Abstract
The Drosophila central brain is largely composed of lineages, units of sibling neurons derived from a single progenitor cell or neuroblast. During the early embryonic period neuroblast generate the primary neurons that constitute the larval brain. Neuroblasts reactivate in the larva, adding to their lineages a large number of secondary neurons which, according to previous studies in which selected lineages were labeled by stably expressed markers, differentiate during metamorphosis, sending terminal axonal and dendritic branches into defined volumes of the brain neuropil. We call the overall projection pattern of neurons forming a given lineage the “projection envelope” of that lineage. By inducing MARCM clones at the early larval stage, we labeled the secondary progeny of each neuroblast. For the supraesophageal ganglion excluding mushroom body (the part of the brain investigated in the present work) we obtained 81 different types of clones, Based on the trajectory of their secondary axon tracts (described in the accompanying paper), we assigned these clones to specific lineages defined in the larva. Since a labeled clone reveals all aspects (cell bodies, axon tracts, terminal arborization) of a lineage, we were able to describe projection envelopes for all secondary lineages of the supraesophageal ganglion. This work provides a framework by which the secondary neurons (forming the vast majority of adult brain neurons) can be assigned to genetically and developmentally defined groups. It also represents a step towards the goal to establish, for each lineage, the link between its mature anatomical and functional phenotype, and the genetic make-up of the neuroblast it descends from.
Keywords: Brain, Development, Drosophila, Lineage, Mapping, SAT
Introduction
In the field of developmental biology, the concepts of cell determination and cell lineage are fundamental to our understanding of the formation of complex tissues and organs. When talking about cell lineage we refer to the genealogy (family tree) of groups of cells. The lineage produced by a progenitor cell is generally used synonymously with the progeny descending from this cell. During early stages of development, a progenitor cell initiates a genetic program that controls the later fate of this cell and its progeny. The genetic program of the progenitor is defined by the expression of cell fate determinants, typically transcription factors, that either remain active in the progeny or trigger the expression of a next tier of factors impacting the fate of the progeny (Guillemot, 2007; Shirasaki and Pfaff, 2002; Skeath and Thor, 2003). Thus, when embarking on the analysis of a complex organ, one of the assumptions that guides our research is that cells which possess a similar phenotype do so because they are part of a lineage produced by a common progenitor which, early on, expresses a set of transcription factors (“intrinsic determinants”) controlling the fate of its lineage. Of course, this assumption has to always be tested against the alternative: extrinsic signals from the environment into which cells are placed trigger a genetic switch in these cells which controls their fate. In this scenario, the family tree of the cells is not important.
The fate of cell lineages in the Drosophila nervous system is heavily influenced by intrinsic determinants. A number of pioneering experiments in which neural progenitors (neuroblasts) were cultured in vitro revealed that, even when removed from their natural environment in the early embryo, neuroblasts are capable of dividing and producing progeny in the same number and cell type (assayed by expression of neurotransmitters) (Huff et al., 1989). Later studies identified many specific transcription factors expressed in neuroblasts (Doe, 1992; Urbach and Technau, 2003). Furthermore, it was shown that the timing of expression of a transcription factor is able to influence the fate of subsets of neurons (“sublineages”) forming part of a lineage (Brody and Odenwald, 2000; Isshiki et al., 2001; Kambadur et al., 1998; Pearson and Doe, 2004). Thus, a neuroblast (N) divides asymmetrically, with one daughter cell (N’) remaining in the state of a dividing neuroblast, whereas the other daughter cell (ganglion mother cell), after an additional round of division, becomes postmitotic and differentiates into two daughter cells (neurons or glia). The asymmetric division allows for a mechanism by which transcription factors are differentially inherited by daughter cells. The general model is that transcription factor (A), expressed during a specific time interval, will be inherited by one daughter cell or sublineage (α). Eventually, (A) is no longer expressed and a second one (B) turns on. All neurons born after (A) is turned off now inherit (B) and become a second sublineage (β). Several transcription factors were identified that are expressed in a sequential manner during neuroblast proliferation in the embryo, and were shown, using molecular markers as a read-out, to influence the fate of embryonic-born (primary) neurons (Isshiki et al., 2001; Kambadur et al., 1998). However, it should be emphasized that many transcription factors are active in neuroblasts from before they are mitotically active through to a later developmental period, and that this window of expression varies for each transcription factor (Doe, 1992; Kumar et al., 2009a-b; Lichtneckert et al., 2008; Urbach and Technau, 2003); these factors would be predicted to have an impact on the fate of an entire lineage.
Analyses of a few select lineages in the larval and/or adult brain support the idea that neuronal fate is controlled by factors inherited by entire lineages and by specific sublineages, which may manifest itself in a lineage's overall structure. Thus, lineages appear as “morphological units,” with all axons forming one or two (in the case of hemilineages; Truman et al., 2010) bundles and terminal arborizations focusing on a discrete neuropil territory. For example, four lineages (MB1-4) form the mushroom body (Crittenden et al., 1998; Ito et al., 1997) and three lineages, (vNB/BAla1, lNB/BAlc and dNB/BAmv3) include the majority of projection neurons connecting the antennal lobe with the protocerebrum (Lai et al., 2008). Endings of all secondary neurons of MB1-4 are confined to the calyx, peduncle, and lobes of the mushroom body; the antennal lobe–associated lineages innervate three compartments, namely the antennal lobe, calyx, and lateral horn (Das et al., 2013; Lai et al., 2008; Stocker et al., 1990). We will use the term “projection envelope” to describe the overall neuropil volume that is innervated by neurons of a lineage. Individual neurons in a lineage form restricted terminal arbors that target smaller volumes within the projection envelope. For example, the neurons produced by MB1-4 in the late embryo/early larva fill the γ-lobe; they are followed by neurons forming the α’/β’ lobes, and finally by neurons of the α/β lobes (Ito et al., 1997; Kunz et al., 2012). In the case of dNB/BAmv3, most neurons innervate a single glomerulus of the antennal lobe and project to discrete regions within the calyx and lateral horn (Jefferis et al., 2001; Yu et al., 2010). It is reasonable to assume that the projection envelope of a lineage, which is shared by all neurons of that lineage, is determined to some extent by transcription factors expressed earlier in development and are common to the neuroblast of that lineage. In addition, other factors expressed later at defined temporal intervals, thereby only reaching neurons born during that interval, may be responsible for more specific structural and functional characteristics that set neurons of a lineage apart from each other.
Whereas both expression of molecular determinants of cell fate and the phenotypic elements of cell fate (e.g., shape of neuronal arbor, choice of pre-and postsynaptic partners, physiological characteristics) can be studied in great detail, the complex cascade of molecular events linking the two levels has remained elusive. What mechanism acts on outgrowing axons and guides/restricts them to a specific compartment? How is this mechanism encoded in the cell fate determinants expressed in the neuroblast? Drosophila offers a favorable system to address these questions: its nervous system is built by a relatively small number of lineages (previous descriptive maps yielded approximately 100 lineages per central brain hemisphere and 28 lineages per ventral nerve cord hemineuromere; Doe, 1992; Urbach and Technau, 2003; Younossi-Hartenstein et al., 1996). Lineages can be globally and/or individually labeled by antibodies for various neuronal proteins (e.g. mushroom body-specific antibodies: Crittenden et al., 1998; neuropeptide pigment-dispersing factor or PDF: Helfrich-Förster et al., 2007; neuropeptide IPNamide: Shafer et al., 2006) and reporter constructs (LacZ and Gal4-based: e.g. en-Gal4, Kumar et al., 2009b; Th-Gal4, Mao and Davis, 2009; Gal4 lines expressed in ellipsoid body neurons, Renn et al., 1999). Maps of the expression of transcription factors in the neuroblasts, as well as the anatomical pattern of lineages at the larval stage, have been generated (Cardona et al., 2010; Pereanu and Hartenstein, 2006; Truman et al., 2004; Urbach and Technau, 2003; Urbach and Technau, 2004). In the accompanying paper (Lovick et al., 2013) we had mapped the association between lineages and neuropil fascicles and followed these fascicles throughout metamorphosis into the adult. In this paper, we have analyzed individual lineages at the adult stage by the MARCM technique (Lee and Luo, 2001), where a GFP reporter gene is activated by somatic recombination in neuroblasts shortly before they enter their larval phase of proliferation. In this manner, all secondary neurons produced by these neuroblasts (the “secondary lineages”) are labeled as “clones.” A clone includes the cluster of cell bodies derived from the larval neuroblast, as well as the axons and terminal arborizations of these cell bodies. Based on the trajectory of their axon bundles, we are able to assign clones to their respective lineages. We analyzed a total of 814 clones located in the supraesophageal ganglion, the largest part of the brain. Excluded from this study are clones in the optic lobes, whose modular (and probably not-lineage based) structure has been described previously (Bausenwein et al., 1992; Fischbach and Dittrich, 1992). Excluded are also clones representing the four well known lineages of the mushroom body (Crittenden et al., 1998; Ito et al., 1997; Kunz et al., 2012), and the lineages of the subesophageal ganglion, which will be analyzed in an upcoming study (Kuert et al., in preparation). Clones fell into 81 groups, where each group corresponded to a known lineage or lineage pair. We provide a brief description of the projection envelopes for all lineages. The complexity of these lineages clearly warrants a much finer level of analysis, taking into account aspects like overlap of terminal arborizations of different lineages, precise relationships between arborizations and compartment boundaries, and variations in the size and location of cell bodies. These investigations, which require that specimens with different clones are digitally registered to a “standard brain”, will be presented in a series of upcoming studies. Note that numerous aspects of lineage analysis has been recently published in two large, independent studies where MARCM clones of secondary lineages were generated (Ito et al., 2013; Yu et al., 2013). The main purpose of the present work is to identify clones with defined lineages, contributing to the ultimate goal of linking the mature anatomical and functional phenotype of a lineage and its constituent neurons with the specific genetic make-up of the embryonic neuroblast that produces the lineage.
MATERIAL AND METHODS
Fly Stocks
Flies were grown at 25°C using standard fly media unless otherwise noted. Fly stocks used are the ones detailed in the Clonal Analysis section below.
Immunohistochemistry
Samples were fixed in 4% paraformaldehyde in phosphate buffer saline (PBS, Fisher-Scientific, pH = 7.4; Cat No. #BP399-4). Tissues were permeabilized in PBT (phosphate buffer saline with 0.3% Triton X-100, pH = 7.4) and immunohistochemistry was performed using standard procedures (Ashburner 1989). The following antibodies were provided by the Developmental Studies Hybridoma Bank (Iowa City, IA): mouse anti-Neurotactin (BP106, 1:10), rat anti-DN-cadherin (DN-EX #8, 1:20), mouse anti-Neuroglian (BP104, 1:30). Secondary antibodies, IgG (Jackson ImmunoResearch; Molecular Probes) were used at the following dilutions: Alexa 546-conjugated anti-mouse (1:500), DynaLight 649-conjugated anti-rat (1:400).
Clonal Analysis
Clones were generated by Flp-mediated mitotic recombination at homologous FRT sites. Larval neuroblast clones were generated by MARCM (Lee and Luo, 2001; see below) or the Flp-out construct (Zecca et al., 1996; Ito et al., 1997).
Mitotic clone generation by Flp-out
To generate secondary lineages clones in the larva using the Flp-out technique; flies bearing the genotype:
hsflp, elavC155-Gal4/+; UAS-FRT-rCD2, y+, stop-FRT-mCD8::GFP
hsflp; Act5C-FRT-stop,y+-FRT-Gal4, UAS-tauLacZ/UAS-src::EGFP
Briefly, early larva with either of the above genotype were heatshocked at 38°C for 30-40 minutes. elavC155-Gal4 is expressed in neurons as well as secondary neuroblasts.
Third instar larval and adult brains were dissected and processed for immunohistochemistry (as described above).
Mitotic clone generation by MARCM
Mitotic clones were induced during the late first instar/early second instar stages by heat-shocking at 38°C for 30 minutes to 1 hour (approximately 12h-44h ALH). GFP-labeled MARCM clones contain the following genotype:
Adult MARCM clones
hsflp/+; FRTG13, UAS-mCD8GFP/FRTG13, tub-GAL80; tub-Gal4/+ or
FRT19A GAL80, hsflp, UAS-mCD8GFP/ elavC155-Gal4, FRT19A; UAS-CD8GFP/+
Larval MARCM clones
hsflp, elavC155-Gal4, FRTG13, UAS-mCD8GFP /Y or hsflp, elavC155-Gal4, FRTG13, UAS-mCD8GFP /; FRT42D, tub-Gal80/FRT42D
Confocal Microscopy
Staged Drosophila larval and adult brains labeled with suitable markers were viewed as whole-mounts by confocal microscopy [LSM 700 Imager M2 using Zen 2009 (Carl Zeiss Inc.); lenses: 40× oil (numerical aperture 1.3)]. Complete series of optical sections were taken at 2-μm intervals. Captured images were processed by ImageJ or FIJI (National Institutes of Health, http://rsbweb.nih.gov/ij/ and http://fiji.sc/) and Adobe Photoshop.
2D registration of clones to standard brain
Brains with MARCM clones were labeled with DN-cad and BP104 to image the SAT and projection envelope relative to the BP104-positive fascicles and DN-cad-positive neuropil compartments. Fasciculation of the SAT of a clone with a fascicle allowed for its identification with a lineage, or lineage pair (see accompanying paper by Lovick et al., 2013). To generate the figure panels of this paper, z-projections of the individual MARCM clones were registered digitally with z-projections of a standard brain (“2D registration”). To this end, the standard brain was subdivided along the antero-posterior axis into six slices of approximately 20 μm thickness. These slices, each one characterized by one or more easily recognized landmark structures (antennal lobe, optic tubercle, ellipsoid body, fan-shaped body, lateral bend of antennal lobe tract, calyx), are introduced in Pereanu et al., 2010, and are depicted throughout the figures of this and the accompanying paper (Lovick et al., 2013). The process of 2D registration involved the following steps:
The confocal stack depicting a given clone was imported into the FIJI program (National Institutes of Health, http://rsbweb.nih.gov/ij/ and http://fiji.sc/) and digitally oriented such that the peduncle was aligned with the z-axis of the stack;
A z-projection of the entire clone (e.g., all sections of the green channel showing label) was generated;
In the case of brains containing more than one clone, background fluorescence and/or fluorescence from other clones were digitally removed to allow visualization of a single clone;
This z-projection was merged with a z-projection of the red channel (BP104 or DN-cad) derived from the confocal sections of one slice. The chosen slice is dependent upon the corresponding clone's SAT location. For example, consider the lineage BAlp4, whose SAT enters the antennal lobe postero-laterally. In terms of antero-posterior brain slices, this SAT forms part of the second slice (“level optic tubercle”);
Both the optic tubercle slice of the standard brain and the brain specimen containing the BAlp4 clone were imported as two layers into a file generated by the Adobe Photoshop program. Using few standard landmarks (location of the peduncle, tips of the MB vertical and medial lobe, vertical midline), the layer containing the clone (rendered temporarily semitransparent) was optimally fitted to the underlying layer representing the standard brain;
The optimally-fitted layer containing the clone was re-opened in FIJI, and then merged with the red channel (BP104 or DN-cad) of the standard brain. For the panels of the figure set depicting clone SATs relative to BP104-positive fascicles (Figs.5, 7, 10, 13, 16, 18), the red channel (BP104) was rendered white in Adobe Photoshop. For the figure set depicting the projection envelopes of clones (Figs.3, 4, 6, 8, 9, 11, 12, 14, 15, 17), the red channel (DN-cad) was rendered magenta by duplicating it in the blue channel.
Figure 5.
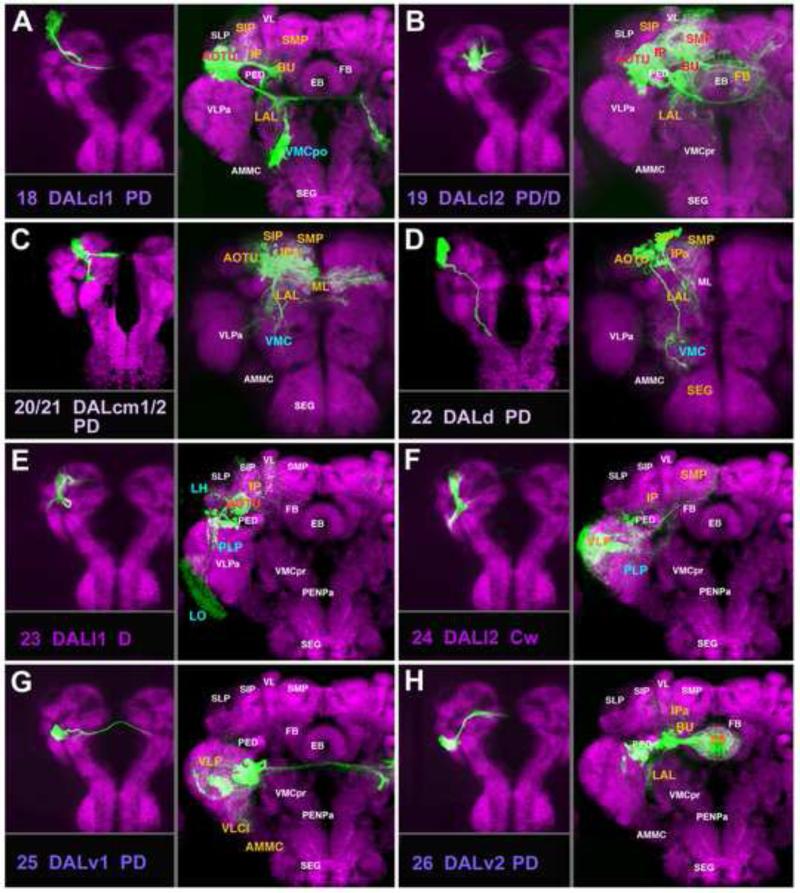
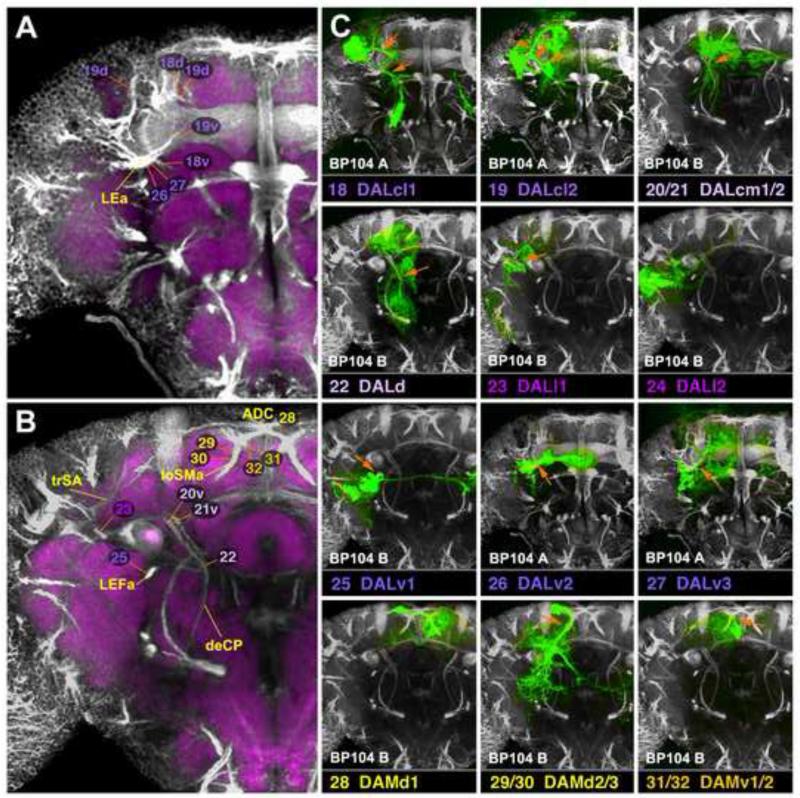
A-H: Clones representing lineages of the DAL group (#18/DALcl1 to #26/DALv2) in the larval and adult brain. For description of how panels are made and displayed, see legend of Fig.2. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 7.
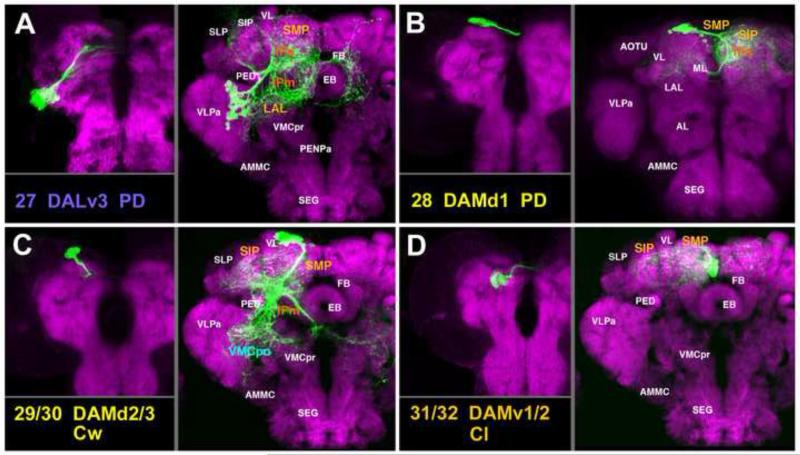
A-D: Clones representing lineages of the DAL group (#27/DALv3) and the DAM group in the larval and adult brain. For description of how panels are made and displayed, see legend of Fig.2. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 10.
