Abstract
Many recent studies have focused on the connection between the composition of specific volatile organic compounds (VOCs) in exhaled breath and various forms of cancer. However, the composition of exhaled breath is affected by many factors, such as lung disease, smoking, and diet. VOCs are released into the bloodstream before they are exhaled; therefore, the analysis of VOCs in blood will provide more accurate results than the analysis of VOCs in exhaled breath. Blood were collected from 16 colorectal cancer patients and 20 healthy controls, then solid phase microextraction–chromatography–mass spectrometry (SPME-GC-MS) was used to analysis the exhaled volatile organic compounds (VOCs). The statistical methods principal component analysis (PCA) and partial least-squares discriminant analysis (PLSDA) were performed to deal with the final dates. Three metabolic biomarkers were found at significantly lower levels in the group of CRC patients than in the normal control group (P < 0.01): phenyl methylcarbamate, ethylhexanol, and 6-t-butyl-2,2,9,9-tetramethyl-3,5-decadien-7-yne. In addition, significantly higher levels of 1,1,4,4-tetramethyl-2,5-dimethylene-cyclohexane were found in the group of CRC patients than in the normal control group (P < 0.05). Compared with healthy individuals, patients with colorectal adenocarcinoma exhibited a distinct blood metabolic profile with respect to VOCs. The analysis of blood VOCs appears to have potential clinical applications for CRC screening.
Keywords: colorectal cancer, volatile organic compounds, biomarkers, solid-phase microextraction, cancer diagnosis
Introduction
Colorectal cancer (CRC) is the third most common malignancy in the world.1 In Europe, CRC is the leading cause of cancer-related death.2 The 5-y relative survival rate for CRC can reach 90.1% for CRC cases that are diagnosed when tumors remain localized; however, the 5-y survival rate for cases of CRC in which the cancer has invaded surrounding tissue and spread to lymph nodes is only 69.2%.3 Early diagnosis and timely intervention improve the quality of life of patients with CRC and reduce cancer-related mortality.4 At present, there are continuing efforts to discover methods for the early detection and diagnosis of CRC. The fecal occult blood test (FOBT) is the most commonly used non-invasive method to screen for CRC. However, the FOBT exhibits low sensitivity and specificity for the detection of early-stage CRC; in fact, the positive predictive value of the FOBT for CRC is only 10%.5 Colonoscopy is a more accurate and reliable screening method for the detection of CRC.6,7 However, the colonoscopy approach also features certain limitations, including high costs, resource-related restrictions, and inconvenience; moreover, a colonoscopy may not only cause discomfort in patients but also produce unanticipated iatrogenic trauma.1,8 Certain specific serum tumor biomarkers, such as carcinoembryonic antigen (CEA) and CA 19-9 (carbohydrate antigen 19-9, also known as cancer antigen 19-9) have been widely used for the clinical diagnosis of CRC. However, neither CEA nor CA 19-9 is able to provide good sensitivity and specificity for CRC diagnoses because both of these antigens also serve as biomarkers for other types of cancer.1,9,10 Therefore, clinical practitioners have long sought to develop an effective screening method for the early detection of CRC that is not only economical and convenient to perform but also exhibits high sensitivity and accuracy. Exhaled breath analysis is a novel approach that has recently been developed for the screening and diagnosis of diseases. Because this approach exhibits the advantages of being convenient and non-invasive, exhaled breath analysis has drawn increasing attention from researchers. Many recent studies have focused on the connection between the composition of specific volatile organic compounds (VOCs) in exhaled breath and various forms of cancer, including lung cancer11,12 and breast cancer.13,14 In 2013, Altomare et al. compared the composition of the exhaled VOCs of a CRC group with the exhaled VOCs of a normal control group and identified CRC-specific VOCs in exhaled breath.15 However, the composition of exhaled breath is affected by many factors, such as lung disease, smoking, and diet.16 VOCs are released into the bloodstream before they are exhaled; therefore, the analysis of VOCs in blood will provide more accurate results than the analysis of VOCs in exhaled breath. In the current study, we intend to utilize gas chromatography/mass spectrometry (GC/MS) and multivariate data analysis to compare the VOCs in blood samples from individuals in a healthy physiological state with the VOCs in blood samples from CRC patients in a pathological state, thereby allowing for the identification of CRC-specific VOCs in the blood.
