (Fig. 3 J–K and M–O). Further we detect vnd expression in the DAL8, DAL9 and DAL10 clusters of the posterior protocerebrum (Table 1; Fig. 3I–P). All of these vnd expressing cell clusters are located ventrally with respect to the neuraxis. In contrast, msh is expressed in the late embryonic brain in the tritocerebral D/T clusters, in the dorsal deuterocerebral BA4, BA5, BA6, BA7, and in a number of clusters at the dorsal deutero-protocerebral boundary region (several DAL, BLA and BLD clusters; Table 1, Fig. 5 J–L).
1.2.5 Genes involved in eye development
Genetic analysis of the formation of the adult eye of Drosophila has lead to the identification of a conserved group of transcriptional regulators collectively known as the retinal determination network (RDN), or early eye genes. This genetic network comprises a hierarchical cascade, where twin of eyeless (toy) activates eyeless (ey), ey in turn activates both eyes absent (eya) and sine oculis (so), and eya and so activate dachshund (dac). Reciprocal positive feedback loops between these genes ensure robust network function (Czerny et al., 1999; Halder et al., 1995; Niimi et al., 1999; Pignoni et al., 1997; Shen and Mardon, 1997). toy, ey, dac and eya are also expressed during embryonic brain development (Kurusu et al., 2000; Noveen, et al., 2000; Kammermeier et al., 2001, Urbach et al., 2003). We have analyzed the expression of toy, ey, dac and eya in the cortex of the late embryonic brain (Table 1). We find that ey and toy are expressed in numerous cell clusters in the protocerebrum, deuterocerebrum and tritocerebrum of the late embryonic brain. dac is expressed in a number of cell clusters in the protocerebrum and deuterocerebrum but not in the tritocerebrum. eya is only expressed in a several primary cell clusters in the protocerebrum and deuterocerebrum. ey, toy and dac but not eya are expressed in all of the developing MB clusters (data not shown). Furthermore ey, dac and eya, but not toy expression is detected the optic lobe primordium.
1.2.6 Other developmental control genes
The expression of four other genes which have play distinct roles during embryonic development was studied. These are the genes labial (lab), extradenticle (exd), Distal-less (Dll), and fork head (fkh). In Drosophila, the homeotic gene lab his expressed in the tritocerebrum, where it is required for proper neuromere differentiation (Hirth et al., 1998). We find that lab expression is restricted to tritocerebral D/T primary cell clusters and cannot be detected in more anterior regions (Table 1). In contrast, the Hox co-factor exd has been reported to be expressed in large parts of the developing tritocerebrum, deuterocerebrum and posterior protocerebrum (Nagao et al., 2000). In accordance with this, we find exd expression in the tritocerebral D/T cluster, in all BA clusters and in P3m and P4m of the deuterocerebrum (but not in the deuterocerebral DAM clusters) and in a number of protocerebral clusters (Table 1). Dll has been found to be expressed in anterior parts of the late embryonic brain, including glia cells in the protocerebrum (Panganiban and Rubenstein, 2002). We analyzed the expression pattern of Dll in the late embryonic brain cortex using the Dll-LacZ reporter line (Kaphingst and Kunes; 1994). We find Dll expression in numerous clusters in the protocerebrum and also in some DAL clusters in the deuterocerebrum but not in tritocerebrum and posterior deuterocerebrum (Table 1). The fkh gene is involved in the development of the embryonic brain, and fkh mutants have a strong defect in the formation of the protocerebral commissure as well as in gut formation (Page, 2002; Jürgens and Weigel, 1988). We find fkh expression in late embryonic brain cortex clusters in the protocerebrum and deuterocerebrum, but not in the tritocerebrum (Table 1).