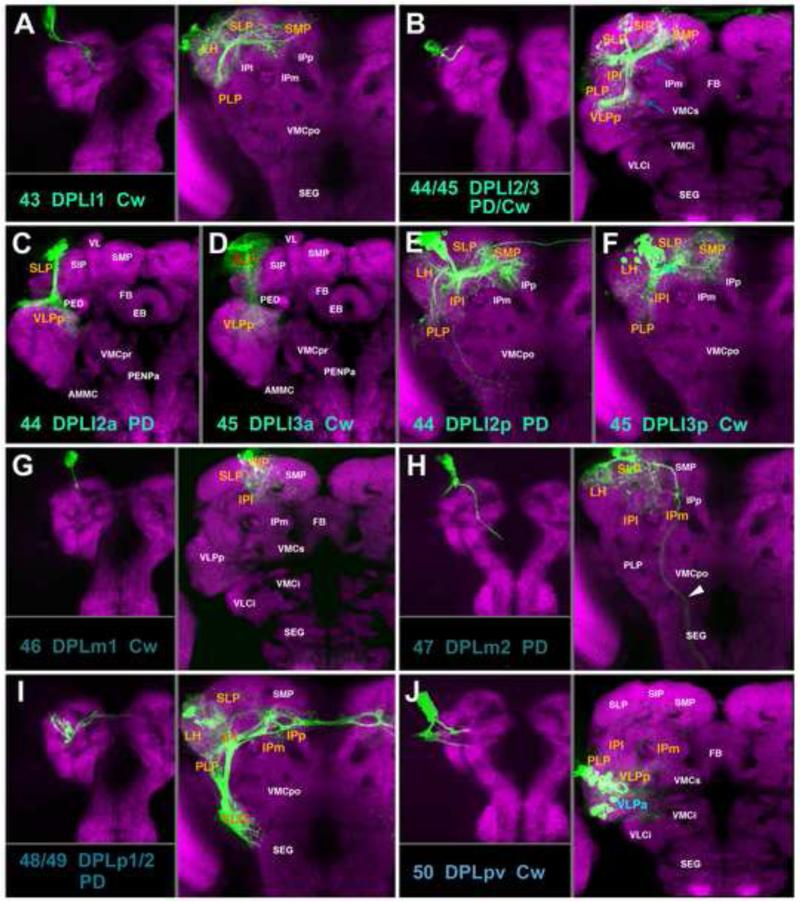
A-J: Clones representing lineages #43/DPLl1 to #50/DPLpv of the DPL group in the larval and adult brain. For description of how panels are made and displayed, see legend of Fig.2. The lineage pair DPLl2/3 is represented by five modified panels. The first of these (B) shows z-projection of entire DPLl2 clone registered to a central slice of the brain (level of fan-shaped body). At the larval stage, only one type of clone, represented at the left of B, exists for the DPLl2/3 pair. Blue arrows point at anterior and posterior HSATs. Panels C and D show z-projections of the anterior hemilineages (#44 DPLl2a and #45/DPLl3a, respectively), registered to an anterior slice. Panels E and F show posterior hemilineages of DPLl2/3, registered to posterior brain slice. White arrows point at an anterior segment of the SAT which, in case of DPLl2, is dense and thin, and in case of DPLl3, wide and diffuse. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 13.
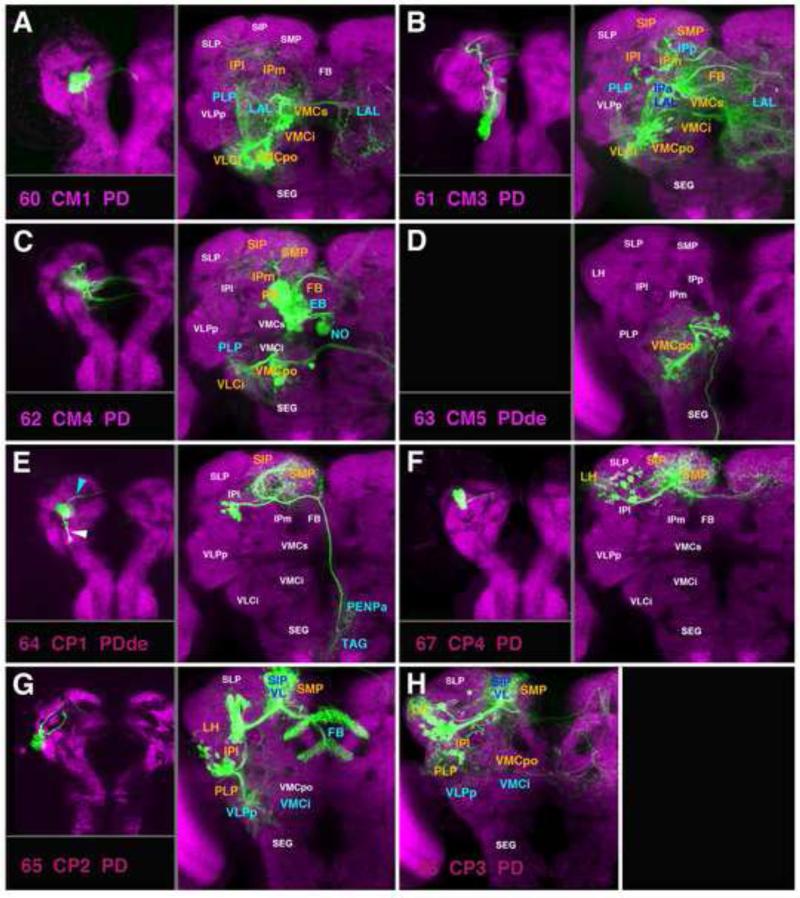
A-H: Clones representing lineages of the CM and CP group in the larval and adult brain. No larval clone for CM5 (D) was isolated. For the pair CP2/3 (G, H), only one larval clone (panel G, left) is shown. For description of how panels are made and displayed, see legend of Fig.2. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 16.
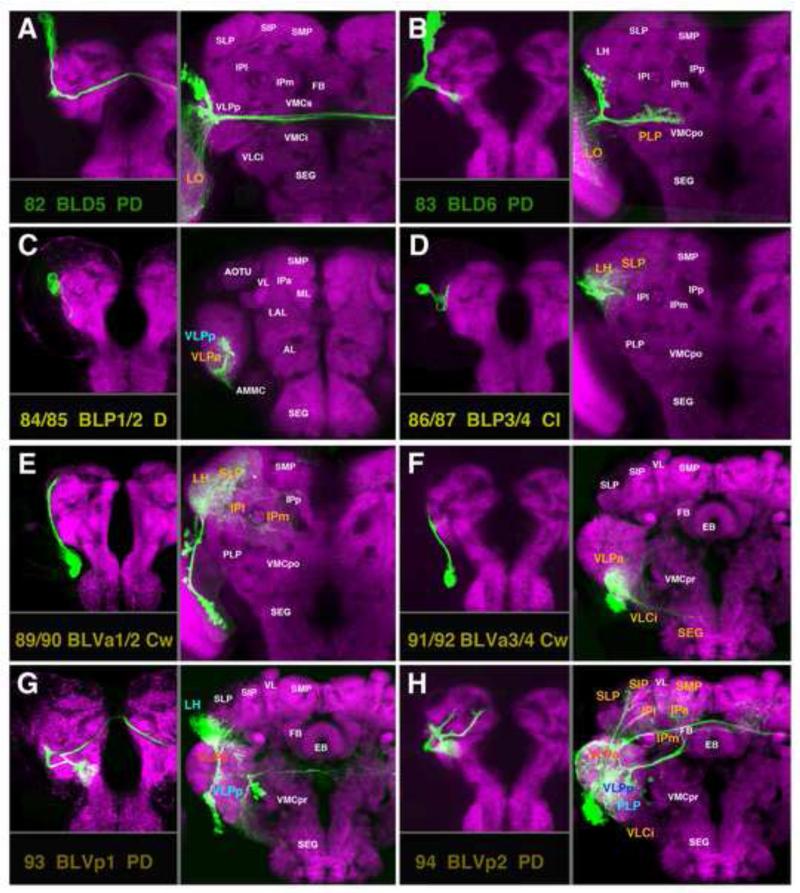
A-H: Clones representing lineages #82/BLD5-#83/BLD6 of the BLD group and lineages of the BLP and BLV group in the larval and adult brain. For description of how panels are made and displayed, see legend of Fig.2. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 3.
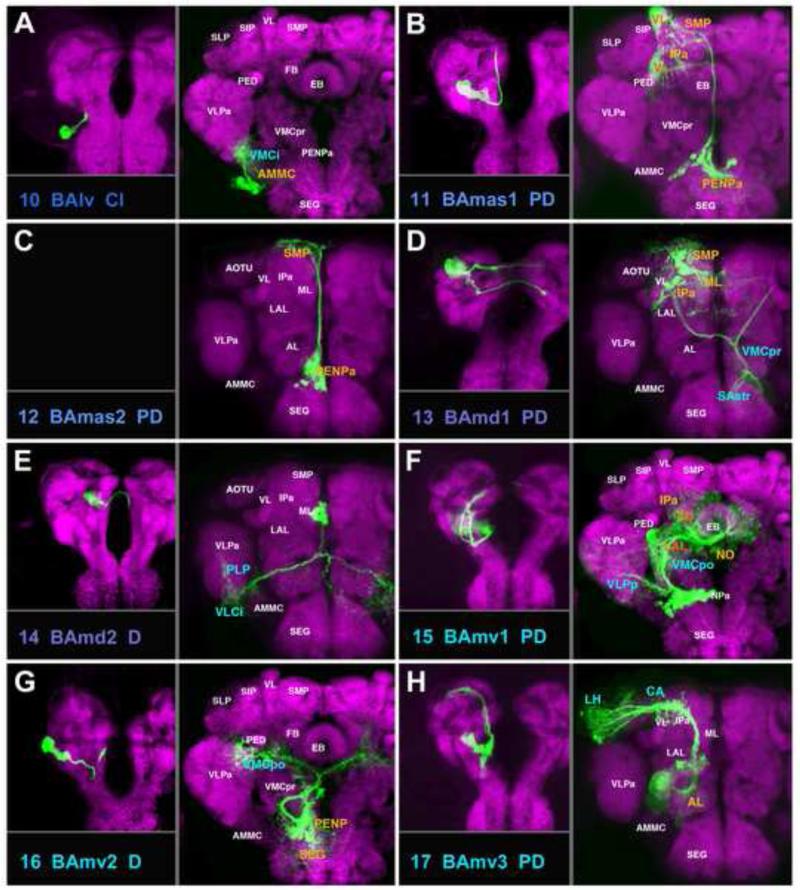
A-H: Clones representing lineages of the BA group (#10/BAlv to #17/BAmv3) in the larval and adult brain. For description of how panels are made and displayed, see legend of Fig.2. Only one larval clone is shown for the pair BAmas1/2 (panels B and C). For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 4.
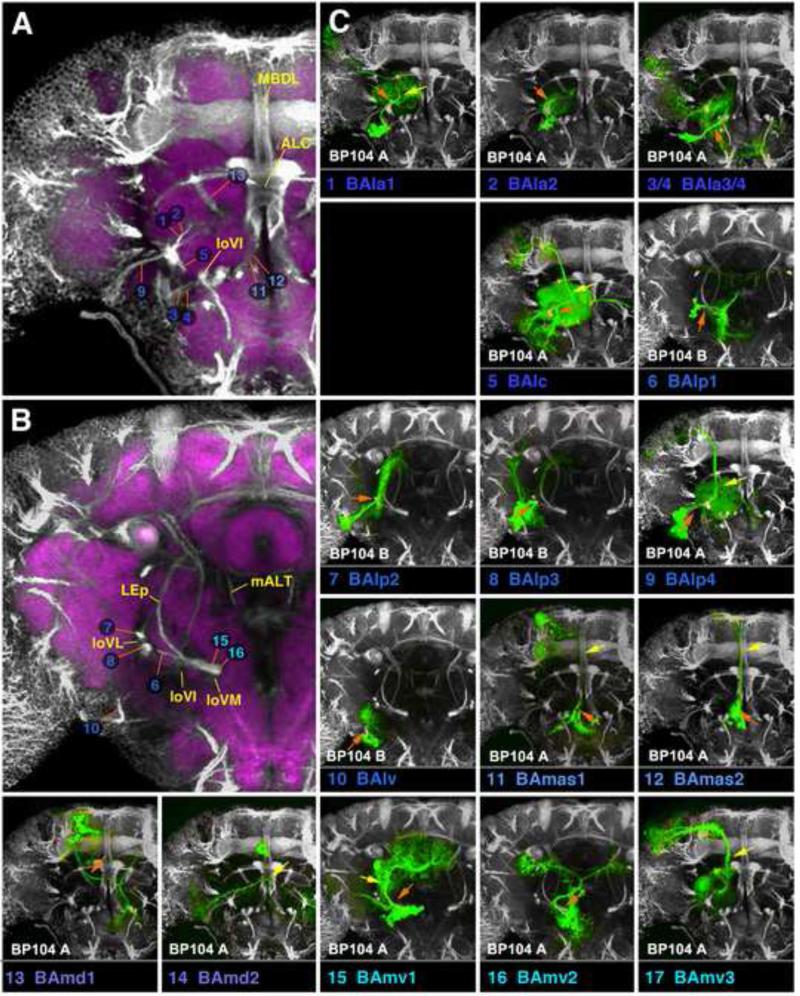
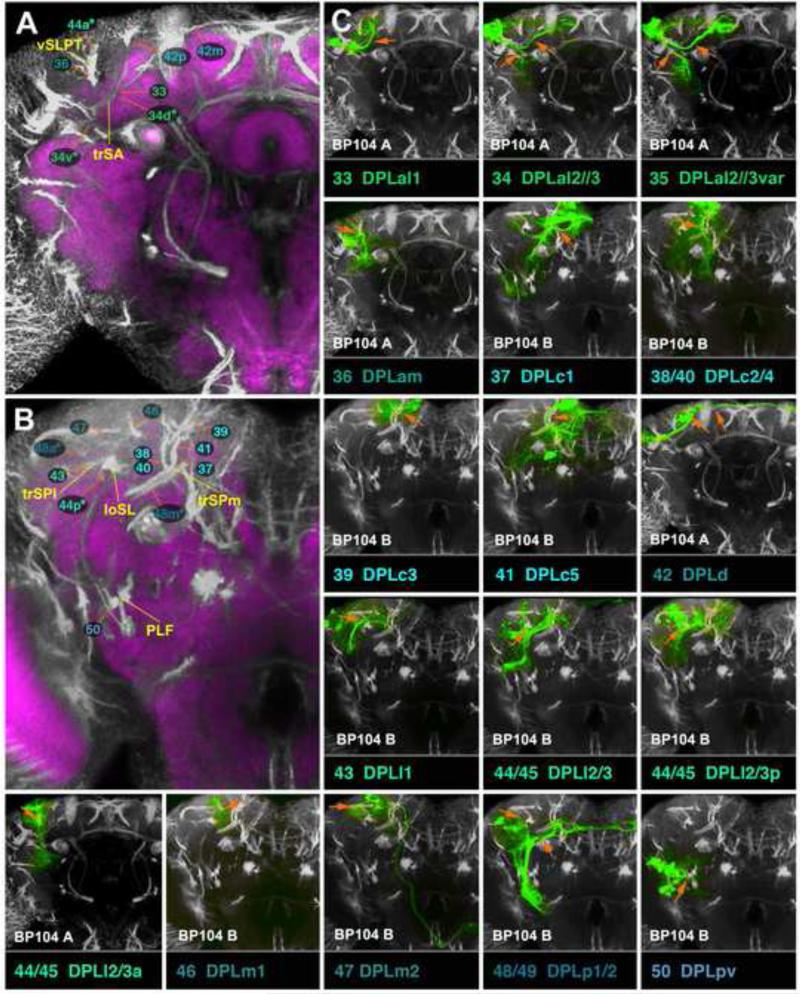
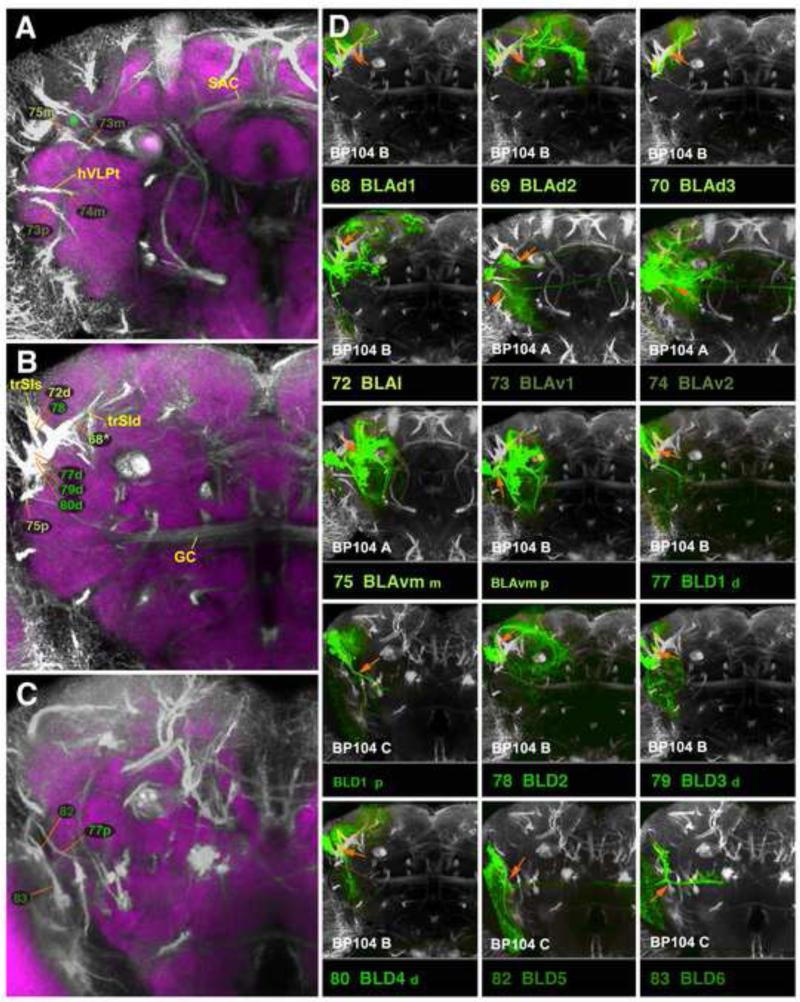
Clones representing lineages of the BA group in the adult brain. This figure, as well as the following figures 6, 9, 12, 15, and 17, are all designed in the same manner. Large panels on the left (A, B in case of Fig.4) show z-projections of frontal confocal sections of adult brain hemisphere labeled with BP104 (white, SATs and neuropil fascicles) and DN-cad (purple; neuropil compartments). Each z-projection represents a brain slice of approximately 15-20µm thickness. Brain slices correspond to different levels along the antero-posterior (“z”) axis. Panels A and B of this figure represent the two slices of the brain where BA lineage tracts enter the neuropil (A: level of optic tubercle and mushroom body lobes; B: posteriorly adjacent slice, marked by ellipsoid body). SATs of lineages are annotated by colored numbers; the same color key is used as in the accompanying paper by Lovick et al. (2013). In cases where two or more SATs or HSATs come very close and cannot be distinguished, the identifying numbers may be contracted into a single number followed by an asterisk; see, for example, the annotation of the HSATs of the DPLal2/3 lineages, #34/35, as “34v*” and “34d*”, in Fig.9A. Fascicles with which SATs are associated are annotated by yellow letters. (C): The small panels in section (C) of this figure show z-projections of clones representing lineages of the BA group. Panels were generated by registering z-projection of clones to a “slice” of the BP104-labeled standard brain, as described in the Materials and Methods section. Each lineage is identified by a number and abbreviation (bottom of its panel), rendered in the same color as that used in panels A and B. For the BP104 channel (white), only slices shown in the left panels (A, B) are used and indicated at the bottom left of the panel (“BP104 A”, “BP104 B”). For example, the first small panel depicts lineage #1/BAla1. The clone is registered to the anterior slice (“optic tubercle/mushroom body lobes”; shown at higher magnification in panel A), because the proximal SAT of BAla1 is contained within this slice. Panels with other lineages (e.g., #10/BAlv) use the more posterior slice (the one shown in panel B; ellipsoid body), because the SAT of BAlv is contained within that slice. In each small panel, the orange-colored arrow points at the proximal SATs by which the clone is identified. In terms of position and orientation, the arrow matches the orange line in panel on the left (A or B) which points at the corresponding BP104-labeled tract. Yellow arrows in the C-panels point at neuropil fascicles joined by the SAT. For example, the yellow arrow in the panel #1/BAla1 points at the beginning of the antennal lobe tract (ALT). For alphabetical list of abbreviations see Table 1. Bar: 25μm
Figure 6.
Clones representing lineages of the DAL and DAM groups in the adult brain. (A, B) z-projections show brain slices at level of optic tubercle/mushroom body lobes (A) and ellipsoid body (B). (C) z-projections of clones representing lineages of the DAL and DAM group. For description of how panels are made and displayed, see legend of Fig.4. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 8.
A-H: Clones representing lineages #33/DPLal1 to #42/DPLd of the DPL group in the larval and adult brain. For description of how panels are made and displayed, see legend of Fig.2. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 9.
Clones representing lineages of the DPL groups in the adult brain. (A, B) z-projections show brain slices at an anterior level (ellipsoid body; A) and posterior level (lateral bend of antennal lobe tract). (C) z-projections of clones representing lineages of the DPL group. For description of how panels are made and displayed, see legend of Fig.4. The lineage pair DPLl2/3 is represented by three panels in C. The first two of these (C44 DPLl2, C45 DPLl3) shows the clone registered to the posterior brain slice, to show association of the posterior HSAT with the obP fascicle (orange arrows; compare to panel B). In the third panel (C 45/DPLl3a), z-projection of the anterior hemilineage of DPLl3 is registered with anterior brain slice, showing entrypoint of anterior HSAT into dorsal SLP (orange arrow). For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 11.
A-H: Clones representing lineages of the DPM group in the larval and adult brain. For description of how panels are made and displayed, see legend of Fig.2. For alphabetical list of all abbreviations see Table 2. Bar: 50μm
Figure 12.