Results
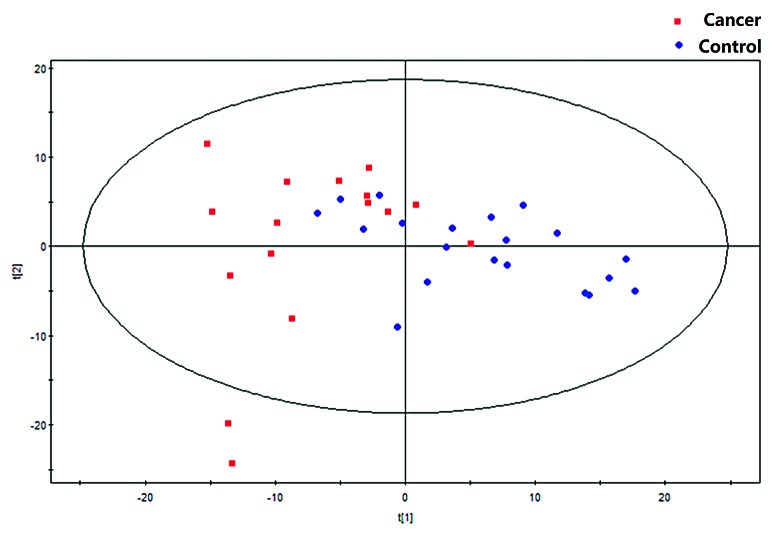
In this study, VOCs in the blood of 16 CRC patients and 20 healthy volunteers were analyzed by GC-MS. Based on the ion peaks in the resulting chromatograms and the results of multivariate analyses, we obtained 341 variables; these variables were then assessed to verify their performance as predictors of CRC. In particular, the PCA approach can separate the main influencing factors from among diverse considerations, revealing the essential aspects of the situation and simplifying complex issues. Thus, we performed PCA analyses of all samples and examined the resulting grouping trends and outliers. A PCA scatterplot revealed the separate trends for the experimental group and the control group; the tight clustering of samples in this plot demonstrated that our approach was effective (Fig. 1).
Figure 1. The PCA model (R2X = 0.642, Q2 = 0.431).
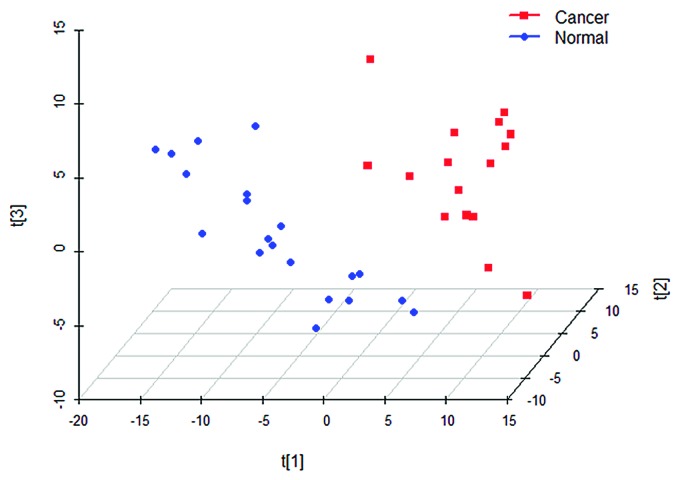
As a supervised analytical technique, partial least squares discriminant analysis (PLS-DA) can be utilized to facilitate the screening of metabolic biomarkers and the exclusion of system variables that are unrelated to pathological conditions.17,18 We established a PLS-DA model that included three potential variables: R2X = 0.445; R2Y = 0.874; and Q2 = 0.678. A PLS-DA scatter plot confirmed that the CRC patients of the cancer group could be clearly separated from the healthy individuals in the normal control group (Fig. 2).
Figure 2. PLS-DA (with the three components of R2X = 0.445, R2Y = 0.874, and Q2 = 0.678).
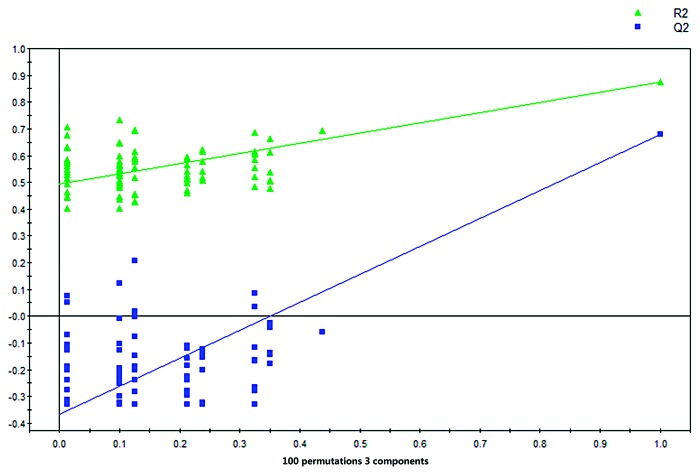
The VIP values of the examined factors in the PLS-DA model were calculated. Based on the standard of a VIP value greater than 1.0, four distinct metabolic biomarkers that could be used to distinguish between the two groups of study participants were selected. These metabolic biomarkers were annotated using the National Institute of Standards and Technology (NIST) 11 database. Finally, the model was validated using a validation plot that was obtained from 100 permutation tests (Fig. 3). All of the R2 and Q2 values for this model were calculated, and the values for both variables were lower in the new model than in the original model. Furthermore, the regression line for Q2 exhibited a negative intercept. Therefore, the effectiveness of the supervised model was fully established.
