Abstract
Candida albicans is an opportunistic pathogen capable of causing life-threatening infections in immunocompromised individuals. Despite its significant health impact, our understanding of C. albicans pathogenicity is limited, particularly at the molecular level. One of the largely understudied enzyme families in C. albicans is small molecule AdoMet-dependent methyltransferases (smMTases), which are important for maintenance of cellular homeostasis by clearing toxic chemicals, generating novel cellular intermediates and regulating intra- and interspecies interactions. Putative smMTase orf19.633 has little homology to any known protein and was previously identified based on its ability to functionally complement a baker’s yeast crg1 mutant in response to protein phosphatase inhibitor cantharidin. In this study, we demonstrated that C. albicans Crg1 (CaCrg1) is a bona fide smMTase that interacts with the toxin in vitro and in vivo. We report that CaCrg1 is important for virulence-related processes such as adhesion, hyphal elongation and membrane trafficking in response to this toxin. Using biochemical and genetic analysis we also found that CaCrg1 plays a role in complex sphingolipid pathway: it binds to exogenous short-chain ceramides in vitro, it interacts genetically with genes of glucosylceramide pathway and the deletion of CaCRG1 leads to significant changes in the abundance of phytoceramides. Finally we found that this novel lipid-related smMTase is required for virulence in the waxmoth Galleria mellonella, a model of infection.
Keywords: Candida albicans, chemical biology, AdoMet-dependent methyltransferase, sphingolipid, cantharidin
INTRODUCTION
The fungus Candida albicans is a normally harmless commensal present in gastrointestinal tracts of the majority of humans, where it exists as a part of healthy microbiome. However, this fungus can cause life-threatening infections in immunocompromised individuals1, 2. Despite being a significant health concern, our current understanding of Candida ’s pathogenicity mechanisms is incomplete. According to the Candida Genome database (www.candidagenome.org) over 70% of C. albicans genes are annotated as uncharacterized, and much of the current characterization relies on homology to genes in the model yeast Saccharomyces cerevisiae.
One of the poorly studied enzyme families in C. albicans are S-adenosylmethionine (AdoMet)-dependent MTases. Small molecule MTases (smMTase) are of a particular interest, because they are involved in biotransformation of endogenous as well as exogenous small molecules (lipids, xenobiotics and secondary metabolites) and maintain cellular homeostasis by clearing toxic chemicals, generating cellular intermediates and regulating intra- and interspecies interactions3–6. Mutations in smMTases can lead to the intracellular accumulation of toxic substrates resulting in cellular dysfunction or altered drug response. Considering the importance of smMTases in response to small molecules, the characterization of these enzymes in C. albicans will enhance our understanding of the fungal drug response and may provide a starting point for the development of novel antifungal drugs. Despite their potential to illuminate basic and applied aspects of Candida growth, smMTase have been refractory to interrogation. The majority of these enzymes do not have an obvious phenotype in standard laboratory conditions and biochemical strategies designed for protein MTases7, 8 are not effective for smMTases because these tests rely on prior knowledge of substrates. Computational analysis can predict functions for smMTases3, 7, 9, yet experimental approaches are required to determine the cellular ligands of smMTases.
We previously used a chemical genetics approach in S. cerevisiae to identify an AdoMet-dependent MTase CRG1 as a gene dose-dependent interactor of cantharidin10, 11. Cantharidin is a secondary metabolite, produced by blister beetles of the Meloidae family. It functions as a precopulatory agent and was also suggested to act as a protection for beetle eggs12, 13. Humans have been also used this natural product as aphrodisiac (aka Spanish fly), a topical therapy for warts and tattoo removal, as well as for the treatment of hepatocellular carcinoma in traditional Chinese medicine14. Additionally, cantharidin analogues have being currently investigated for their applications in anticancer therapy15.
Although the primary targets of cantharidin are type I and type II protein phosphatases16, 17, we showed that in baker’s yeast cantharidin interacts with MTase Crg1 in vitro and Crg1 maintains lipidome homeostasis in response to the drug in non-pathogenic fungi11. In C. albicans putative MTase orf19.633 is also a gene-dose modulator of cantharidin response. At the primary sequence level ScCRG1 and CaCRG1 have a limited homology within their putative MTase domains (19.2% identity and 38.5% similarity), indicating that the function of orf19.633 in its response to the toxin could not be inferred solely from its sequence. Despite this evolutionary divergence, our functional tests show that CaCrg1 robustly rescues Saccharomyces crg1 deletion mutant. BLASTp analysis reveals that CaCRG1 shares homology with other human fungal pathogens genes with unknown functions: Candida dubliniensis (CD36_30360, 77.9% identity, 85.2% similarity), Candida tropicalis (CTRG_00537, 65.4% identity, 80.3% similarity), Candida parapsilosis (CPAR2_204610, 57.9% identity, 74.3% similarity), Candida orthopsilosis (CORT_0D04720, 57.8% identity, 74% similarity). Given the steady increase in non-albicans infections18 these observations suggest that the study of CaCrg1 can provide insight into these related pathogens.
In the present study, we used cantharidin as a small molecule probe to characterize the putative MTase CaCRG1 in C. albicans. Our biochemical and genetic analysis provides an evidence that CaCrg1 is a lipid-binding smMTase essential for cellular defense against chemical stress and for maintenance of virulence-related processes in the response to cantharidin. We also demonstrated that CaCRG1 is important for virulence of C. albicans in the waxworm G. mellonella.