Clones representing lineages of the DPM group in the adult brain. (A, B) z-projections show brain slices at a posterior level (lateral bend of antennal lobe tract; A) and central level (fan-shaped body, great commissure; B). (C) z-projections of clones representing lineages of the DPM group. For description of how panels are made and displayed, see legend of Fig.4. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 14.
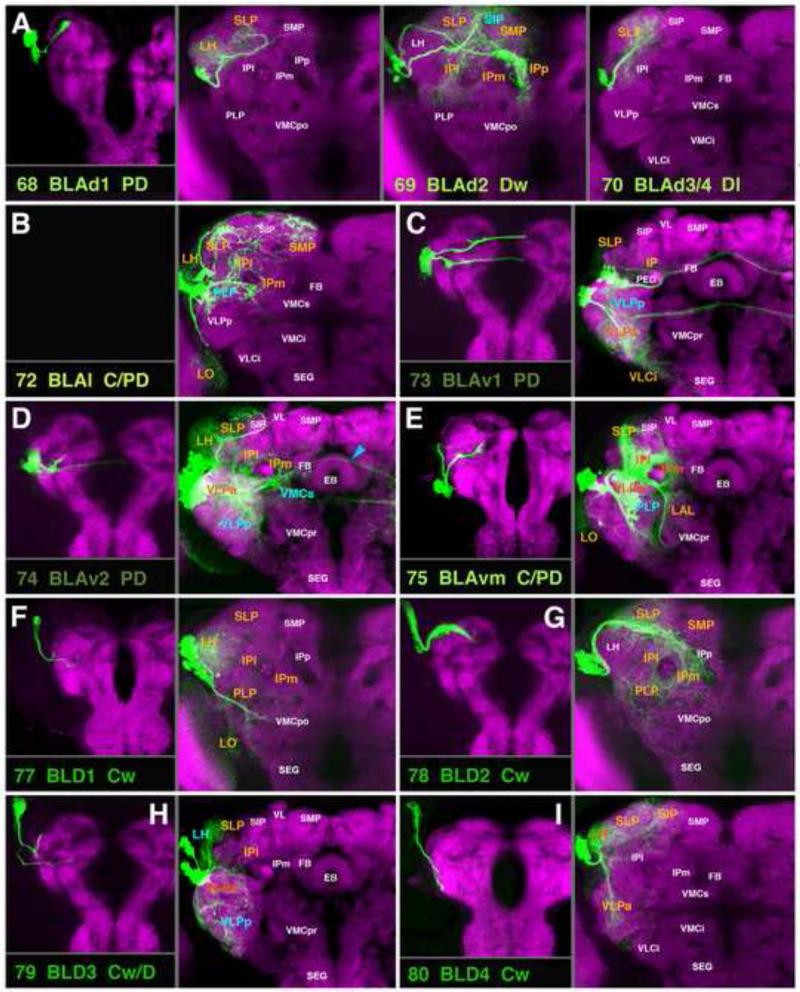
A-I: Clones representing lineages of the BLA group and lineages #77/BLD1-#80/BLD4 of the BLD group in the larval and adult brain. No larval clone for BLAl (B) was isolated. For the quartet BLAd1-4 (A) only one larval clone (panel A, left) is shown. For description of how panels are made and displayed, see legend of Fig.2. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 15.
Clones representing lineages of the BLA and BLD group in the adult brain. (A-C) z-projections show slices of the brain at an anterior level (A; level ellipsoid body), central level (B; fan-shaped body and great commissure) and posterior level (C; lateral bend of antennal lobe tract). (D) z-projections of clones representing lineages of the BLA and BLD group. For description of how panels are made and displayed, see legend of Fig.4. The lineages BLAvm and BLD1are represented by two panels each in D. The first of the BLAvm panels (D75 BLAvm m) shows the clone registered to the anterior brain slice, to show projection of the medial HSAT of BLAvm between VLPa and SLP (orange arrow). In the second BLAvm panel (BLAvm p) the posterior HSAT of BLAvm along the surface of the VLPp is indicated (orange arrow). Similarly, separate entry points of the dorsal and posterior HSATs of BLD1 are shown in panels D77 BLD1 d and BLD1 p, respectively. For alphabetical list of all abbreviations see Table 1. Bar: 50μm
Figure 17.
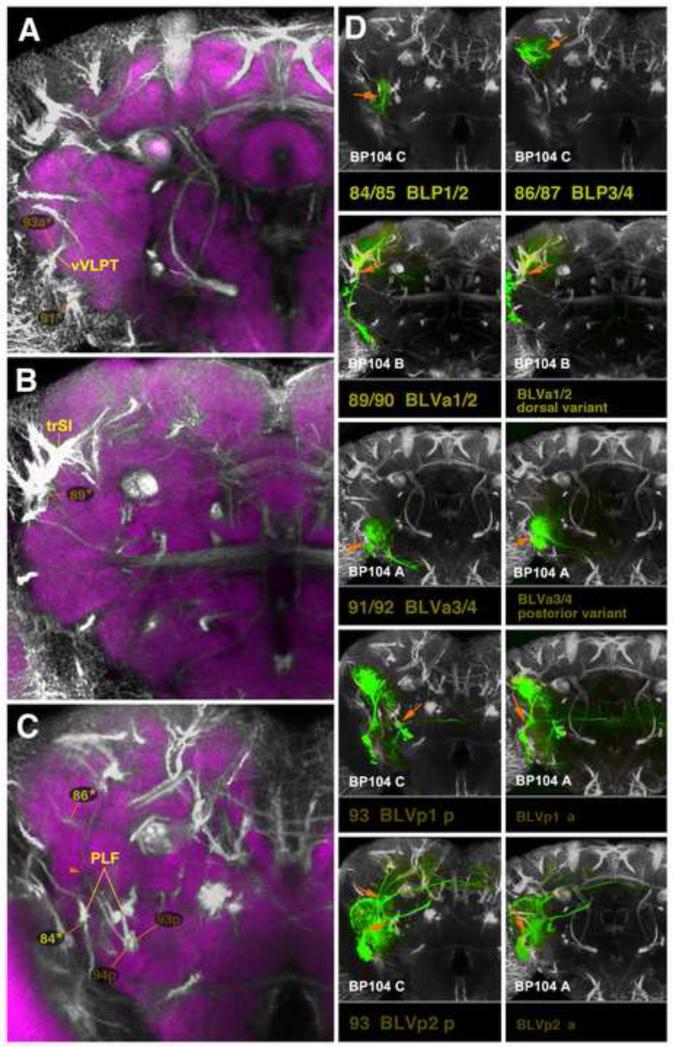
Clones representing lineages of the BLP and BLV group in the adult brain. (A-C) z-projections show slices of the brain at an anterior level (A; level ellipsoid body), central level (B; fan-shaped body and great commissure) and posterior level (C; lateral bend of antennal lobe tract). (D) z-projections of clones representing lineages of the BLP and BLV group. The lineage pairs BLVa1/2 (#89/90) and BLVa3/4 (#93/94) each are represented by two panels which show differences in location of cell body clusters. Lineages BLVp1 (#93) and BLVp2 (#94) are shown in two panels each. The left panels (D93 BLVp1 p; D94 BLVp2 p) shows the clone registered to the posterior brain slice, to show projection of the posterior HSAT of BLVp1/2 along the PLF fascicle (orange arrows). In the right panels (BLVp1 a, BLVp2 a) the anterior HSATs of these lineages, penetrating the VLPa, are shown (orange arrow). For alphabetical list of all abbreviations see Table 1. Bar: 50μm
RESULTS
MARCM clones reveal the projection envelope of secondary lineages
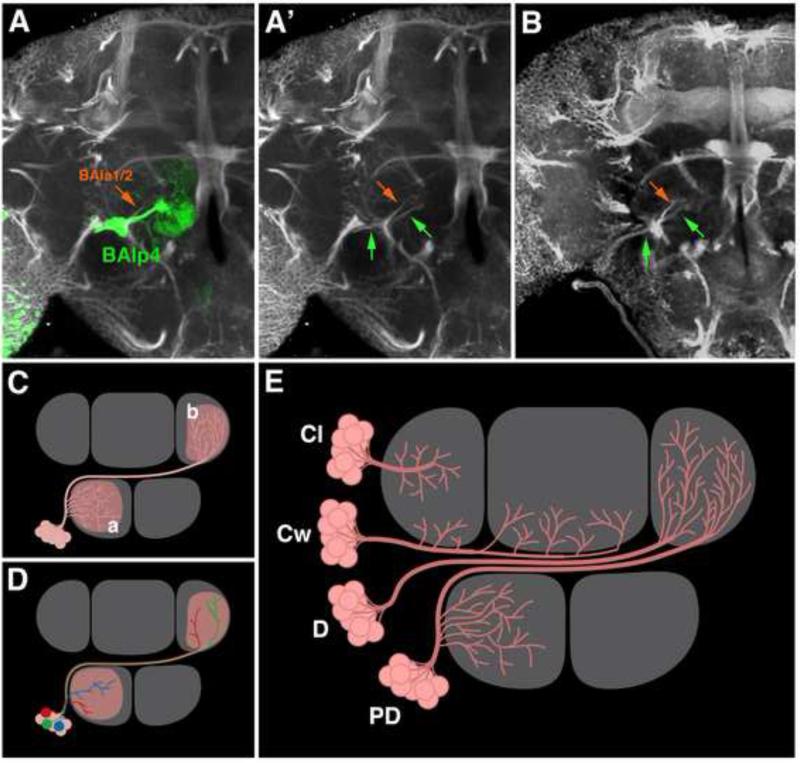
We analyzed a total of 814 secondary clones, distributed over 499 brains. About half of the brains had a single clone, the other half had two or more (up to five; brains containing in excess of five clones were discarded). Aside from clonal labeling of individual secondary lineages, most brains also contained labeled-endings of afferents from the optic lobe and/or antennal nerve. All clones could be assigned to a specific secondary lineage (or lineage pairs) based on the entry point and trajectory of the SAT, defined as the fiber bundle that directly emerges from the cell body cluster and enters the neuropil (Fig.1A-B; BAlp4 clone assigned to BP104-labeled SAT). Given the number of clones analyzed, most secondary lineages were represented by more than one clone. We observed a wide range, with some lineages represented more than 20 times and others less than five times (average: ten clones per lineage; Table 2).
Figure 1.
Secondary lineages: SATs and projection envelope. (A-C) Assignment of clones to their respective secondary lineages is based on the entry point and trajectory of the SAT. Panel (A) shows a z-projection of an adult brain labeled with BP104 (white), containing a MARCM clone of the BAlp4 lineage (green). The SAT of BAla1/2 is shown in orange. Panel (A’) shows BP104 channel of the same z-projection. Green arrows in (A’-B) point at the SAT of the BAlp4 clone; as visible by green arrows in (A’), the SAT follows one of the BP104-positive tracts that can be followed back through metamorphosis to lineage BAlp4. Panel (B) represents a z-projection of the standard brain used in this paper, at an antero-posterior level corresponding to the one used for (A/A’). Note invariant pattern of BP104-positive fiber bundles in (A/A’) and (B) (green arrows: BAlp tract; red arrow: BAla1/2 tract). (C-D) Relationship of individual neurons and projection envelope. Schematic representation of a lineage (pink) with a projection envelope including compartments “a” and “b”. Individual neurons of the lineage could (all or in part) form arborizations throughout all compartments of the projection envelope [represented by red neuron in panel (D)], or could project to one or the other compartment [blue and green neuron in (D)]. (E) Classification of lineages based on the contour of projection envelope. Shown are four classes of lineages. In “PD” (“proximal distal”) lineages, a long segment of the SAT connects proximal to distal arborizations. In “C” (continuous) lineages, branches emerge at more or less regular intervals along the entire length of the SAT. Two subtypes of continuous lineages, local (“Cl”) and widefield (“Cw”), are illustrated. In “D” (“distal”) lineages, terminal arborizations are delimited to the distal end of the SAT. Bar: 25μm.
Table 2.
List of abbreviations of neuropil fascicles (left), compartments (center), and entry portals of lineage-associated tracts (right).
| Fascicles | Abbr. | Compartments | Abbr. | Entry portals | Abbr. |
|---|---|---|---|---|---|
| Anterior-dorsal commissure | ADC | Antennal lobe | AL | Anterior entry portal of the ML | ptML a |
| Antennal lobe commissure | ALC | Antenno-mechanosensory and motor center | AMMC | Anterior portal of the lateral horn | ptLH a |
| Antennal lobe tract | ALT | Anterior optic tubercle | AOTU | Anterior superior lateral protocerebrum portal | ptSLP a |
| Inner antennal lobe tract | iALT | Anterior periesophageal neuropil | PENPa | Antero-dorsal entry portal of the VLP | ptVLP ad |
| Medial antennal lobe tract | mALT | Bulb | BU | Dorrso-lateral superior ventro-lateral protocerebrum portal | ptVLP dls |
| Outer antennal lobe tract | oALT | Ellipsoid body | EB | Dorsal antennal lobe portal | ptAL d |
| Anterior optic tract | AOT | Fan-shaped body | FB | Dorsal spur portal | ptSP d |
| Anterior superior transverse fascicle | trSA | Inferior protocerebrum | IP | Dorso-lateral entry portal of the ML | ptML dl |
| Central protocerebral descending fascicle | deCP | Anterior IP | IPa | Dorso-lateral inferior ventro-lateral protocerebrum portal | ptVLP dli |
| Cervical Connective | CCT | Lateral IP | IPl | Dorso-lateral portal of protocerebral bridge | ptPB dl |
| Commisure of the lateral accessory lobe | LALC | Medial IP | IPm | Dorso-lateral vertical lobe portal | ptVL dl |
| Dorsal commissure of anterior subesophageal ganglion | DCSA | Posterior IP | IPp | Dorso-medial entry portal of the ML | ptML dm |
| Dorsolateral root of the fan-shaped body | dlrFB | Lateral accessory lobe | LAL | Dorso-medial portal of protocerebral bridge | ptPB dm |
| Fronto-medial commissure | FrMC | Lateral horn | LH | Dorso-medial ventro-lateral protocerebrum portal | ptVLP dm |
| Great commissure | GC | Mushroom body | MB | Dorso-medial vertical lobe portal | ptVL dm |
| Horizontal ventrolateral protocerebral tract | hVLPT | Calyx | CA | Lateral antennal lobe portal | ptAL l |
| Intermediate superior transverse fascicle | trSI | Medial lobe | ML | Lateral portal of calyx | ptCA l |
| Deep bundle of irSI | trSI d | Peduncle | PED/P | Lateral portal of the posterior lateral protocerebrum | ptPLP l |
| Superficial component of trSI | trSI s | Spur | SP | Lateral portal of the superior lateral protocerebrum | ptSLP l |
| Lateral ellipsoid fascicle | LE | Vertical lobe | VL | Medial portal of calyx | ptCA m |
| Anterior LE | LEa | Noduli | NO | Posterior inferior portal of the posterior lateral protocerebrum | ptPLP pi |
| Posterior LE | LEp | Posterior lateral protocerebrum | PLP | Posterior portal of superior lateral protocerebrum | ptSLP p |
| Lateral equatorial fascicle | LEF | Protocerebral bridge | PB | Posterior portal of the lateral horn | ptLH p |
| Anterior LEF | LEFa | Subesophageal ganglion | SEG | Posterior superior portal of the posterior lateral protocerebrum | ptPLP ps |
| Posterior LEF | LEFp | Superior protocerebrum | SP | Posterior ventro-medial cerebrum portal | ptVMCpo |
| Medial equatorial fascicle | MEF | Superior intermediate protocerebrum | SIP | Postero-lateral portal of superior lateral protocerebrum | ptSLP pl |
| Medial root of the fan-shaped body | mrFB | Superior lateral protocerebrum | SLP | Postero-medial portal of superior lateral protocerebrum | ptSLP pm |
| Median bundle | MBDL | Anterior SLP | SLPa | Ventral antennal lobe portal | ptAL v |
| Oblique posterior fascicle | obP | Posterior SLP | SLPp | Ventral entry portal of the VLCi | ptVLCi v |
| Posterior commissure of the posterior lateral protocerebrum | pPLPC | Superior medial protocerebrum | SMP | Ventral portal of calyx | ptCA v |
| Posterior lateral fascicle | PLF | Ventro-lateral cerebrum | VLC | Ventral portal of protocerebral bridge | ptPB v |
| External component of PLF | PLFe | Anterior VLC | VLCa | Ventral spur portal | ptSP v |
| Dorsolateral component of PLF | PLFdl | Inferior VLC | VLCi | Ventro-lateral antennal lobe portal | ptAL vl |
| Dorsomedial component of PLF | PLFdm | Lateral VLC | VLCl | Ventro-lateral inferior ventro-lateral protocerebrum portal | ptVLP vli |
| Ventral component of PLF | PLFv | Ventro-medial cerebrum | VMC | Ventro-lateral portal of calyx | ptCA vl |
| Posterior superior transverse fascicle | trSP | Anterior VMC | VMCa | Ventro-lateral superior ventro-lateral protocerebrum portal | ptVLP vls |
| Lateral trSP | trSPl | Inferior VMC | VMCi | Ventro-lateral vertical lobe portal | ptVL vl |
| Medial trSP | trSPm | Post-commissural VMC | VMCpo | Ventro-medial antennal lobe portal | ptAL vm |
| Sub-ellipsoid commissure | SuEC | Pre-commissural VMC | VMCpr | Ventro-medial ventro-lateral protocerebrum portal | ptVLP vm |
| Subesophageal-protocerebral system | SPS | Superior VMC | VMCs | Ventro-medial vertical lobe portal | ptVL vm |
| Superior arch commissure | SAC | Ventro-lateral protocerebrum | VLP | ||
| Superior commissure of the posterior lateral protocerebrum | sPLPC | Anterior VLP | VLPa | ||
| Superior lateral longitudinal fascicle | loSL | Posterior VLP | VLPp | ||
| Anterior loSL | loSLa | ||||
| Posterior loSL | loSLp | ||||
| Superior medial longitudinal fascicle | loSM | ||||
| Anterior loSM | loSMa | ||||
| Posterior loSM | loSMp | ||||
| Supra-ellipsoid body commissure | SEC | ||||
| Ventral fibrous center | VFC | ||||
| Ventral longitudinal fascicle | loV | ||||
| Intermediate loV | loVIa | ||||
| Lateral loV | loVLa | ||||
| Medial loV | loVMa | ||||
| Posterior-lateral loV | loVP | ||||
| Vertical posterior fascicle | vP | ||||
| Vertical tract of the superior lateral protocerebrum | vSLPT | ||||
| Vertical tract of the ventro-lateral protocerebrum | vVLPT |
Based on our analysis of SAT development (see accompanying paper by Lovick et al., 2013), 56 lineages defined in the late larva have SATs that can be individually followed within the neuropil throughout development (Table 2). Within this group, we could identify clones in all cases except one, DALv3. The projection pattern of DALv3 has been characterized previously (Kumar et al., 2009a). A second group of 30 lineages (e.g., BAmas1/2; DALcm1/2) have SATs that form pairs or form a quartet (the four BLAd lineages). In these cases, it is not possible to predict whether the two lineages forming the pair (or quartet, in the case of BLAd1-4) will have projection envelopes that are identical or different. Within the group of 30 lineages, four pairs (BAmas1/2, DPMpl1/2, CP2/3, BLP1/2) were obtained that had clones with significantly different arborization patterns. This suggests that paired lineages with identical SATs form distinct arborization patterns (e.g. BAmas1/2; Fig.4C11-12). In three pairs within this group (DPLl2/3, BLVa1/2, BLVa3/4) the patterns were very similar, but the trajectory of part of the SATs differed consistently (e.g. DPLl2/3 in Fig.10B-F). In the case of the BLAd1-4 quartet we isolated three different classes of clones. In the eight remaining pairs (BAla3/4, DALcm1/2, DAMd2/3, DAMv1/2, DPLal2/3, DPLc2/4, DPLp1/2, BLP3/4) only one type of clone was recovered, suggesting that these lineages form identical arborization patterns. Alternatively, it is possible that we could recover clones for only one member of the pair, which is unlikely given the fact that an average of ten clones per lineage was obtained for all other lineages.
A significant fraction of lineages form more than one SAT. In cases where these tracts separate from the very beginning where axon tracts enter the neuropil we tentatively assume that they represent two separate hemilineages (or sublineages, in case of type II lineages); ultimate proof for their status as “true” hemilineages would have to come from experimental studies such as those done for thoracic lineages (Truman et al., 2010) or a small number of engrailed-positive brain lineages (Kumar et al., 2009a). As described in the accompanying paper by Lovick et al. (2013), hemilineages move apart during metamorphosis in a number of cases. GFP labeled clones provided confirmation for this movement of hemilineages. All except one (BLAvm) of the lineages in question, notably BAlc, BAmd1, DPLl2/3, CP2/3, BLAl, BLAv1, BLVp1/2, were represented by more than five clones; for BLAvm we have three clones. In all cases, GFP labeling invariably marked both hemilineages simultaneously, whereas other, independent lineages could be represent by a clone in some cases, but not in others. In case of several lineages for which the movement of SATs and HSATs was difficult to follow (BLD1, BLD3, BLAl, DPLc5), the existence of two separating hemilineages was confirmed. In three cases, BAmd2, DPLm2 and DPMl1, the analysis of MARCM clones made it possible to identify the proper lineage in the adult brain. Thus, the SAT of BAmd2 cannot be followed beyond P24 because its entry and proximal SAT is masked by the arrays of antennal afferent surrounding it. A clone with the characteristic SAT entry point and crossing in the antennal lobe commissure confirmed BAmd2 for the adult brain. The same applied for DPMl1, whose characteristic descending SAT is not visible beyond P24. DPLm2 represents a unique case where the MARCM clone united two clusters of cells that had been previously considered to represent two separate lineages. Thus, in our larval analysis, DPMl2 with a characteristic centrifugal axon bundle projecting to the ring gland was considered as a separate lineage (Pereanu et al., 2006). MARCM clones showed that the ring gland associated axons form part of DPLm2 instead.