Figure 3.y-intercepts: R2 = (0.0, 0.492), Q2 = (0.0, −0.366). Validation plot obtained from 100 permutation tests.
The results of the present study were as follows: Three metabolic biomarkers were found at significantly lower levels in the group of CRC patients than in the normal control group (P < 0.05): phenyl methylcarbamate, ethylhexanol, and 6-t-butyl-2,2,9,9-tetramethyl-3,5-decadien-7-yne. In addition, significantly higher levels of 1,1,4,4-tetramethyl-2,5-dimethylene-cyclohexane were found in the group of CRC patients than in the normal control group (P < 0.05). Additional information is provided in Table 1.
Table 1. Specific VOC biomarkers found at abnormal levels in the blood of CRC patients.
| Potential biomarker | P value | FC | VIP |
|---|---|---|---|
| Phenyl methylcarbamate | 0.00417 | −0.77 | 1.1085 |
| Ethylhexanol | 0.00618 | −1.04 | 1.7287 |
| 1,1,4,4-Tetramethyl-2,5-dimethylene-cyclohexane | 0.00867 | 4.93 | 1.0635 |
| 6-t-Butyl-2,2,9,9-tetramethyl-3,5-decadien-7-yne | 0.00377 | −0.88 | 1.0884 |
The staging of patients was included in the multi-variate analysis. No separation trend between early stage (I and II) CRC patients and late stage (III) CRC patients was observed in the PCA score plot. The PLS-DA model with one component automatically built in the SIMCA-P software showed a classification tendency (R2X = 0.262; R2Y = 0.493; and Q2 = 0.147), while the validation plot from 100 permutation tests could not assure the validity of this supervised PLS-DA model as not all of the R2 and Q2 values were lower in the new models than in the original model. Therefore, our data could not robustly support the classification of different stages in CRC patients, which might be limited by the relative small sample size. Further investigations consisting of larger sample sizes are needed to validate our findings.
Discussion
In this study, the composition of VOCs in the blood of patients with CRC and those of healthy individuals were analyzed and compared with identify CRC-specific VOCs for the screening and diagnosis of CRC. The experimental design is theoretically reasonable; metabolic changes have occurred in patients with CRC that inevitably lead to the production of abnormal metabolites. These compounds are released into the bloodstream before being transported to the alveoli via blood circulation; through alveolar gas exchange, abnormal VOCs are discharged into the air as components of each exhaled breath.19 In theory, the VOC components in an individual’s blood should be consistent with the VOC components that this individual exhales; however, components of an individual’s exhaled breaths may be affected if this individual suffers from lung disease or has a history of smoking.16 In the current study, to eliminate the effects of food and other factors on experimental results, the research subjects were strictly required to fast for 8 h prior to the acquisition of samples.