RESULTS AND DISCUSSION
A Functional CaCrg1 is Important for Cantharidin Resistance
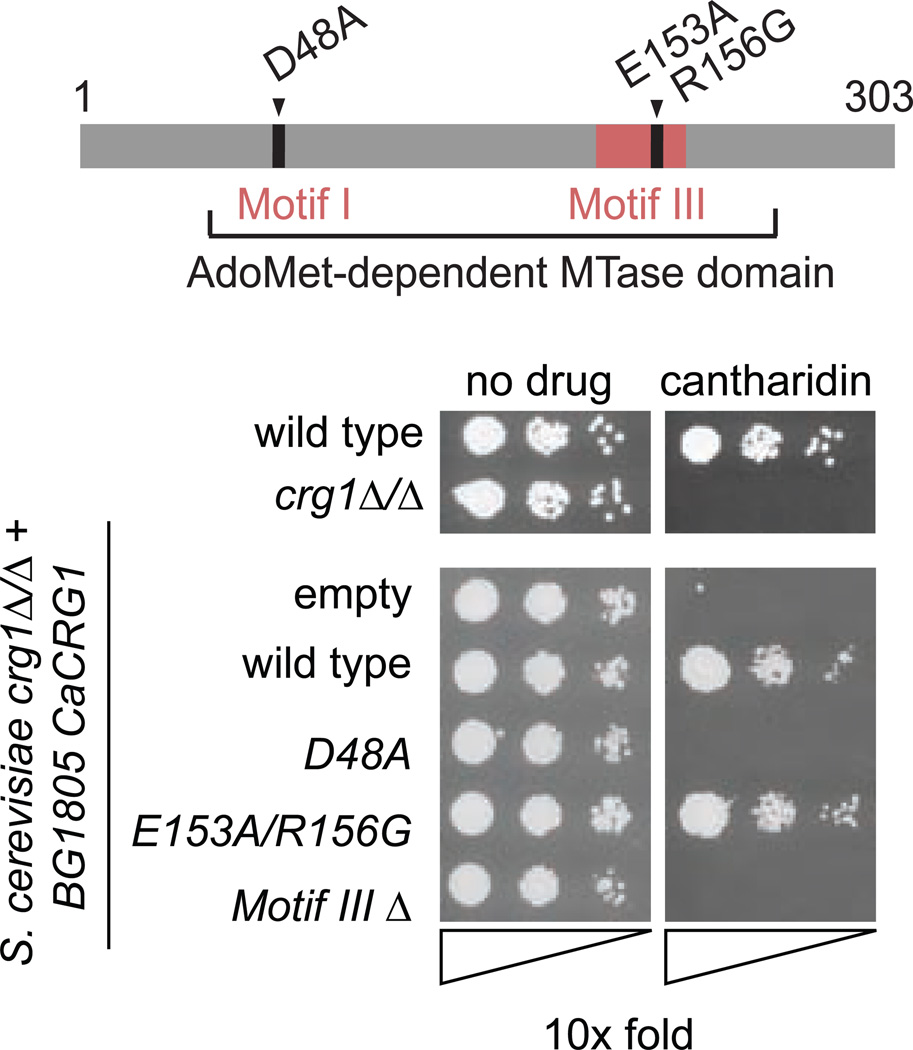
The aim of this work was to characterize a putative MTase orf19.633 (CaCRG1) in response to small molecule cantharidin and uncover biological functions for this enzyme. Orf19.633 (hereafter CaCrg1) was annotated as a putative MTase with diagnostic AdoMet-binding motifs10, 11. To test if it is a functional MTase, we synthesized a codon-optimized CaCRG1 sequence (Bio Basic Inc) and expressed it via a galactose-inducible promoter from a plasmid in S. cerevisiae (Supplementary Figure 1A). The synthesized gene was further used as a template to produce mutant alleles (D48A, E153A-R156G, and motif IIIΔ), using S. cerevisiae crg1Δ/Δ null mutant as the expression host. Galactose-induced overexpression of wt CaCRG1 completely rescued crg1Δ/Δ sensitivity to cantharidin whereas the mutant alleles (D48A and Motif IIIΔ) failed to confer cantharidin resistance (Figure 1). The failure to complement was not due to reduced expression of the mutated CaCrg1 proteins (Supplementary Figure 1B), indicating that the MTase domain of CaCrg1 is both necessary and sufficient for cellular survival in the presence of cantharidin.
Figure 1. A functional MTase domain of CaCrg1 is required for cantharidin resistance.
The diagram of MTase domain with the point mutations and the deletion of Motif III (top). Growth of S. cerevisiae wt and crg1Δ cells overexpressing empty vector BG1805, wt and mutated CaCRG1 alleles (D48A, E153A/R156G, and Motif IIIΔ) in the presence of cantharidin (80 µM) (bottom).
Cantharidin is Methylated by CaCrg1
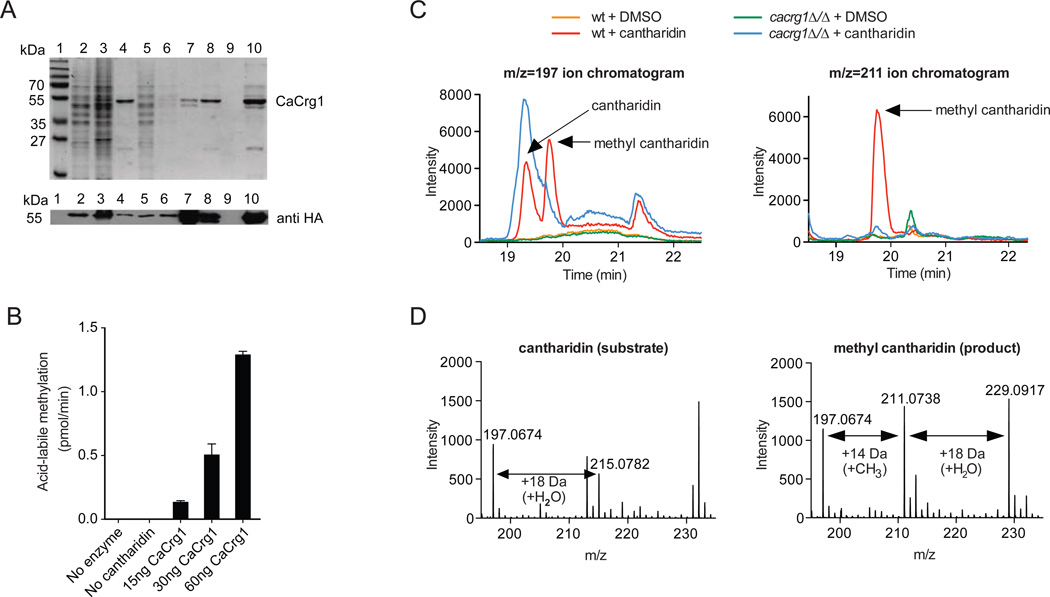
Because CaCrg1 is required for cantharidin resistance, we tested if CaCrg1 catalyzes a methylation reaction on cantharidin similar to that of ScCrg110, 11. We purified the Candida enzyme expressed in baker’s yeast (Figure 2A) and found that an acid-hydrolyzed reaction mixture of the purified CaCrg1, cantharidin and S-adenosyl-[methyl-14C]-L-methionine results in the formation of volatile radioactive methyl ester (as methanol) (Figure 2B). This activity was dependent on the presence of both the protein and cantharidin (Supplementary Figure 2) demonstrating that CaCrg1 is a functional MTase methylating cantharidin in vitro.
Figure 2. CaCrg1 is a small molecule MTase.
A. Coomassie-stained 12% SDS-PAGE of purified His-tagged CaCrg1. lane 1, molecular weight standards; lane 2, soluble cell extract; lane 3, insoluble fraction; lane 4, Ni2+ Sepharose beads after wash 1; lane 5, unbound to beads cell extract; lane 6, wash 1; lane 7, beads after three washes; lane 8, non-concentrated elute; lane 9, flow-through; lane 10, concentrated and desalted elute. The expression of CaCrg1 was assessed with a mouse monoclonal anti-HA antibody (bottom).
B. CaCrg1 shows robust MTase activity with cantharidin as the substrate in vitro. The reactions containing varying amounts of CaCrg1 enzyme and cantharidin were tested on production acid-labile methylated ester. The error bars represent the standard deviation of two separate experiments each performed in duplicate.
C. CaCrg1 is required for a formation of methyl cantharidin in vivo. Wt and cacrg1Δ/Δ cells were cultured in the presence and absence of cantharidin before extraction of intracellular metabolites and analysis by LC-MS/MS. Single-ion chromatograms of various cellular extracts are shown for the mass ranges corresponding to cantharidin (m/z=197±100 ppm) (left panel) and methyl cantharidin (m/z=211±100 ppm) (right panel). Arrows mark the elution patterns for cantharidin and methyl cantharidin.
D. Averaged spectra of the cantharidin (left panel) and methyl cantharidin (right panel). Chromatographic peaks from the cantharidin-treated wt are shown and ions of interest are indicated.