Classification of lineages based on the geometry of their projection envelope
The GFP-labeled clone, when superimposed on a backdrop of an adult brain labeled with a neuropil marker (anti DN-cadherin; from here on called DN-cad) or axonal marker (anti-Neurotactin, from here on referred to as BP104), allows one to map the neurite arborizations of all neurons of a single lineage (the “projection envelope”) with respect to brain neuropil compartments (Fig.1C). Note that the relationship between the projection envelope of a lineage and the projection of an individual neuron forming part of a lineage is not simple (Fig.1D). For example, when an envelope includes two neuropil compartments, a and b, there are two possibilities: (1) each individual neuron may also have arborizations in a and b (Fig.1D, red neuron); or alternatively, a subset of neurons might only project to a or to b (Fig.1D, green neuron and blue neuron, respectively). Nonetheless, documenting the projection envelope for each lineage represents a significant step towards describing brain circuitry. In this paper we will provide an overview of the projection envelopes for each lineage of the supraesophageal ganglion, following the same topology-based ordering used in the accompanying paper (Lovick et al., 2013).
Representative clones and lineage restricted markers used in previous studies suggested that, aside from their topology (spatial relationship of a SAT entry point and projection relative to neuropil tracts and compartments), lineages can also be classified according to the “geometry”, defined as the distribution of axonal/dendritic branches relative to the main SAT (Larsen et al., 2009). It should be noted that unlike vertebrate neurons, where dendritic branches connected to the cell body are separated from axonal branches, insect neurons have a neurite tree on which dendritic and axonal branches are frequently intermingled (Hartenstein et al., 2008; Watson and Schürmann, 2002). The degree of intermingling, nonetheless, varies for different neurons and presumably different lineages. For example, in the case of the well characterized lineages of antennal lobe (AL) projection neurons (e.g. BAmv3, BAlc, and BAla1), dendritic branches are concentrated along the proximal region of the SAT in the AL; whereas axonal branches are close to the distal tip of the SAT in the calyx (CA) and lateral horn (LH). The long segment of the SAT connecting proximal to distal, called the antennal lobe tract, is devoid of branches. Lineages with this geometry were classified as “PD” (“proximal distal”) lineages (Larsen et al., 2009; Fig.1E). In other lineages, branches emerged at more or less regular intervals along the entire length of the SAT (“continuous” or “C” lineages); or were all concentrated at its distal tip (“distal” or “D” lineages). Further analysis using fluorescent reporters differentially localized to either dendritic or axonal branches (e.g. UAS-ICAM5ΔECD::mCherry and UAS-GFP-KDEL; Nicolai et al., 2010 and Okajima et al., 2005, respectively) can be used to compare their distribution in the C, D, and PD lineages.
In the following, clones representing individual lineages will be described in the order established for the lineage tracts in the accompanying paper (Lovick et al., 2013). Clones are documented in three sets of figures. In one set of figures, we show z-projections of clones with spatial respect to the BP104-labeled scaffold of neuropil fascicles, starting with lineages entering the anterior brain surface (BA: Fig.4; DAL and DAM: Fig.6), followed by those of the dorsal surface (DPL: Fig.9), posterior surface (DPM, CM, CP: Fig.12), and finally, lateral surface (BLA, BLD, BLP, BLV: Figs.15 and 17). These figures illustrate the identification of clones with their corresponding lineages. The second set of figures show z-projections of clones registered to DN-cad-labeled brain slices, in the order as described above (BA: Figs.2, 3; DAL: Fig.5; DAM: Fig.7; DPL: Figs.8, 10; DPM: Fig.11; CM and CP: Fig.13; BLA and BLD: Fig.14; BLP and BLV: Fig. 16), illustrating the projection envelopes of all lineages. In all panels of all these figures, the adult MARCM clone is paired with a larval clone (MARCM or Flp-out; see Materials and Methods for more details) representing the corresponding lineage, documenting the similarity in SAT projection patterns between larval and adult stages. The third set of figures (supplementary figures S1-S5) show schematic renderings of SATs and main locations of terminal arborizations for all lineages. Lineages sharing important aspects of their projection are combined together, such that Fig.S1 shows lineages with SATs connecting ventral to dorsal compartments, Fig.S2 has lineages interconnecting ventral compartments, lineages in Fig.S3 are associated strongly with the central complex and mushroom body, Fig.S4 presents lineages of the superior protocerebrum projecting ventrally (including the subesophageal ganglion and thoracico-abdominal ganglion), and Fig.S5 shows lineages interconnecting dorsal compartments of the brain.
Figure 2.
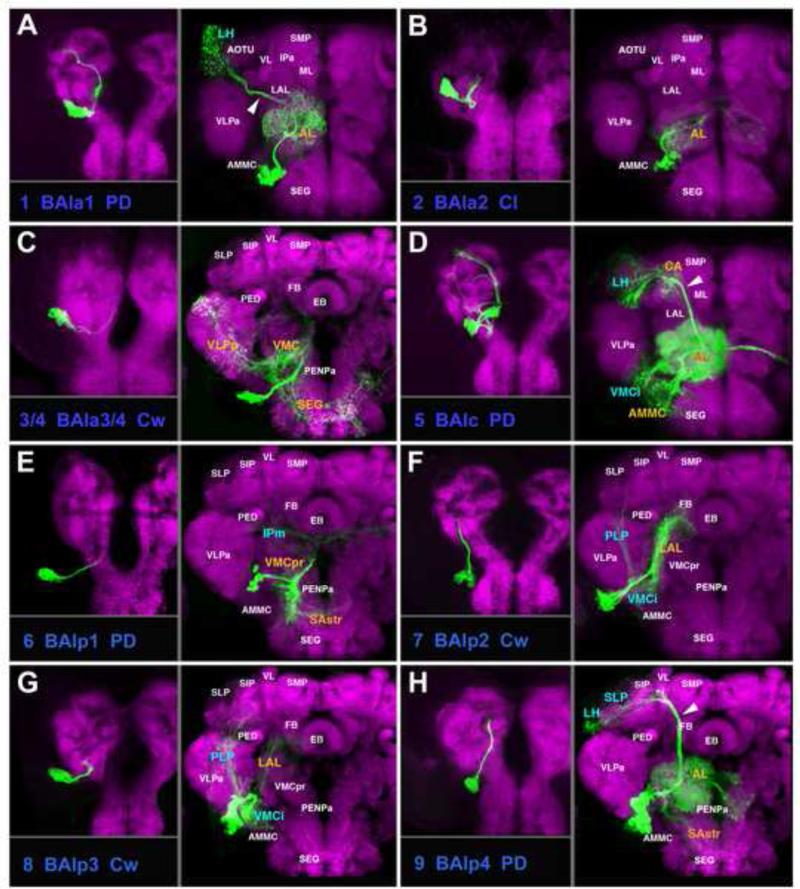
A-H: Clones representing lineages of the BA group (#1/BAla1 to #9/BAlp4) in the larval and adult brain. This and the following figures 3, 5, 7, 8, 10, 11, 13, 14, 16 are designed in the same manner: Each lineage is represented by a panel showing z-projection of a larval brain hemisphere on the left, and of an adult hemisphere on the right. Number and abbreviation of the lineage is given at the bottom left of the panel. Next to the abbreviation, the type of lineage (C, P, PD) is indicated. Images were generated by registering full z-projection of clones (that is, a z-projection containing all sections of a stack showing GFP label) to a z-projection of a “slice” of the DN-cad-labeled standard brain, as described in the Materials and Methods section. DN-cad visualizes neuropil compartments, annotated by white letters on part of panels showing adult brain. Compartments receiving major innervation by the lineage shown in a given panel are annotated by colored letters. Innervated compartments contained within the brain slice shown by DN-cad are in orange; compartments located significantly anterior or posterior to the slice shown appear in blue letters. For example, #1/BAla1 (A) is represented by a clone registered to an anterior brain slice [level optic tubercle (OTU)/mushroom body medial lobe (ML)]. Major compartments visible at that level are annotated by small white letters. Within this slice, only the antennal lobe is innervated by BAla1; it is annotated by orange letters. BAla1 projects postero-laterally towards the lateral horn, which is located in a slice posterior to the one shown. The lateral horn (LH) is therefore annotated by blue letters. For alphabetical list of all abbreviations see Table 2. Bar: 50μm.
BA Lineages
The BA group comprises PD, C, and D lineages whose neurons are mostly associated with the ventral brain compartments (Figs.2-4; Figs.S1, S2). Four PD lineages, BAla1 (#1; Fig.2A), BAlc (#5; Fig.2D; dorsal hemilineage), BAlp4 (#9; Fig.2H), and BAmv3 (#17; Fig.3H) include all of the projection neurons connecting the antennal lobe (AL) and superior protocerebrum. After forming proximal (dendritic) branches in the AL, neurites of all neurons of these lineages converge and exit the AL posteriorly as the common antennal lobe tract (ALT; yellow arrows in Fig.4C1, C5, C9, C17). Axons of BAla1 soon leave this bundle and directly head for the lateral horn (LH) via the medial-lateral ALT (mlALT; Fig.2A, white arrowhead; Fig.S1). The remaining three SATs (#5, 9, 17) stay together as the medial ALT (mALT) which continues dorso-posteriorly towards the calyx (CA), before bending laterally towards the LH (arrowheads in Figs.2D, H; 3H; Fig.S1). As published previously (Das et al., 2013), the lineage BAlp4 (#9) forms proximal branches that not only reach the AL, but also part of the ventrally adjacent subesophageal ganglion (abbreviated as SAstr in Fig.2H; possibly a domain with gustatory input), and projects to the superior lateral protocerebrum, rather than the CA and LH (Fig.2H; Fig.S1). The ventral hemilineage of BAlc (#5v) includes complex projection neurons which are mostly unrelated to the AL. They project ventro-posteriorly, forming the loVI fascicle (Fig.4A, B; Fig.S2; see accompanying paper by Lovick et al., 2013). Proximal branches of the loVI arborize in the antenno-mechanosensory and motor center (AMMC; Fig.2D). Distally, this tract forms a T-junction, with one branch projecting laterally into the inferior domain of the ventro-lateral cerebrum (VLCi) and the other branch crossing the midline via the great commissure to reach the contralateral inferior ventro-lateral cerebrum (VLCi; Fig.2D; Fig.S2). In addition, a thin branch continues dorso-laterally towards the LH; this constitutes the lateral antennal lobe tract (not resolved in Fig.2D; Fig.S1).
Two additional PD lineages, BAmas1 and BAmas2 (#11, #12; Fig.3B, C), form a connection between the tritocerebrum and the superior medial protocerebrum and mushroom body, respectively. Thus, proximal branches of BAmas1/2 form dense arborizations in the tritocerebrum. The tritocerebrum is also called, in a segment-neutral manner, anterior peri-esophageal neuropil (PENPa; Kumar et al., 2009b; Pereanu et al., 2010; Fig.3B, C). The SATs of BAmas1/2 then project dorsally through the median bundle and reach the superior medial protocerebrum (Fig.4C11, C12). Short distal terminal branches of BAmas2 end here; BAmas1 bends laterally and forms terminal arbors in the dorsal lobe of the mushroom body (Fig.3B, C; Fig.S1).
BAmd1 (#13) and BAmd2 (#14) are complex lineages with commissural tracts. The dorsal HSAT of BAmd1 (#13d) projects medially directly behind the mushroom body medial lobe and crosses in the fronto-dorsal commissure; terminal branches innervate the medial lobe of both hemispheres, as well as the anterior inferior protocerebrum on the ipsilateral side (Figs.3D; 4A, C13; Fig.S3). The ventral HSAT of BAmd1 (#13v) projects diagonally through the AL, crosses in the antennal lobe commissure (ALC), and then bifurcates into a dorsal branch directed towards the superior lateral protocerebrum (SLP) and a ventral branch with a large terminal domain in the lateral accessory lobe (LAL), ventro-medial cerebrum (VMC), and subesophageal ganglion (SEG) (Figs.3D; 4C13; Fig.S4). BAmd2 (#14) enters near the midline, in between the antennal lobes of either side. The SAT bifurcates, with one branch crossing in the antennal lobe commissure (Fig.4C14; Fig.S2). The ipsi- and contralateral branches project in a nearly symmetrical fashion postero-laterally, innervating the ventrolateral cerebrum (VLCi) and posterior lateral protocerebrum (PLP) (Fig.3E; Fig.S2).
Another complex PD lineage, BAmv1 (#15), is marked by the per-Gal4 driver line and has been documented previously (Spindler and Hartenstein, 2010; Spindler and Hartenstein, 2011). The large proximal SAT of BAmv1 (#15p) forms a major component of the loVM that passes underneath the AL into the ventromedial cerebrum (VMC) (Fig.4B, C15). The SAT splits into three major branches: one curving dorsally and medially towards the central complex; the second continuing posteriorly into the VMC; the third one extending laterally towards the ventro-lateral protocerebrum (VLPa/p) (Fig.S2). Terminal branches innervate the lateral accessory lobe (LAL), the fan-shaped body (FB), the noduli (NO), the VMC, and the VLP (shown in orange letters in Fig.3F; Fig.S2). BAmv2 (#16) has a distally branching (type D) single SAT that accompanies BAmv1 in the loVM fascicle (#15; Fig.4B, C16). At the level of the great commissure the tract turns medially and dorsally and splits into an ipsilateral and contralateral component that innervate the VMC surrounding the great commissure (Fig.3G; Fig.S2).
BAlp2 and BAlp3 (#7, #8) are lineages with long C-type SATs that contribute to the lateral component of the ventral longitudinal fascicle (loV; Fig.4B, C7, C8). The BAlp2 tract splits into a dorsal branch with dense terminal fibers in the lateral part of the LAL compartment and a posterior branch that continues posteriorly (Fig.2F), innervating VLCi and PLP (Fig.S2). BAlp3 has a single tract that follows BAlp2 towards the VLCi and PLP (Fig.2G; Fig.S2).
BAla3/4 (#3/4) and BAlv (#10) have C-type SATs, and BAlp1 (#6) has a PD-type SAT that enter from a position lateral of the AL (Fig.4A, B, C). The first three of these lineages (#3/4, 6) project medially towards the ventromedial cerebrum: BAlp1 crosses the loVM fascicle at its dorsal surface and BAla3/4 at its ventral surface (Fig.4A, B, C3/4; C6). BAlp1 has terminal arborizations within the VMC compartment, with ventral branches reaching into the SEG/PENPa (Fig.2E; Fig.S2). BAla3, marked by the driver en-Gal4 (Kumar et al., 2009b), has widespread terminals in the VMC (Fig.2C; Fig.S2). BAla4 extends alongside BAla3; only a single type of clone was recovered for the BAla3/4 pair. BAlv (#10) contacts the inferior VLC (VLCi) from ventral (Fig.4B, C10) and has terminal fibers confined to the VLCi and neighboring AMMC (Fig.3A; Fig.S2).
DAL Lineages
Projections of the DAL lineages are predominantly associated with the medial and vertical lobes of the mushroom body (ML, VL), the central complex, and the adjacent protocerebral compartments (AOTU, SMP, IPa). Most of the DAL lineages, including the DALcl1/2, DALcm1/2, DALd, DALl1, and DALv1-3 are PD lineages with long tracts, many of which are commissural.
DALcl1 and DALcl2 (#18, #19; Figs.5A, B; 6C18, C19), located laterally of the mushroom body vertical lobe (VL), form a pair of PD lineages associated with the anterior optic tubercle (AOTU), central complex, and adjoining compartments; each one consists of two hemilineages whose diverging HSATs in a “pincer-like” manner enclose the mushroom body spur (SP; Fig.6A, C18, C19). Dense proximal branches of DALcl1 and DALcl2 innervate the AOTU (Fig.5A, B; Fig.S3). The ventral hemilineage tract of DALcl1 (#18v) passes underneath the SP and continues medially, crossing the midline in the subellipsoid commissure (SEC; Fig.5A; Fig.S3). Terminal arborizations of this tract end bilaterally in the LAL. In addition, on each side, a posteriorly directed branch of DALcl1v projects along the MEF fascicle towards the posterior VMC compartment (VMCpo; Figs.5A; 6A). The ventral DALcl2 hemilineage projects as part of the lateral ellipsoid fascicle (LEa) towards the central complex (Figs.5B, 6A, C19; Fig.S3). Dorsal hemilineage tracts of DALcl1/2 (#18d/19d) curve over the dorsal surface of the spur (SP) and peduncle. DALcl1 projects in a fairly restricted manner to the bulb (BU, a small compartment relaying information towards the ellipsoid body, EB; Fig.5A; Fig.S3); DALcl2 projects in a more widespread manner, including the BU, adjacent LAL, IP, and SMP (Fig.5B; Fig.S3).
DALd (#22) and the DALcm1/2 pair (#20/21) are located medially of the mushroom body vertical lobe (VL; Figs.5C, D; 6B, C20-22). DALd (#22) constitutes a PD lineage with dense proximal arborizations in the IPa and the superior intermediate protocerebrum (SIP), which surround the medial lobe and vertical lobe, respectively (Fig.5D; Fig.S4). The SAT of DALd (#22) projects ventro-medially, crossing the peduncle, and continues as part of the deCP tract (Fig.6B, C22). Distal arborizations are found in the VMC and SEG (Fig.5D; Fig.S4). The lineage pairs DALcm1/2 each have two PD hemilineages with very similar projection patterns in all recovered clones. This suggests that both lineage pairs possess the same projection envelope. The medial hemilineage tracts pass behind the medial lobe (ML) and enter the fronto-medial commissure (FrMC; Fig.6C20-21; Fig.S3). In terms of projection, arbors are found bilaterally in the ML and the surrounding IPa, AOTU, SIP, and SMP compartments (Fig.5C; Fig.S3). The ventral hemilineage tracts of DALcm1/2 pass through the elbow formed by the VL and peduncle before turning ventrally (Fig.5C; Fig.S4). This projection initially forms part of the deCP, but then separates from the fascicle, extending into the ventral brain as a separate, loose bundle. The proximal terminal branches are found throughout the anterior domain of the SMP, IP, and the distal branches in the VMC (Fig.5C; Fig.S4).
Two DAL lineages, DALl1 and DALl2 (#23, #24), are located laterally of the DALcl group, flanking the anterior VLP compartment (VLPa; Figs.5E, F; 6C23, C24). DALl1 (#23) is a PD lineage with a conspicuous recurrent projection. The SAT projects posteriorly, splits into a ventral branch with arborizations in the PLP and adjacent lobula (LO), and a recurrent branch that turns dorsally and anteriorly forming arborizations in the anterior SLP, IP and AOTU (Figs.5E, 6B; Fig.S5). DALl2 (#24; Fig.5F) represents a C lineage. Its short SAT projects into the anterior VLPa where it splits into several groups of terminal arborizations filling much of the anterior and posterior VLP compartment (VLPa/p). A few “outlier” branches continue dorso-medially into the IP and SMP compartments (Fig.5F).
The DALv group, comprising three PD lineages with commissural connections, is located ventral of the spur. DALv1 (#25) has a long unbranched proximal SAT that forms the LEFa fascicle (Figs.5G; 6B, C25) and then bifurcates into an ipsilateral and commissural branch that crosses in the great commissure. Terminal arborizations fill the ipsilateral and contralateral posterior VLP and neighboring VLCi compartments (Fig.5G). Ipsilaterally, there is a projection to the posterior SEG (not shown). DALv2 (#26) and DALv3 (#27) are marked by the expression of per-Gal4 and en-Gal4, as described previously (Kumar et al., 2009b; Spindler and Hartenstein, 2010; Spindler and Hartenstein, 2011); proximal SATs form the lateral ellipsoid fascicle (LEa) (Figs.5H; 6A, C26-27; 7A). The DALv2 (#26) lineage forms large proximal arborizations in the BU, as well as distal, ring-shaped branches of the EB (Fig.5H; Fig.S3), that represent the ring (R)-neurons of the EB (Spindler and Hartenstein, 2011). Additional terminal arborizations of DALv2 are found in the adjacent LAL and IPa (Fig.5H). DALv3 (#27), marked by the expression of en (Kumar et al., 2009b), projects alongside DALv2 in the LEa fascicle, which then splits into a dorsal and ventral commissural branch (Fig.6A, C27; Fig.S3). DALv3 terminal arborizations are confined to the ipsilateral and contralateral inferior protocerebrum (IPa/m) and the SMP (Fig.7A; see Kumar et al., 2009b for detail).
DAM Lineages
The small group of DAM lineages is located in the anterior dorso-medial cortex and has arborizations predominantly associated with the SMP/SIP and adjacent IPm/IPa compartments. DAMd1 (#28), a PD lineage with a unique recurrent commissural projection, first crosses the midline in the anterior dorsal commissure (ADC; Figs.6B, C28; 7B). It forms profuse arborizations in the contralateral SMP, SIP, and IPa; and crosses back via the fronto-medial commissure (FrMC) to form distal arbors in the ipsilateral SMP and IPa (Figs.6B, C28; 7B; Fig.S5). The DAMd2/3 pair (#29/30) comprises large C-type lineages (Figs.6B, 7C). Among the clones recovered for this pair, only a single type of projection envelope could be observed. The DAMd2/3 tract forms the anterior longitudinal superior medial fascicle (loSMa), continuously giving off terminal arborizations throughout the SIP, SMP, and IPm compartments (Figs.6B, C29-30; 7C; Fig.S5). Posteriorly, projections of DAMd2/3 extend ventrally to fill regions of the ipsilateral and contralateral VMCpo (Fig.7C; Fig.S5). The DAMv1/2 (#31/32) paired lineages also possess an indistinguishable projection envelope. The short SAT enters the SMP from anterior (Fig.6B, C31-32) and splays out into dense terminal arborizations, filling much of the SMP compartment (Fig.7D; Fig.S5).