Certain studies have demonstrated that different pathological types of cancer tissue produce distinct combinations of VOCs during abnormal metabolic processes.20,21 Orna et al. performed exhaled breath analyses of patients with different lung cancer pathologies and found that the composition of exhaled VOCs was different among these patients; in particular, the concentrations of the three compounds of 2-ethyl-1-hexanol, 1,3-dimethyl-benzene, and 1,3-bis(1,1-dimethylethyl)-benzene were significantly higher in patients with adenocarcinoma than in patients with squamous cell carcinoma.22 Chen et al.23 examined four types of lung cancer, including adenocarcinoma, squamous cell carcinoma, bronchial carcinoid tumor, and non-small-cell lung cancer, and determined that different VOCs are exhaled by patients with different lung cancer types. Because exhaled metabolites are derived from metabolites in the blood that are released into the air via the pulmonary circulation, pathological differences among patients may also correlate with differences in the metabolites in patients’ blood. Given the potential influence of pathological type on the experimental results, we only included patients who were diagnosed with colorectal adenocarcinoma in this study. For data analysis, we used PCA, PLSDA with permutation tests, and multivariate data analysis techniques to validate the effectiveness of our model. In combination, these analyses demonstrated that our model was valid and could clearly distinguish CRC patients from healthy individuals with an accuracy that was significantly above chance.20
In recent years, many investigations have focused on analyzing CRC by determining the urinary metabolic profiles, blood levels of water-soluble metabolites, and exhaled VOCs of CRC patients.8,15,21 To the best of our knowledge, no study has investigated the specific volatile metabolic biomarkers that exist in the blood of CRC. Chen performed a metabolomics analysis of the effects of CRC on urine and determined that relative to the urine of a control group of healthy individuals, the urine of CRC patients exhibited significantly higher levels of lactic acid but lower levels of histidine and methionine.21 This increase in the levels of lactic acid and other substances is caused by the elevated metabolic state of cancer cells. In particular, glycolysis increases in these cells; as a result, large quantities of glucose are converted into lactic acid even under aerobic conditions, a phenomenon that is known as the “Warburg effect”.24,25 Low levels of histidine occur in CRC patients due to increases in the activity of histidine decarboxylase, which catalyzes the decarboxylation of histidine to produce histamine.7 Reduced methionine levels in CRC patients reflect increases in methylation that occur during tumor development, which result in declines in methyl donor levels.26 Nishiumi et al. established a prediction model for serum-soluble metabolites in the context of CRC and demonstrated that higher serum levels of 2-hydroxybutyrate and kynurenine are found in a group of CRC patients than in a normal control group.8 This increase in 2-hydroxybutyrate levels may relate to oxidative stress.27 The production of kynurenine from tryptophan is catalyzed by indoleamine 2,3-dioxygenase; thus, because higher levels of tryptophan metabolism are observed in tumor cells than in normal cells, tumor cells will generate abnormally elevated levels of kynurenine.8 The aforementioned research primarily addresses two types of pathological mechanisms of CRC, namely, increases in both oxidative stress and methylation.
Peng et al. analyzed the composition of exhaled breaths to create a model of CRC-associated metabolic biomarkers, which include ([1,1-dimethylethyl]thio) acetic acid; 4-(4-propylcyclohexyl)-4’-cyano(1,1’-biphenyl)-4-yl ester benzoic acid; 1,3-dimethyl benzene; 2-amino-5-isopropyl-8-methyl-1-azulenecarbonitrile; and 1,1-(1-butenylidene)bis benzene.28 Altomare et al. also identified various CRC-specific VOCs, including 2-methylbutane, cyclohexane, decanal, 4-methyl-2-pentanone, 1,2-pentadiene, 2-methylpentane, 4-methyloctane, methylcyclopentane, 3-methylpentane, methylcyclohexane, 1,3-dimethylbenzene, 1,4-dimethylbenzene, and nonanal.15 These volatile biomarkers are mostly alkanes, ketones, aldehydes, and substituted benzene compounds. The mechanisms through which these metabolites are generated remains under investigation, although the majority of the extant studies support the hypothesis that these metabolites are produced in response to oxidative stress.15,29 Tumor tissue demonstrates vigorous growth and consequently exhibits high energy demands. The malignant growth of cancer cells can lead to genetic mutations and the abnormal expression of proteins, causing the excessive oxidation of polyunsaturated fatty acids in cell membranes and/or the production of excessive reactive oxygen species (ROS). The available evidence indicates that in cases of various types of cancers, there are high levels of antioxidant enzyme activity and deficiencies in antioxidant vitamins.30-34
The results of the current study revealed that CRC-specific VOCs in blood included the following compounds: phenyl methylcarbamate, ethylhexanol, 6-t-butyl-2,2,9,9-tetramethyl-3,5-decadien-7-yne, and 1,1,4,4-tetramethyl-2,5-dimethylene-cyclohexane. The methylated alkanes, such as 1,1,4,4-tetramethyl-2, 5-dimethylene-cyclohexane, that were identified as CRC-specific VOCs in our results were similar to the metabolic biomarkers of melanoma that were identified by Abaffy et al., which include 4-methyl decane, dodecane, and undecane. Abaffy et al. has speculated that in melanoma, methylated alkanes are produced by in vivo methylation.35 Abnormal DNA methylation is a common mechanism that occurs during the development of many types of cancer; for instance, increased levels of DNA methyltransferase have been detected in certain cancer patients.36-38 Hassanein et al. reported that the abnormal proliferation of cancer cells is the fundamental principle underlying the component analysis of metabolites in cancer patients; in particular, these researchers propose that cancer cell proliferation may lead to abnormal protein expression, thereby causing the peroxidation of cell membranes and the release of VOCs.39 In blood samples from CRC patients, we detected VOCs, such as 1,1,4,4-tetramethyl-2,5-dimethylene-cyclohexane, that were consistent with the CRC-specific VOCs that Altomare reported in 2013. One of the metabolites detected by Altomare was cyclohexane, which has the same fundamental chemical structure as the 1,1,4,4-tetramethyl-2,5-dimethylene-cyclohexane that was identified in our study but differs only with respect to substituents. These results validate the principle that a correlation exists between blood metabolites and the VOCs of exhaled breath.
