Abstract
The discovery of Helicobacter pylori overturned the conventional dogma that the stomach was a sterile organ and that pH values < 4 were capable of sterilizing the stomach. H. pylori are an etiological agent associated with gastritis, hypochlorhydria, duodenal ulcers, and gastric cancer. It is now appreciated that the human stomach supports a bacterial community with possibly 100s of bacterial species that influence stomach homeostasis. Other bacteria colonizing the stomach may also influence H. pylori-associated gastric pathogenesis by creating reactive oxygen and nitrogen species and modulating inflammatory responses. In this review, we summarize the available literature concerning the gastric microbiota in humans, mice, and Mongolian gerbils. We also discuss the gastric perturbations, many involving H. pylori, that facilitate the colonization by bacteria from other compartments of the gastrointestinal tract, and identify risk factors known to affect gastric homeostasis that contribute to changes in the microbiota.
Keywords: Helicobacter pylori, gastric, stomach, microbiota, cancer, hypochlorhydria, bacterial colonization
Introduction
The human microbiota consists of approximately 100 trillion microbial cells that outnumber our human cells by 10 to 1.1 Through the efforts of the Human Microbiome Project,2,3 the human oral and fecal microbiota have been more extensively studied than other sites in the gastrointestinal (GI) tract. However, given the recognition that each site of the GI tract has its unique microbiota,4 it is necessary to further investigate each of these ecosystems to identify their role in health and disease states.
Conventional wisdom espoused the dogma that the stomach was a sterile organ and that pH values < 4 were able to sterilize the stomach,5 but in the past 30 years with the discovery of Helicobacter pylori,6,7 it is now known that the stomach supports a bacterial community with hundreds of phylotypes (approximate species-level taxa),8-10 and while pH values < 4 prevent bacterial overgrowth, the acidic milieu is not capable of sterilizing the stomach.11 Data suggest that the microbial density of the stomach is 101-103 CFU/g.12-14 The stomach, along with the esophagus and the duodenum, are the least colonized regions of the GI tract, in contrast to the high bacterial counts (1010 to 1012 CFU/g) observed in the colon.12-14 The low bacterial densities within this portion of the GI tract are due to the effects of rapid peristalsis, low pH and/or high bile concentration.15 As H. pylori are directly implicated as an etiological agent in several gastric diseases, including gastric atrophy and cancer,16 it is important to determine the contributions made by other bacteria in gastric health and disease.
Stomach Anatomy
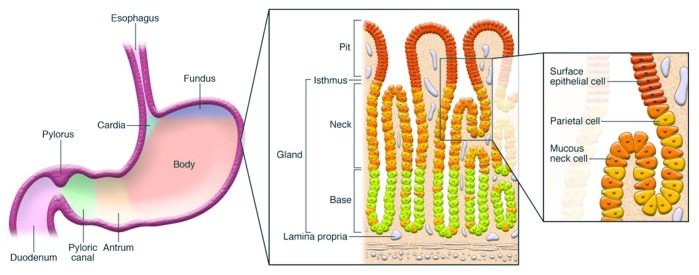
The human stomach is divided into three anatomic regions: the cardia, the fundus/corpus, and the antrum. The cardia is distal to the gastresophageal junction, and its glands primarily secrete mucus. The fundus/corpus comprises close to 80% of the organ and contains the oxyntic glands. The antrum is proximal to the pyloric sphincter, which separates the stomach from the duodenum, and contains pyloric glands (Fig. 1). Both oxyntic and pyloric glands possess mucous neck cells, D cells, and Enterochromaffin cells. They differ as oxyntic glands possess parietal (oxyntic) cells, chief (zymogenic), and enterochromaffin-like cells that produce HCl, pepsinogen, and histamine, respectively. The main feature of pyloric glands is the presence of G cells used to generate gastrin, a key hormone in the regulation of acid secretion.17
Figure 1. Diagram depicting anatomy of the stomach and histological representation of the oxyntic glands of the body of the stomach. It is these glands, which include parietal cells, that are lost in gastric atrophy. Reproduced with permission from Fox and Wang 2007.16
The role of the stomach is 2-fold. First, the stomach secretes gastric juice, composed mainly of proteolytic enzymes and hydrochloric acid, which provide the environment necessary for denaturing of proteins and facilitates the absorption of nutrients. Second, gastric acid plays a role in suppressing the density of ingested microorganisms and assists in preventing infection by pathogens.18 The intragastric pH of 1–2 is the primary restrictive component of the stomach, and severely limits bacterial colonization and survival.15 To prevent damage to the mucosa from HCl and pepsinogen, mucous neck cells throughout the stomach generate mucus that lines the gastric epithelium. While the human gastric lumen has a pH of 1–2, the mucus layer establishes a pH gradient that increases the pH to 6–7 at the surface of the mucosa.19 This is achieved by unique properties of the mucus which permit acid to flow from parietal cells into crypts which communicate with the lumen, but do not allow acid at pH < 4 from penetrating the mucus layer.19 The mucus layer consists of several mucins, such as MUC1, MUC5AC, MUC5AB, and MUC6, and forms two sublayers, an inner mucus layer that is firmly attached to the epithelia and a loose mucus layer interfacing with the lumen.20,21 In the context of understanding the dynamics of the gastric microbiota, it is necessary to consider the site of isolation, as bacteria (and importantly bacterial DNA) may be isolated from the gastric juice, which is too formidable a barrier for colonization (isolated DNA may reflect transient bacteria), compared with the mucosa, which presents a more hospitable environment for microbial colonization. However, during abnormal or disease states, this balance may be perturbed, leading to bacterial colonization. Reduction of gastric acid secretion, whether by parietal cell loss or drug-induced inhibition, can lead to hypochlorhydria (pH between 4–7) or even achlorhydria (pH 7), and increases the risk of bacterial overgrowth and possible deleterious infections throughout the GI tract.18
Gastric Perturbations by Helicobacter pylori
Helicobacter pylori are gram-negative bacteria that successfully colonize the human stomach, infecting 50% of the world’s population. H. pylori are uniquely adapted to colonize the gastric niche. This process has extensively reviewed by others.22,23 Upon infection, H. pylori utilize urease and α-carbonic anhydrase to generate ammonia and HCO32- which mitigate the effects of low pH.24,25 The local increases in pH facilitate the bacteria's passage through acidic gastric fluid and the pH-sensitive mucous layer. Using chemotaxis, the bacteria navigate the pH gradient to their niche near the host epithelium.26,27 Infection with H. pylori, or the closely related pathogen H. felis, have been shown to alter the mucus barrier by affecting the expression of mucins Muc1, Muc4, and Muc5b.28,29 Once established in the inner mucus layer, H. pylori can utilize diverse adhesins (e.g., SabA and BabA) to attach to epithelial cells. Once attached, bacterial effector molecules, both secreted [vacuolating cytotoxin (VacA) and cytotoxin-associated gene A (CagA)] or attached [components of the type IV secretion system (CagL)], modulate gastric epithelial cell behavior leading to loss of cell polarity, release of nutrients and chemokines (e.g., IL-8), and of particular interest for this review, regulation of acid secretion via control of gastrin and H+/K+ ATPase.22,30,31
In response to H. pylori infection, the host mounts an acute inflammatory response characterized by infiltration of neutrophils and mononuclear cells which leads to a chronic, active gastritis. H. pylori protect themselves from reactive oxygen and nitrogen species (RONS) via detoxifying enzymes (catalase and superoxide dismutase) and arginase which limits nitric oxide production from immune cells.22 Furthermore, H. pylori lipopolysaccharide (LPS) and flagellin do not elicit strong inflammatory responses, which limit specific immune responses to the bacteria.22 The ineffectual, acute response leads to the establishment of a chronic inflammatory state. The adaptive immune response to H. pylori is mainly mediated by cellular (T cell), rather than humoral (B cell), immunity and is comprised of proinflammatory and regulatory T cell responses.32 Broadly, proinflammatory T helper 1 (TH1) and TH17 cells secrete cytokines (e.g., interleukin-2 (IL-2), IL-17, IL-22, and IFN-γ) that increase proinflammatory cues, and promote both neutrophil recruitment and macrophage activation. TH1 and TH17 cells play an important role in controlling H. pylori infection, but also mediate infection-associated immunopathology.33,34 Regulatory T (TREG) cells mediate immune tolerance that allows the persistence of H. pylori and minimizes host damage caused by excessive immunopathological T cell responses.35 A recently proposed mechanism demonstrates how H. pylori can modulate both proinflammatory and regulatory T-cell responses via the release of both IL-1β and IL-18, following inflammasome activation.23 IL-1β promotes the induction of T-box transcription factor (T-bet)-dependent T helper 1 (TH1) and RAR-related orphan receptor γt (RORγt)-dependent TH17 cells, and the expression of IFN-γ and IL-17, while IL-18 promotes FOXP3-dependent CD4+CD25+ TREG cells.34 Therefore, the host's attempts to eradicate H. pylori increase gastric immunopathology (gastritis, epithelial damage such as atrophy and intestinal metaplasia), which alters the gastric compartment and its microbiota, and may subsequently progress to gastric cancer. Due to its role in gastric cancer, Helicobacter pylori was one of the first infectious agents recognized by the International Agency for Research on Cancer (IARC) as a class I, or definite, carcinogen.36