DPL Lineages
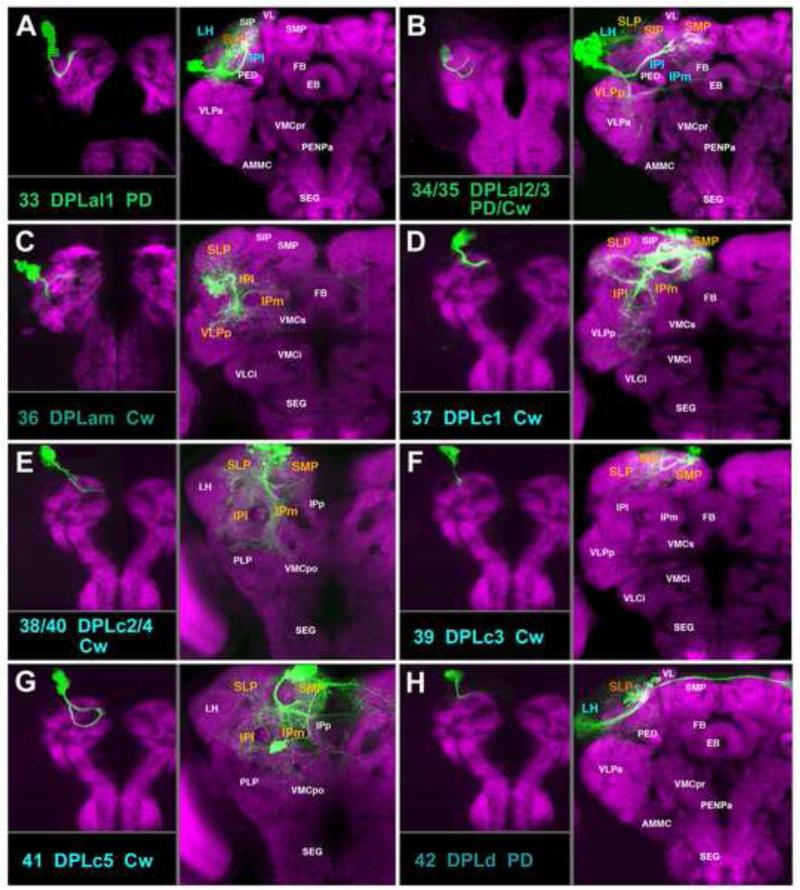
DPL lineages predominantly innervate the lateral domains of the superior and inferior protocerebrum. Five lineages, DPLal1-3, DPLam, and DPLd represent the anterior subgroups, located dorso-laterally of the anterior optic tubercle (AOTU). DPLal1-3 (#33-35) are PD lineages recognizable by their crescent shaped SATs which form the anterior transverse fascicle of the superior protocerebrum (trSA; Figs.8A, B; 9A, C33-35; Fig.S5). Proximal arborizations of DPLal1 (#33) fill the deep regions of the SLP, LH, and the adjacent lateral IP (IPl); distal arbors innervate the dorso-anterior SLP (Fig.8A; Fig.S5). The DPLal2/3 (#34/35) pair has an indistinguishable projection envelope, each with two hemilineages (Figs.8B; 9C34, C35). The dorsal hemilineage (#34/35d) resembles DPLal1 (#33), forming part of the trSA (Fig.9A), but arborizing more widely than DPLal1 in the LH, SIP, SLP, SMP, and much of the IP (IPm/l, Fig.8B). The ventral HSAT forms projections in the medial IP and the adjacent posterior VLP (VLPp) (Fig.8B; Fig.S5). We recovered one clone where the two HSATs extended at a moderate distance from each other (Fig.8C); this could represent a random variant, or indicate that DPLal2/DPLal3 do differ in regard to their exact HSAT pathfinding. DPLam (#36) is a C-type lineage marked by the expression of engrailed and has been described previously (Kumar et al., 2009b). Projecting its single SAT ventro-posteriorly via the vSLPT fascicle (Figs.8C, 9A, C36), DPLam arborizes widely in the anterior SLP and the central part of the IPl/m and the VLPp (Fig.8C; Fig.S4). DPLd (#42) forms sparse proximal arborizations in the SIP and part of the adjacent SLP (Figs.8H, 9C42). The lineage has two HSATs, a medial one crossing the midline in the anterior-dorsal commissure (ADC) and projecting to the contralateral SIP (#42m; Figs.8H, 9A), and a posterior one that extends posteriorly along the anterior part of the loSL fiber system, forming terminal arborizations in the LH and lateral SLP (#42p; Figs.8H; Fig.9C42; FigS5).
The remainder of the DPL group, including DPLc1-5, DPLl1-3, DPLm1-2, DPLp1-2, and DPLpv are located in the posterior brain cortex. DPLc1-5 (#37-41; Fig.8D-G) enter through a common portal located at the junction between the SLP and SMP compartments (Fig.9B; C37-41) and have arborizations focused on the superior and inferior protocerebrum. DPLc1 (#37) is a C lineage with a characteristic crescent-shaped tract that forms part of the medial trSP fiber system (trSPm, Figs.8D; 9B, C37). Arborizations fill much of the SLP/SMP and the posterior part of the IPl/m (Fig.8D; Fig.S5). DPLc2/4 (#38, #40) is a C-type lineage pair that also forms part of the trSP fascicle (Fig.9B, C38-40). Unlike DPLc1, DPLc2/4 do not curve dorso-medially into the more anterior and dorsal part of the SMP; rather the pair remains close to the IPl/m, filling the compartment with widespread terminal arborizations and additional branches in the deep SLP/SMP compartments (Fig.8E; Fig.S5). DPLc3 (#39), another C-type lineage, has a short, anteriorly-directed SAT and arborizes in the central parts of the SLP, SIP, and SMP (Fig.8F; Fig.S5). DPLc5 (#41) possesses two hemilineages (#41a/p) which, in the adult are spaced relatively far apart from one another. The anterior hemilineage produces a curved SAT that enters alongside DPLc1-4 (Fig.9B, C41), extending antero-medially into the anterior SMP and part of the SLP; its dense terminal arborizations fill this compartment and the adjacent domains of the IP (Fig.8G; Fig.S5). The posterior DPLc5 hemilineage (#41p) is located at the ventro-posterior brain surface; the HSAT projects antero-dorsally, joining the loSM fascicle and crossing the midline in the ADC commissure (Fig.9C41). Terminal arborizations overlap with those of the anterior hemilineage in the SMP and IPm (Fig.8G).
DPLl1 (#43) and DPLl2/3p (#44/45p) enter the postero-lateral neuropil surface at the junction between the SLP/LH (Fig.9C43-45). The DPLl2/3p pair projects anteriorly, forming the loSL fascicle (Fig.9B). From the loSL fascicle, terminal branches sprout off and innervate the superior brain compartments (LH, SIP, SLP, and SMP) and ventrally directed branches also reach into the PLP, VLPp, and IPl (Fig.10B, E, F; Figs.S4, S5). While the DPLl2/3p hemilineages innervate identical compartments, they have distinct fasciculation patterns. Only one of the hemilineages, DPLl2p (#44p), forms a tight tract; fibers of the other hemilineage (DPLl3p, #45p) are more loosely aggregated (Fig.10E, F). This same characteristic holds true for the anterior hemilineages (#44/45a). As described in detail in the accompanying paper (Lovick et al., 2013), the anterior hemilineage tracts of DPLl2/3 (#44/45a) shift forward during metamorphosis and enter the anterior surface of the SLP (Fig.9A, C44, C45); they project ventrally into the upper part of the VLPp compartment (Fig.10C,D; Fig.S4). In contrast to DPLl2a (#44a), which forms a thin, compact tract with dense endings in a narrowly defined subdomain of VLPp (Fig.10C), DPLl3a (#45a) axons form a loose tract and extend diffuse terminal arborizations along their entire trajectory from the SLP to the VLPp (Fig.10D). DPLl1 (#43) enters the brain at the same point as DPLl2/3 (Fig.9B, C43), but sends its tract medially via the trSP fascicle, arborizing in the posterior SLP/SMP; a lateral branch innervates the LH/PLP (Fig.10A; Figs.S4, S5).
DPLm1 and DPLm2 (#46, #47; Fig.10G, H) are located lateral of the DPLc cluster, dorsal of the mushroom body calyx. DPLm1 (#46) is a C-type lineage and projects anteriorly in the SLP (Fig.9B, C46), producing branches in the SLP as well as the adjoining IPl/ SIP compartments (Fig.10G; Fig.S5). DPLm2 (#47) also innervates the SLP and adjacent IP; in addition, it sends a short SAT laterally (Fig.9B, C47) to form a terminal arbor in the lateral horn (LH; Fig.10H; Fig.S5). A long, thin fiber bundle of DPLm2 leaves the brain and projects to the ring gland (Fig.10H, arrowhead).
For the pair DPLp1/2 (#48/49), we were only able to isolate a single clonal type (Fig.10I). The paired tract enters the postero-lateral neuropil surface at the base of the lateral horn. A long, medial branch extends in the oblique posterior (obP) fascicle, across the peduncle and the brain midline, forming terminal arborizations along its trajectory in the posterior IP/SLP of both hemispheres (Figs.9C48, C49; 10I; Fig.S4). The anteriorly-directed HSAT of DPLp1/2 penetrates into the LH and forms profuse terminal branches in this compartment (Figs.9B; 11I; Fig.S4). A massive projection of DPLp1/2 is directed ventrally (#48v) along the vertical posterior tract (vP) into the PLP and posterior VLCi compartments (Fig.10I; Fig.S4). The posterior-most of the DPL lineages, DPLpv (#50) enters the posterior neuropil surface ventro-laterally; its SAT follows the postero-lateral fascicle anteriorly (PLF; Fig.9B, C50). Terminal branches appear along the entire length of the SAT and innervate the PLP/VLPp compartments and the adjacent IPl/m (Fig.10J; Fig.S2).
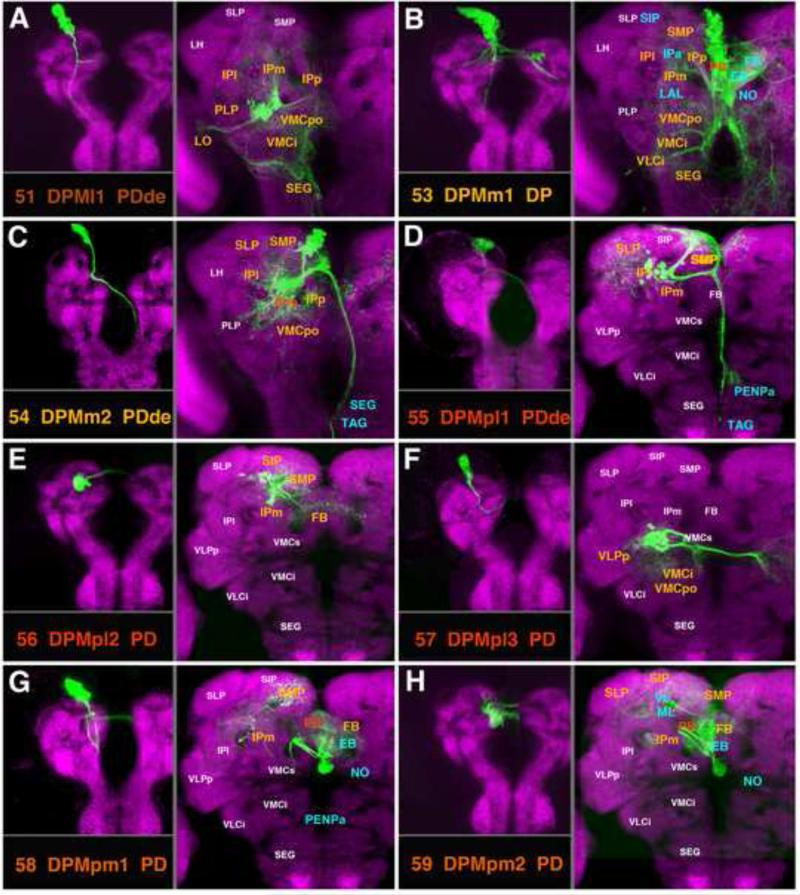
DPM Lineages
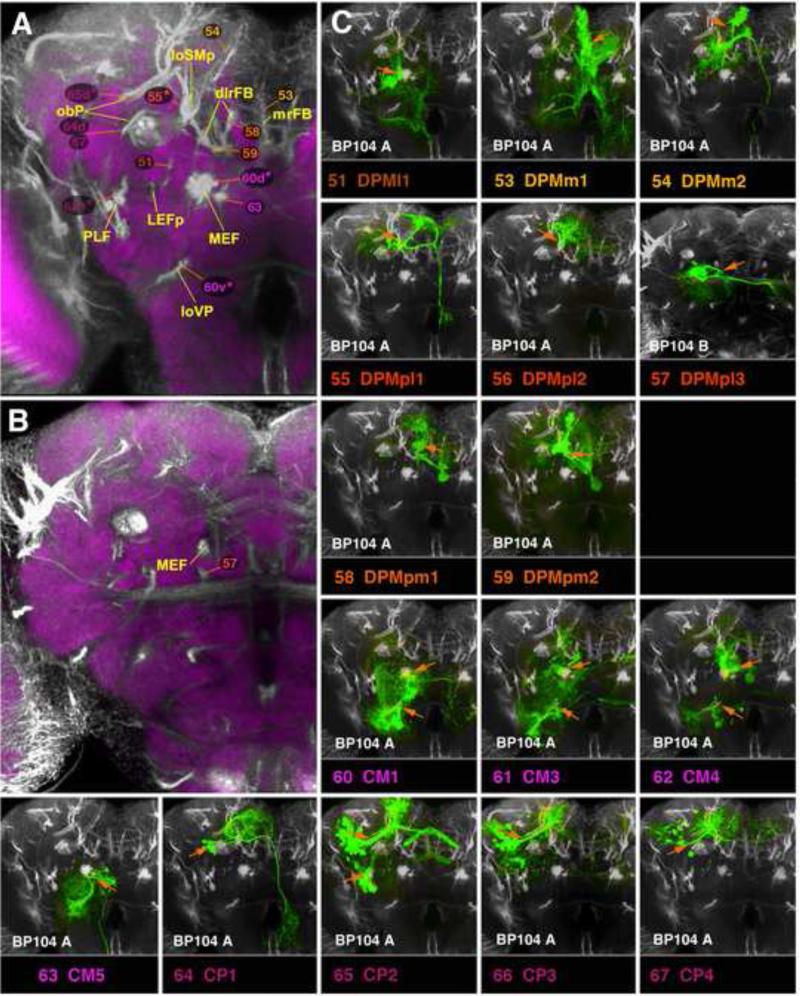
Located in the postero-medial brain, DPM lineages are primarily connected with the compartments of the central complex and the medial protocerebrum (SMP, SIP, IP). Three of the DPM lineages (DPMm1, DPMpm1, and DPMpm2) are type II lineages which have been recently described (Bayraktar et al., 2010; Bello et al., 2008; Boone and Doe; 2008; Ito and Awasaki, 2008; Izergina et al., 2009), where they were termed DM1 (DPMm1), DM2 (DPMpm1), and DM3 (DPMpm2), respectively. Expression of two genes, distalless (Izergina et al., 2009) and earmuff (Bayraktar et al., 2010), mark the type II lineages. Along with another type II lineage, CM4 (#62, see below; called DM4 in Bello et al., 2008), DPMm1, DPMpm1, and DPMpm2 (#53, #58, #59) include sub-lineages whose SATs characteristically enter through the dorso-lateral and the medial roots of the fan-shaped body (dlrFB, mrFB; Fig.12A, C53, C58, C59). They form the columnar neurons of the central complex, connecting specific small domains of the protocerebral bridge (PB) in a topographical manner with segments and sectors of the FB and EB, respectively (Ito and Awasaki, 2008; Yang et al., 2013; Fig.11B, G, H; Fig.S3). In addition, these type II lineages have other sub-lineages with widespread terminal arborizations outside the central complex. The most prominent arborizations of DPMm1 (#53) are found in the (1) medial IP and deep layers of the adjacent SMP/SLP (via SSAT #53a following the loSM), (2) posterior VMC of both hemispheres, (via SSAT #53d), (3) in the LAL, IPa, ventromedial cerebrum, SEG (via the anterior and descending SSATs #53c/e; Figs.11B; 12A, C53;.Figs.S4, S5). DPMpm1, via its long forward-directed SSAT #58a, has terminal arborizations in the anterior SMP, IPm, and PENPa (tritocerebrum; Fig.11G; Fig.S4). DPMpm2 (#59) arborizes more widely in the superior protocerebrum (SLP, SIP, SMP) and mushroom body lobes via its loSM-associated SSAT, #59a (Fig.11H; Fig.S5).
Lineages DPMpl1 and 2 (#55, #56) enter the posterior neuropil as the most lateral component of the posterior loSM fascicle. The tract extends into the superior protocerebrum, with branches all along its length (SMP, SIP, SLP; Figs.11D, E; 12A, C55, C56; Figs.S4, S5). DPMpl1 is one of the lineages with a long descending fiber bundle, which leaves the loSM, crosses in the chiasm of the median bundle (MBDLchi), follows the median bundle ventrally, and forms terminal arborizations in the PENPa (tritocerebrum), SEG, and TAG (Figs.11D; 12A, C55; Fig.S4). The SAT formed by DPMpl2 (#56) has no descending projections, but, after leaving the loSM, it continues medially into the FB where it forms wide-field arborizations (Fig.11E; Fig.S3). DPMpl3 (#57), whose cell bodies are initially located close to those of DPMpl1/2 (hence inclusion of this lineage in the same subgroup), but shift ventrally during metamorphosis, project along the MEF fascicle (Fig.12B, C57) and innervate specific ventral compartments, including the VMCpo, VLPp, and VLCi. This lineage also has a strong commissural component, reaching, via the great commissure, the contralateral VLCi and VLPp (Fig.11F; Fig.S2).
Two DPM lineages, DPMl1 (#51) and DPMm2 (#54), innervate the posterior brain compartments and send a descending tract towards the SEG and TAG (Figs.11A, C; 12C51, C54; Fig.S4). DPMl1 (#51) arborizes more ventrally than DPMm2 (#54), including in the IP (IPl; IPm/p), VMCi, VMCpo, and SEG (Fig.11A; Fig.S4). DPMm2 also branches in the posterior realm of the IP (IPl; IPm/p), as well as the adjoining SMP, SLP, and VMCpo (Fig.11C; Fig.S4). DPMl1 also has a laterally-directed branch which reaches the lobula (LO).
CM Lineages
Three of the four CM lineages (CM1, #60; CM3, #61; CM4, #62; labeled DM5, DM6, and DM4, respectively, in Izergina et al., 2009) are large type II lineages with multiple sub-lineages. Each of the three has one major ventral SSAT; the three ventral SSATs of CM1-4), forming the loVP fascicle (Lovick et al., 2013; #60v* in Fig.12A), arborize in the postero-ventral brain, including the VCMpo, VMCs, PLP, and VLCi compartments (Fig.13A-C; Fig.S2). The ventral SSATs of CM3/4 have a commissural component crossing in the pPLPC commissure and reaching the postero-ventral compartments of the contralateral hemisphere (Fig.13B-C; Fig.S2). The intermediate and dorsal SSATs of the lineages CM1-4 (#60d* in Fig.12A) connect the postero-ventral brain to more anterior and dorsal regions of the neuropil. CM1 and CM3 each have one SSAT (#60d and 61d2) that travels with the MEF fascicle and arborizes posteriorly (VMCpo), as well as more anteriorly (VMCs, IPl/m, LAL; Figs.12A; 13A, B; Fig.S2). CM1 (#60) has a conspicuous commissural component that interconnects the LAL compartments of either side (Fig.13A; Fig.S2). CM3 (#61d1) also arborizes throughout the entire FB Fig.13B; Fig.S3). As described in the previous section, CM4 (#62) is one of the four lineages (beside DPMm1, DPMpm1, and DPMpm2) which produces columnar neurons of the central complex: the CM4 SSAT (#62d) forming these arborizations is uncrossed and innervates the most lateral part of the PB and FB (Fig.13C; Fig.S3). CM3 and CM4 have one other major SSAT (#61/62a) that projects dorsally along the loSM and interconnects dorsal protocerebral compartments along their antero-posterior axis (SIP/SMP; Fig.13B, C; Fig.S5).
CM5 (#63), the most medial member of the CM group, has an SAT that enters the posterior neuropil medially of the MEF fascicle (Fig.12A, C63). CM5 is the third lineage (beside DPMl1 and DPMm2) which has a long SAT descending posteriorly towards the thoracico-abdominal ganglion (TAG); its proximal arbors are focused on the VMCpo compartment (Fig.13D; Fig.S4).
CP Lineages
The four CP lineages (CP1-4) are located laterally adjacent to the CM group and form mostly projection neurons associated with the superior and inferior protocerebrum. The CP2/3 pair (#65/66) each produces a dorsal and ventral HSAT (HSATd, HSATv) that have a characteristic spatial relationship to the mushroom body peduncle (PED; Fig.12A, C65, C66). Even though the two lineages innervate similar neuropil compartments, each shows distinctive differences. The lineage defined as CP2, with its dorsal HSAT (#65d), forms arborizations in the LH, SIP, and SMP and also projects to the mushroom body vertical lobe and fan-shaped body where it forms wide-field arborizations (Fig.13G; Figs.S3; S5). The dorsal component of CP3 (#66d) has denser innervations in the LH, but misses the projection to the fan-shaped body (Fig.13H). The ventral HSATs of CP2/3 (#65/66v) project along the PLF fascicle that converges upon the peduncle from ventrally (Fig.12A, C65, C66). They form terminal arbors along their axons in the ventro-lateral and inferior protocerebrum (PLP, VLPp, IPm/l; Fig.13G, H; Fig.S2).