In our results, lower blood concentrations of phenylmethylcarbamate and ethylhexanol were observed in the cancer group than in the normal control group. A potential mechanism underlying this phenomenon is the consumption that occurs during tumor cell proliferation. Filipiak et al. cultured the Calu-1 lung cancer cell line and analyzed the composition of the headspaces of these cell cultures; from this analysis, these researchers determined that during cancer cell growth, there are reductions in the levels of certain volatile biomarkers, such as butyl acetate and 2-methylpropanal, indicating that these compounds are consumed during the process of tumor cell proliferation.40 Our experimental results indicating CRC-specific reductions in certain metabolites, including phenylmethylcarbamate, ethylhexanol and 6-t-butyl-2,2,9,9-tetramethyl-3,5-decadien-7-yne, could also be explained by tumor consumption mechanisms, although these compounds were different from the compounds reported by Filipiak et al. This difference likely reflects the fact that the current study and the investigation by Filipiak et al. examined different types of cancer; therefore, the metabolites and the consumption mechanisms of these cancers would be expected to differ.
Our experiments exhibited certain limitations. For instance, this study examined a relatively small sample. In addition, although the CRC patients who participated in the study demonstrated the same pathology, the effect of clinical stage on exhaled VOCs was not investigated. In future research, we will increase the sample size and analyze the impact of cancer stage on exhaled VOCs. In addition, new techniques will be tested in an attempt to render the exhaled breath analysis approach a more convenient and cost-effective technique for the screening of potential CRC patients.28 Compared with healthy individuals, patients with colorectal adenocarcinoma exhibited a distinct blood metabolic profile with respect to VOCs. The analysis of blood VOCs appears to have potential clinical applications for CRC screening.
Materials and Methods
Human subjects
This protocol of this study was approved by the Ethics Committee of Harbin Medical University (No. 201314). A total of 16 patients who were admitted to the First Affiliated Hospital of Harbin Medical University between May 2011 and October 2012 for the resection of CRC were selected for this study (the cancer group). All of the selected study participants were recruited in accordance with the strict inclusion and exclusion criteria established by our research institution. In particular, the following inclusion criteria were utilized with respect to CRC patients: (1) an age of 25 to 70 y, (2) the presence of pathologically confirmed colorectal adenocarcinoma, and (3) agreement to participate in the study, as indicated by a signed informed consent form. The following exclusion criteria were utilized with respect to study participants: (1) pregnancy, lactation, or the possibility of pregnancy, (2) the presence of a known congenital disease, (3) a family history of mental illness, (4) the presence of a chronic inflammatory disease, (5) symptoms of an acute disease during the 2 weeks prior to study enrollment, (6) a history of infectious disease, and (7) patients who received the prior therapeutic regimen (chemotherapy, radiotherapy or complementary therapies). In addition to the cancer group, 20 healthy volunteers were also included in this study. The inclusion criteria with respect to healthy volunteers were colonoscopy results indicating the absence of CRC and no history of malignancies or infectious disease.
As detailed in Table 2, the 16 CRC patients who were selected for the cancer group included 11 males and 5 females. The mean age of the patients in the cancer group was 55.6 y, with a standard deviation (SD) of 14.0 y, and 5 of these patients were smokers, early stages I and II: 11; late stages III: 5. The normal control group of 20 patients included 8 males and 12 females. The mean age of individuals in the normal control group was 49.6 y, with an SD of 9.0 y.
Table 2. Demographic characteristics of the study subjects.
| Colon cancer | Rectal cancer | Colorectal cancer | Normal controls | |
|---|---|---|---|---|
| Subjects (n) | 8 | 8 | 16 | 20 |
| Age (mean ± SD) | 55.8 (14.7) | 55.4 (14.0) | 55.6 (14.0) | 49.6 (9.0) |
| Sex | ||||
| Male | 6 | 5 | 11 | 8 |
| Female | 2 | 3 | 5 | 12 |
| Smokers (n) | 3 | 2 | 5 | 7 |
Solid-phase microextraction (SPME) procedures
A manual SPME holder with carboxen/polydimethylsiloxane (CAR/PDMS) fibers of 75 μm in thickness was purchased from Supelco. The SPME fiber was inserted into the vial and exposed to the headspace of a blood sample (2 ml, taken from the ulnar vein) for 20 min at 40 °C. Subsequently, the desorption of volatiles occurred in a hot GC injector at 200 °C for 2 min.