1.3 Anteroposterior, dorsoventral and neuromeric subdivisions of the embryonic brain cortex
During early neurogenesis the brain anlage consists of a single-layered epithelium, the procephalic neurectoderm, from which neuroblasts delaminate. Similar to the neuroblasts of the ventral nervous system, individual brain neuroblasts can be assigned coordinates in a two dimensional Cartesian coordinate system (Urbach et al., 2003). The demarcation of anteroposterior and dorsoventral domains during these early embryonic stages (stages 8–11) is reflected by the expression of specific patterning genes, such as the columnar genes, segment polarity genes or head gap genes. In the late embryonic brain (stage 15), the subdivision into clear anteroposterior and dorsoventral domains is more complicated since each brain hemisphere is a three dimensional structure consisting of the developing neuropile surrounded by the primary clusters of the cell cortex. Furthermore, the neuraxis of the late embryonic brain manifests a prominent hook-like backward flexure. Thus at this developmental stage anterior according to body axis is posterior according to neuraxis, while dorsal according to body axis is ventral according to neuraxis (compare Hirth et al., 1995). Previously, neuromeric boundary regions of the late embryonic brain have been estimated by using en as marker (Hirth et al., 1995). A more precise determination of neuromeric boundaries and other anteroposterior subdivisions in the late embryonic brain cortex will require additional information such as the combinatorial expression patterns of the molecular markers described here. Although this information is still incomplete, we find that a tentative assignment of neuronal clusters to neuromeric domains in the late embryonic brain is possible. A summary scheme of this tentative assignment of neuronal projection clusters to brain neuromeres is shown in Figure 6. On the basis of anteroposterior position and relation to en-stripes, we propose that the D/T clusters are tritocerebral and that the DAM and BA clusters are deuterocerebral. We assign the DAL1–DAL7 clusters to the deuterocerebrum and DAL8–10 (which express the protocerebral marker tll and/or vnd as a marker for posterior segmental boundary; see below) to the protocerebrum. The remaining cell clusters in more anterior (according to neuraxis; see above) regions are all assigned to the protocerebrum.
Figure 6.
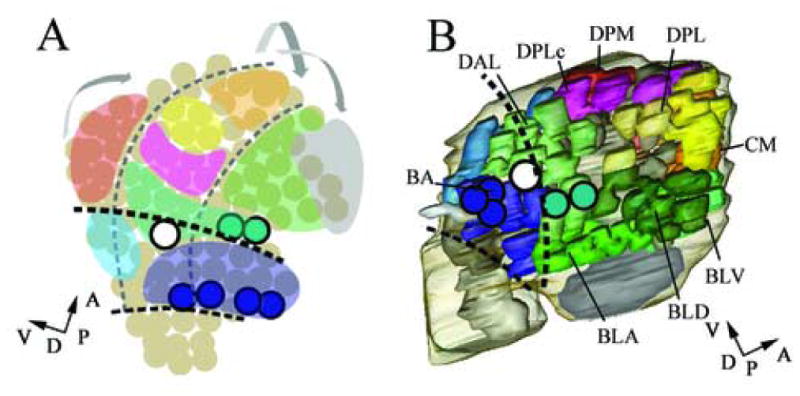
A summary model of tentative assignment of neuronal projection clusters to brain neuromeres in comparison to the neuromeric subdivision of the early brain and late stage embryonic brain. (A) Neuromeric and dorsoventral subdivision of neuroblasts of the stage 11 embryonic brain according to Urbach et al., 2003. The marked expression of engrailed delimits neuronal segments (dark circles). The b1 en-stripe (or en head spot) delimits the posterior protocerebrum (green, B), the b2 en-stripe (or en antennal stripe) delimits the posterior deuterocerebrum (blue, B). (A) Correspondingly engrailed expressing neuroblasts in the antennal neuromere and ocular neuromere delimit neuromeric boundaries during earlier stages. (B) Tentative subdivision of the stage 15 late embryonic brain. We assign the DAL1-DAL7 clusters to the deuterocerebrum and DAL8-10 to the protocerebrum. The remaining cell clusters in more anterior are assigned to the protocerebrum. (B) According to marker gene expression the brain cortex clusters BA, BLA, BLD and BLV define a dorsal domain while the DPM; DPL, DPLc, CP, CM clusters define a ventral domain. (A, B) Perpendicular arrows show the anteroposterior and dorsoventral orientation of the brain according to neuraxis. (A, B) Dark dashed lines indicated neuromeric boundaries between protocerebral (anterior), deuterocerebral (middle) and tritocerebral (posterior) domains. (A) dashed lines indicate columnar domains of the embryonic neurectoderm. (A) Morphogenetic deformation of the neuroblast map between early and late embryonic stages indicated by arrows; Potential progenitor domains are color coded accordingly (compare Table 1).