CP1 and CP4 (#64, #67) have similar SATs to the HSATds of CP2/3, crossing over the peduncle along the obP fascicle. Characteristically, the tracts of CP1 and 4 are closer to the peduncle than those of CP2/3 (compare Fig.12C64/C67 to C65/C66). Both CP1 and CP4 have dense terminal arborizations in the LH, SIP, and SMP compartments (Fig.13E, F). CP1 (#64), in the late larval brain, has a dorsal (blue arrowhead in Fig.13E) and ventral hemilineage (white arrowhead in Fig.13E): the HSATv projects along the posterior LEF fascicle. In the adult, the tract of the dorsal hemilineage (#64) can be followed along the loSM towards the MBDLchi, where it joins DPMm2 and DPMpl1 to descend towards the SEG/TAG (Fig.12C64; Fig.S4). We identified a total of four clones in different brains for CP1. However, none of them had a ventral hemilineage component, even though a BP104-positive LEFp bundle could be clearly distinguished in the adult (Fig.12A; see accompanying paper by Lovick et al., 2013). One possible explanation is that the ventral CP1 hemilineage undergoes apoptosis during metamorphosis. CP4 has only a dorsal component, both in the larva and adult (Fig.13F; Fig.S5).
BLA Lineages
BLA lineages are located at the antero-lateral neuropil surface. A subgroup of five dorsal BLAs, BLAd1-4 (#68-71) and BLAl (#72), form SATs that converge on one neuropil fascicle, the trSI (Figs.14A, B; 15B, D68-72; Fig.S5), which primarily interconnects domains of the superior protocerebrum. For the four BLAds defined in the larva, three types of clones with different projection envelopes were recovered (Fig.14A); these were assigned arbitrarily to the lineages BLAd1 (#68), BLAd2 (#69), and BLAd3/4 (#70/71). BLAd1 arborizes in the LH and SLP (Fig.14A); BLAd2 is focused more on the SMP and SIP, but has an additional branch that follows the superficial trSI (trSIs), and innervates the posterior SLP, IPp, and IPm/l (Fig.14A; Fig.S5); BLAd3/4 has restricted arborizations in the medial SLP (Fig.14A; Fig.S5). The BLAl lineage (#72) has two separate hemilineages. The dorsal hemilineage (#72d) projects along the trSIs (Figs.14B; 15B, D72) and arborizes in the posterior regions of the LH, SLP, and SMP compartments (Fig.14B; Fig.S5). The medial hemilineage tract (#72m) follows the surface of the VLP, close to the anterior optic tract (green asterisk in Fig.15A), and arborizes in the IPl/m and PLP, respectively (Fig.14B; Fig.S2).
The three ventral BLA lineages, BLAv1 (#73), BLVa2 (#74), and BLAvm (#75), have two hemilineages each and interconnect ventro-lateral compartments of both hemispheres, also forming projections to the superior protocerebrum (Fig.14C-E). The medial hemilineage of BLAv1 (#73m) projects over the anterior surface of the VLPa compartment, following the anterior optic tract (Fig.15A, D73); the tract then extends underneath the peduncle and crosses the midline in the superior arch commissure (SAC). Branches innervate (ipsi- and contralaterally) the VLPa/p and the anterior SLP/IP compartments (Fig.14C; Fig.S1). The medial hemilineage of BLAv2 (#74m) projects medially through the VLPa compartment along the hVLPT tract (Fig.15A, D74). Although the HSATm of the BLAv2 lineage arborizes in a similar ipsilateral territory as BLAv1m (IPl/m, Fig.14D), the hemilineage lacks a strong commissural component across the SAC, but forms a bundle which crosses posteriorly of the central complex towards the contralateral IP (Fig.14D, blue arrowhead). The posterior hemilineages of BLAv1/2 (#73/74p) are directed through the ventro-lateral protocerebrum (Fig.15A) and across the great commissure, arborizing bilaterally in the VLPa/p (Figs.14C, D; 15D73, C74; Fig.S2). The posterior hemilineage of BLAv2 (#74p) has a strong dorsally-directed branch towards the LH and SLP compartments (Fig.14D; Fig.S1). The HSATm of BLAvm (#75m) is located at the antero-dorsal surface of the VLP, where it projects dorsally, passing the anterior optic tract (green asterisk in Fig.15A, D75). The HSATm of BLAvm has widespread terminal arborizations in the dorsal VLPa and dorso-posterior adjacent compartments: the SLP, IPl/m, and PLP (Fig.14E; Fig.S1). The posterior hemilineage of BLAvm (#75p) is located at the lateral surface of the ventrolateral protocerebrum. Its tract, similar to that of BLAl, follows the trajectory of the anterior optic tract (Fig.15B, D75). Anteriorly, it sends arborizations into the LAL compartment (Fig.14E); posteriorly, it innervates the PLP (Fig.14E; Fig.S2).
BLD Lineages
Six BLD lineages were defined in the larva. The anterior four of these, BLD1-4 (#77-80), lie posteriorly adjacent to the dorsal BLAs and, like those, mostly innervate the superior protocerebrum, with tracts forming the superficial component of the trSI (Figs.14F-I; 15B, D77-80; Fig.S5). The terminal arborizations of BLD1 (#77) are fairly restricted to the LH (Fig.14F; Fig.S5); BLD3 (#79) and BLD4 (#80) also innervate the LH, but have more widespread arborizations in adjacent areas of the superior protocerebrum (SLP, SMP, SIP; Fig.14H, I; Fig.S5). Ventrally-directed branches of BLD3/4 arborize in the VLP compartment (Fig.14H, I; Fig.S4). BLD2 (#78) follows the trSIs towards the posterior neuropil surface (Fig.15B), arborizing in the posterior SLP/SMP, the IPp, IPl/m, and PLP (Fig.14G; Fig.S5). BLD1 and BLD3 each has an additional hemilineage. In case of BLD1 these neurons (#77p) are located further posteriorly and project into the PLP (Fig.15C, D77). Terminal arborizations are to be found in the PLP, IP, and the lobula (Fig.14F; Fig.S4). In case of BLD3 one finds an anterior hemilineage (#79a) with projections to the ventral tip of the VLPa compartment (Fig.14H).
Two additional BLD lineages, BLD5 and BLD6 (#82 and 83), are located at the postero-lateral neuropil surface and form connections with the lobula (LO) (Fig.16A-B). BLD5, marked by the expression of the gene atonal (Hassan et al., 2000; Spindler and Hartenstein, 2010; Spindler and Hartenstein, 2011), has a characteristic straight commissural tract interconnecting the ipsi- and contralateral lobula (Figs.15C, D82; 16A). BLD6 is located further ventro-posteriorly (Fig.15C, D83); it has widespread arborizations in the LO, but innervates a restricted “focus” located in the posterior VLPp (Fig.16B).
BLP Lineages
BLP lineages form two pairs in the postero-lateral brain. BLP1/2 (#84/85) are located ventrally, projecting along the PLF fascicle that innervates ventral domains of the VLP compartments (Figs.16C; 17C, D84, D85; Fig.S2). With the exception of a single clone (Fig.S7), all other clones assigned to the BLP1/2 pair have the same, fairly restricted projection envelope (Fig.16C). The clone belonging to the exception (Fig.S7) possesses much more widespread arborizations in the PLP and posterior domains of the IP. This clone suggests that BLP1 and 2 possess different envelopes. BLP3/4 (#86/87) lineages are located dorsally; the pair sends a short dorso-anteriorly directed tract dorso-anteriorly, innervating the LH and the adjacent SLP compartment (Figs.16D; 17C, D86, D87; Fig.S5).
BLV Lineages
BLV lineages, similar to the BLA lineage group, innervate the ventro- and dorso-lateral protocerebrum. BLVa3/4 (#91/92) form a pair whose short SAT penetrates into the VLPa from ventrally; the lineage pair forms restricted terminal arborizations in the VLPa and the neighboring VLCi (Figs.16F; 17A, D91, D92; Fig.S2). Even though the projection envelope of all clones recovered for this lineage is similar, it appears as if in some clones, the cell body cluster and SAT entry point is located more posteriorly than most other clones (Fig.17D92/93, “posterior variant” of BLVa3/4”). BLVa1/2 (#89/90) form another pair with SATs that enter at the ventral side of the trSI fascicle (Fig.17B) and terminates shortly thereafter (at the junction of the IP/SLP; Figs.16E; 17D89/90; Fig.S5). Terminal arborizations of the BLVa1/2 pair are focused on the LH, SLP, and adjacent IPl/m compartments. Recovered clones for the BLVa1/2 pair had very similar projection envelopes but differed with respect to the location of the cell body clusters. In four out of thirteen clones, the cluster was located dorso-anteriorly of the VLPa, at the level of the BLAd lineages (“dorsal variant” of BLVa1/2; Fig.17D). The BLVa1/2 reporter, so-Gal4 is expressed in the larval lineages (Chang et al., 2003) and remains on in the “dorsal variant” in the adult (VH, unpublished). In the remaining eight BLVa1/2 clones, the cell body clusters are spread out within an elongated volume of the cortex in the cleft between the optic lobe and VLP (Fig.16E and left panel of Fig.17D89/90). We speculate that these two variants (one with a dorsal position; the other with a ventral or spread-out dorsal-to-ventral position) represent the two lineages of the BLVa1/2 pair.
BLVp1/2 (#93, 94) each have two hemilineages that migrate apart during metamorphosis. The posterior hemilineages of BLVp1/2 (#93/94p; HSATp) project along the PLF (Fig.17C, D93, 94). The BLVp1 HSATp innervates the VLPp and VLCi compartments and, via a commissural tract crossing as part of the great commissure, innervates the contralateral VLPp (Figs.16G; 17D94; Fig.S2). The HSATp of the BLVp2 lineage has a different trajectory: two branches project dorsally to the superior protocerebrum (SLP, SIP, and SMP) and the anterior IP (IPa and IPl/m; Fig.16H; Fig.S1). The anterior hemilineages of BLVp1/2 (#93a/94a) are located at the ventro-anterior brain surface and laterally adjacent to BLVa3/4 (Fig.17A, D93, D94). Terminal arborizations branch in the VLP, SLP, and LH (Fig.16G, H; Fig.S1). BLVp2 has a strong commissural component which crosses the SAC and projects to the contralateral VLP (Fig.16H; Fig.S1).
Discussion
Identification of MARCM clones with secondary lineages
We show in this paper that MARCM clones induced in the early larva, labeling post-embryonically derived (e.g. secondary) neurons, can be assigned to specific lineages based on the stereotyped trajectory of their axonal tracts. These tracts are formed in the larva and, as documented in the accompanying paper, remain visible as coherent, BP104-positive fiber bundles throughout metamorphosis (Lovick et al., 2013). 56 lineages form tracts with unique properties; all but one of these 56 lineages (e.g. DALv3, marked by en-Gal4; Kumar et al., 2009b) has been represented in our collection of MARCM clones. All other lineages were represented at least twice, with many of them occurring at high frequencies (n>25). We currently have no explanation for the existence of such “hot” and “cold” spots of clone induction. Given that the lineages begin to proliferate at slightly different time points at the first-to-second larval instar transition (Ito and Hotta, 1992; Lovick et al., in preparation), we speculate that the exact timing of the heat pulse may play an important role for the large variations in the frequencies of clone generation.
30 lineages are paired (tracts of two adjacent clusters form one composite bundle) and eight form two “quartets” (four tracts coalesce into a single thick bundle). One of the “quartets” represents lineages of the mushroom body (MB), while the others are the four BLAd lineages (BLAd1-BLAd4). Based on the individually-labeled lineage MARCM clones, it is clear that within these composite bundles, axons of the different lineages do not intermingle. In brains labeled only with global markers (e.g. BP104), one cannot separate individual SATs within the pairs and quartets, and cannot predict how many clones with different projection envelopes to expect. All members of a pair/quartet could either form clones with identical projection envelopes or they could form two/four anatomically distinct clones. In nine cases, each member of the pair has a unique arborization pattern. For example, BAmas1/2, both of which project along the median bundle, appear to have distinct fields of arborization: proximal branches of BAmas1 project in the ventral PENPa (tritocerebrum) and the SEG, projecting towards the VL/SMP; while BAmas2 form proximal arborizations dorsally in the PENPa and distally and bilaterally in the SMP. Importantly, although these projection envelopes are clearly different, they include adjacent brain territories. This generally holds true for most lineages: lineages with neurons (and, at an earlier stage, neuroblasts) located close to each other also typically innervate adjacent neuropil territories (see schematic representations of projection envelopes in the Fig.S1-S5). Another case in point is the quartet BLAd1-4, for which we could identify three different types of clones whose projection envelopes were all confined to the superior protocerebrum where they targeted contiguous territories (BLAd1: lateral horn; BLAd2: lateral SLP, SIP, SMP; BLAd3: medial SLP).
The fact that in case of the BLAd lineages we identified three, and not four, types of clones could mean that “the fourth” BLAd lineage has a projection envelope that is indistinguishable from one of the other three BLAd members; alternatively, we might have missed the clone, since that particular lineage (like DALv3 mentioned above) represents a cold spot of inducibility. The same reasoning applies to six pairs of lineages (see Table 2) for which also a single clone type was noted. It is unlikely that for all of these pairs one of the members was missed, given their good overall representation (e.g., 10 clones for the BAla3/4 pair, 11 for DALcm1/2, 14 for DAMd2/3, and 13 for DAMv1/2). In these cases, we favor the interpretation that a lineage might have been ‘duplicated” to increase the overall number of neurons sharing the same projection envelope.
Lineage-based analysis of brain macrocircuitry
With respect to the overall shape of their projection envelopes, lineages fall into several classes. A more in depth discussion of these different classes will have to await the detailed analysis of projection envelopes relative to each other, and relative to the boundaries of neuropil compartments. To this end, clones are being registered to one “model brain”, in which their spatial relationships can be established. We anticipate that this work (Wong et al., in prep) promises to yield further insight regarding the validity of compartment boundaries (do projection envelopes of multiple lineages respect the boundaries defined on the basis of synapse density? Do projection envelopes reveal additional subdivisions of compartments?), as well as regarding brain macrocircuitry (how strongly are compartments connected on the basis of sharing in a certain number of projection envelopes?).
Some of these questions have been already addressed in two recent papers where, employing a MARCM-based approach, Ito et al. (2013) and Yu et al. (2013) have published a comprehensive atlas of secondary lineages for the adult Drosophila brain. They chose a terminology in which the term for a lineage was based upon one of the neuropil compartments heavily innervated by that lineage. Both studies concur with our conclusion that the pattern of projection and arborization of secondary lineages is highly invariant. Based on their characteristic SAT projection and projection envelope, most lineages depicted in Ito et al. (2013) and Yu et al., (2013) can be identified with the secondary lineages described here, even on the basis of z-projections alone. Note for example the close correspondence between the projection envelopes shown for the well documented type II DPM, CM and CP lineages (DPMm1-DM1; DPMpm1 – DM2; DPMpm2 – DM3; CM4 – DM4; CM3 – DM6; CM1 – DM5; CP2 – DL1; CP3 – DL2), but also other newly described lineages with long, characteristic SAT trajectories (e.g., BAmas1 – FLAa2; BAmas2 – FLAa3; BAmd2 – WEDd2; DALcl1 – AOTUv3; DALv1 – VLPa2; BLAv1 – VLPl&d1). However, given that clones described in Ito et al. (2013) and Yu et al. (2013) are presented in the absence of labeled fascicles, the unambiguous matching of their nomenclature with ours should await the careful comparison of confocal stacks. Nonetheless, three aspects concerning the overall coverage of lineages applying different driver lines to visualize clones deserve mentioning.
First, “cold spots”, i.e., lineages, known to exist on the basis of independent data, which are not represented by MARCM clones are apparent in all three studies. Interestingly, DALv3 (independently documented by its expression of engrailed-Gal4; Kumar et al., 2009) seems to be a “universal cold spot”, since it is not represented in our study, neither in that of Ito et al. (2013) or Yu et al. (2013). Other cold spots may depend on the driver line used; for example, the characteristic lineage of local antennal lobe interneurons, BAla2 (Das et al., 2013) is represented by multiple clones in our study, as well as in Yu et al. (2013), where it is called ALv2, but not in Ito et al. (2013). Similarly, BAmas1 (FLAa2) and BAmv2 (VESa2) are represented by clones in Yu et al. (2013), but not Ito et al. (2013).
Secondly, the mapping of lineages in our study is restricted to clones that could be clearly assigned to the BP104-positive fiber bundles corresponding to lineage associated axon tracts traceable from the larva into the adult. Following this approach, we could assign a type of clone to each lineage originally defined in the larva based on possessing a neuroblast and a unique SAT; the only exceptions were the DALv3 “cold spot”, and the uncertainty concerning the six paired lineages for which we recovered only clone (see above). The study by Ito et al. (2013) lists a sizeable number of clones (e.g., the majority of their VPN clones), located in the lateral brain and including projections between optic lobe and central brain, that are most likely derived from neuroblasts that originate from the inner optic anlage. These lineages were not included in our larval catalog of central brain lineages (Pereanu and Hartenstein, 2006; Cardona et al., 2010), and are not considered in this paper, even though we also recovered frequent examples of VPN-type clones. The developmental definition of a boundary between central brain and optic lobe, in particular lobula, is complex, and will require further work.
Finally, the participation of secondary lineages in the production of glia also needs further clarifications. All classes of glia increase in number during the larval period, and part of this increase is due to the proliferation of dedicated glial progenitors (or glial cells themselves, which continue to divide), whereas another part results from the generation of glial cells from within secondary (neural) lineages (Pereanu et al., 2005). More recent studies have shown that several of the type II lineages are responsible for the generation of much of the ensheathing glia of the central complex (Viktorin et al., 2011), as well as some of the optic lobe associated glia (Viktorin et al., 2013). The use of elav-Gal4 as a driver precluded us from visualizing glial progeny among the clones described in this study. However, Yu et al. (2013), utilizing flip-out techniques (actin5CP-FRT-stop-FRT-GAL4 and actin5CP-loxP-stop-loxP-lexA::p65) and twin spot MARCM (nSyb-GAL4) describe a small number of lineages that included glia among their progeny. One of these, DM5 (CM1 in our nomenclature) produced both ensheathing glia and astrocyte like glia; another one, DL1 (CP2 in our nomenclature) generated a population of optic lobe-associated ensheathing glia that most likely corresponded to the set of glia described in Viktorin et al. (2013). However, none of the large number of central complex-associated ensheathing glia, derived from the type II DM lineages according to Viktorin et al. (2011) were labeled in Yu et al. (2013) or Ito et al. (2013). Aside from the possibility that this is due to a property of the driver line used (), one may explain this discrepancy by the timing of clone induction. Thus, if the very first sublineages generated by the DM neuroblasts are dedicated glial progenitors, one might miss their progeny if clonal induction occurs at a slightly later time point.
A lineage-based approach to study mechanisms controlling Drosophila brain development
In previous studies, a number of secondary lineages marked by Gal4-reporters had been identified in the larval brain and linked to their adult counterparts. The best characterized lineages are the four lineages of the mushroom body (Ito et al., 1997; Ito and Awasaki, 2008; Lee et al., 1999; Zhu et al., 2003), and the five which form the projection and local interneurons of the antennal lobe (Das et al., 2013; Das et al., 2008; Komiyama et al., 2003; Lai et al., 2008; Yu et al., 2010). Additional lineages have been characterized based on the restricted expression pattern of transgenic reporters and protein markers (e.g., en-Gal4; ato-Gal4; per-Gal4; empty spiracles, ems; Hassan et al., 2000; Kumar et al., 2009b; Lichtneckert et al., 2008; Spindler and Hartenstein, 2010; Spindler and Hartenstein 2011; Srahna et al., 2006). The identification of projection envelopes of adult MARCM clones for all central brain lineages presented in this paper will aid in the identification of additional lineage-specific markers from among the numerous existing collections of Gal4 enhancer-trap lines (Hayashi et al., 2002; Jenett et al., 2012; Pfeiffer et al., 2008).
Taking advantage of the fact that lineages form structural units whose individual neurons share a common trajectory and terminal arborization, a selected number of genes (encoding for members of developmentally-relevant molecular pathways or important cell-cell interactions, such as adhesion molecules) have been analyzed using a lineage-based approach. This type of approach was pioneered in a series of studies that revolve around the MB lineages. In these studies, the roles of many crucial players important for proliferation, cytoskeletal dynamics, and cell-to-cell adhesion were dissected through conditional loss- and gain-of-function experiments, using MB-specific drivers under Gal4/UAS control (Billuart et al., 2001; Lee et al., 2000a,b; Ng et al., 2002; Reuter et al., 2003; Scott et al., 2001). Additional lineage-specific (e.g. non-MB) Gal4 drivers have been identified and similar approaches (like the MB studies) have been taken to identify critical players for secondary lineage morphogenesis (through conditional knock-outs and gain-of-function, as described above; Bello et al., 2003; Kuert et al., 2012; Marin et al., 2012; Maurange et al., 2008; Spindler and Hartenstein, 2011; Zheng et al., 2006).