GC/MS analyses
The analysis was performed on a GC/MS instrument (Shimadzu GC-MS QP 2010) equipped with a DB-5MS (length 30 min × inner diameter (ID) 0.250 mm × film thickness 0.25 µm) (Agilent Technologies) porous-layer open-tubular PLOT column. Injections were performed in the splitless mode with a splitless time of 1 min. The temperature of the injector was 200 °C. The flow rate of the helium (99.999%) carrier gas was maintained at a constant 2 ml min−1. The column temperature was held at 40 °C for 2 min to concentrate the hydrocarbons at the head of the column; subsequently, the column temperature was increased by 70 °C min−1 to 200 °C for 1 min, ramped by 20 °C min−1 to 230 °C, and held at 230 °C for 3 min. The MS analyses were performed in full scan mode, with a scan range of 35–200 amu. The ion source was maintained at 200 °C, and an ionization energy of 70 eV was used for each measurement.
The extraction and pretreatment of raw GC/MS data
Raw GC/MS data were converted into CDF-format (NetCDF) files by the Shimadzu GCMS Postrun Analysis software package and subsequently processed by the XCMS toolbox (http://metlin.scripps.edu/download/). The default XCMS parameters were utilized, with the following exceptions: xcmsSet (fwhm = 8, snthresh = 6, max = 200); retcor (method = “linear”, family = “gaussian”, plottype = “mdevden”); and a bandwidth of 8 for the first grouping command and of 4 for the second grouping command.41,42 The data set of aligned mass ions was exported from XCMS for further processing by Microsoft Excel, which was used to normalize the data prior to the multivariate analyses.
Statistical analyses
The normalized data were exported to SIMCA-P 11.5 for principal component analysis (PCA) to detect grouping trends and outliers. Partial least-squares discriminant analysis (PLSDA) was then performed, using the default seven-round cross-validation approach, and the corresponding variable importance in the projection (VIP) values in the PLSDA model were calculated. To prevent overfitting, permutation tests with 100 iterations were performed to validate the supervised model. In addition, the nonparametric Kruskal–Wallis rank sum test was performed to determine the significance of each metabolite. Potential metabolic biomarkers were selected based on VIP values and nonparametric P values, using thresholds of 1.0 and 0.01, respectively.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Financial support by grants from the National Natural Science Foundation of China (30972839), China Postdoctoral Science Foundation (2013M531069), Foundation of Heilongjiang Educational Committee (12531245), and Doctoral Fund of the First Affiliated Hospital of Harbin Medical University (2012B006) are gratefully acknowledged.
Glossary
Abbreviations:
- VOCs
volatile organic compounds
- PLSDA
partial least-squares discriminant analysis
- SPME-GC-MS
solid phase microextraction–chromatography–mass spectrometry
- PCA
principal component analysis
- CRC
colorectal cancer
- FOBT
fecal occult blood test
- CEA
carcinoembryonic antigen
- GC/MS
gas chromatography/mass spectrometry
- SD
standard deviation
- CAR/PDMS
carboxen/polydimethylsiloxane
- PCA
principal component analysis
- VIP
variable importance in the projection
- NIST
National Institute of Standards and Technology
- ROS
reactive oxygen species
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/26723
References
- 1.Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep. 2008;41:685–92. doi: 10.5483/BMBRep.2008.41.10.685. [DOI] [PubMed] [Google Scholar]
- 2.Bingham S, Riboli E. Diet and cancer--the European Prospective Investigation into Cancer and Nutrition. Nat Rev Cancer. 2004;4:206–15. doi: 10.1038/nrc1298. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 4.Montrose DC, Zhou XK, Kopelovich L, Yantiss RK, Karoly ED, Subbaramaiah K, Dannenberg AJ. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev Res (Phila) 2012;5:1358–67. doi: 10.1158/1940-6207.CAPR-12-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandel JS, Church TR, Bond JH, Ederer F, Geisser MS, Mongin SJ, Snover DC, Schuman LM. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000;343:1603–7. doi: 10.1056/NEJM200011303432203. [DOI] [PubMed] [Google Scholar]