Gastric Cancer
Gastric cancer is the fourth most common cancer and the second leading cause of cancer-related death worldwide.37 Both incidence and mortality rates are about twice as high in males as in females.37 Over 70% of cases occur in developing nations, concentrated in Eastern Asia, Eastern Europe, and Central and South America. In contrast, Australia, Africa, Southern Asia, Western Europe and North America are areas of low risk. Not surprisingly, in 2008, Eastern Asia has the highest mortality rates (28.1 and 13.0 per 100,000 men and women, respectively), while North America has the lowest (2.8 and 1.5 per 100,000 men and women, respectively).38 The high mortality to incidence ratio is due in part to the lack of clinical symptoms in most cases of early gastric cancer, which makes early detection difficult.39 Improvements in sanitation (resulting in reduced H. pylori infections), nutrition (greater access to fresh food and decreased dietary salt intake), use of endoscopies and antimicrobial eradication of H. pylori have contributed to the decrease in gastric cancer rates worldwide.40
Approximately 90% of gastric cancers are adenocarcinomas, malignant epithelial tumors that arise from the gastric glandular epithelium.41 Anatomically, gastric cancers are categorized as proximal and distal. Proximal adenocarcinomas are more similar to esophageal adenocarcinomas and may be associated with the absence of H. pylori,40 while distal adenocarcinomas originate in the antrum and are commonly associated with H. pylori infection. Histologically, gastric adenocarcinomas are classified as either diffuse-type or intestinal-type.42 Diffuse-type tumors are characterized by pan-gastritis but no atrophy, are potentially familial in distribution, are present in younger populations and are found uniformly throughout the world in both men and women.43,44 Intestinal-type tumors are characterized by a corpus-dominated gastritis with gastric atrophy and intestinal metaplasia, are associated with regions of high gastric cancer risk and H. pylori infection, and occur more frequently in elderly men.16
Helicobacter pylori, the Gastric Microbiota and Progression to Gastric Cancer
As the host's chronic inflammatory response is incapable of eradicating H. pylori, chronic gastritis ensues, which over decades can progress through a series of discrete steps known as the Correa pathway which involve atrophy, intestinal metaplasia, dysplasia, and intestinal-type gastric adenocarcinoma.16,45 In the context of the stomach, atrophic gastritis is the loss of specialized glandular tissue, such as the oxyntic glands, which impairs acid secretion and the differentiation of gastric progenitor cells.46-48 The loss of parietal cells, which creates a state of hypochlorhydria (pH > 4), facilitates the colonization of the stomach by various bacteria, including those with nitrosating ability which are not regularly cultured from a normal, healthy stomach.45
Since the advent of H2 receptor antagonists (H2RA) in the mid-1970s, there has been an ongoing clinical interest in the microbiota colonizing the stomach. Hypochlorhydria induced by acid suppression is associated with higher levels of gastric nitrites and an increased risk of gastric cancer.49-51 Chronic H2RA therapy or atrophic gastritis promote overgrowth of nitrosating bacteria that convert nitrite and other nitrogen compounds in gastric juice to produce carcinogenic N-nitroso compounds (NOC).45 These chemical reactions are favored in hypochlorhydric stomachs where pH > 4 allows the persistence of nitrites by reducing the antioxidant activity of vitamin C, a powerful nitrosation inhibitor.52,53 The introduction of proton pump inhibitors (PPI) elevated and sustained gastric pH levels even further.54 Studies found a logarithmic relationship between intragastric pH and median bacterial counts in the gastric juice and mucosa and increased risks for enteric infections and bacterial diarrhea.55,56 A review of the literature notes that multiple non-H. pylori organisms have been isolated from the stomach in hypochlorhydric patients, including Lactobacillus spp, Streptococcus spp, Pseudomonas spp, Xanthomonas spp, Proteus spp, Klebsiella spp, Neisseria spp, Escherichia coli, and Campylobacter jejuni.54
Acid suppressive drugs also affect the progression of H. pylori pathogenesis. In stomachs with normal or high acid production, H. pylori gastritis is limited to the antrum and this pattern is usually associated with the development of duodenal ulcers, and not gastric cancer.16 In stomachs with lower acid secretion, as caused by acid suppression or atrophy, H. pylori shifts to a corpus predominant gastritis, which drives parietal cell loss and is associated with increased gastric cancer risk.56,57 Furthermore, increased pH may enhance H. pylori-induced lesions to the gastric mucosa mediated by RONS.58 Another harmful effect of acid suppression is the deregulation of gastrin. Both H. pylori infection and high pH induce hypergastrinemia (to stimulate parietal cells), but prolonged hypergastrinemia can be deleterious due to gastrin's trophic effects on the oxyntic mucosa, which promotes gastric stem cell proliferation and increase the risk of enterochromaffin-like cell hyperplasia.56,59 Eradication of H. pylori did not lead to full recovery of acid secretion in patients with profound hypochlorhydria but did reduce hypergastrinemia.60,61 One study indicated that in H. pylori–infected patients, the high serum levels of gastrin prior to PPI therapy were associated with the most marked progression in gastric atrophy during acid suppression therapy.62 At the same time, H. pylori has been shown to enhance the acid suppressive effects of both H2RAs and PPIs, as well as increasing the risk of atrophic gastritis, bacterial levels and elevation of N-nitrosamines.56,63 The presence of both H. pylori and non-H. pylori bacteria also increased atrophy observed in patients under acid suppressive regimes.56 Animal studies support the hypothesis that Helicobacter infection might accelerate atrophy in hypergastrinemic individuals or patients undergoing acid suppression therapy.64-66 In Mongolian gerbils, omeprazole treatment of H. pylori infected animals led to increased neutrophil and lymphoid infiltration, higher corpus atrophy scores and increased adenocarcinomas.66 H. felis–infected hypergastrinemic mice treated with omeprazole manifested a more rapid progression to dysplasia.64 The pathological changes to the stomach can become so profound that the niche inhabited by Helicobacter spp changes, as evidenced by the decline in Helicobacter spp colonization levels observed in cases with severe achlorhydria and gastric cancer in humans16,67 and mice.68 The loss of H. pylori may also facilitate the colonization of other bacterial populations into this niche. As such, gastric atrophy is considered a critical step in the progression to intestinal-type gastric cancer, and is a strong marker of gastric cancer risk.69
Methods for Determining the Gastric Microbiota
Given culture conditions have not been established for the majority of microbes colonizing the GI tract, culture-based methods provide an incomplete and biased picture of the biodiversity of intestinal microbiota. Therefore, culture-independent molecular methods based on 16S rRNA genes, such as fluorescent in situ hybridization (FISH),70 dot-blot hybridization with rRNA-targeted probes,71 targeted qPCR,72 traditional or sequence-aided community fingerprinting [including denaturing gradient gel electrophoresis (DGGE),73 temperature gradient gel electrophoresis (TGGE),74 and terminal restriction fragment length polymorphism (T-RFLP)75], sequencing of cloned 16S rDNA,74 microarrays (PhyloChip),76 and next-generation sequencing77 (NGS) have been used to determine the gut microbiota in diverse regions of the GI tract. While new technologies (e.g., API78 and MALDI-TOF mass spectrometry79) have improved the identification of cultured organisms, culture remains limited by the inability to culture all the organisms of interest, but also by time consuming technical demands. It has been argued that culture-based methods provide the advantage of distinguishing viable microorganisms, which DNA-based assays cannot.80 However, in the context of decreased bacteriocidal activity in the stomach (e.g., hypochlorhydria), bacteria in transit can also be cultured.