More recently, it has been demonstrated that neurons/lineages, which may express a common set of molecular factors, react very differently to the loss of these factors. This has been made possible by the identification and characterization of lineage-specific Gal4 lines and highlights the importance of utilizing a multi-lineage approach when studying neural development. One example is the role of the Par-complex proteins, Bazooka (Baz)/Par-3/Par-6, in determining the shape of secondary neurons (Spindler and Hartenstein, 2011). Outside the nervous system, the Par complex plays an essential role for epithelial cell polarity and migration (Cong et al., 2010; Ellenbroek et al., 2012; Hurd et al., 2003; Ohno, 2001; Wang et al., 2006; Wang et al., 2012). In some vertebrate neurons, Par appears to be required for the differentiation of nascent neuronal processes into axons and dendrites (Chen et al., 2006; Nishimura et al., 2004; Shi et al., 2004). In the Drosophila post-embryonic brain, Baz is expressed by neuronal progenitors and postmitotic neurons. A Gal4-inducible Baz::GFP fusion reporter (under the control of UAS enhancer sequences; Sánchez-Soriano et al., 2005) driven by per-Gal4 (marking BAla1, BAmv1, DAlv2) and ato-Gal4 (marking BLD5) revealed that the Baz protein accumulates at positions along the SATs where terminal branches will appear (Spindler and Hartenstein, 2011). For example, in the PD-type lineages BAla1 (using per-Gal4) and BLD5 (using ato-Gal4), Baz::GFP is both concentrated at the cortex-neuropil boundary and the distal ending of the SAT. Loss of function of Baz by MARCM, results in ectopic terminal branches, either along the SAT (DALv2 and BAmv1) or at its distal tip (BLD5). In the case of BAla1, loss of baz results in aberrant pathway choices, forming additional SATs into the iACT (Spindler and Hartenstein, 2011). Interestingly, loss-of-function of another member of the Par family, par6, phenocopies baz loss-of-function clones in case of the DALv2 and BLD5 lineage, but not BAla1 and BAmv1, further supporting the notion that the requirement of different Par-complex members varies from one lineage to the next. A previous study, demonstrating that the Par complex is not required for the development of mushroom body lineages (Rolls and Doe, 2004), also supports this idea. Although the mushroom body lineages have been traditionally used as “test lineages” to understand gene function, it is clear that such an approach may not be sufficient. Since gene function (in the case of baz and par6) may be lineage-dependent, a “multi-lineage approach” is more favorable and will provide a clearer picture of gene function in developing neurons.
Dissecting lineages: hemilineages, sub-lineages, neurons
Many type I lineages consist of two hemilineages (Truman et al., 2010). To generate hemilineages, a NB divides asymmetrically to produce an intermediate cell (ganglion mother cell, GMC). The GMC divides symmetrically to produce two post-mitotic sibling neurons. Typically, cell fate determinants (eg. Numb, a repressor of Notch signaling) are asymmetrically segregated into the two neurons, such that one cell acquires an ‘A’ fate (inherits Numb and represses active Notch signaling) and the other acquires a ‘B’ fate (does not inherit Numb and has active Notch signaling; Lin et al., 2012; Truman et al., 2010). In some cases, one hemilineage is fated to undergo programmed cell death (Kumar et al., 2009a; Truman et al., 2010); in others, both hemilineages survive, but are morphologically and most likely, functionally unique. It was suggested that in cases where both hemilineages survive, two separate bundles or HSATs, emerge from the cell body cluster. Upon entering the neuropil, the two HSATs diverge and target different neuropil compartments. In this work and the accompanying paper (Lovick et al., 2013), we have identified 20 lineages possessing these properties (see Table 1). In the majority of cases, both the hemilineage cell body clusters and the neuropil entry points of their HSATs move apart to some extent during metamorphosis. This morphogenetic shift is extreme for several lineages (e.g. DPLl2/3, DPLc5, BLAl, BLAv1, BLAvm, BLVp1/2). For eight of these lineages, although there is a single cell body cluster and SAT; the tract splits into two components with different trajectories (Table 1). In these cases, it remains unclear whether the existence of more than one tract suggests that there are two separate hemilineages; an in-depth analysis of individual neurons forming parts of these lineages may help answer this question.
Table 1.
List of secondary lineages of the Drosophila brain.
| A Lineage Name | B Lineage SAT Number | C Number Clones | D Lineage Type | E Fascicle Joined by Lineage | F Main Ipsilateral Compartments Innervated by Lineage | G Commissure Joined by Lineage | H Main Contralateral Compartments Reached by Lineage |
|---|---|---|---|---|---|---|---|
| BAla1 | 1 | 16 | PD | mlALT | AL - LH | ||
| BAla2 | 2 | 9 | Cl | 0 | AL | ||
| BAla3 | 3 | 10 | Cw | 0 | VMC VLP SEG | ||
| BAla4 | 4 | ||||||
| Balc | 5 d | 12 | PD | mALT | AL - CA LH | ||
| 5 v | PDco | loVI | AMMC - VLCi | GC | VLCi | ||
| BAlp1 | 6 | 9 | PD | 0 | VMCpr < SAsbtri Ipm | ||
| BAlp2 | 7 | 6 | Cw | loVL | LAL VLCi PLP | ||
| BAlp3 | 8 | 9 | Cw | loVL | LAL VLCi PLP IP l | ||
| BAlp4 | 9 | 7 | PD | mALT | AL SAsbtri - LHa SLPp SIP | ||
| Balv | 10 | 7 | Cl | 0 | VLCi AMMCp | ||
| BAmas1 | 11 | 7 | PD | MBDL | PENPa SAsbtri-VL SMP SIP | ||
| BAmas2 | 12 | 4 | PD | MBDL | PENPad - SMPa | ||
| BAmd1 | 13 d | 12 | PDco | 0 | SMP IP a ML | FrMC | ML |
| 13 v | PD | 0 | ML? IPa? - | ALC | VMCpr SAsbtri | ||
| BAmd2 | 14 | 9 | Dl | 0 | 0 - VLCi PLP | ALC | VLCi PLP |
| BAmv1 | 15 d | 16 | PD | loVM LEp | LAL IPa SMPa - FB | ||
| 15 p | Dw? | loVM | 0? - VMCpo PLP | ||||
| 15 dn | Dw? | 0? - VLPp | |||||
| BAmv2 | 16 | 12 | Dw | loVM | 0-VMCpo PENPa/p SAsbtri | ||
| BAmv3 | 17 | 24 | PD | mALT | AL - CA LH | ||
| DALcl1 | 18 d | 14 | PD | 0 | AOTU SLP SIP - BU IP | ||
| 18 v | PDco | 0 | 0 - LAL | SuEC | LAL | ||
| 18 vn | MEF | LAL - VMCpo bilat | |||||
| DALcl2 | 19 d | 9 | PDco | 0 | AOTU SIP SMP - LAL | SuEC | SMP SIP LAL |
| 19 v | Dw | LEa | 0 - FB SMP | ||||
| 19 dn | Cw | 0 | AOTU SIP SMP | ||||
| DALcm1 | 20/21 m | PDco | 0 | IPa LAL SIP SMP VL ML | FrMC | VL ML | |
| DALcm2 | 20/21 v | PDde | deCP | AOTU SIP IP - VMC | |||
| DALd | 22 | 8 | PDde | deCP | IPa AOTU LAL SIP SMP - VMC | ||
| DALl1 | 23r | 12 | Dw | trSId | LH IP AOTU | ||
| 23v | 0 | PLP Lo | |||||
| DALl2 | 24 | 7 | Cw | 0 | VLPap PLP SMP IP | ||
| DALv1 | 25 | 8 | PDco | LEFa | VLP VLCi AMMC SEG | GC | VLP |
| DALv2 | 26 | 27 | PD | LE a | BU IPa LAL - EB | ||
| DALv3 | 27 d | PDeo | LE a | SMP IPa/m/l | SEC | SMP IPa | |
| 27 v | PDco | LE a | LAL IP | SuEC | LAL IP | ||
| DAMd1 | 28 | 2 | PDco | ADC | SMP - SIP SMP IPa | FrMC | IPa SMP |
| DAMd2 | 29 | 14 | Cw | loSMa | SMP SIP IPm VMCpo | ||
| DAMd3 | 30 | ||||||
| DAMv1 | 31 | 13 | Cw | 0 | SMP SIP | ||
| DAMv2 | 32 | ||||||
| DPLal1 | 33 | 15 | PD | trSA | SLPp LH IPl - SLPa | ||
| DPLal2 | 34/35 d | 16 | PD | trSA | SLP LH IP - SIP SMP IP | ||
| DPLal3 | 34/35 v | Cw | 0 | SLP IPm/l VLPp | |||
| DPLam | 36 | 12 | Cw | vSLPT | SLP VLPp IPm/l | ||
| DPLc1 | 37 | 10 | Cw | trSPm | SLP IPm/l SMP | ||
| DPLc2 | 38 | 9 | Cw | trSPm | SLP LH IPm/l SMP | ||
| DPLc4 | 40 | ||||||
| DPLc3 | 39 | 3 | Cw | 0 | SLP SIP SMP | ||
| DPLc5 | 41 a | 14 | Cw | trSPm | IPm/l SLP SMP | ||
| 41 p | PDco | 0 | IP p/m SMP | ADC | SMP | ||
| DPLd | 42 m | 3 | PDco | 0 | LH? SLP | ADC | SLP |
| 42 p | PD | trSId | SLP - LH | ||||
| DPLl1 | 43 | 2 | Cw | trSPl | PLP SLP LH SMP | ||
| DPLl2 | 44 p | 21 | PD | loSL | LH PLP IPl SLP (-) SIP SMP | ||
| 44 a | PD | vSLP | SLP VLPp | ||||
| DPLl3 | 45 p | 8 | Cw | loSL | LH IP l SLP SIP SMP | ||
| 45 a | Cw | vSLP | SLP? VLPp | ||||
| DPLm1 | 46 | 5 | Cw | 0 | SLP SIP IP l | ||
| DPLm2 | 47 | 2 | PD | 0 | IP m/l SLP (-) LH ncc | ||
| DPLp1 | 48/49 m | 23 | PDco | obP | SLP LH - SMP | sPLPC | IPp/m/l PLP VLCi |
| DPLp2 | 48/49 v | Cw | (veP) | LH SLP IP l PLP VLCi | |||
| 48/49 a | PD | 0 | LH (-) SLP | ||||
| DPLpv | 50 | 7 | Cw | PLF | PLP IPm/l VLPa/p | ||
| DPMl1 | 51 | 4 | PDde | DPPT | VMCpo/s PLP IP Lob-SEG | ||
| DPMm1 | 53 a | 20 | Cw | loSMp | IPp IP m/l SMP SIP | ||
| 53 b | PD | mrFB | PB - FB EB NO co | ||||
| 53 c | PD?de | mrFB | PB? - SEG VMCi PENPp VLCi | IPa LAL | |||
| 53 d | PDco | 0 | VMCpo | 0 | VMCpo | ||
| IP SLP SMP VMCpo VLPa (-) | |||||||
| DPMm2 | 54 | 6 | PDde | 0 | SMP | MBDLchi | SEG TAG |
| DPMpl1 | 55 | 4 | PD co/de | loSMp | IP SLP SMPp - SMPa | MBDLchi | PENPa TAG |
| DPMpl2 | 56 | 3 | PD | loSMp | SMP SIP IPp/m - FB | ||
| DPMpl3 | 57 | 5 | PDco | MEF | VMCpo (-) VLCi VLPp | GC | VMCpo VMCi VLPp |
| mACT | |||||||
| DPMpm1 | 58 a | 24 | PD? | MBDL | PB? - SMP IP m PENPa | ||
| 58 b | PD | dlrFB | PB - FB EB NO co | ||||
| DPMpm2 | 59 a | 21 | PDco | loSMp | SMP IPm SIP SLP VL ML | SAC | SMP SIP |
| 59 b | PD | dlrFB | PB - FB EB NO | ||||
| CM1 | 60 d | 15 | PD | MEF | VMCpo - IPm/l VMCs LAL | SuEC | LAL VMCi |
| 60 v | Cw | loVP | PLP VMCpo VMCi VLCi | ||||
| CM3 | 61 a | 8 | PDco | loSMp | SIP SMP IPm | SEC | SMP |
| 61 d1 | PD? | MEF | VMCpo? IP? - FB | ||||
| 61 d2 | PD? | MEF | VMCpo? IP? - LAL IPa | ||||
| 61 v | PDlco | loVP | PLP VMCpo - VLCi IPm/l/p | pPLPC | PLP | ||
| CM4 | 62 a | 2a | Cw | loSMp | SMP SIP | ||
| 62 d | PD | MEF | PB - FB EB NO | ||||
| 62 v | PDco | loVP | PLP VMCpo VLCi | pPLPC | PLP | ||
| CM5 | 63 | 9 | PDde | 0 | VMCpo - SEG TAG | ||
| CP1 | 64 d | 4 | PDco/de | obP loSMp | SIP SMPp - SMPa | MBDL chi | PENPa TAG |
| 64 v | N | ||||||
| obP loSM | |||||||
| CP2 | 65 d | 13 | PD | OE | LH SIP SMP VL - FB | ||
| 65 v | PD | PLF | PLP IPl - VLP VMC | ||||
| CP3 | 66 d | 4 | PDco | obP loSMp | LH PLP - SIP SMP VL | SEC | SMP SIP VL |
| 66 v | PD | PLF | PLP IPl - VMC VLP | ||||
| CP4 | 67 | 4 | PDco | obP loSM | LH SLP - SIP VL SMP | SEC | SMP |
| BLAd1 | 68 | 15 | PD | trSId | LH (-) SLP | ||
| BLAd2 | 69 d | 5 | Dw | trSId | SIP SMP SLP IP | ||
| 69 s | Dw | trSIs | SLP IP | ||||
| BLAd3 | 70 | 3 | Dl | trSId | SLP | ||
| BLAd4 | 71 | 0 | N | ||||
| BLAl | 72 d | 6 | Cw | trSIe | LH SLP SMP | ||
| 72 m | PD | 0 | PLP - IP | ||||
| BLAv1 | 73 m | 17 | PDco | 0 | VLPa/p - SLP IPm/l | SAC | IPm/l VLPa |
| 73 p | PDco | 0 | VLPa/p | GC | VLPa/p | ||
| 73 pn | PD | 0 | VLPad (-) VLPav VLCi | ||||
| BLAv2 | 74 m | 10 | PD | 0 | VLPap SLPa IP m/l VMCs | pCCXc | IPm/l |
| 74 p | PDco | 0 | VLPp IPm/l | GC | IP VLP | ||
| 74 pn | Cl | 0 | LH | ||||
| BLAvm | 75 m | 3 | Cw | 0 | VLPa SLP PLP IPm | ||
| 75 p | PD | 0 | PLP - LAL PLP | ||||
| BLD1 | 77 d | 5 | Cw | trSIe | LH SLP | ||
| 77 p | Cw | 0 | PLP IPm/l Lob | ||||
| BLD2 | 7a | 12 | Cw | trSIe | SLP PLP SMP IPm/l | ||
| BLD3 | 79 d | 9 | Cw | trSIe | SLP LH | ||
| 79 vn | Cw | 0 | VLPp/a IPl | ||||
| 79 a | Dl | 0 | VLPa/p | ||||
| BLD4 | 80 d | 10 | Cw | trSIe | LH SLP SIP | ||
| 80 v | Cl | 0 | VLPp | ||||
| BLD5 | 82 | 16 | PDco | 0 | Lob | GC | Lob |
| BLD6 | 83 | 5 | PD | 0 | Lob - PLP | ||
| BLP1 | 84 | 1 | Cw | PLF | PLP VLP IP Lob | ||
| BLP2 | 85 | 13 | Dl | PLF | VLPa VLPp | ||
| BLP3 | 86 | 29 | Cl | 0 | LH SLP | ||
| BLP4 | 87 | ||||||
| BLVa1 | 89 | 4 | Cw | 0 | LH SLP IPm/l | ||
| BLVa2 | 90 | 9 | Cw | 0 | LH SLP IPm/l | ||
| BLVa3 | 91 | 8 | Cw | 0 | VLPa VLCi SEG | ||
| BLVa4 | 92 | 11 | Cw | 0 | VLPa VLCi SEG | ||
| BLVp1 | 93 p | 10 | PDco | PLFv | VLPp VLCi | GC | VLPp |
| 93 a | PDco | vVLPT | VLPa/p < LH basal | ||||
| BLVp2 | 94 p | 6 | PDco | PLF | SIP SMP IPa | SEC | SIP |
| 94 a | PDco | vVLPT | VLPa SLPa IPm/l | SAC | VLPa |
Column A: Lineage names.
B: Number identifying lineage-associated tracts (SATs). In lineages with multiple hemilineage tracts or sublineage tracts, these are individually listed (e.g., dorsal hemilineage tract of BAlc is identified as “5d”, ventral hemilineage tract as “5v”).
C: Number of MARCM clones isolated for lineage
D: Main class of lineage based on contour of projection envelope. “PD”: lineage with separate proximal and distal arborizations; “C”: lineage with terminal arborizations emerging at more or less regular intervals along the entire length of the SAT; “D”: lineages where arborizations are concentrated at distal tip of SAT. Lower case “l” (“local”) stands for small volume filled by arborization; “w” (“wide”) indicates large volume; “co” indicates commissural SAT; “de” signifies descending tract.
E: Neuropile fascicle joined by lineage-associated tract. For abbreviations of fascicle names, see Table 2. “0” indicates that tract does not form part of any designated fascicle
F: Ipsilateral neuropile compartments receiving strong innervation by lineage. In cases of PD lineages where proximal and distal terminal arborizations can be distinguished based on labeled clone, a hyphen separates compartments receiving proximal arbors (left of hyphen) from those receiving distal arbors (right of hyphen). For abbreviations, see Table 2.
G: Commissure joined by lineage associated tract. For abbreviations, see Table 2.
H: Contralateral neuropile compartments receiving strong innervation by lineage. For abbreviations, see Table 2.
Lineages and hemilineages are comprised of sequentially-born neurons, which may all share in the common projection envelope; however, they can be grouped into sub-classes which differ among each other in regards to their detailed phenotype (e.g. projection patterns). This has been investigated for the four lineages of the MB and for some of the lineages associated with the AL (Jefferis et al., 2001, 2004; Kunz et al., 2012; Lai et al., 2008; Lee et al., 1999; Yu et al., 2009 and 2010; Zhu et al., 2003). The MB lineages undergo four sequential phases of proliferation, producing sub-lineages with different properties (Lee et al., 1999; Zhu et al., 2003; Kunz et al., 2012). In the early embryo, MB neuroblasts give rise to a small set of non-intrinsic neurons (as opposed to the intrinsic neurons or Kenyon cells) that do not contribute to the neuropil of the mushroom body (Kunz et al., 2012). From mid-embryonic stages onward, the MB lineages switch to a mode that generates γ-neurons, followed by α’/β’ neurons (most of third instar larva), pioneer α/β neurons (at the start of metamorphosis), and α/β neurons (during mid-to-late phases of metamorphosis). Within these sub-lineages, neurons might form even smaller groupings, defined by the specific territory inside the calyx or MB lobes they innervate. For example, α/β neurons born at different times in the pupa appear to send terminal arbors to different domains of the calyx (Zhu et al., 2003).
The correlation between birth order and neuronal projection has been elucidated in great detail for the adNB/BAmv3 lineage. The projection envelope of this lineage includes the antennal lobe, calyx and lateral horn (Lai et al., 2008). The antennal lobe is formed by over 50 specific glomeruli, each glomerulus characterized by the endings of olfactory neurons sharing a common olfactory receptor (Jefferis et al., 2001; Vosshall et al., 1999). Single-cell clonal analysis of the adNB/BAmv3 lineage (Yu et al., 2010) indicated that dendritic branches of neurons born at a certain time point always innervated a single, invariant glomerulus. In other words, dendrites innervate the antennal lobe in a largely non-overlapping, “tiled” manner. The same applies to axonal terminals which form an “odour map” in the calyx (Jefferis et al., 2001; Jefferis et al., 2004).
The projection envelopes shown for the central brain lineages will help to manage the large number of individual neurons that emerge in past and future studies of fly brain development and brain function. The shape of a large fraction of adult central brain neurons, imaged as single-cell clones, has recently become available (Chiang et al., 2011). Many of these neurons are readily identifiable as members of specific secondary lineages. A few examples are shown in supplementary figure S6. Panels A1-A5 show individual neurons that project along the median bundle, shared by lineages BAmas1 and BAmas2, and fall within the projection of BAmas2 (panel A; proximal arborization in dorsal PENPa compartment, distal arbors bilaterally in antero-dorsal SMP). Aside from this shared envelope, neurons shown in A1-A5 clearly differ in regard to the details of their distal arborization. For example, A1 has widespread, sparse endings bilaterally in the SMP; A2 projects only ipsilaterally; A3 has bilateral terminations which are denser and more restricted than those of A3. It is likely that these different projections, similar to those of adNB/BAmv3 neurons discussed above, are correlated to the birth dates of the corresponding neurons. Panels B1, C1, D1, and E1/E1’ show single cell clones of neurons which follow the projection envelopes of lineages BLVa3/4 (B), DAMv1/2 (C), BLD2 (D), and vNB/BAla1 (E), respectively.
An interesting case is presented in E2/E2’: the neuron shown has proximal arborizations in the antennal lobe and follows the trajectory of the vNB/BAla1 SAT, passing underneath the peduncle towards laterally. However, distal arborizations are not in the lateral horn (as in the case of secondary vNB/BAla1 neurons), but in the ventrolateral protocerebrum. There are three other features that set the neuron shown in E2/E2’ apart from secondary vNB/BAla1 neurons: the large cell body, located slightly more dorsally than secondary BAla1 cells, as well as the thick axon (compare E2 with E1). We propose that E2 shows a primary vNB/BAla1 neuron. A recent study (Das et al., 2013) indicated that many primary neurons of this lineage enter the antennal lobe through the same portal later used by the secondaries, form proximal terminal branches in the antennal lobe, and follow (in part) the medial antennal lobe tract. However, most of these primaries do not reach the larval counterpart of the lateral horn, but terminate in other (adjacent) territories (Das et al., 2013). These are all properties of the neuron shown in E2/E2’. We are currently preparing clones of brain lineages that include primary neurons (Omoto et al. in preparation), which will allow us to visualize the “primary projection envelope” of lineages, and thereby help to identify primary neurons. Ultimately, suitable Gal4 drivers have to be identified for all lineages, to then engage on a single-cell clonal analysis as the one pioneered for adNB/BAmv3.