- 6.Winawer SJ, Zauber AG, Ho MN, O’Brien MJ, Gottlieb LS, Sternberg SS, Waye JD, Schapiro M, Bond JH, Panish JF, et al. The National Polyp Study Workgroup Prevention of colorectal cancer by colonoscopic polypectomy. N Engl J Med. 1993;329:1977–81. doi: 10.1056/NEJM199312303292701. [DOI] [PubMed] [Google Scholar]
- 7.Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y, Ni Y, Zhao A, Cai S, Xu LX, et al. Urinary metabonomic study on colorectal cancer. J Proteome Res. 2010;9:1627–34. doi: 10.1021/pr901081y. [DOI] [PubMed] [Google Scholar]
- 8.Nishiumi S, Kobayashi T, Ikeda A, Yoshie T, Kibi M, Izumi Y, Okuno T, Hayashi N, Kawano S, Takenawa T, et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS One. 2012;7:e40459. doi: 10.1371/journal.pone.0040459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher RH. Carcinoembryonic antigen. Ann Intern Med. 1986;104:66–73. doi: 10.7326/0003-4819-104-1-66. [DOI] [PubMed] [Google Scholar]
- 10.Kronborg O, Fenger C, Olsen J, Jørgensen OD, Søndergaard O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348:1467–71. doi: 10.1016/S0140-6736(96)03430-7. [DOI] [PubMed] [Google Scholar]
- 11.Phillips M, Cataneo RN, Cummin AR, Gagliardi AJ, Gleeson K, Greenberg J, Maxfield RA, Rom WN. Detection of lung cancer with volatile markers in the breath. Chest. 2003;123:2115–23. doi: 10.1378/chest.123.6.2115. [DOI] [PubMed] [Google Scholar]
- 12.Gordon SM, Szidon JP, Krotoszynski BK, Gibbons RD, O’Neill HJ. Volatile organic compounds in exhaled air from patients with lung cancer. Clin Chem. 1985;31:1278–82. [PubMed] [Google Scholar]
- 13.Hietanen E, Bartsch H, Béréziat JC, Camus AM, McClinton S, Eremin O, Davidson L, Boyle P. Diet and oxidative stress in breast, colon and prostate cancer patients: a case-control study. Eur J Clin Nutr. 1994;48:575–86. [PubMed] [Google Scholar]
- 14.Phillips M, Cataneo RN, Ditkoff BA, Fisher P, Greenberg J, Gunawardena R, Kwon CS, Rahbari-Oskoui F, Wong C. Volatile markers of breast cancer in the breath. Breast J. 2003;9:184–91. doi: 10.1046/j.1524-4741.2003.09309.x. [DOI] [PubMed] [Google Scholar]
- 15.Altomare DF, Di Lena M, Porcelli F, Trizio L, Travaglio E, Tutino M, Dragonieri S, Memeo V, de Gennaro G. Exhaled volatile organic compounds identify patients with colorectal cancer. Br J Surg. 2013;100:144–50. doi: 10.1002/bjs.8942. [DOI] [PubMed] [Google Scholar]
- 16.Gordon SM, Wallace LA, Brinkman MC, Callahan PJ, Kenny DV. Volatile organic compounds as breath biomarkers for active and passive smoking. Environ Health Perspect. 2002;110:689–98. doi: 10.1289/ehp.02110689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469–79. doi: 10.1021/pr060594q. [DOI] [PubMed] [Google Scholar]
- 18.Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115–22. doi: 10.1021/ac0713510. [DOI] [PubMed] [Google Scholar]
- 19.Miekisch W, Schubert JK, Noeldge-Schomburg GF. Diagnostic potential of breath analysis--focus on volatile organic compounds. Clin Chim Acta. 2004;347:25–39. doi: 10.1016/j.cccn.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Pasikanti KK, Esuvaranathan K, Ho PC, Mahendran R, Kamaraj R, Wu QH, Chiong E, Chan EC. Noninvasive urinary metabonomic diagnosis of human bladder cancer. J Proteome Res. 2010;9:2988–95. doi: 10.1021/pr901173v. [DOI] [PubMed] [Google Scholar]
- 21.Chen JL, Fan J, Yan LS, Guo HQ, Xiong JJ, Ren Y, Hu JD. Urine Metabolite Profiling of Human Colorectal Cancer by Capillary Electrophoresis Mass Spectrometry Based on MRB. Gastroenterol Res Pract. 2012;2012:125890. doi: 10.1155/2012/125890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barash O, Peled N, Tisch U, Bunn PA, Jr., Hirsch FR, Haick H. Classification of lung cancer histology by gold nanoparticle sensors. Nanomedicine. 2012;8:580–9. doi: 10.1016/j.nano.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Xu F, Wang Y, Pan Y, Lu D, Wang P, Ying K, Chen E, Zhang W. A study of the volatile organic compounds exhaled by lung cancer cells in vitro for breath diagnosis. Cancer. 2007;110:835–44. doi: 10.1002/cncr.22844. [DOI] [PubMed] [Google Scholar]