There is some evidence that the deuterocerebral/protocerebral domains of the late embryonic brain cortex can be further subdivided into dorsal and ventral regions. The columnar gene vnd, which in the early embryonic brain and VNC is expressed in ventrally located NBs, is expressed in the late embryonic brain cortex in some of the cell clusters that are located ventral according to neuraxis (see above). Moreover, the columnar gene msh, which in the early embryonic brain and VNC is expressed in dorsally located NBs, is detected in the late embryonic brain cortex in some of the cell clusters that are located dorsal according to neuraxis (see above). If we can take these findings as indicative of DV patterning in the deuterocerebral/protocerebral regions of the cortex, we would, based on position of primary cell clusters relative to the neuraxis, propose that the brain cortex clusters BA, BLA, BLD and BLV define a dorsal domain while the DPM; DPL, DPLc, CP, CM clusters define a ventral domain. However, it should be stressed that this assignment of cortex clusters to dorsoventral domains is highly preliminary and requires verification by substantial experimental data.
1.4. Developmental control gene expression in brain development of Drosophila and vertebrates
Even though the embryonic brain of Drosophila is very different in neuroanatomical and morphological properties from its vertebrate counterpart, many of the gene homologs expressed in the developing brain of Drosophila can also be detected in the embryonic vertebrate brain. In the developing mouse neocortex the Emx2 and Emx1 as well as the Pax6 genes have been proposed to be genetic regulators of arealization (O’Leary and Nakagawa, 2002). Emx1 and Emx2 are expressed in the embryonic mouse neocortex in a rostrolateral (low) to caudomedial (high) gradient, whereas Pax6 expression follows an opposite concentration gradient (Gulisano et al., 1996; Mallamaci et al., 1998). The Drosophila homologs of these genes, ems and ey/toy, also show some degree of regionalized expression in the developing brain. ems is expressed predominantly in posterior parts (deuterocerebrum). In contrast, ey and toy are expressed throughout the brain largely overlapping in the protocerebrum. Thus similar to vertebrates ey/toy are co-expressed in anterior regions, whereas ems is present in more posterior regions.
In Drosophila, the homeobox genes vnd, intermediate neuroblasts defective (ind) and msh are expressed and required in longitudinal domains that form a ventral (vnd), intermediate (ind) and dorsal (msh) column in the ventral neurectoderm (Isshiki et al., 1997; McDonald et al., 1998; Chu et al., 1998; Weiss et al., 1998). The vertebrate homologs of these genes, Nkx2 (vnd), Gsh (ind) and Msx (msh) are similarly involved in dorsoventral patterning of the spinal cord (Qiu et al., 1998; Pera et al., 1998; Pabst et al., 1998; Shimamura et al., 1995; Wang et al., 1996). The dorso-ventrally ordered expression of vnd/Nkx2 and msh/Msx continues anteriorly into the anlage of the brain (Urbach and Technau 2003b; 2004). Similarly, expression of vnd and msh in the late embryonic brain cortex displays dorso-ventral specificity, where vnd is expressed in more postero-ventral portions and msh in more antero-dorsal domains (according to neuraxis).
In vertebrate CNS development the opposing action of signaling mechanisms from the ventral side (floorplate) and from the dorsal side (roofplate) are essential to specify neuronal identity in the neural tube (for review see Chizhikov and Millen, 2005; Marti and Bovolenta, 2002). Sonic hedgehog (Shh), expressed in the floorplate, is involved in inducing ventral fate in neurons and neuronal precursors of the neural tube, while suppressing dorsal fates. Wnt signaling from the dorsally located roof plate acts to inhibit the Shh response in developing neural tissue. Interestingly in the late embryonic brain cortex of the fly, we find that Drosophila wg is specifically expressed in dorso-anterior cell clusters whereas hh, the Drosophila homolog of Shh, is expressed in a spatially opposite manner in ventro-posterior cell clusters.