For all technologies dependent on hybridization, amplification, identification or sequencing of the 16S rRNA gene, the quality of DNA extractions is critically important as it may bias the results, due to varying degrees of microbial resistance to processing by enzymes, chaotropic agents, or bead beating.81 FISH, dot-blot hybridization and qPCR are highly specific and useful techniques when a defined set of organisms are being studied. However, given the need to design and test specific probes for each queried organism and the low-throughput nature of the assays, these techniques are not as useful in surveying large collections of microbes. Newer technologies, such as microarrays/PhyloChip and high-throughput qPCR arrays, easily address the concerns of the low-throughput nature of dot-blot hybridization and qPCR and allow the capacity to query 100s to 1000s of organisms in a single run. Nevertheless, a significant investment has to be made to design and test comprehensive qPCR probe sets or microarrays like the PhyloChip. In the case of the PhyloChip, the array can distinguish > 50,000 different operational taxonomic units (OTUs) and incorporates bioinformatic tools to dissect the generated data.76,82 The high number of detectable OTUs effectively allows most users to use the PhyloChip without a priori knowledge of the sample composition, which is not possible with qPCR or FISH. However, the limitation of the PhyloChip lies in its inability to multiplex samples, which makes it unfeasible for most labs to process more than a few samples.
Techniques that allow unbiased surveillance of the entire microbial community without a priori knowledge of the composition rely on 16S rRNA gene analysis and include community fingerprinting, Sanger-sequencing of 16S rDNA libraries and next generation sequencing (NGS) of 16S rRNA genes. The 16S rRNA gene is homologous in all bacteria, highly conserved in overall structure, not readily transferred between species, and contains 9 variable regions that allow phylogenetic identification of species or the definition of operative taxonomic units (OTUs).83 Community fingerprinting techniques are capable of surveying unknown microbial communities and are flexible in post-processing. As bands can be analyzed visually on the gel, and subsequently confirmed using PCR or sequencing methodologies, the user can customize the degree of confidence in the assay's results. The drawbacks for these techniques are the low resolution between bands (i.e., multiple organisms can have similar bands) and the high level of expertise needed for execution. Sanger-sequencing of 16S rDNA libraries can be used in conjunction to community fingerprinting methods or direct PCR amplification from the sample. Earlier microbiome studies relied on library sequencing.71,74 The advent of better sequencing technologies, which process more reads and do not require cloning, and better bioinformatics tools have rendered this technology more obsolete. While not free of biases in PCR amplification, massively parallel sequencing with NGS removes selection biases that could occur with 16S rDNA clonal libraries, provide easier processing, increase sequencing coverage and provide better resolution than other methods. Technically, many of the skills needed for processing NGS samples are familiar to molecular biologists. Its current limitations are primarily complexity of bioinformatic analysis, and secondarily, access to equipment and cost. The secondary concerns are being addressed as the equipment becomes less expensive and more readily available, and the cost of sequencing continues to decrease. The primary obstacle for most labs has been processing millions of relatively short reads effectively, but considerable resources have been allocated to resolve these limitations. Applications like 16S profiling have become quite standard. Briefly, programs take raw data generated by the NGS machine and remove low-quality sequences and further processing, such as trimming barcodes and adaptor sequences, prepare sequences for comparison. Software aligns sequences against reference databases such as SILVA,84 GREENGENES,85 or the Ribosomal Database Project (RDP)86 to identify microbes most closely associated to a given sequence. Currently NGS may be the best method in terms of balancing ease of use, accessibility and cost for microbiome studies. A more complete discussion of methods, DNA isolation and bioinformatic analysis can be found in this review.81
The Gastric Microbiota
Despite the declining prevalence of H. pylori infection worldwide, H. pylori still infects 50% of the world's population.22 We and others would argue that H. pylori are indeed an autochthonous species in the gastric niche.1,74,87 As such, this review will describe the gastric microbiota in both humans and key rodent models, with the inclusion and absence of H. pylori, while evaluating the effects of altered gastric states on the microbiota. In this review, we will include higher and lower taxonomic information, such as phylum and genus, for ease of comparison (Fig. 2). For older publications, we have updated classification systems to better reflect current nomenclature. The studies reviewed have been conducted in populations worldwide, but as samples for gastric microbiota analysis are more difficult to obtain than the oral or fecal microbiota samples, many studies rely on patients undergoing an endoscospy. This may bias studies as these subjects may not be reflective of an asymptomatic population. Collections of gastric juices have been used in the past, but may be compromised because the samples also reflect the transient populations of the stomach.