To determine if CaCrg1 is required for in vivo methylation of cantharidin , we investigated the metabolism of cantharidin in wt and a cacrg1Δ/Δ mutant. Mid-exponentially grown cells were treated with cantharidin (100 µM) or DMSO for 90 min. Intracellular metabolites were rapidly extracted and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). In the m/z=197 single-ion chromatogram, we observed the peak corresponding to cantharidin (m/z=197) in wt and cacrg1Δ/Δ cells grown in the presence of the drug (Figure 2C, left panel). These chromatographic peaks eluting at 19.4 min with m/z ratios matching cantharidin were absent in cells treated only with DMSO. Next, we examined the m/z=211 single-ion chromatogram, which corresponds to the mass range of methyl cantharidin (m/z=211) (Figure 2C, right panel). We observed a large peak eluting at 19.9 min in wt cells treated with cantharidin in the mass range matching methylated cantharidin. In contrast, no peak in this mass range was observed in cantharidin-treated cacrg1Δ/Δ cells or in cells treated with DMSO alone. The spectra of the 19.4-min cantharidin peak in the drug-treated wt cells corresponded to cantharidin (m/z=197) and cantharidin water adduct or hydrated cantharidin derivative (m/z=215) (Figure 2D, left panel). When we analyzed the spectra of the CaCrg1-dependent 19.9-min methyl cantharidin peak in the drug-treated wt, we saw ions corresponding to methyl cantharidin (m/z=211), hydrated methyl cantharidin (m/z=229), as well as unmodified cantharidin (m/z=197), a possible product of in-source fragmentation (Figure 2D, right panel). Our findings indicate that cantharidin is methylated in vivo in C. albicans, and that CaCrg1 is the small molecule AdoMet-dependent MTase responsible for this activity.
CaCRG1 is Important for Candida Morphogenesis in Response to the Drug
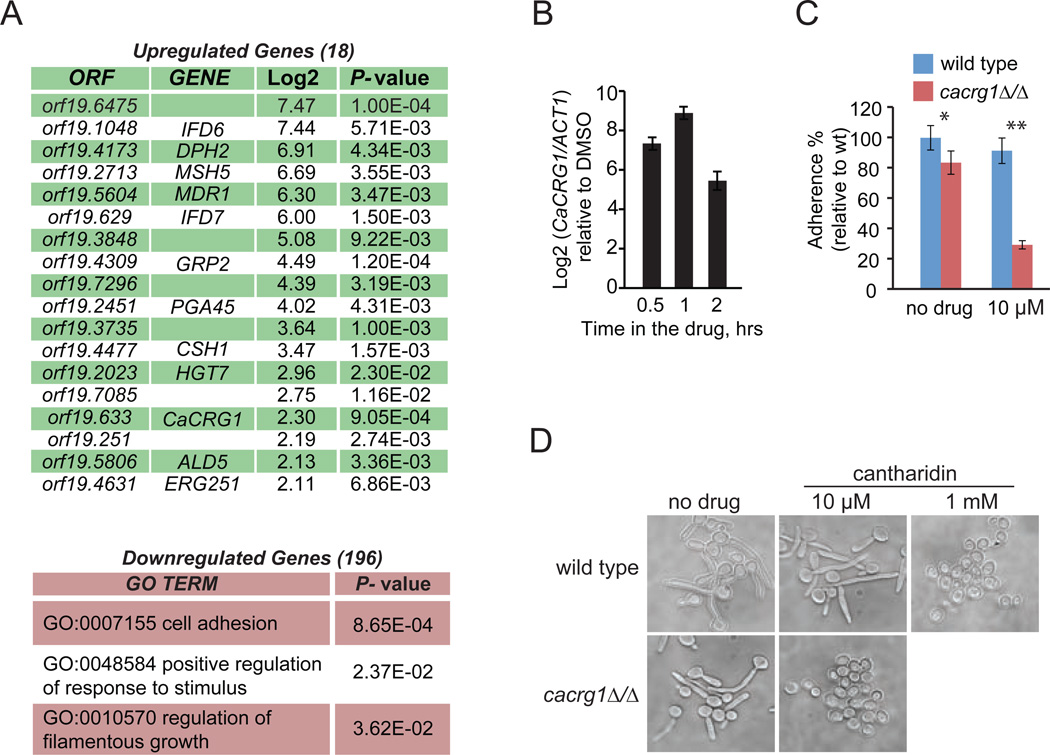
Despite its long history of use, the antifungal activity of cantharidin has not been characterized in detail19. To define its molecular mechanism in C. albicans, we profiled its transcriptional response using Affymetrix gene expression arrays (stCANDIDA 1a). Exponentially grown cells were treated with cantharidin at its IC50 (2 mM) in YPD and with DMSO for 30 min. Analysis of the transcriptome revealed 235 differentially expressed genes (log2 (cantharidin/DMSO)<|2|, P-value <0.05; Figure 3A and Supplementary Table 1), 91% of which were downregulated. These genes were significantly enriched in the following Gene Ontology (GO) term processes: “cell adhesion” (P-value <8.65×10−4), “positive regulation of response to stimulus” (P-value <2.37×10−2), and “regulation of filamentous growth” (P-value <3.6×10−2). CaCRG1 was among the significantly upregulated genes (log2 >2.3, P-value <9.0×10−4), and qRT-PCR analysis confirmed that the relative abundance of CaCRG1 transcript increases in the response to cantharidin in a time-dependent manner (Figure 3B). Because, cantharidin is a potent protein phosphatase inhibitor17, it likely perturbs gene expression by interfering with phosphorylation-dependent signaling events in a cell.
Figure 3. CaCRG1 is important for cantharidin-perturbed morphogenesis and membrane trafficking in C. albicans.
A. Transcriptional profile of wt grown in the presence of cantharidin. GO Biological Term Enrichment was applied to significantly (P-value<0.05) downregulated genes (log2 (drug/DMSO) >|2|).
B. qRT-PCR analysis demonstrates that CaCRG1 is a cantharidin-responsive gene. Data are means of at least three independent experimental replicates, and error bars are SD.
C. cacrg1Δ/Δ has reduced adherence to plastic surface in the presence and absence of cantharidin. *P-value <0.05, ** <0.01.
D. cacrg1Δ/Δ fails to form hyphae in the presence of cantharidin.
Because cantharidin treatment leads to downregulation of genes involved in two phenotypes that are directly related to C. albicans virulence (adhesion and filamentation)20, we assessed these phenotypes in cacrg1Δ/Δ mutant in the presence of the drug. cacrg1Δ/Δ showed reduced adherence to plastic (P-value <1.0×10−3; Figure 3C and Supplementary Figure 3) and completely failed to adhere to plastic in the presence of non-growth inhibitory doses of cantharidin (10µM) (P-value <8.0×10−10). The mutant also failed to germinate when exposed to a non-growth inhibitory dose (10 µM) (Figure 3D), whereas under standard conditions (without the drug at 37°C) cacrg1Δ/Δ underwent hyphal elongation similarly to wt. These results indicate that cantharidin treatment affects germination of fungi and CaCrg1 is required to maintain fungal morphogenesis in response to the drug.