Taken together, the expression pattern of several molecular markers, such as the ey/toy/Pax6 genes; the dorsoventral genes vnd/Nkx2 and msh/Msx genes; the wg/Wnt1 genes and the hh/Shh genes display an unexpected degree of similarity during embryonic brain development in insects and vertebrates. Even though the mode of embryonic brain development in these two animal groups differs in many aspects and results in largely different neuroanatomy and brain morphology, a wealth of publications show that many genes that control the development of the brain are conserved, in both expression and function.
2. MATERIAL AND METHODS
2.1 Fly strains and genetics
Wild-type flies are of the strain Oregon R. Further, the following fly strains were used: UAS::Synaptobrevin-GFP (Estes et al., 2000; kindly provided by V. Hartenstein and R. Stocker); C155 elav::Gal4 (Lin and Goodman 1994; Sprecher et al., 2004; an enhancer trap line that expresses Gal4 in postmitotic neurons) hedgehog-lacZ (16E) (Mohler et al., 1995), wingless-lacZ (Broadus et al.,1995) and Distal-less-lacZ (kindly provided by G. Boekhoff-Falk). All experiments reported here were carried out at 25°C. Egg collections were done on yeasted apple juice agar plates (Ashburner, 1989) Embryos were staged according to Campos-Ortega and Hartenstein (1997).
2.2 Immunocytochemistry
Embryos were dechorionated, fixed and immunostained, according to Therianos et al. 1995. Primary antibodies were rabbit anti-HRP (FITC-conjugated) 1:100 (Jan and Jan, 1982) (Jackson Immunoresearch), rabbit anti-βGAL 1:200–1:400 (Milan Analytika), mouse anti-βGAL 1:20 (DSHB), mouse anti-FAS II 1:5 (Lin and Goodman, 1994, DSHB), rat anti-ELAV 1:30 (DSHB), mouse anti-REPO 1:20 (DSHB), rabbit anti-GFP 1:200 (Torrey Pines Biolabs), mouse anti-GFP at 1:20 (Roche), rabbit anti-LAB at 1:100 (F. Hirth and H. Reichert, unpublished), rat anti-LAB at 1:500 (F. Hirth and H. Reichert, unpublished), mouse anti-NRT at 1:20 (BP106 antibody, DSHB), rabbit anti VND at 1:500 (Mc Donald et al., 1998; kindly provided by C.Q. Doe), mouse anti-EN at 1:1 (Patel et al., 1989; DSHB), mouse anti-EYA 1:50 (Bonini et al., 1993, DSHB), mouse-anti-DAC at 1:50 (Mardon et al., 1994; DSHB), rabbit anti-HTH at 1:200 (Pai et al., 1998), and monoclonal mouse anti-EXD at 1:2 (Aspland and White, 1997), rat anti-EMS at 1:100 (Walldorf and Gehring, 1992), guinea pig anti-FKH 1:500 (Lehmann and Korge, 1996), rabbit anti-EY at 1:300, rabbit anti-TOY at 1:300 (both kindly provided by U. Walldorf), guinea pig a ODD 1:100 (Kosman et al., 1998); rabbit anti-OTD at 1:200 (Hirth et al., 2003). As secondary antibodies we used the respective Alexa-488, Alexa-568, and Alexa-647 antibodies generated in goat (Molecular probes), all 1:150.