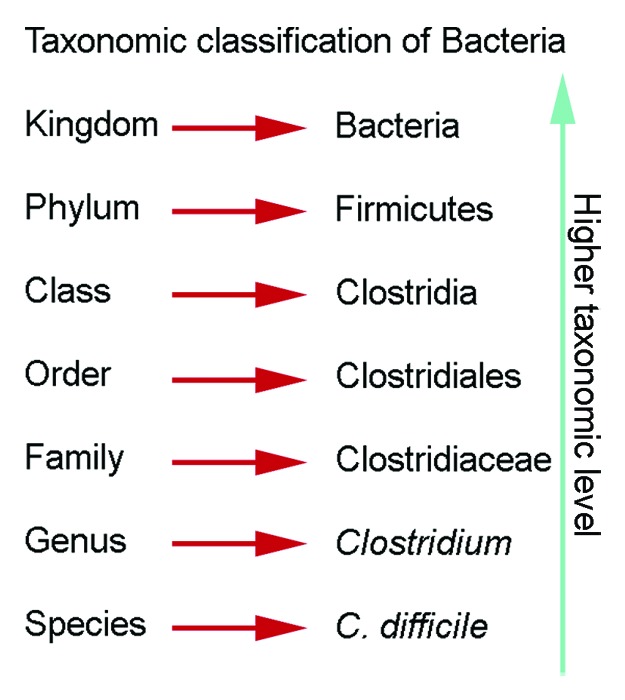
Figure 2. Taxonomic classification of bacteria. Descriptions of the gastric microbiota focus on the levels of phylum and genus.
Gastric Microbiota in Humans
The major constituent of the gastric microbiota in more than half of all humans is the Proteobacteria H. pylori.22 As discussed above, the bacteria's effects on the gastric mucosa affect the ecological niches in the stomach, which allow the colonization of other bacteria. H. pylori are fastidious, microaerophilic bacteria, which have influenced earlier reports utilizing culture as the primary means of H. pylori identification.
Culture-Based Identification of Gastric Microbiota
Historically, the low intragastric pH (pH < 2) of the stomach was considered a barrier to gastric microbial colonization. As such, the stomach was historically considered a sterile organ, and the bacteria present were considered transient species. In a review prior to the discovery of H. pylori, the bacteria isolated from the stomach (at > 103 CFU/g) included Firmicutes (genera Lactobacillus, Streptococcus, Clostridium, and Veillonella), Actinobacteria (genus Bifidobacterium), and Proteobactearia (coliforms), and at a lower frequency other bacteria (Firmicutes (genera Peptostreptococcus and Staphylococcus), Bacteroidetes (genus Bacteroides), and Actinobacteria (genus Actinobacillus)) and yeasts (Candida and others).1 We have summarized several representative studies that use culture-based techniques to assess the microbiota in the stomach in Table 1. In the literature using culture methodologies, the most prevalent or abundant phylum, regardless of H. pylori status, is Firmicutes, followed by Proteobacteria and Bacteroidetes. Depending on the study, Actinobacteria may be the second or third most prevalent phylum. The most commonly found genera were Streptococcus, Lactobacillus, Bacteroides, coliforms, Staphylococcus, Veilonella, Corynebactieum and Neisseria, which may reflect both the interest of the investigators and what bacteria are more easily cultivable.49,79,88-94 Comparing studies that assess H. pylori status, H. pylori status did not alter the prevalence ranking with Firmicutes, Proteobacteria and Bacteroidetes being the top three phyla, when the quantification of H. pylori was not included.49,79,88-94 A study that evaluated the effects of gastric cancer found increases in Proteobacteria along with Firmicutes (genera Veilonella and Streptococcus) and species from the Bifidobacterium/Lactobacillus group.91 Of note, a study using culture and MALDI-TOF mass spectrometry was able to detect a Proteobacteria called Acinetobacter lwoffii,79 which experimentally caused gastritis and hypergastrinemia in mice.95 These findings suggest that non-H. pylori species can promote chronic inflammatory conditions. It is interesting that the literature does have examples of culturable organisms, but the scientific community refused to accept the presence and importance of gastric microbiota. The culture studies also demonstrated the fastidiousness of H. pylori and the limitations of culture, as more recent studies, have found that Proteobacteria are the dominant phylum in H. pylori infected subjects due to the high levels of gastric H. pylori.9,10
Table 1. Studies analyzing the human gastric microbiota using culture-based methods.
| Study | Method | # of samples | Age range | Gender | Ethnicity or Location | HP status | Phyla | Genera observed | Genus information | Clinical presentation | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Savage 1977 | N/A | N/A | N/A | N/A | Various | N/A | Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes, and yeasts | 9+ | Firmicutes (Lactobacillus), Firmicutes (Streptococcus), Actinobacteria (Bifidobacterium), Firmicutes (Clostridium), Firmicutes (Veillonella), Proteobacteria (Coliforms), other bacteria (Firmicutes (Peptostreptococcus, Staphylococcus) Bacteroidetes (Bacteroides), and Actinobacteria (Actinobacillus)) and yeasts | N/A | Review |
| Sharma et al. 1984 | Gastric juice culture | 10 | 19–26 (mean 22.6) | Male | England | N/A | Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria | 9 | Actinobacteria (Corynebacterium), Firmicutes (Staphylococcus), Proteobacteria (Neisseria), Firmicutes (Lactobacillus, Veilonella, Streptococcus,Bacillus), Bacteroidetes (Bacteroides), Proteobacteria (Acinetobacter) | Omeprazole daily for 2 weeks on healthy patients | OMP increased bacterial load and nitrite levels but returned to normal after Tx discontinued |
| Stock-bruegger 1985 | N/A | N/A | N/A | N/A | Various | N/A | Firmicutes, Proteobacteria, Bacteroidetes | 7 | Proteobacteria (Enterobacteriaceae), Firmicutes (Enterococcus), Firmicutes (Streptococcus), Firmicutes (Staphylococcus), Firmicutes (Lactobacillus), Firmicutes (Clostridium), Bacteroidetes (Bacteroides) | Hypochlorhydria | Review |
| Sjostedt et al. 1985 | Gastric juice culture | 10 | Pool: 28–74 | Pool: 39 males, 21 females | Sweden | N/A | Firmicutes, Proteobacteria, Actinobacteria | 6 | Firmicutes (Staphylococcus), Proteobacteria (Neisseria), Firmicutes (Streptococcus), Actinobacteria/Firmicutes (Bifidobacterium/Lactobacillus), Firmicutes (Veilonella), Bacteroidetes (Bacteroides) | Healthy | Only 1–4 of 10 patients had cultures of each |
| Gastric juice culture | 10 | Pool: 28–74 | Pool: 39 males, 21 females | Sweden | N/A | Proteobacteria, Firmicutes, Actinobacteria | 9 | Firmicutes (Streptococcus), Actinobacteria/Firmicutes (Bifidobacterium/Lactobacillus) and Firmicutes (Veilonella). Also observed Proteobacteria (Klebsiella, Escherichia, Pseudomonas), Firmicutes (Bacillus), Candida spp | Gastric cancer | 2–6 of 10 patients had cultures of each | |
| Thorens et al. 1996 | Gastric juice culture | 37 | 20–60 (mean 42) | 39 males, 8 females | Switzerland | N/A | Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes | 11 | Firmicutes (Streptococcus), Actinobacteria (Corynebacterium), Proteobacteria (Neisseria), Actinobacteria (Micrococcus), Bacteroidetes (Bacteroides), Firmicutes (Staphylococcus), Proteobacteria (Escherichia coli), Proteobacteria (Klebsiella), Actinobacteria (Rothia (previously Stomatococcus)), Firmicutes (Enterococcus, Lactobacillus) | Patients with routine endoscopy with Omeprazole and Cimetidine | Bacterial overgrowth on acid suppression. Oral and intestinal microbes seen |