CaCrg1 Maintains Membrane Trafficking during Cantharidin Exposure
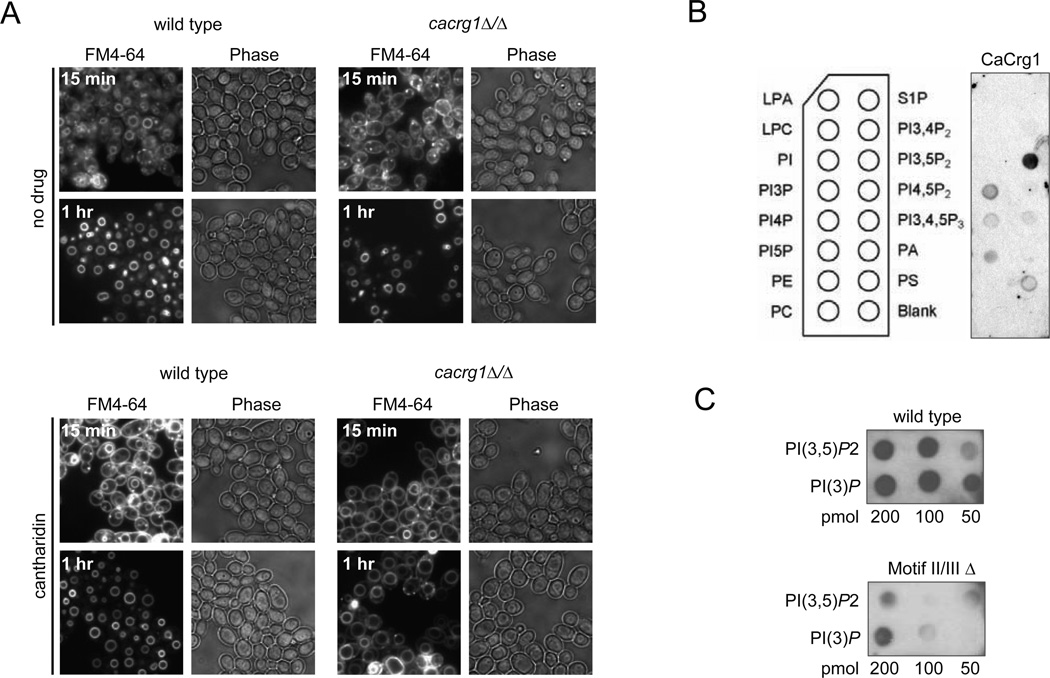
To better understand how response to cantharidin is manifested on a cellular level we examined C. albicans wt and cacrg1Δ/Δ cells microscopically. Because we previously found that cantharidin treatment affected the formation of actin patches11 (the sites of endocytosis in S. cerevisiae crg1Δ/Δ mutant), we tested if endocytosis is perturbed by the drug in C. albicans. Using the lipophilic styryl dye FM 4–64 to follow the dynamics of membrane internalization and transport via endosomal intermediates to the vacuole21, we found that cantharidin interferes with endocytosis or membrane trafficking in cacrg1Δ/Δ mutant (Figure 4A). After 15 min of cantharidin treatment (250 µM), both wt and the mutant demonstrated brightly stained plasma membrane and vacuolar membranes. Within 60 min of the drug exposure, wt had exclusively vacuolar membrane staining (Figure 4A) whereas in cacrg1Δ/Δ mutants the plasma membrane staining remained as small puncta, and vacuoles were enlarged. Because endosome system is essential for trafficking of membrane components (e.g. lipids) and is a point of sorting cargo either for degradation or recycling it back to plasma membrane, our observations suggest that CaCrg1 is important for membrane trafficking in response to cantharidin. Consistent with this finding, CaCrg1 also preferentially binds in vitro to established biomarkers of early and late endosomes (Supplementary Figure 4A), the membrane phosphoinositides phosphatidylinositol phosphate PI(3)P and phosphatidylinositol bisphosphate PI(3,5)P222, 23, and the binding was dependent on the MTase domain of CaCrg1 (Figure 4B and 4C). In the baker’s yeast we found that in addition to its cytoplasmic distribution GFP-tagged ScCrg1 is co-localized with the vacuolar membrane (Supplementary Figure 4B). Taken together, our findings suggest that CaCrg1 is involved in fungal morphogenesis and the concomitant changes in membrane trafficking that occur during this shift.
Figure 4. CaCrg1 maintains membrane trafficking during cantharidin exposure.
A. Visualization of endosome dynamics in wt and cacrg1Δ/Δ after 15 min and 60 min in the presence and absence of cantharidin.
B. Lipid-protein overlay assay of CaCrg1. Lipids: lysophosphatidic acid (LPA), lysophosphocholine (LPC), phosphatidylinositol (PtdIns), PtdIns phosphate (PI(n)P), phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine-1-phosphate (S1P), phosphatidic acid (PA), phosphatidylserine (PS).
C. Validation of the lipid-overlay experiment with PI(3)P and PI(3,5)P 2.
Affinity-purified CaCrg1 Binds Exogenous Short-Chain Ceramides in Vitro
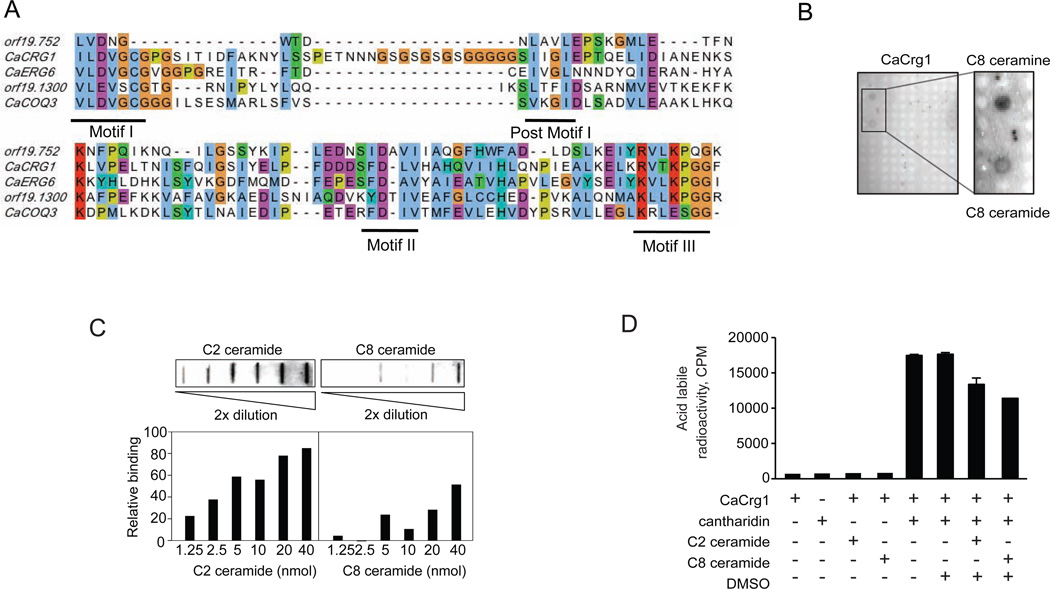
In its MTase domain CaCrg1 demonstrates limited similarity to small molecule MTases, such as orf19.300 (27.2% identity, 54.3% similarity), orf19.752 (28% identity, 46.6% similarity), CaCOQ3 (27.3% identity, 51.2% similarity), CaERG6 (28% identity, 44.1% similarity) (Figure 5A). Having established that CaCrg, a small molecule MTase, that binds to membrane lipids and it has partial homology to lipid-related MTases we tested whether CaCrg1 interacts with bioactive lipids. To address this, we prepared a microarray comprising 195 bioactive lipids spotted on a nitrocellulose-coated slide (see Supplementary Table 2) and assessed binding of CaCrg1 by a lipid-overlay assay. We found that purified CaCrg1 binds specifically to C8-ceramide (N-octanoylsphingosine) and its analogue C8-ceramine (N-octylsphingosine) (Figure 5B and 5C). CaCrg1 also binds to C2-ceramide and it does not bind to C16-ceramide (N-palmitoylsphingosine) and other sphingolipid species (Supplementary Figure 5A and 5B). Although, the observed binding of CaCrg1 to ceramides is of particular interest (because these bioactive molecules are involved in stress response, cell growth, senescence, apoptosis, and autophagy24), these results must be interpreted with caution because endogenous short-chain ceramides (C2, C6 and C8) are not found in yeast. As such, the biological relevance of observed binding of CaCrg1 to these molecules in vitro remains to be determined. We also observed that co-incubation of CaCrg1with C2- and C8-ceramides and cantharidin resulted in significant reduction in the formation of methyl esters in acid-labile methylation assay (Figure 5D). This suggests that, in vitro, short-chain ceramides may compete with cantharidin for the binding site as a substrate. In our mass spectrometry analysis, however, we did not detect the formation of methyl derivatives of ceramides, indicating that in the tested conditions short chain ceramides are not substrates of CaCrg1.