2.3 Laser confocal microscopy and generation of 3D digital models
For laser confocal microscopy, a Leica TCS SP was used. Optical sections ranged from 0.2 – 1 μm recorded in line average mode with picture size of 512 × 512 pixels, or 1024 × 1024 pixels. Captured images from optical sections were arranged and processed using IMARIS (Bitplane), either directly exporting merged stacks for figures or for further use in ImageJ and AMIRA. Complete series of optical sections of the Leica confocal software were imported and processed using ImageJ. The raw tiff stacks were further imported into the AMIRA program, where user-defined “materials” were drawn manually around the labeled structures which were to be included in the model. Subsequently, the program synthesizes a surface by triangulation around the defined “materials”, which was further processed and smoothened. The process of warping was performed as follows: one pre-defined model was imported into a second model; to achieve the most precise overlay of the two models, the surface of one model was three dimensionally adapted by transforming; thereafter expression domains were assigned to specific cortex-clusters (Figure 3C, exemplifies the overlay of the specific projection clusters of two morphed models). For 2D representations in figures, screen shots were generated of appropriate angle, virtual lighting and transparency of individual material surfaces (For review see Pereanu and Hartenstein, 2004). Final figures were arranged and labeled using Adobe Photoshop.
2.4 Nomenclature
The nomenclature chosen for the 72 primary clusters observed at late embryonic stage 15 corresponds to that given in Younossi-Hartenstein et al. (Younossi-Hartenstein et al., 2006). Briefly, this nomenclature takes into account the location of a primary clusters in the cortex and the position where its primary axon bundle enters the neuropile. Specifically, D/T groups are tritocerebral, DAM and P2 groups are anteromedial deutocerebral, BA and P1 groups are anterolateral deuterocerebral, P4m groups are posteromedial deuterocerebral, DAL and P3c groups are at the proto-deuterocerebral boundary, CM, BLA/D/P/V and P5m groups are basal protocerebral, DPL, DPLc, P4Iv, and P3I groups are dorso-lateral protocerebral, DPM and P3m groups are dorsomedial protocerebral, MB (mushroom body), CP and P4Id groups are posteromedial protocerebral. According to Younossi-Hartenstein et al. many of the neuronal clusters that make up these groups are likely to reflect lineages of single neuroblast clones; these are designated in the nomenclature by a single number. However, in other cases the clusters become subdivided into morphologically discrete subclusters (i.e. neuroblast clones) after late stage 15; in these cases the clusters are designated by a compound nomenclature that combines the numbers of the subclusters (for instance BLD1/2, which will give rise later in embryogenesis to BLD1 and BLD2).
Acknowledgments
We thank G. Boekhoff-Falk, the Developmental Studies Hybridoma Bank, C.Q. Doe, B. Hartmann, K. Matthews, D.M. Mellerick, J. Reinitz, R. Stocker, G.M. Technau and R. Urbach, U. Walldorf for flies and antibodies. This work was supported by the Swiss NSF to H.R. and S.G.S and NIH Grant NS 29367 to V.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. pp. 214–217. [Google Scholar]
- Aspland SE, White RAH. Nucleocytoplasmic localisation of extradenticle protein is spatially regulated throughout development in Drosophila. Development. 1997;124:741–747. doi: 10.1242/dev.124.3.741. [DOI] [PubMed] [Google Scholar]
- Bhat KM. The patched signaling pathway mediates repression of gooseberry allowing neuroblast specification by wingless during Drosophila neurogenesis. Development. 1996;122:2921–2932. doi: 10.1242/dev.122.9.2921. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The Eyes Absent Gene - Genetic-Control of Cell-Survival and Differentiation in the Developing Drosophila Eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ. New Neuroblast Markers and the Origin of the Acc/Pcc Neurons in the Drosophila Central-Nervous-System. Mechanisms of Development. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. Springer; Heidelberg: 1997. [Google Scholar]