| Adamsson et al. 1999 | Culture - Biochemistry, Gas chromatography | 30 | 35–75 (mean 58.5) | 14 males, 16 females | Sweden | Pos. | Firmicutes (58%), Proteobacteria (22.2%), Actinobacteria (14.3%), Bacteroidetes (3.8%) and Fusobacterium (1.5%). | 12 to 13 | Firmicutes (Streptococcus), Proteobacteria (Neisseria), Proteobacteria (Haemophilus), Firmicutes (Staphylococcus), Firmicutes (Peptostreptococcus), Firmicutes (Veilonella), Actinobacteria (Actinomyces), Actinobacteria (Micrococcus), Bacteroidetes (Prevotella-Bacteroides), Proteobacteria (Enterobacteriaceae), Yeasts, Firmicutes / Actinobacteria (Lactobacillus and Bifidobacterium), Fusobacteria (Fusobacterium) | Non-ulcer dyspepsia treated with anti-HP triple therapies (omeprazole, metronidazole and either amoxicillin or clarithromycin) | Data presented prior to antibiotics. HP eradicated. |
| Mowat et al. 2000 | Gastric juice culture | 18 | HP neg 20–45 (mean 27) | N/A | Scotland | Neg. | Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria | 11 | Firmicutes (Streptococcus), Firmicutes (Staphylococcus), Proteobacteria (Neisseria), Firmicutes (Lactobacillus), Proteobacteria (Haemophilus), Bacteroidetes (Bacteroides), Firmicutes (Clostridium), Firmicutes (Veilonella), Proteobacteria (Coliform), Actinobacteria (Corynebacterium), Proteobacteria (Moraxella (previously Branhamella)), Yeast | Healthy | Samples collected during omeprazole treatment (pH 3). |
| Gastric juice culture | 11 | HP pos 21–45 (mean 29) | N/A | Scotland | Pos. | Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria. HP decreased Firmicutes and increases in Proteobacteria/Bacteroidetes | 11 | Firmicutes (Streptococcus), Proteobacteria (Neisseria), Firmicutes (Staphylococcus), Bacteroidetes (Bacteroides), Firmicutes (Veilonella), Proteobacteria (Haemophilus), Firmicutes (Clostridium), Proteobacteria (HP), Proteobacteria (Moraxella (previously Branhamella)), Firmicutes (Lactobacillus), Actinobacteria (Corynebacterium).# | Healthy | Samples collected during omeprazole treatment (pH 8). Nitrosating bacteria found. HP presence was not high. | |
| Kato et al. 2006 | Biopsy, Gastric juice culture with qPCR | 10 | 9 to 16 | 3 males, 7 females | Japan | Pos. | Firmicutes, Actinobacteria, Bacteroidetes | 5 | Firmicutes (Lactobacillus), Actinobacteria (Bifidobacterium), Firmicutes (Eubacterium), Bacteroidetes (Bacteroides), Firmicutes (Faecalibacterium (previously Fusobacteria) prausnitzii) | Children (gastritis, duodenal ulcer, reflux esophagitis, non-ulcer dyspepsia). | Did not culture HP. Only recovered bacteria from 2 children |
| Biopsy, Gastric juice culture with qPCR | 10 | 9 to 16 | 3 males, 7 females | Japan | Neg. | 1 | Firmicutes (Lactobacillus) | Did not culture HP. Only recovered from 1 child with highest pH | |||
| Biopsy, Gastric juice culture with qPCR | 10 | 33–79 | 4 males, 6 females | Japan | Pos. | Firmicutes, Proteobacteria, Bacteroidetes | 9 | Firmicutes (Streptococcus), Firmicutes (Bacillus), Firmicutes (Veilonella), Proteobacteria (Neisseria), Firmicutes (Lactobacillus), Bacteroidetes (Bacteroides), Firmicutes (Peptostreptococcus), Firmicutes (Staphylococcus) | Adults (gastritis, gastric ulcer, early gastric cancer, gastric adenoma, non-ulcer dyspepsia) | Did not culture HP. Higher bacterial load (100 fold) correlated with higher pH | |
| Biopsy, Gastric juice culture with qPCR | 10 | 33–79 | 4 males, 6 females | Japan | Neg. | Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria | 9 | Firmicutes (Bacillus), Firmicutes (Streptococcus), Firmicutes (Staphylococcus), Proteobacteria (Neisseria), Firmicutes (Veilonella), Bacteroidetes (Bacteroides), Fusobacteria (Fusobacterium) | Adults (gastritis, gastric ulcer, early gastric cancer, gastric adenoma, non-ulcer dyspepsia) | Did not culture HP | |
| Zilberstein et al. 2007 | Biopsy, Culture | 20 | Mean age 42 | 8 males, 12 females | Brazil | N/A | Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria, Bacteroidetes | 18 | Firmicutes (Lactobacillus) (41.6%), Firmicutes (Veilonella) (41.6%), Firmicutes (Clostridium) (33.3%), Actinobacteria (Corynebacterium) (25%), Proteobacteria (Escherichia) (16%), Bacteroidetes (Bacteroides) (16%), Proteobacteria (Klebsiella) (16%), Firmicutes (Peptococcus) (16%). Other genera observed were Fusobacteria (Fusobacterium), Actinobacteria (Propionibacterium), Firmicutes (Peptostreptococcus, Staphylococcus, and Streptococcus), Proteobacteria (Proteus and Enterobacter) and yeast | Healthy | Found acid resistant strains. Proteobacteria likely from lower bowel |
| Hu et al. 2012 | Biopsy, Culture + MALDI-TOF mass spectrometry | 67 samples from 103 patients | 81 patients < 50 y old | 51 males, 52 females | China | Pos. | Firmicutes, Proteobacteria, Actinobacteria | Proteobacteria (Neisseria), Firmicutes (Streptococcus), Actinobacteria (Rothia), Firmicutes (Staphylococcus) | Upper GI diseases (duodenal ulcers, gastric ulcers, gastritis, dyspepsia, esophagitis) | Detected Acinetobacter lwoffi | |
| Delgado et al. 2013 | Biopsy, Culture | 12 | Mean age 60.5 | 6 males, 6 females | Spain | Neg. | Firmicutes, Actinobacteria, Proteobacteria | 9 | Actinobacteria (Propionibacterium), Firmicutes (Lactobacillus), Firmicutes (Staphylococcus) | Dyspeptic without PPI | P. acnes was surprising but it has shown in other studies (Monstein et al.) |
N/A, not available; Pos., positive; Neg., negative; #, underlined values are altered by HP infection compared to negative controls
16s rRNA Based Identification of the Human Gastric Microbiota
Culture-independent studies use a variety of molecular methods (Table 2). Of the eight studies reviewed, four different molecular methods were used to survey the human gastric microbiota based on the analysis of a gastric biopsy sample. Three studies utilized NGS technologies,10,80,96 two studies used Sanger sequencing of a 16S rDNA library,8,9 two studies used a community fingerprinting method to define a library for Sanger sequencing,74,75 and one study utilized a PhyloChip.82 There is considerable variation in the gastric microbiome between individuals at the genus level, and perhaps future standardization of technologies to survey the gastric microbiota will facilitate more robust comparisons.