Figure 5. CaCrg1 binds short-chain ceramides in vitro.
A. Alignment of protein sequences of CaCrg1 MTase domain and its closest homologues. The protein sequences are aligned using the MUSCLE software with EMBL-EBI Alignment program. Conserved motifs in the MTase domain are underlined.
B. CaCrg1 binds to C8-ceramide and C8-ceramine molecules in vitro.
C. Lipid-CaCrg1 overlay assay with biologically active ceramide analogs (C2- and C8-ceramides spotted on the nitrocellulose membrane. Quantification of relative binding of CaCrg1 to ceramides was performed with ImageJ software.
D. The addition of ceramides decreases acid-labile methylation of cantharidin by CaCrg1 in vitro.
CaCRG1 Interacts with Genes of Sphingolipid Biosynthesis Pathway
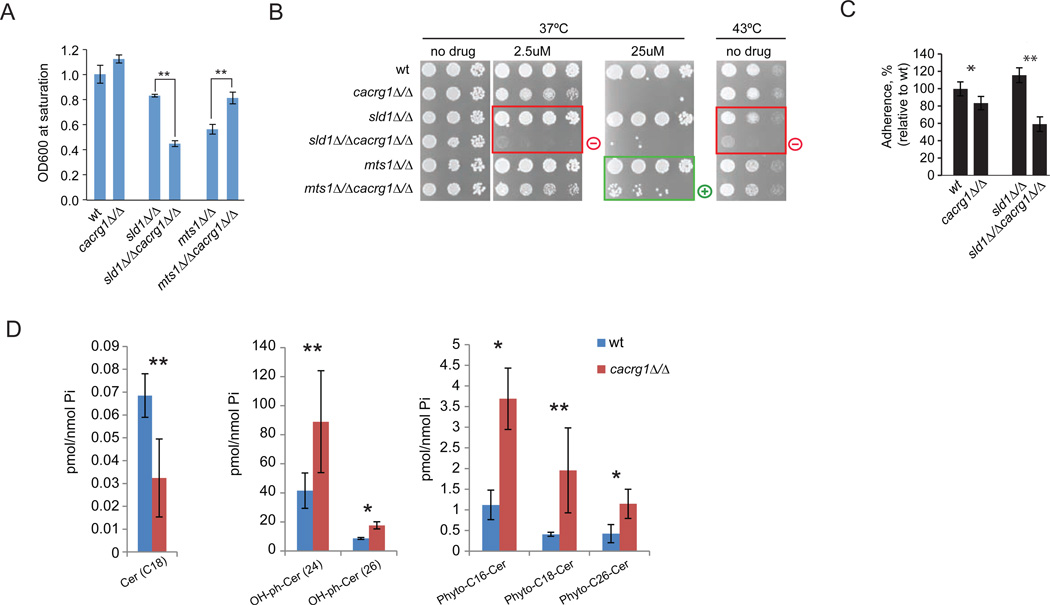
Ceramides serve as both bioactive molecules and as structural elements required for the biosynthesis of complex sphingolipids (e.g. glucosylceramides (GlcCer))24. Complex sphingolipids, generated by an addition of sugar group to otherwise cytotoxic ceramides, have been recognized as participatory in host-pathogen interactions25–28. Phylogenetic analysis also suggested that CaCrg1 is a putative GlcCer MTase29. To investigate biological significance of the observation that CaCrg1 interacts with ceramides in vitro and to test if CaCRG1 is related to complex sphingolipid biosynthesis, we assessed genetic interactions between CaCRG1 and GlcCer genes by constructing isogenic double deletion mutants in C. albicans with a cacrg11Δ/Δ strain. Considering that the occurrence of a genetic interaction between two genes is extremely rare event (~0.5%), genetic analysis performed by assessing fitness of constructed double deletion mutations is a powerful method to investigate gene function as it was successfully demonstrated in baker’s yeast. For example, if fitness of the double mutant is worse than the combination of the corresponding single mutants (known as synthetic sick or lethal), the genes are likely to act in overlapping pathways or in the same essential pathway30. In contrast, positive or suppressive interactions between two genes characterized by an enhanced fitness of a double mutant compared to singles suggest that these genes are likely to associate physically or act in the same pathway.
The fitness of the generated double mutants was analyzed at elevated temperatures and in the presence of cantharidin. Both cacrg1Δ/Δsld1Δ/Δ and cacrg1Δ/Δmts1Δ/Δ double mutants showed drastically altered fitness when grown in liquid medium at 39°C compared to the corresponding single mutants and wt (Figure 6A). Specifically, the double homozygous deletion strain cacrg1Δ/Δsld1Δ/Δ is synthetically sick (negative genetic interaction) at 39°C in liquid media and at 43°C on solid SD media. cacrg1Δ/Δmts1Δ/Δ mutant showed alleviating (positive) interactions at 39°C and in the presence of cantharidin (Figure 6A and 6B). Because neither of these genes were sensitive as single deletion mutants, the effect is specific to the double mutant combination. CaCRG1 also had a negative genetic interaction with HSX11 and HET1 in the presence of cantharidin (50 µM) at 30°C (Supplementary Figure 6A). Additionally, we observed synthetic lethality in cacrg1Δ/Δsld1Δ/Δ mutant grown at 37°C and cantharidin (2.5 µM), and an enhanced fitness of cacrg1Δ/Δmts1Δ/Δ mutant at 37°C and cantharidin (25 µM) compared to the single mutants. The observed genetic interactions detected between CaCrg1 and sphingolipid-modifying enzymes in the presence of the compound may reflect a critical role of sphingolipids in sustaining the barrier function of the plasma membrane towards compounds such as cantharidin.
Figure 6. CaCrg1 is important for sphingolipid biosynthesis.
A. Fitness of double deletion mutants in liquid SC media at 39°C.
B. CaCRG1 interacts with GlcCer genes in a condition-dependent manner. The unexpected phenotypes for double mutants (relative to wt and crg1Δ/Δ mutant) are highlighted: “+” denotes positive genetic interactions, “-“ denotes negative genetic interaction.
C. Adherence of cacrg1Δ/Δsld1Δ/Δ to abiotic surface.
D. Abundance of ceramide-related species in cacrg1Δ/Δ and wt. Statistically significant difference in the abundance (Student’s t-test) is shown in red. Error bars are standard deviation. *P-value<0.05, **P-value<0.1, Student’s t-test.
Phenotypically, cacrg1Δ/Δsld1Δ/Δ had significantly reduced adherence to plastic (Figure 6C), and had a drastically different morphological appearance compared to wt and the corresponding single deletion mutants (Supplementary Figure 6B). The observation of negative and positive genetic interactions between CaCRG1 and GlcCer genes (e.g. sphingolipid delta-8 desaturase SLD1 ) detected via measuring growth fitness and other phenotypes, such as adhesion and colony morphology, of the double mutant cacrg1Δ/ΔsldΔ/Δ suggests that CaCRG1 is required to buffer the absence of these genes in the stress conditions. Thus, CaCRG1 may act in a parallel pathway to GlcCer pathway. The positive or suppressive interaction observed between CaCRG1 and MTS1 indicates that the product of Mts1 may be toxic in the absence of CaCrg1 (e.g. membrane integrity is compromised), and therefore, the absence of both MTS1 and CaCRG1 results in an increased fitness of a mutant.