- Chizhikov VV, Millen KJ. Roof plate-dependent patterning of the vertebrate dorsal central nervous system. Dev Biol. 2005;277:287–95. doi: 10.1016/j.ydbio.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Chu H, Parras C, White K, Jimenez F. Formation and specification of ventral neuroblasts is controlled by vnd in Drosophila neurogenesis. Genes Dev. 1998;12:3613–24. doi: 10.1101/gad.12.22.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RA, Ohlen TV. Vnd/nkx, ind/gsh, and msh/msx: conserved regulators of dorsoventral neural patterning? Curr Opin Neurobiol. 2000;10:63–71. doi: 10.1016/s0959-4388(99)00049-5. [DOI] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Molecular Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Estes PS, Ho GLY, Narayanan R, Ramaswami M. Synaptic localization and restricted diffusion of a Drosophila neuronal synaptobrevin - Green fluorescent protein chimera in vivo. Journal of Neurogenetics. 2000;13:233. doi: 10.3109/01677060009084496. [DOI] [PubMed] [Google Scholar]
- Gulisano M, Broccoli V, Pardini C, Boncinelli E. Emx1 and Emx2 show different patterns of expression during proliferation and differentiation of the developing cerebral cortex in the mouse. Eur J Neurosci. 1996;8:1037–50. doi: 10.1111/j.1460-9568.1996.tb01590.x. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of Ectopic Eyes by Targeted Expression of the Eyeless Gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hartmann B, Hirth F, Walldorf U, Reichert H. Expression, regulation and function of the homeobox gene empty spiracles in brain and ventral nerve cord development of Drosophila. Mechanisms of Development. 2000;90:143–153. doi: 10.1016/s0925-4773(99)00237-3. [DOI] [PubMed] [Google Scholar]
- Hirth F, Hartmann B, Egger B, Reichert H. Homeotic gene function in embryonic brain development of Drosophila. European Journal of Neuroscience. 1998;10:384–384. [Google Scholar]
- Hirth F, Kammermeier L, Frei E, Walldorf U, Noll M, Reichert H. An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development. 2003;130:2365–2373. doi: 10.1242/dev.00438. [DOI] [PubMed] [Google Scholar]
- Hirth F, Therianos S, Loop T, Gehring WJ, Reichert H, Furukubotokunaga K. Developmental Defects in Brain Segmentation Caused by Mutations of the Homeobox Genes Orthodenticle and Empty Spiracles in Drosophila. Neuron. 1995;15:769–778. doi: 10.1016/0896-6273(95)90169-8. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Takeichi M, Nose A. The role of the msh homeobox gene during Drosophila neurogenesis: implication for the dorsoventral specification of the neurectoderm. Development. 1997;124:3099–109. doi: 10.1242/dev.124.16.3099. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. Antibodies to Horseradish-Peroxidase as Specific Neuronal Markers in Drosophila and in Grasshopper Embryos. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Watts RJ, Luo L. Development of neuronal connectivity in Drosophila antennal lobes and mushroom bodies. Curr Opin Neurobiol. 2002;12:80–6. doi: 10.1016/s0959-4388(02)00293-3. [DOI] [PubMed] [Google Scholar]
- Jurgens G, Weigel D. Terminal Versus Segmental Development in the Drosophila Embryo - the Role of the Homeotic Gene Fork Head. Rouxs Archives of Developmental Biology. 1988;197:345–354. doi: 10.1007/BF00375954. [DOI] [PubMed] [Google Scholar]
- Kammermeier L, Leemans R, Hirth F, Flister S, Wenger U, Walldorf U, Gehring WJ, Reichert H. Differential expression and function of the Drosophila Pax6 genes eyeless and twin of eyeless in embryonic central nervous system development. Mechanisms of Development. 2001;103:71–78. doi: 10.1016/s0925-4773(01)00328-8. [DOI] [PubMed] [Google Scholar]