Table 2. Studies analyzing the human gastric microbiota using 16S rRNA identification methods.
| Study | Method | # of samples | Age range | Gender | Ethnicity or Location | HP status | Phyla | Genera (OTUs) observed | Genus information | Clinical status | Comments |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Monstein et al. 2000 | Biopsy/TTGE/Sanger-sequenced 16S rDNA library | 13 of 22 | Pool: male (44–79), female (43–76) | Pool: 10 males, 12 females | Sweden | Pos. | Proteobacteria, Firmicutes, Actinobacteria | 11 | Proteobacteria (Pseudomonas), Firmicutes (Enterococcus, Staphylococcus, Streptococcus), and Actinobacteria (Rothia (prev. Stomatococcus)). Low abundance Proteobacteria (Acinetobacter, Brevundimonas, Enterobacter, Helicobacter (non HP), and Rhizobium) and Actinobacteria (Propionibacterium) | No gastritis(5) or no clinical gastritis (8) | Gastritis increased HP numbers and reduced detection of other bacteria. Detected other Helicobacters |
| Bik et al. 2006 | Biopsy/Sanger-sequenced 16S rDNA library | 23 | 42–78 | 22 males, 1 female | USA (13 Caucasian, 5 Hispanic, 5 African-American) | 12 of 23 Pos., but 7 of 11 Neg. had counts of HP | Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria | 128 | HP = 72% of reads in Pos. patients, but still 11% in Neg. patients. Non-HP genera in the overall library were Firmicutes (Streptococcus, Veilonella), Bacteroidetes (Prevotella), Actinobacteria (Rothia), Fusobacteria (Fusobacterium) |
Medically indicated gastroscopies. 8/23 on acid blockers. pH range from 2–7 | Only detected H. pylori from genus Helicobacter. H. pylori is dominant but does not affect underlying structure and diversity. HP samples had relative lack of Bacteroidetes phylotypes |
| Andersson et al. 2008 | Biopsy/454 pyrosequencing of V6 of 16S rRNA amplicons | 3 | 61–76 | N/A | Sweden | Neg. | Actinobacteria (46.8%), Firmicutes (29.6%), Bacteroidetes (11.1%), Proteobacteria (10.8%), Fusobacteria (1.1%) | 262 | Firmicutes (Streptococcus, Gemella), Actinobacteria (Actinomyces), Bacteroidetes (Prevotella) | Healthy | Only 33 of 262 seen in all three Neg. samples. H. pylori reduced diversity. Found many throat organisms, so may be transients. Non-throat organisms were Proteobacteria |
| 3 | 61–76 | N/A | Sweden | Pos. | Proteobacteria (96%), Firmicutes (1.8%), Actinobacteria (1.1%), Bacteroidetes (0.8%) and Fusobacteria (0.1%) | 33 | HP 93–97% | Healthy | HP dominates with 93–97% of reads | ||
| Dicksved et al. 2009 | Biopsy/T-RFLP/Sanger-sequenced 16S rRNA library | 10 | 52–88 | 8 males, 2 females | Sweden | 8 of 10 | Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria | 102 | Firmicutes (Streptococcus, Lactobacillus and Veilonella) and Bacteroidetes:Prevotella | Gastric cancer (intestinal- and diffuse-type) | Low abundance of HP. Did not find disease related association from clustering of T-RFLP data from dyspepsia and GC samples |
| Biopsy/T-RFLP | 5 | 52–88 | 2 males, 3 females | Sweden | 0 of 5 | Firmicutes, Bacteroidetes, Actinobacteria | Firmicutes (Veilonella, Streptococcus) and Bacteroidetes (Prevotella) | Dyspepsia | |||
| Li et al. 2009 | Biopsy/Sanger-sequenced 16S rDNA library | 5 | Pool - 40–87 | Female | Hong Kong | Neg. | Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, Fusobacteria | 133 | Proteobacteria (Neisseria), Bacteroidetes (Prevotella), Firmicutes (Streptococcus), Proteobacteria (Haemophilus), Bacteroidetes (Porphyromonas) | Asymptomatic | Found 1–2 dominant genera per phyla. Neisseria and Haemophilus higher in asymptomatic patients |
| 5 | Pool - 40–87 | Female | Hong Kong | Neg. | Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Fusobacteria | 133 | Firmicutes (Streptococcus), Bacteroidetes (Prevotella), Proteobacteria (Neisseria, Haemophilus), Bacteroidetes (Porphyromonas) | Gastritis | Streptococcus higher in gastritis. Streptococcus and Prevotella levels correspond with Bik et al. | ||
| Maldonado-Contreras et al. 2010 | Biopsy/PhyloChip | 4 | 37–80 | 2 males, 2 females | 4 Amerindians | Neg. | Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes | 154 (family) | Gastritis | ||
| 8 | 21–59 | 4 males, 4 females | 6 Amerindians, 2 USA (South Asia and Africa) | Pos. | Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes | 137 (family) | Dyspepsia, GERD, Gastritis | No differences on taxonomic complexity but HP increases relative abundance of non-Helicobacter bacteria from Proteobacteria, Spirochetes, and Acidobacteria. Decreased abundance of Actinobacteria, Bacteroidetes and Firmicutes. | |||
| Stearns et al. 2011 | Biopsy/V3 16S rRNA sequencing (Sanger and Illumina) (data for males and females) | 4 | > 50 | 2 males, 2 females | Canada | Unk. | Overall antrum Proteobacteria (50.66%), Firmicutes (35.7%), Bacteroidetes (13.47%). Overall corpus Firmicutes (69.0%), Proteobacteria (16.7%), Fusobacteria (8.1%) | 23 antrum 24 body | Healthy undergoing gastroscopy | High variability between subjects and between antrum and corpus. Data hard to interpret as they only report phyla and genera present in all 4 samples. | |
| Same data (only males) | 2 | > 50 | Male | Canada | Unk. | Antrum: Male (avg) Proteobacteria 99.6%. Corpus Male 1 Proteobacteria (45.0%), Fusobacteria (22.1%), Firmicutes (16.0%), Bacteroidetes (14.9%) and SR1 (2.0%). Corpus Male 2 Firmicutes (Parvimonas) (99.44%) | Antrum: Males = 99% Proteobacteria - no genus data (HP?) Corpus: Male 1 = Firmicutes (Parvimonas) (99.4%), Male 2 = Proteobacteria (no data) (45%), Fusobacteria (no data) (22.1%), Firmicutes (Streptococcus, Veilonella, other) (16%), Bacteroidetes (Prevotella, Porphyromonas, other) (14.9%), SR1 (no data) (2%) | Healthy undergoing gastroscopy | n = 2 per site | ||
| Same data (only females) | 2 | > 50 | Female | Canada | Unk. | Female antrum: Firmicutes, Bacteroidetes. Female corpus: Firmicutes. | Antrum: Female = Firmicutes (Streptococcus) (72.6%) and Bacteroidetes (Prevotella) (27.3%). Corpus: Female = Firmicutes [Veilonella, Streptococcus and Oribacterium] (99.9%) | Healthy undergoing gastroscopy | n = 1 per site | ||
| Delgado et al. 2013 | Biopsy/454 pyrosequencing of 16S rDNA amplicons | 4 sequenced | Mean age 60.5 | Pool: 6 males, 6 females | Spain | Neg. | Firmicutes, Proteobacteria, Actinobacteria with small proportions of Deinococcus-Thermus, Bacteroidetes and Gemmatimonadetes | 19 core. 69 total | Firmicutes (Streptococcus, Lactobacillus), Actinobacteria (Propionibacterium), and Proteobacteria (Methylobacterium) were dominant. Dominant genera varied by sample. | dyspeptic without PPI | High variability between subjects |
N/A, Not available, Pos., positive, Neg., negative, Unk., unknown H. pylori status
In the studies surveyed, the most prominent phyla commonly detected in the stomach are Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria and Fusobacteria (Table 2).9,10,74,75,80,82,96 The most abundant phyla in H. pylori positive subjects are Proteobacteria, Firmicutes and Actinobacteria. In the absence of H. pylori, the most abundant phyla are Firmicutes, Bacteroidetes and Actinobacteria. In humans, H. pylori are by far the most dominant species in the stomach, comprising 72 to 99% of sequencing reads.9,10 In the absence of H. pylori, analysis of the known H. pylori negative subjects consistently shows the presence of Streptococcus spp, which seems to be the most abundant genus in these subjects.8,75,80,96 In the gastric microbiota, the non-Helicobacter genera commonly detected are Streptococcus, Prevotella, Veillonella, and Rothia.