Deletion of CaCRG1 Results in the Accumulation of Phytoceramides
To further investigate the effect of deletion of CaCRG1 on the levels of sphingolipids, we measured the abundance of ceramides, GlcCer and precursors for inositol-containing sphingolipids in wt and cacrg1 deletion mutants. The deletion of CaCRG1 resulted in a significant increase in OH-ceramides and phytoceramides compared to the levels in wt (Figure 6D): OH-phytoCer (26) (P-value<0.04), OH-phytoCer (24) (P-value< 0.1), phyto-C16-Cer (P-value<0.05), phyto-C18-Cer (P-value< 0.06) and phyto-C26-Cer (P-value< 0.04). This accumulation of specific precursors of inositol-containing sphingolipid species in the mutant indicates that CaCrg1 functions in this branch of the complex sphingolipid pathway for the generation of GlcCer. Furthermore, these results are consistent with our genetic interaction analysis which demonstrated that CaCRG1 may act in either parallel or overlapping pathways with GlcCer biosynthesis. For example, CaCrg1 may play a role either in a conversion of phytoceramides to complex inositol-containing sphingolipids (IPC and MIPC) by methylating specific phytoceramides or in a negative regulation of the breakdown of the complex inositol-containing sphingolipids (via a salvage pathway). Combined, these data show that CaCrg1, along with these other gene products is important in complex sphingolipid biosynthesis in vivo. The mechanistic relationships between these pathway components will require detailed follow-up studies.
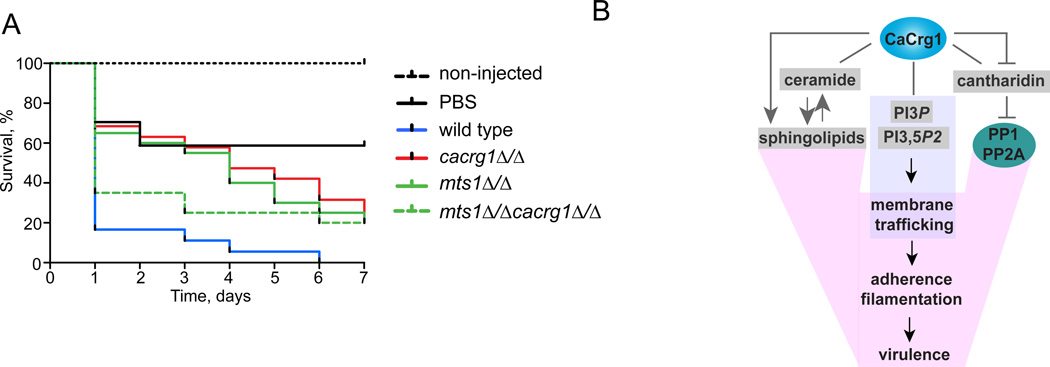
CaCrg1 is Important for C. albicans Virulence in a Galleria mellonella
Sphingolipid biosynthesis has been implicated in the virulence of pathogenic fungi25–28. Therefore, to test the role of CaCrg1 in the pathogenicity of C. albicans, we examined the effect of deletion of CaCRG1 on infectivity of the greater wax moth G. mellonella, an established invertebrate model of infection31, 32. At least 16 larvae were used for each treatment and controls using a single blind design. Each larvae was injected with 5×105 stationary phase cells, incubated at 37°C and assessed for viability every 24 hrs. We found that mtsΔ/Δ has decreased virulence in the infected waxmoth larvae (Figure 7A), in accordance with the previous infection experiments performed in mice28. A survival analysis of the infected larvae revealed that the deletion of CaCRG1 also significantly attenuated the virulence of C. albicans compared to wt injected larvae (P- value <0.0001, log-rank test). We also found that cacrg1Δ/Δmts1Δ/Δ has increased virulence relative to the single mutants suggesting that the condition-dependent positive genetic interactions we observed between CaCRG1 and MTS1 in vitro can be recapitulated in the infection model. Our findings demonstrate that CaCrg1 plays a role in host-pathogen interactions. One plausible explanation of these observations is that CaCrg1 is important for fungal virulence via the regulation of the levels of phytoceramides, yet additional supporting experimental evidence would be required to make this conclusion. Furthermore, as-yet-unidentified endogenous substrates of CaCrg1 are likely involved in pathogenesis and may be revealed under these conditions.
Figure 7. CaCrg1 is important for C. albicans virulence in Galleria mellonella.
A. Kaplan-Meier survival plot demonstrating that the deletion of CaCRG1 results in increased survival (relative to wt) of G. mellonella larvae injected with C. albicans.
B. A model demonstrating how CaCrg1 and it functional interactions play a role in drug response and fungal virulence . CaCrg1 interacts with toxic cantharidin that inhibits protein phosphatases involved in multiple biological processes. Upon the drug exposure CaCRG1 maintains membrane trafficking, adhesion and hyphal elongation, the processes required for fungal virulence.
In summary, we demonstrated that C. albicans CaCrg1 is a bona fide smMTase that interacts with the cytotoxic cantharidin in vitro and in vivo, and other lipid molecules contributing to its biological role (Figure 7B) . We found that CaCrg1 is important for virulence-related processes such as adhesion, hyphal elongation and membrane trafficking in the response to cantharidin. CaCrg1 is related to complex sphingolipid biosynthesis: it binds to exogenous short-chain ceramides in vitro, it interacts genetically with genes of the GlcCer pathway and the deletion of CaCRG1 leads to significant changes in the abundance of OH-ceramides and phytoceramides required for the biosynthesis of complex sphingolipids. Finally we found that this novel lipid-related smMTase is required for virulence in the waxmoth Galleria mellonella, model of infection.
METHODS
Strains and Growth Conditions
Yeast strains and plasmids used in this study are described in Supplementary Table 3 and 4, respectively. Cantharidin from Sigma Aldrich was dissolved in DMSO and stored at −20°C. Ceramides (N-acetylshphingosine, N-octanoylsphingosine, N-palmitoylsphingosine) were from Avanti Polar Lipids, Inc., dissolved in DMSO or ethanol. Cells analyzed by spot dilutions were normalized to an equal OD600, 10-fold diluted, spotted onto solid media and incubated at 30°C for 2 days.
Microarray Analysis
Cells grown to mid-exponential phase in YPD were incubated with cantharidin (2 mM) for 30 min and harvested by centrifugation. Isolation of RNA and hybridization to the microarrays was performed as described11. Three independent replicates were used for the analyses. Hybridization to Affymetrix custom expression array (stCANDIDA 1a) (Affymetrix) was followed by the extraction of intensity values for the probes using the GeneChip Operating Software (Affymetrix). The resulting files containing probe position and intensities were further analyzed by aligning the probes that match the position of the Candida Genome Database list of defined ORFs. Quantile normalized datasets were further analyzed (Supplementary Table 1). The significance for a differential expression was set as log2 (drug/DMSO) >|2|, P-value <0.05 as determined by Student’s t test. Significantly up- and downregulated transcripts were further tested for Gene Ontology (GO) Biological process term enrichment using AmiGo (http://amigo.geneontology.org) with P-value cutoff of 0.05 and multiple testing corrections (Bonferroni).