- Kaphingst K, Kunes S. Pattern-Formation in the Visual Centers of the Drosophila Brain - Wingless Acts Via Decapentaplegic to Specify the Dorsoventral Axis. Cell. 1994;78:437–448. doi: 10.1016/0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Kosman D, Small S, Reinitz J. Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev Genes Evol. 1998;208:290–4. doi: 10.1007/s004270050184. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development. 2002;129:409–19. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–52. [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–76. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lehmann M, Korge G. The fork head product directly specifies the tissue-specific hormone responsiveness of the Drosophila Sgs-4 gene. Embo Journal. 1996;15:4825–4834. [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and Increased Expression of Fasciclin-Ii Alters Motoneuron Growth Cone Guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Mallamaci A, Iannone R, Briata P, Pintonello L, Mercurio S, Boncinelli E, Corte G. EMX2 protein in the developing mouse brain and olfactory area. Mech Dev. 1998;77:165–72. doi: 10.1016/s0925-4773(98)00141-5. [DOI] [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. Dachshund Encodes a Nuclear-Protein Required for Normal Eye and Leg Development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development. 2005;132:725–37. doi: 10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- Marti E, Bovolenta P. Sonic hedgehog in CNS development: one signal, multiple outputs. Trends Neurosci. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Holbrook S, Isshiki T, Weiss J, Doe CQ, Mellerick DM. Dorsoventral patterning in the Drosophila central nervous system: the vnd homeobox gene specifies ventral column identity. Genes & Development. 1998;12:3603–3612. doi: 10.1101/gad.12.22.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinertzhagen IA. The Synaptic Populations of the Flys Optic Neuropil and Their Dynamic Regulation - Parallels with the Vertebrate Retina. Progress in Retinal Research. 1993;12:13–39. [Google Scholar]
- Mohler J. Spatial Regulation of Segment Polarity Gene-Expression in the Anterior Terminal Region of the Drosophila Blastoderm Embryo. Mechanisms of Development. 1995;50:151–161. doi: 10.1016/0925-4773(94)00332-h. [DOI] [PubMed] [Google Scholar]
- Morante J, Desplan C. Building a projection map for photoreceptor neurons in the Drosophila optic lobes. Semin Cell Dev Biol. 2004;15:137–43. doi: 10.1016/j.semcdb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Nagao T, Endo K, Kawauchi H, Walldorf U, Furukubo-Tokunaga K. Patterning defects in the primary axonal scaffolds caused by the mutations of the extradenticle and homothorax genes in the embryonic Drosophila brain. Development Genes and Evolution. 2000;210:289–299. doi: 10.1007/s004270050316. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Embryonic development of the Drosophila brain. I. Pattern of pioneer tracts. J Comp Neurol. 1998;402:10–31. [PubMed] [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Noveen A, Daniel A, Hartenstein V. Early development of the Drosophila mushroom body: the roles of eyeless and dachshund. Development. 2000;127:3475–88. doi: 10.1242/dev.127.16.3475. [DOI] [PubMed] [Google Scholar]
- O’Leary DD, Nakagawa Y. Patterning centers, regulatory genes and extrinsic mechanisms controlling arealization of the neocortex. Curr Opin Neurobiol. 2002;12:14–25. doi: 10.1016/s0959-4388(02)00285-4. [DOI] [PubMed] [Google Scholar]
- Pabst O, Herbrand H, Arnold HH. Nkx2-9 is a novel homeobox transcription factor which demarcates ventral domains in the developing mouse CNS. Mech Dev. 1998;73:85–93. doi: 10.1016/s0925-4773(98)00035-5. [DOI] [PubMed] [Google Scholar]
- Page DT. Inductive patterning of the embryonic brain in Drosophila. Development. 2002;129:2121–2128. doi: 10.1242/dev.129.9.2121. [DOI] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, Sun YH. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila. Genes & Development. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JLR. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Pera EM, Kessel M. Demarcation of ventral territories by the homeobox gene NKX2.1 during early chick development. Dev Genes Evol. 1998;208:168–71. doi: 10.1007/s004270050170. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Digital three-dimensional models of Drosophila development. Current Opinion in Genetics & Development. 2004;14:382–391. doi: 10.1016/j.gde.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by Decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Qiu M, Shimamura K, Sussel L, Chen S, Rubenstein JL. Control of anteroposterior and dorsoventral domains of Nkx-6.1 gene expression relative to other Nkx genes during vertebrate CNS development. Mech Dev. 1998;72:77–88. doi: 10.1016/s0925-4773(98)00018-5. [DOI] [PubMed] [Google Scholar]