The effects of H. pylori on the gastric microbiota are not fully understood. Numbers of H. pylori increase with the onset of gastritis,74 which may reflect changes in the gastric niche that allow H. pylori to outcompete other bacteria and increase H. pylori levels.97 While H. pylori made up 72% of reads observed and decreased the overall diversity and evenness of the gastric microbiota, Bik et al. determined that the underlying diversity and richness is higher in H. pylori positive samples than H. pylori negative samples when H. pylori reads are removed.9 However, the authors did note a relative lack of Bacteroidetes in H. pylori infected patients.9 Other studies disagree, finding a strong effect of H. pylori on the composition of the gastric microbiota.10,82 In one study, H. pylori accounted for 93–97% of all reads in the infected stomach, and substantially decreased the diversity as only 33 phylotypes were observed in H. pylori positive individuals while 262 phylotypes were observed in H. pylori negative subjects.10 In a separate study, PhyloChip data was analyzed using non-metric multidimensional scaling, and demonstrated that H. pylori infection accounted for 28% of the variation seen in the analysis. This was despite the fact that no differences in taxonomic complexity were seen in terms of abundance of different phyla or the numbers of families identified.82 However, the authors determined that H. pylori infection increased the relative abundance of Proteobacteria (non-Helicobacter bacteria), Spirochaetes and Acidobacteria and decreased the relative abundance of Actinobacteria, Bacteroidetes and Firmicutes when compared with H. pylori negative individuals.82
The effects of the absence of H. pylori on the microbiota has also been the focus of other studies.8,80 An important consideration when evaluating this literature is to note that multiple studies report the ability to detect H. pylori sequences at extremely low levels in subject who were H. pylori negative by other diagnostic means.9,74,80,82 This may reflect a host response that led to significant reduction of H. pylori or the presence of non-H. pylori helicobacters.22 Li et al. sequenced 16S rRNA clones from H. pylori-uninfected patients with gastritis and without gastritis.8 Asymptomatic patients had a much lower level of Firmicutes (genus Streptococcus) but instead had a higher proportion of Proteobacteria (genera Neisseria and Haemophilus) similar to Monstein et al. who observed more Proteobacteria (genus Pseudomonas) in asymptomatic H. pylori infected patients.8,74 Their results share similarity with the Bik et al. study;9 both groups found that Streptococcus spp and Prevotella spp accounted for ~40% of reads in H. pylori-uninfected subjects presenting with gastric disease.8 A second study focusing on H. pylori-negative subjects demonstrated that the gastric microbiota is extremely variable at lower taxonomic levels; the four subjects sampled shared the same ranking in terms of phyla (Firmicutes, Proteobacteria and Actinobacteria, from most abundant to least). But upon closer inspection, the four subjects had different abundances of each phylum, being more evident at the genus level.80 While the study identified 69 different genera, a core set of 19 was observed in all four samples with Firmicutes (genus Streptococcus), Actinobacteria (genus Propionibacterium), Firmicutes (genus Lactobacillus) and Proteobacteria (genus Methylobacterium) being of importance, in spite of the fact that the dominant genus and proportions varied from sample to sample.80 The authors also compared their sequencing results from the results of bacterial culture from the same gastric samples and found robust concordance as the four dominant cultivable genera were Propionibacterium, Lactobacillus, Streptococcus, and Staphylococcus. While three of the four genera match the sequenced data, the inclusion of Staphylococcus, which was not a strong contributor to the sequencing data, reflects bias toward cultivable organisms.
Regarding the uniformity of the microbiota within the stomach, Bik et al. and Li et al. found no differences in the microbiota of the antrum and corpus in their populations, with the exception of decreased Prevotella in the antrum of gastritis patients by Li et al.8,9 In contrast, Stearns et al. documented anatomical differences between subjects and between the antrum and corpus (Fig. 3).96 The antrum was dominated by Proteobacteria, Firmicutes and Bacteroidetes while the corpus microbiota was predominantly Firmicutes, Proteobacteria, Fusobacteria and Bacteroidetes. The focus of the study surveyed the bacterial microbiota along the GI tract, and unfortunately, did not assess stomach data in depth. In their effort to find commonality in the microbiota of the four subjects, the authors have focused on the genera shared between subjects and omitted useful information from supplemental tables. For example looking at the phyla represented in the three antral samples, it is evident that the two male samples were composed of > 99% of Proteobacteria, while the single female sample did not have significant amounts of Proteobacteria, as the microbiota was composed of Firmicutes (72.6%) and Bacteroidetes (27.3%) (Fig. 3).96 Hence the representation of a Proteobacteria dominated antrum is misleading. As Streptococcus and Prevotella were present in all four samples, it was possible to determine that these constitute the two most abundant genera in the female antrum, as seen in other H. pylori negative samples in other studies.8,9 However, the sequence reads corresponding to Helicobacter spp were not reported, and it is not possible to determine whether Helicobacter spp were in fact the dominant species in the antrum of the male subjects, as noted in other studies.9,10 Another interesting finding is that Proteobacteria were the most abundant phyla in the corpus of one of the two male subjects, while the corpus of the other male subject is exclusively colonized by Firmicutes of the genus Parvimonas.96 Unfortunately, the sample size is too small and the inter individual variability is too high to determine if there are effects due to gender in this study. Dicksved et al. explored the effect of gastric cancer on H. pylori and the gastric microbiota.75 The authors found