Cloning and Purification of CaCrg1 Fusion Protein
The sequence of CaCRG1 was optimized for expression in S. cerevisiae and synthesized with sequences for restriction enzyme digestion sites BsrGI in the universal vector pUC57. The synthesized CaCRG1 was cut out with BsrGI, SAP-treated and co-transformed with BsrGI-linearized BG1805 vector into a S. cerevisiae crg1Δ/Δ mutant. CaCRG1 was cloned downstream of a GAL1 inducible promoter and in frame with a triple affinity tag at C-terminal (His6-HAepitope-3Cprotease site-ZZprotein A). Transformants selected in SD media lacking uracil (SD-Ura) were screened by PCR and for cantharidin resistance. Clones were sequence-verified. To express CaCrg1, cells were grown to mid-exponential phase in SD-Ura containing 2% raffinose then induced with 2% galactose. Cells were harvested after overnight induction, and CaCrg1 expression was verified by with anti-HA antibodies. Induction and purification of CaCrg1 was performed as described previously11.
Site-Directed Mutagenesis
CaCRG1 missense and deletion mutants were prepared using the Phusion Site-directed mutagenesis kit (Finnzymes – Thermo Fisher Scientific) with the primers listed in Supplementary Table 5. Clones were sequence-verified. To express mutated CaCrg1, transformants were grown to mid-exponential phase in SD-Ura and 2% raffinose, and induced by the addition of 2% galactose. Cantharidin (30 µM) was used to test sensitivity of mutants. Cells were harvested after 3 hrs of induction, and CaCrg1 expression was verified with anti-HA antibodies.
Metabolomic Profiling of C. albicans Cellular Extracts
Wt and cacrgΔ/Δ mutants were cultured in the presence of cantharidin, and cellular extracts were prepared for metabolomic analysis by mass spectrometry based on methods described previously11. Briefly, cells were cultured in SC medium overnight at 30 °C. Mid-exponential cells were treated with cantharidin (100 µM) or DMSO alone (1%). After 90 min of growth at 30 °C, cells were rapidly isolated onto 45-mm diameter Millipore nylon filter membranes (0.45-µm pore size) via vacuum filtration. The filter was then transferred to a petri dish containing 800 µL 80:20 acetonitrile:H2O and the dish was incubated at 4°C for 15 min before the extract was transferred to a tube. The filters were washed again with 200 µl of extraction buffer. The extract was centrifuged at 20,800 rcf for 5 min and the supernatant was isolated. The pellet was re-extracted with 200 µL of extraction buffer and incubated at 4 °C for 15 min. After centrifugation at 20,800 rcf for 5 min, the supernatants from both extraction steps were pooled, neutralized with 120 µL 15% ammonium bicarbonate, dried by vacuum centrifugation, and frozen. The samples were resuspended in 100 µL of H2O before analysis by LC-MS/MS using methods that we described previously11.
In Vitro Methylation Reactions
Reaction mixtures containing 0.2 mM cantharidin (prepared as a stock solution of 10 mM cantharidin in DMSO) and 20 µM S-adenosyl-[methyl-14C]-L-methionine (48.8 mCi/mmol; PerkinElmer Inc.) in a buffer of 0.1 M sodium phosphate, pH 7.4, were mixed with either 0.015 µg, 0.03 µg, or 0.06 µg of recombinant C. albicans CaCrg1 protein in a final volume of 50 µL. Control reactions were performed in the absence of protein (no enzyme) or in the absence of cantharidin (DMSO solvent alone) with 0.09 µg of the CaCrg1 protein. Samples were incubated for 120 min at 30 °C and the reaction quenched by the addition of 40 µL of 2 M HCl. Methylation of cantharidin was determined by acid-labile volatility as described previously11. A portion of the quenched reaction mixture (80 µL) was spotted on a filter paper which was then placed in a scintillation vial containing 5 mL of Safety-Solve cocktail (Research Products International) and incubated for 4 hrs at room temperature. Radioactivity released as 14C-methanol was measured by counting the vial after removal of the paper.
Lipid-Protein Overlay Assay
The Screen-Well Bioactive lipid library containing 195 bioactive lipids (Supplementary Table 2) were obtained from Enzo Life Sciences, Inc via Cedarlane Laboratories. Lipids were spotted onto FAST glass slides covered with nitrocellulose polymer (Whatman Ltd, GE Healthcare) and the binding between CaCrg1 and lipids was analyzed by standard lipid-overlay assay. Briefly, lipids dissolved in chloroform/methanol/water (1:2:0.8) were spotted on PVDF membrane. Dried membranes were blocked for 1 hr in 3% fatty acid-free BSA in TBST (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 0.1% Tween20). The arrays were incubated with affinity purified HA-tagged CaCrg1 (2 µg mL−1) overnight at 4°C with gentle stirring. The membrane was rigorously washed six times for 30 min in TBST, incubated with mouse anti-HA monoclonal antibody for 1 hr, washed again as before, incubated with anti-mouse-horseradish peroxidase conjugate. Finally, the membrane was washed 12 times for 1 hr in TBST, and the membrane-bound HA-fusion CaCrg1 was detected by ECL.
FM4–64 Labeling for Vacuolar Membrane Dynamics
Wt and cacrg1Δ/Δ cells were grown overnight in SC. Cells grown to mid-exponential phase in YPD a 30°C were concentrated to OD600 of 20, and stained with lipophilic dye FM4–64 (40 µM) for 45 min at 25°C. Cells were washed twice and resuspended in 200 µL YPD. Cells were treated with 250 µM cantharidin and incubated at 30°C for 1 hr with shaking. Cells were observed after 15 min and 1 hr of cantharidin treatment with 63x objective, and fluorescence images (Cy3 filter) were acquired using AxioVision software on an Axiovert 200M fluorescence microscope (Zeiss).
C. albicans Adhesion Assay
Wt and cacrg11Δ/Δ cells grown in YPD at 30°C overnight were washed with PBS pH 7.4 two times. Cells were inoculated into SC media to final OD600 of 0.5. After 2 hr incubation at 37°C, non-adherent cells were removed by three washes with PBS pH 7.4. Adherent cells stained with 0.1% crystal violet for 5 min, then washed with PBS three times, 0.25% SDS one time, PBS two times. To resolubilize crystal violet, 150 µL isopropanol-0.04N HCl and 50 µL of 0.25% SDS were added to each well. The absorbance of each well was measured using a microplate reader at A590.
Quantitative Real-Time PCR Analysis
Cells grown to mid-exponential phase in YPD medium were incubated with cantharidin for varying amounts of time, harvested by centrifugation, frozen in liquid N2 and stored at −80 °C. RNA extraction and QRT-PCR analysis was performed as described previously11.
Construction of Double Mutants
The double knockout strains were generated using SAT technology33. SAT was PCR amplified from pJK863 (pLC49) using specific primers (Supplementary Table 5), containing sequence homologous to SAT and a gene of interest. PCR-amplified product was transformed into wt and cacrg11Δ/Δ mutants using standard transformation protocol. Nourseothricin (NAT)-resistant transformants were PCR tested for a proper integration of the construct. The SAP2 promoter was induced to drive expression of FLP recombinase to excise the NAT marker cassette for a subsequent reuse. The same procedure was repeated until all alleles were knocked out with SAT cassette. This strain was additionally tested for the absence of any wt alleles by PCR.
Mass spectrometry Analysis of Lipids
Total lipids were extracted as described previously34. Briefly, wt and cacrg1 null mutants were grown at 39°C for 48 hrs in SC media. Cells were washed twice with PBS and counted. 5×108 cells were placed in a single glass tube and lipids were extracted using Mandala followed by Bligh and Dyer extraction. A quarter of the lipid samples were used for inorganic phosphate determination. The remaining lipids were analyzed by MS and MS/MS scans using a TSQ7000 triple quadruple mass spectrometer with electrospray ionization as described.