- Ramaekers A, Magnenat E, Marin EC, Gendre N, Jefferis GS, Luo L, Stocker RF. Glomerular maps without cellular redundancy at successive levels of the Drosophila larval olfactory circuit. Curr Biol. 2005;15:982–92. doi: 10.1016/j.cub.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, Taghert PH. Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J Neurobiol. 1999;41:189–207. [PubMed] [Google Scholar]
- Richter S, Hartmann B, Reichert H. The wingless gene is required for embryonic brain development in Drosophila. Development Genes and Evolution. 1998;208:37–45. doi: 10.1007/s004270050151. [DOI] [PubMed] [Google Scholar]
- Rudolph KM, Liaw GJ, Daniel A, Green P, Courey AJ, Hartenstein V, Lengyel JA. Complex regulatory region mediating tailless expression in early embryonic patterning and brain development. Development. 1997;124:4297–4308. doi: 10.1242/dev.124.21.4297. [DOI] [PubMed] [Google Scholar]
- Shen WP, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martinez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–33. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Zhang Y, Holmgren R, Carroll SB, Doe CQ. Specification of Neuroblast Identity in the Drosophila Embryonic Central-Nervous-System by Gooseberry-Distal. Nature. 1995;376:427–430. doi: 10.1038/376427a0. [DOI] [PubMed] [Google Scholar]
- Sprecher SG, Muller M, Kammermeier L, Miller DF, Kaufman TC, Reichert H, Hirth F. Hox gene cross-regulatory interactions in the embryonic brain of Drosophila. Mech Dev. 2004;121:527–36. doi: 10.1016/j.mod.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- Strausfeld NJ, Hansen L, Li Y, Gomez RS, Ito K. Evolution, discovery, and interpretations of arthropod mushroom bodies. Learn Mem. 1998;5:11–37. [PMC free article] [PubMed] [Google Scholar]
- Strauss R, Heisenberg M. A higher control center of locomotor behavior in the Drosophila brain. J Neurosci. 1993;13:1852–61. doi: 10.1523/JNEUROSCI.13-05-01852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R, Schnabel R, Technau GM. The pattern of neuroblast formation, mitotic domains and proneural gene expression during early brain development in Drosophila. Development. 2003;130:3589–606. doi: 10.1242/dev.00528. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003a;130:3621–37. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Segment polarity and DV patterning gene expression reveals segmental organization of the Drosophila brain. Development. 2003b;130:3607–20. doi: 10.1242/dev.00532. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Neuroblast formation and patterning during early brain development in Drosophila. Bioessays. 2004;26:739–51. doi: 10.1002/bies.20062. [DOI] [PubMed] [Google Scholar]
- Walldorf U, Gehring WJ. Empty Spiracles, a Gap Gene Containing a Homeobox Involved in Drosophila Head Development. Embo Journal. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen X, Xu H, Lufkin T. Msx3: a novel murine homologue of the Drosophila msh homeobox gene restricted to the dorsal embryonic central nervous system. Mech Dev. 1996;58:203–15. doi: 10.1016/s0925-4773(96)00562-x. [DOI] [PubMed] [Google Scholar]
- Weiss JB, Von Ohlen T, Mellerick DM, Dressler G, Doe CQ, Scott MP. Dorsoventral patterning in the Drosophila central nervous system: the intermediate neuroblasts defective homeobox gene specifies intermediate column identity. Genes Dev. 1998;12:3591–602. doi: 10.1101/gad.12.22.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Hartenstein V. Pattern, time of birth, and morphogenesis of sensillum progenitors in Drosophila. Microscopy Research and Technique. 1997;39:479–491. doi: 10.1002/(SICI)1097-0029(19971215)39:6<479::AID-JEMT3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–29. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nguyen B, Shy D, Hartenstein V. Embryonic origin of the Drosophila brain neuropile. J Comp Neurol. 2006;497:981–98. doi: 10.1002/cne.20884. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ungar A, Fresquez C, Holmgren R. Ectopic Expression of Either the Drosophila Gooseberry-Distal or Proximal Gene Causes Alterations of Cell Fate in the Epidermis and Central-Nervous-System. Development. 1994;120:1151–1161. doi: 10.1242/dev.120.5.1151. [DOI] [PubMed] [Google Scholar]