Supplementary Material
Supplementary Figure S1 Schematic diagram of lineages of the ventral brain with strong projections connecting ventral and dorsal neuropil domains. Each lineage is represented by a numbered sphere (location of SAT entry point into neuropil), a line (SAT trajectory in neuropil), and small circles (location of terminal arborizations). SAT trajectories are based upon BP104 labeled brains (see accompanying paper by Lovick et al., 2013), as well as MARCM clones of corresponding lineages. Location of terminal arborizations are based on MARCM clones (see Figs.3-18 of this paper). To show lineage projections in relationship to neuropil structures, lines and circles, representing SATs and terminal arborizations, respectively, are superimposed upon a set of four z-projections of confocal sections of a brain labeled with DN-cad (synapses), in which fascicles and compartment boundaries appear as signal-negative/signal-poor domains. Z-projections represent the brain slices at the antero-posterior levels used throughout this paper (bottom: optic tubercle and mushroom body medial lobe; second from bottom: ellipsoid body; third: fan-shaped body and great commissure; top: lateral bend of antennal lobe tract towards calyx). Z-projections are compressed (50%) along the y-axis, such as to give the set of these images the appearance of a cut-away diagram of the neuropil in antero-dorsal view. SATs entering the neuropil from anterior to associate with a specific fascicle are shown as opaque lines, converging on the signal-negative domain corresponding to that fascicle. Note, for example, the SATs of BAla1 (#1), BAlc d (#5d), BAlp4 (#9) and BAmv3 (#17), whose SATs form the antennal lobe tract ACT). The colored lines representing these SATs target the signal-negative “hole” formed by the ACT in the posterior antennal lobe. Proximal arborizations (small circles) are formed in the antennal lobe. After passing through that “hole” (and thereby disappearing “behind” the first z-projection), the lines representing the SATs are rendered semitransparent; once they “reappear” in the space between the first (bottom) and second z-projection (one above bottom), lines turn opaque again, and so on. Reaching the posterior level (top), SATs turn laterally and form distal terminal arborizations, indicated by small circles. Gray lines indicate HSATs or SSATs of lineages whose projection does not conform with the “theme” of this figure, i.e., connections between ventral and dorsal neuropil compartments. For example, the posterior hemilineage of BLAv1 (#73p) is confined to ventral neuropil domains, and is rendered gray. Neuropil compartments (white lettering) are annotated on the right side of the z-projections. See Table 2 for complete list of abbreviations.
Supplementary Figure S2 Schematic diagram of lineages with strong projections interconnecting ventral neuropil domains. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S3 Schematic diagram of lineages with strong projections involving the central complex and mushroom body lobes. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S4 Schematic diagram of lineages of the dorsal brain with strong projections towards the ventral neuropil, subesophageal ganglion and thoracic-abdominal ganglion. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S5 Schematic diagram of lineages with strong projections interconnecting dorsal neuropil domains. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S6 Tentative matching of single brain neurons to their respective lineages. Panels A-E show z-projections of clones representing five lineages or lineage pairs, BAmas2, BLVa3/4, DAMv1/2, BLD2, and BAla1. Numbered panels (A1-A5, B1, C1, D1, E1/E1’) show z-projections of single cell clones, published by Chiang et al. (2011). Based on their neuropil entry point and contour of the projection envelope, neurons correspond to the lineages shown. E2/E2’ is an example of a putative primary neuron. For detail, see Discussion. Single-cell clones were generated by Chiang et al., 2011 and are available for viewing at FlyCircuit: A Database of Drosophila Brain Neurons (http://www.flycircuit.tw/). Panels correspond to clone Neuron ID's as follows: A1, fru-F-700004; A2, fru-F-700003; A3, fru-F-700028; A4, 5-HT1B-F-200000; A5, fru-X-200003; B1, VGlut-F-100001; C1, VGlut-F-200024; D1, VGlut-F-200058; E1/E1`, VGlut-F-200315; E2/E2`, VGlut-F-200000. Bar: 50μm
Supplementary Figure S7 Clone representing lineage BLP1. (A) z-projections show slice of the brain at a posterior level (level posterior to fan-shaped body). (B) z-projection of clone whose axons fasciculate with the SAT of BLP1/2 (#83/84) (arrowhead). Whereas all other clones representing the BLP1/2 pair have restricted projections to the VLP compartment (see Fig.18C), this one clone possesses additional projections to the PLP and IP.
Highlights.
Lineages form fiber tracts (SATs) innervating discrete sets of brain compartments
Based on SAT trajectory, we assigned MARCM clones to secondary lineages
Characterization of supraesophageal ganglion secondary lineage projection envelopes
Framework linking adult-specific neurons with genetically distinct secondary lineages
ACKNOWLEDGEMENTS
We thank the members of the Hartenstein laboratory for critical discussions during the preparation of this manuscript. We thank S. Fung, F. Wang, J.D. Nguyen, and K. Wang for their involvement in generating larval and adult MARCM clones. We are grateful to the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for y strains and antibodies. This work was supported by NIH grant (R01 NS29357-15). K.T.N. is supported by the NSF Graduate Research Fellowship Program (No. DGE-0707424). J.J.O. is supported by the Ruth L. Kirschstein National Research Service Award (No. GM007185).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M. A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. Drosophila. pp. 214–217. [Google Scholar]
- Bausenwein B, Dittrich AP, Fischbach KF. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 1992;267:17–28. doi: 10.1007/BF00318687. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Boone JQ, Drummond ML, Doe CQ. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 2010;5:26. doi: 10.1186/1749-8104-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billuart P, Winter CG, Maresh A, Zhao X, Luo L. Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell. 2001;107:195–207. doi: 10.1016/s0092-8674(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 2008;68:1185–1195. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev. Biol. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Arganda I, Pereanu W, Schindelin J, Hartenstein V. Identifying neuronal lineages of Drosophila by sequence analysis of axon tracts. J. Neurosci. 2010;30:7538–7553. doi: 10.1523/JNEUROSCI.0186-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T, Younossi-Hartenstein A, Hartenstein V. Development of neural lineages derived from the sine oculis positive eye field of Drosophila. Arthropod Struct. Dev. 2003;32:303–317. doi: 10.1016/j.asd.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Chen YM, Wang QJ, Hu HS, Yu PC, Zhu J, Drewes G, Piwnica-Worms H, Luo ZG. Microtubule affinity-regulating kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc. Natl. Acad. Sci. U S A. 2006;103:8534–8539. doi: 10.1073/pnas.0509955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Cong W, Hirose T, Harita Y, Yamashita A, Mizuno K, Hirano H, Ohno S. ASPP2 regulates epithelial cell polarity through the PAR complex. Curr. Biol. 2010;20:1408–1414. doi: 10.1016/j.cub.2010.06.024. [DOI] [PubMed] [Google Scholar]
- Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn. Mem. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- Das A, Gupta T, Davla S, Prieto-Godino LL, Diegelmann S, Reddy OV, Raghavan KV, Reichert H, Lovick J, Hartenstein V. Neuroblast lineage-specific origin of the neurons of the Drosophila larval olfactory system. Dev. Biol. 2013;373:322–337. doi: 10.1016/j.ydbio.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Sen S, Lichtneckert R, Okada R, Ito K, Rodrigues V, Reichert H. Drosophila olfactory local interneurons and projection neurons derive from a common neuroblast lineage specified by the empty spiracles gene. Neural Dev. 2008;3:33. doi: 10.1186/1749-8104-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Ellenbroek SI, Iden S, Collard JG. Cell polarity proteins and cancer. Semin. Cancer Biol. 2012;22:208–215. doi: 10.1016/j.semcancer.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Guillemot F. Cell fate specification in the mammalian telencephalon. Prog. Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Cardona A, Pereanu W, Younossi-Hartenstein A. Modeling the developing Drosophila brain: rationale, technique and application. BioScience. 2008;58:823–836. [Google Scholar]
- Hassan BA, Bermingham NA, He Y, Sun Y, Jan YN, Zoghbi HY, Bellen HJ. atonal regulates neurite arborization but does not act as a proneural gene in the Drosophila brain. Neuron. 2000;25:549–61. doi: 10.1016/s0896-6273(00)81059-4. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Ito K, Sado Y, Taniguchi M, Akimoto A, Takeuchi H, Aigaki T, Matsuzaki F, Nakagoshi H, Tanimura T, Ueda R, Uemura T, Yoshihara M, Goto S. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- Huff R, Furst A, Mahowald AP. Drosophila embryonic neuroblasts in culture: autonomous differentiation of specific neurotransmitters. Dev. Biol. 1989;134:146–157. doi: 10.1016/0012-1606(89)90085-7. [DOI] [PubMed] [Google Scholar]
- Hurd TW, Fan S, Liu CJ, Kweon HK, Hakansoon K, Margolis B. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr. Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. Adv. Exp. Med. Biol. 2008;628:137–158. doi: 10.1007/978-0-387-78261-4_9. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Izergina N, Balmer J, Bello B, Reichert H. Postembryonic development of transit amplifying neuroblast lineages in the Drosophila brain. Neural Dev. 2009;4:44. doi: 10.1186/1749-8104-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Vyas RM, Berdnik D, Ramaekers A, Stocker RF, Tanaka NK, Ito K, Luo L. Developmental origin of wiring specificity in the olfactory system of Drosophila. Development. 2004;131:117–130. doi: 10.1242/dev.00896. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TB, Shepherd D, Murphy H, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Johnson WA, Luo L, Jefferis GS. From lineage to wiring specificity: POU domain transcription factors control precise connections of Drosophila olfactory projection neurons. Cell. 2003;112:157–167. doi: 10.1016/s0092-8674(03)00030-8. [DOI] [PubMed] [Google Scholar]
- Kuert PA, Bello BC, Reichert H. The labial gene is required to terminate proliferation of identified neuroblasts in post embryonic development of the Drosophila brain. Biol. Open. 2012;1:1006–1015. doi: 10.1242/bio.20121966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bello B, Reichert H. Lineage-specific cell death in postembryonic brain development of Drosophila. Development. 2009a;136:3433–3442. doi: 10.1242/dev.037226. [DOI] [PubMed] [Google Scholar]
- Kumar A, Fung S, Lichtneckert R, Reichert H, Hartenstein V. Arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropil. J. Comp. Neurol. 2009b;517:87–104. doi: 10.1002/cne.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz T, Kraft KF, Technau GM, Urbach R. Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development. 2012;139:2510–2522. doi: 10.1242/dev.077883. [DOI] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Larsen C, Shy D, Spindler SR, Fung S, Pereanu W, Younossi-Hartenstein A, Hartenstein V. Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Dev. Biol. 2009;335:289–304. doi: 10.1016/j.ydbio.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000a;28:807–818. doi: 10.1016/s0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- Lee T, Winter C, Marticke SS, Lee A, Luo L. Essential roles of Drosophila RhoA in the regulation of neuroblast proliferation and dendritic but not axonal morphogenesis. Neuron. 2000b;25:307–316. doi: 10.1016/s0896-6273(00)80896-x. [DOI] [PubMed] [Google Scholar]
- Lichtneckert R, Nobs L, Reichert H. Empty spiracles is required for the development of olfactory projection neuron circuitry in Drosophila. Development. 2008;135:2415–2424. doi: 10.1242/dev.022210. [DOI] [PubMed] [Google Scholar]
- Lin S, Kao CF, Yu HH, Huang Y, Lee T. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS Biol. 2012;10:E1001425. doi: 10.1371/journal.pbio.1001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto J, Wong DC, Nguyen JD, Hartenstein V. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. doi: 10.1016/j.ydbio.2013.07.008. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front. Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin EC, Dry KE, Alaimo DR, Rudd KT, Cillo AR, Clenshaw ME, Negre N, White KP, Truman JW. Ultrabithorax confers spatial identity in a context-specific manner in the Drosophila postembryonic ventral nervous system. Neural Dev. 2012;7:31. doi: 10.1186/1749-8104-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C, Cheng L, Gould AP. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–447. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. PNAS. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Kato K, Yamaguchi T, Fukata Y, Ohno S, Kaibuchi K. Role of the PAR-3-KIF3 complex in the establishment of neuronal polarity. Nat. Cell Biol. 2004;6:328–334. doi: 10.1038/ncb1118. [DOI] [PubMed] [Google Scholar]
- Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 2001;13:641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- Okajima T, Xu A, Lei L, Irvine KD. Chaperone activity of protein OFucosyltransferase 1 promotes Notch receptor folding. Science. 2005;307:1599–1603. doi: 10.1126/science.1108995. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 2005;283:191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Kumar A, Jennett A, Reichert H, Hartenstein V. Development-based compartmentalization of the Drosophila central brain. J. Comp. Neurol. 2010;518:2996–3023. doi: 10.1002/cne.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, Taghert P. Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J. Neurobiol. 1999;41:189–207. [PubMed] [Google Scholar]
- Reuter JE, Nardine TM, Penton A, Billuart P, Scott EK, Usui T, Uemura T, Luo L. A mosaic genetic screen for genes necessary for Drosophila mushroom body neuronal morphogenesis. Development. 2003;130:1203–1213. doi: 10.1242/dev.00319. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Doe CQ. Baz, Par-6 and aPKC are not required for axon or dendrite specification in Drosophila. Nat Neurosci. 2004;7:1293–1295. doi: 10.1038/nn1346. [DOI] [PubMed] [Google Scholar]
- Sánchez-Soriano N, Bottenberg W, Fiala A, Haessler U, Kerassoviti A, Knust E, Löhr R, Prokop A. Are dendrites in Drosophila homologous to vertebrate dendrites? Dev. Biol. 2005;288:126–138. doi: 10.1016/j.ydbio.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Scott EK, Lee T, Luo L. enok encodes a Drosophila putative histone acetyltransferase required for mushroom body neuroblast proliferation. Curr Biol. 2001;11:99–104. doi: 10.1016/s0960-9822(01)00020-3. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Helfrich-Förster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster's neuronal circadian pacemakers reveals new neuronal classes. J. Comp. Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Cheng T, Jan LY, Jan YN. APC and GSK-3beta are involved in mPar3 targeting to the nascent axon and establishment of neuronal polarity. Curr. Biol. 2004;14:2025–2032. doi: 10.1016/j.cub.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Thor S. Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Hartenstein V. The Drosophila neural lineages: a model system to study brain development and circuitry. Dev. Genes Evol. 2010;220:1–10. doi: 10.1007/s00427-010-0323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler SR, Hartenstein V. Bazooka mediates secondary axon morphology in Drosophila brain lineages. Neural Dev. 2011;6:16. doi: 10.1186/1749-8104-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srahna M, Leyssen M, Choi CM, Fradkin LG, Noordermeer JN, Hassan BA. A signaling network for patterning of neuronal connectivity in the Drosophila brain. PLoS Biol. 2006;4:E348. doi: 10.1371/journal.pbio.0040348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF, Lienhard MC, Borst A, Fischbach KF. Neuronal architecture of the antennal lobe in Drosophila melanogaster. Cell Tissue Res. 1990;262:9–34. doi: 10.1007/BF00327741. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development. 2010;137:53–61. doi: 10.1242/dev.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Early steps in building the insect brain: neuroblast formation and segmental patterning in the developing brain of different insect species. Arthropod Struct Dev. 2003;32:103–123. doi: 10.1016/S1467-8039(03)00042-2. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Neuroblast formation and patterning during early brain development in Drosophila. Bioessays. 2004;26:739–751. doi: 10.1002/bies.20062. [DOI] [PubMed] [Google Scholar]
- Viktorin G, Riebli N, Popkova A, Giangrande A, Reichert H. Multipotent neural stem cells generate glial cells of the central complex through transit amplifying intermediate progenitors in Drosophila brain development. Dev. Biol. 2011;356:553–65. doi: 10.1016/j.ydbio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Viktorin G, Riebli N, Reichert H. A multipotent transit-amplifying neuroblast lineage in the central brain gives rise to optic lobe glial cells in Drosophila. Dev. Biol. 2013;379:182–94. doi: 10.1016/j.ydbio.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- Wang Y, Du D, Fang L, Yang G, Zhang C, Zeng R, Ullrich A, Lottspeich F, Chen Z. Tyrosine phosphorylated Par3 regulates epithelial tight junction assembly promoted by EGFR signaling. EMBO J. 2006;25:5058–5070. doi: 10.1038/sj.emboj.7601384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Khan Z, Kaschube M, Wieschaus EF. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature. 2012;484:390–393. doi: 10.1038/nature10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson AH, Schürmann FW. Synaptic structure, distribution, and circuitry in the central nervous system of the locust and related insects. Microscopy Research and Technique. 2002;56:210–226. doi: 10.1002/jemt.10031. [DOI] [PubMed] [Google Scholar]
- Yang JS, Awasaki T, Yu HH, He Y, Ding P, Kao JC, Lee T. Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. J. Comp. Neurol. 2013;521(12):Spc1. doi: 10.1002/cne.23339. doi: 10.1002/cne.23366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J. Comp. Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Yu HH, Chen CH, Shi L, Huang Y, Lee T. Twin-spot MARCM to reveal the developmental origin and identity of neurons. Nat. Neurosci. 2009;12:947–953. doi: 10.1038/nn.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Kao CF, He Y, Ding P, Kao JC, Lee T. A complete developmental sequence of a Drosophila neuronal lineage as revealed by twin-spot MARCM. PLoS Biol. 2010;8:E1000461. doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Basler K, Struhl G. Direct and long-range action of a wingless morphogen gradient. Cell. 1996;87:833–844. doi: 10.1016/s0092-8674(00)81991-1. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zugates CT, Lu Z, Shi L, Bai JM, Lee T. Baboon/dSmad2 TGF-beta signaling is required during late larval stage for development of adult-specific neurons. EMBO J. 2006;25:615–627. doi: 10.1038/sj.emboj.7600962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Chiang AS, Lee T. Development of the Drosophila mushroom bodies: elaboration, remodeling and spatial organization of dendrites in the calyx. Development. 2003;130:2603–2610. doi: 10.1242/dev.00466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Schematic diagram of lineages of the ventral brain with strong projections connecting ventral and dorsal neuropil domains. Each lineage is represented by a numbered sphere (location of SAT entry point into neuropil), a line (SAT trajectory in neuropil), and small circles (location of terminal arborizations). SAT trajectories are based upon BP104 labeled brains (see accompanying paper by Lovick et al., 2013), as well as MARCM clones of corresponding lineages. Location of terminal arborizations are based on MARCM clones (see Figs.3-18 of this paper). To show lineage projections in relationship to neuropil structures, lines and circles, representing SATs and terminal arborizations, respectively, are superimposed upon a set of four z-projections of confocal sections of a brain labeled with DN-cad (synapses), in which fascicles and compartment boundaries appear as signal-negative/signal-poor domains. Z-projections represent the brain slices at the antero-posterior levels used throughout this paper (bottom: optic tubercle and mushroom body medial lobe; second from bottom: ellipsoid body; third: fan-shaped body and great commissure; top: lateral bend of antennal lobe tract towards calyx). Z-projections are compressed (50%) along the y-axis, such as to give the set of these images the appearance of a cut-away diagram of the neuropil in antero-dorsal view. SATs entering the neuropil from anterior to associate with a specific fascicle are shown as opaque lines, converging on the signal-negative domain corresponding to that fascicle. Note, for example, the SATs of BAla1 (#1), BAlc d (#5d), BAlp4 (#9) and BAmv3 (#17), whose SATs form the antennal lobe tract ACT). The colored lines representing these SATs target the signal-negative “hole” formed by the ACT in the posterior antennal lobe. Proximal arborizations (small circles) are formed in the antennal lobe. After passing through that “hole” (and thereby disappearing “behind” the first z-projection), the lines representing the SATs are rendered semitransparent; once they “reappear” in the space between the first (bottom) and second z-projection (one above bottom), lines turn opaque again, and so on. Reaching the posterior level (top), SATs turn laterally and form distal terminal arborizations, indicated by small circles. Gray lines indicate HSATs or SSATs of lineages whose projection does not conform with the “theme” of this figure, i.e., connections between ventral and dorsal neuropil compartments. For example, the posterior hemilineage of BLAv1 (#73p) is confined to ventral neuropil domains, and is rendered gray. Neuropil compartments (white lettering) are annotated on the right side of the z-projections. See Table 2 for complete list of abbreviations.
Supplementary Figure S2 Schematic diagram of lineages with strong projections interconnecting ventral neuropil domains. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S3 Schematic diagram of lineages with strong projections involving the central complex and mushroom body lobes. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S4 Schematic diagram of lineages of the dorsal brain with strong projections towards the ventral neuropil, subesophageal ganglion and thoracic-abdominal ganglion. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S5 Schematic diagram of lineages with strong projections interconnecting dorsal neuropil domains. Lineages are identified by numbers. The diagram is designed in the same manner as explained for Figure S1. See Table 2 for complete list of abbreviations.
Supplementary Figure S6 Tentative matching of single brain neurons to their respective lineages. Panels A-E show z-projections of clones representing five lineages or lineage pairs, BAmas2, BLVa3/4, DAMv1/2, BLD2, and BAla1. Numbered panels (A1-A5, B1, C1, D1, E1/E1’) show z-projections of single cell clones, published by Chiang et al. (2011). Based on their neuropil entry point and contour of the projection envelope, neurons correspond to the lineages shown. E2/E2’ is an example of a putative primary neuron. For detail, see Discussion. Single-cell clones were generated by Chiang et al., 2011 and are available for viewing at FlyCircuit: A Database of Drosophila Brain Neurons (http://www.flycircuit.tw/). Panels correspond to clone Neuron ID's as follows: A1, fru-F-700004; A2, fru-F-700003; A3, fru-F-700028; A4, 5-HT1B-F-200000; A5, fru-X-200003; B1, VGlut-F-100001; C1, VGlut-F-200024; D1, VGlut-F-200058; E1/E1`, VGlut-F-200315; E2/E2`, VGlut-F-200000. Bar: 50μm
Supplementary Figure S7 Clone representing lineage BLP1. (A) z-projections show slice of the brain at a posterior level (level posterior to fan-shaped body). (B) z-projection of clone whose axons fasciculate with the SAT of BLP1/2 (#83/84) (arrowhead). Whereas all other clones representing the BLP1/2 pair have restricted projections to the VLP compartment (see Fig.18C), this one clone possesses additional projections to the PLP and IP.