Virulence Assay
The C. albicans virulence assay was performed on waxworm larvae of G. mellonella. Larvae were obtained from Port Credit Pet Center. C. albicans cells grown overnight in YPD at 30 °C, were washed with PBS, and 5×105 cells were injected into the larvae in 20 µL of PBS and incubated at 37 °C. Dead larvae were scored daily. Kaplan-Meier plots were generated using GraphPad Prism software and significant difference in survival was analyzed by log-rank test.
Supplementary Material
Acknowledgments
We thank M. Gebbia, T. Durbic and A. Surendra for the experimental assistance, and L. Cowen for providing the plasmids. We also thank P. Yau for assistance with printing lipids onto FAST slides.
Funding
EL was supported by Ontario Graduate Scholarship and University of Toronto Open Fellowship. CN and GG are supported by grants from the NHGRI and the Canadian Cancer Society (Grant # 020380), SC is supported by NIH grant GM026020. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions
Conceived and designed the experiments: EL SGC GG CN
Performed the experiments: EL DW AR BY
Analyzed the data: EL DW MP
Contributed reagents/materials/analysis tools: KCO
Wrote the paper: EL CN
REFERENCES
- 1.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 2.Odds FC. Candida infections: an overview. Crit Rev Microbiol. 1987;15:1–5. doi: 10.3109/10408418709104444. [DOI] [PubMed] [Google Scholar]
- 3.Petrossian TC, Clarke SG. Multiple Motif Scanning to identify methyltransferases from the yeast proteome. Mol Cell Proteomics. 2009;8:1516–1526. doi: 10.1074/mcp.M900025-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley R, Chasteen TG. Microbial methylation of metalloids: arsenic, antimony, and bismuth. Microbiol Mol Biol Rev. 2002;66:250–271. doi: 10.1128/MMBR.66.2.250-271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach SJ. Biological methylation. Biol Rev Camb Philos Soc. 1945;20:158–176. doi: 10.1111/j.1469-185x.1945.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 6.Fujioka M. Mammalian small molecule methyltransferases: their structural and functional features. Int J Biochem. 1992;24:1917–1924. doi: 10.1016/0020-711x(92)90287-b. [DOI] [PubMed] [Google Scholar]
- 7.Wlodarski T, Kutner J, Towpik J, Knizewski L, Rychlewski L, Kudlicki A, Rowicka M, Dziembowski A, Ginalski K. Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome. PLoS One. 2011;6:e23168. doi: 10.1371/journal.pone.0023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo M. Current chemical biology approaches to interrogate protein methyltransferases. ACS Chem Biol. 2012;7:443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrossian T, Clarke S. Bioinformatic Identification of Novel Methyltransferases. Epigenomics. 2009;1:163–175. doi: 10.2217/epi.09.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, Giaever G, St Onge RP, Nislow C. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat Chem Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 11.Lissina E, Young B, Urbanus ML, Guan XL, Lowenson J, Hoon S, Baryshnikova A, Riezman I, Michaut M, Riezman H, Cowen LE, Wenk MR, Clarke SG, Giaever G, Nislow C. A systems biology approach reveals the role of a novel methyltransferase in response to chemical stress and lipid homeostasis. PLoS genetics. 2011;7:e1002332. doi: 10.1371/journal.pgen.1002332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisner T, Smedley SR, Young DK, Eisner M, Roach B, Meinwald J. Chemical basis of courtship in a beetle (Neopyrochroa flabellata): Cantharidin as "nuptial gift". Proc Natl Acad Sci U S A. 1996;93:6499–6503. doi: 10.1073/pnas.93.13.6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eisner T, Smedley SR, Young DK, Eisner M, Roach B, Meinwald J. Chemical basis of courtship in a beetle (Neopyrochroa flabellata): cantharidin as precopulatory "enticing" agent. Proc Natl Acad Sci U S A. 1996;93:6494–6498. doi: 10.1073/pnas.93.13.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laidley CW, Cohen E, Casida JE. Protein phosphatase in neuroblastoma cells: [3H]cantharidin binding site in relation to cytotoxicity. J Pharmacol Exp Ther. 1997;280:1152–1158. [PubMed] [Google Scholar]
- 15.Efferth T, Rauh R, Kahl S, Tomicic M, Bochzelt H, Tome ME, Briehl MM, Bauer R, Kaina B. Molecular modes of action of cantharidin in tumor cells. Biochemical pharmacology. 2005;69:811–818. doi: 10.1016/j.bcp.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Li YM, Casida JE. Cantharidin-binding protein: identification as protein phosphatase 2A. Proc Natl Acad Sci U S A. 1992;89:11867–11870. doi: 10.1073/pnas.89.24.11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honkanen RE. Cantharidin, another natural toxin that inhibits the activity of serine/threonine protein phosphatases types 1 and 2A. FEBS Lett. 1993;330:283–286. doi: 10.1016/0014-5793(93)80889-3. [DOI] [PubMed] [Google Scholar]
- 18.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korting HC, Schlaf-Maier U, Schafer-Korting M. [Antimicrobial effects of in vitro-simulated ketoconazole-cantharidin blister fluid levels on candida albicans] Mykosen. 1986;29:297–305. [PubMed] [Google Scholar]
- 20.Zakikhany K, Thewes S, Wilson D, Martin R, Albrecht A, Hube B. From attachment to invasion: infection associated genes of Candida albicans. Nihon Ishinkin Gakkai Zasshi. 2008;49:245–251. doi: 10.3314/jjmm.49.245. [DOI] [PubMed] [Google Scholar]
- 21.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michell RH, Heath VL, Lemmon MA, Dove SK. Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions. Trends Biochem Sci. 2006;31:52–63. doi: 10.1016/j.tibs.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 23.Gillooly DJ, Raiborg C, Stenmark H. Phosphatidylinositol 3-phosphate is found in microdomains of early endosomes. Histochem Cell Biol. 2003;120:445–453. doi: 10.1007/s00418-003-0591-7. [DOI] [PubMed] [Google Scholar]
- 24.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 25.Heung LJ, Luberto C, Del Poeta M. Role of sphingolipids in microbial pathogenesis. Infect Immun. 2006;74:28–39. doi: 10.1128/IAI.74.1.28-39.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramamoorthy V, Cahoon EB, Thokala M, Kaur J, Li J, Shah DM. Sphingolipid C-9 methyltransferases are important for growth and virulence but not for sensitivity to antifungal plant defensins in Fusarium graminearum. Eukaryot Cell. 2009;8:217–229. doi: 10.1128/EC.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oura T, Kajiwara S. Candida albicans sphingolipid C9-methyltransferase is involved in hyphal elongation. Microbiology. 2010;156:1234–1243. doi: 10.1099/mic.0.033985-0. [DOI] [PubMed] [Google Scholar]
- 28.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ternes P, Sperling P, Albrecht S, Franke S, Cregg JM, Warnecke D, Heinz E. Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J Biol Chem. 2006;281:5582–5592. doi: 10.1074/jbc.M512864200. [DOI] [PubMed] [Google Scholar]
- 30.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 31.Fallon J, Kelly J, Kavanagh K. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol Biol. 2012;845:469–485. doi: 10.1007/978-1-61779-539-8_33. [DOI] [PubMed] [Google Scholar]
- 32.Brennan M, Thomas DY, Whiteway M, Kavanagh K. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol Med Microbiol. 2002;34:153–157. doi: 10.1111/j.1574-695X.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 33.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 34.Singh A, Qureshi A, Del Poeta M. Quantitation of cellular components in Cryptococcus neoformans for system biology analysis. Methods in molecular biology. 2011;734:317–333. doi: 10.1007/978-1-61779-086-7_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.