- 24.Mal M, Koh PK, Cheah PY, Chan EC. Metabotyping of human colorectal cancer using two-dimensional gas chromatography mass spectrometry. Anal Bioanal Chem. 2012;403:483–93. doi: 10.1007/s00216-012-5870-5. [DOI] [PubMed] [Google Scholar]
- 25.Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, Chen JL. Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World J Gastroenterol. 2011;17:727–34. doi: 10.3748/wjg.v17.i6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevedomskaya E, Ramautar R, Derks R, Westbroek I, Zondag G, van der Pluijm I, Deelder AM, Mayboroda OA. CE-MS for metabolic profiling of volume-limited urine samples: application to accelerated aging TTD mice. J Proteome Res. 2010;9:4869–74. doi: 10.1021/pr100634d. [DOI] [PubMed] [Google Scholar]
- 27.Gall WE, Beebe K, Lawton KA, Adam KP, Mitchell MW, Nakhle PJ, Ryals JA, Milburn MV, Nannipieri M, Camastra S, et al. RISC Study Group alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng G, Hakim M, Broza YY, Billan S, Abdah-Bortnyak R, Kuten A, Tisch U, Haick H. Detection of lung, breast, colorectal, and prostate cancers from exhaled breath using a single array of nanosensors. Br J Cancer. 2010;103:542–51. doi: 10.1038/sj.bjc.6605810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight JA. Free radicals: their history and current status in aging and disease. Ann Clin Lab Sci. 1998;28:331–46. [PubMed] [Google Scholar]
- 30.Skrzydlewska E, Kozuszko B, Sulkowska M, Bogdan Z, Kozlowski M, Snarska J, Puchalski Z, Sulkowski S, Skrzydlewski Z. Antioxidant potential in esophageal, stomach and colorectal cancers. Hepatogastroenterology. 2003;50:126–31. [PubMed] [Google Scholar]
- 31.Terry P, Lagergren J, Ye W, Nyrén O, Wolk A. Antioxidants and cancers of the esophagus and gastric cardia. Int J Cancer. 2000;87:750–4. doi: 10.1002/1097-0215(20000901)87:5<750::AID-IJC19>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 32.Polat MF, Taysi S, Gul M, Cikman O, Yilmaz I, Bakan E, Erdogan F. Oxidant/antioxidant status in blood of patients with malignant breast tumour and benign breast disease. Cell Biochem Funct. 2002;20:327–31. doi: 10.1002/cbf.980. [DOI] [PubMed] [Google Scholar]
- 33.Choi MA, Kim BS, Yu R. Serum antioxidative vitamin levels and lipid peroxidation in gastric carcinoma patients. Cancer Lett. 1999;136:89–93. doi: 10.1016/S0304-3835(98)00312-7. [DOI] [PubMed] [Google Scholar]
- 34.Bakan E, Taysi S, Polat MF, Dalga S, Umudum Z, Bakan N, Gumus M. Nitric oxide levels and lipid peroxidation in plasma of patients with gastric cancer. Jpn J Clin Oncol. 2002;32:162–6. doi: 10.1093/jjco/hyf035. [DOI] [PubMed] [Google Scholar]
- 35.Abaffy T, Duncan R, Riemer DD, Tietje O, Elgart G, Milikowski C, DeFazio RA. Differential volatile signatures from skin, naevi and melanoma: a novel approach to detect a pathological process. PLoS One. 2010;5:e13813. doi: 10.1371/journal.pone.0013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 37.De Marzo AM, Marchi VL, Yang ES, Veeraswamy R, Lin X, Nelson WG. Abnormal regulation of DNA methyltransferase expression during colorectal carcinogenesis. Cancer Res. 1999;59:3855–60. [PubMed] [Google Scholar]
- 38.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–8. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, Grogan EL, Massion PP. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5:992–1006. doi: 10.1158/1940-6207.CAPR-11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Filipiak W, Sponring A, Mikoviny T, Ager C, Schubert J, Miekisch W, Amann A, Troppmair J. Release of volatile organic compounds (VOCs) from the lung cancer cell line CALU-1 in vitro. Cancer Cell Int. 2008;8:17. doi: 10.1186/1475-2867-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin HM, Edmunds SI, Helsby NA, Ferguson LR, Rowan DD. Nontargeted urinary metabolite profiling of a mouse model of Crohn’s disease. J Proteome Res. 2009;8:2045–57. doi: 10.1021/pr800999t. [DOI] [PubMed] [Google Scholar]
- 42.Gao X, Pujos-Guillot E, Martin JF, Galan P, Juste C, Jia W, Sebedio JL. Metabolite analysis of human fecal water by gas chromatography/mass spectrometry with ethyl chloroformate derivatization. Anal Biochem. 2009;393:163–75. doi: 10.1016/j.ab.2009.06.036. [DOI] [PubMed] [Google Scholar]