few differences in the microbiota of 10 gastric cancer patients and five H. pylori-negative dyspeptic controls.75 While 8 of 10 gastric cancer patients were H. pylori positive, the abundances of H. pylori were very low, perhaps reflecting the changes in the gastric niche that occur with gastric cancer.75 The altered stomach was colonized by multiple Streptococcus spp, including S. mitis, S. parasanguinis and S. bovis (currently S. infantarius which has been associated with colorectal cancer98). However, this study relied on T-RFLP to determine the microbiota, which lacks the resolution to determine subtle shifts in abundance or species composition that may influence gastric pathogenesis.75 Currently studies are lacking that systematically evaluate the gastric microbiota in clinically defined populations to enable distinguishing differences in microbial numbers or diversity related to atrophy, intestinal metaplasia and gastric cancer (intestinal- vs. diffuse-type cancers). It is noteworthy that three studies found a relationship between increased Streptococcus spp and gastric disease.9,75,82
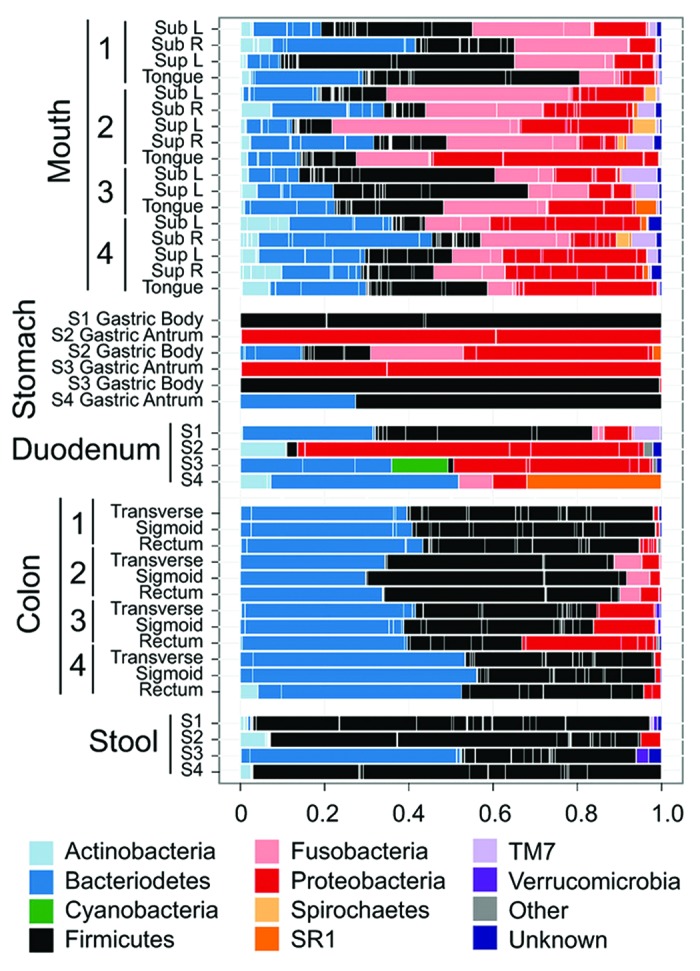
Figure 3. Human microbiota composition in multiple sites of the GI tract, including mouth, stomach, duodenum, colon and stool. Note the high variability between individuals and between the antrum and corpus in the stomach. The stomach microbiota also differs significantly from other sites in the GI tract. Reproduced with permission from Stearns et al. 2011.96
While it has been conjectured that the indigenous microbiota might be a reflection of transient bacteria from the mouth and esophagus, three separate studies demonstrated that in spite of high inter-subject variability, the gastric microbiota were distinguishable from microbiota found in the mouth, nose, and distal GI tract.10,80,96 Comparing the general trends observed in this review with data from other similar sites, the human gastric microbiota is different from the microbiota of the oropharynx,99 but in the absence of H. pylori, the structure and composition most resembles the microbiota reported for the distal esophagus with unique differences due to the makeup of Proteobacteria.10,100
Gastric Microbiota in Mice and Mongolian Gerbils
When considering the gastric microbiota of rodents in the context of H. pylori-induced disease, it is important to recognize several key differences: (1) H. pylori is not an autochthonous member of the microbiota and mouse-adapted strains are needed to infect the mouse,101-103 (2) mice have relatively high intragastric pHs of 3–4,71,104 while Mongolian gerbils have a pH < 2105, more similar to humans, (3) the gastric anatomy differs between humans and rodents, as there is a considerable non-glandular forestomach composed of squamous epithelium (Fig. 4),106 and (4) transient bacteria in the stomach may be due to coprophagia, which is common in mice but not in Mongolian gerbils.106 Using culture methods, it has been noted that there is a relatively simple, but indigenous, gastric microbiota in rats and mice, consisting of mainly of Firmicutes (genera Lactobacillus, Streptococcus, Clostridium, Veilonella), coliforms from Proteobacteria, anaerobic bacteria like Bifidobacterium spp and yeasts.1,107
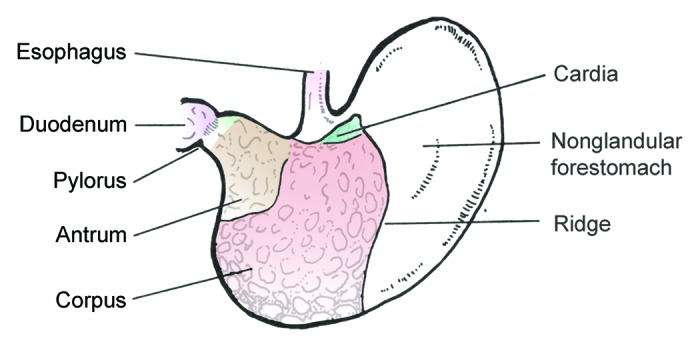
Figure 4. Illustration depicting anatomy of the mouse stomach. The anatomy of the gerbil stomach is similar. The nonglandular forestomach is the site of dense colonization by lactobacilli, which substantially contribute to the differences in the gastric microbiota of humans and rodents.
Mice
Using diverse sampling methods, mouse strain backgrounds and vendor sources, the normal gastric microbiota has been shown to be predominantly dominated by Firmicutes (genus Lactobacillus)(Table 3).71,77,103,104 Using culture and T-RFLP analysis, Lactobacillus represented > 99% of the bacteria in the stomach, with the presence of the remaining bacteria (Proteobacteria (genera Escherichia, Moraxella, Pasteurella, Enterobacter, and Actinobacillus), Firmicutes (genera Staphylococcus and Enterococcus), and Actinobacteria (genus Micrococcus)) at < 1%).103,104 Other studies have Lactobacillus spp as the most abundant genus in the gastric microbiota, but detect significant contributions (35–45%) from other bacterial phyla.71,77 However, while differences in levels of Lactobacillus spp prompted further investigation into the gastric microbiota, Rolig et al. found that the dominant phyla Firmicutes (74% of reads) was mainly composed of the class Clostridia (44% of reads) and not Lactobacillus spp in their control mice.76 In studies where Lactobacillus spp did not compose > 99% of the stomach microbiome, Bacteroidetes was the second most abundant phylum, and significant contributions were made by Cyanobacteria, Verrumicrobia, Proteobacteria and Actinobacteria76,77 The variability in results was highlighted by Rolig et al., who showed that C57BL/6 mice from different vendors had different levels of two different Lactobacillus spp and had different responses to H. pylori infection, highlighting the importance of husbandry and the environment on the gastrointestinal microbiota profile.76 Another possible explanation for the reported variability of Lactobacillus spp levels in the stomach is the inclusion of the squamous epithelium forestomach during sectioning of the stomach. In our studies, the squamous forestomach, which plays a limited role in H. pylori-induced pathogenesis, is routinely removed during necropsy.72,77 However, it has been noted that the squamous epithelium is the primary site of colonization of lactobacilli.108 Further standardization of methodologies is required to compare equivalent data.
Table 3. Gastric microbiota studies in mice.
| Study | Method | # of Samples | Age | Sex | Species | HP status | Phyla (%) | Genus information | HP effect |
|---|---|---|---|---|---|---|---|---|---|
| Kabir et al. 1997 | Culture | 3 | 5 wk-old + up to 9 wk infection | Male | BALB/c mice (Japan) | HP infected/no colonization | Firmicutes (99.9%), Proteobacteria (< 0.01%) | Firmicutes (Lactobacillus) = 99.9% and Firmicutes (Enterococcus and Staphylococcus) and Proteobacteria (Enterobacter) = < 0.01% | Blocked by high number of Lactobacillus spp |
| Aebischer et al. 2006 | Dot-blot hybridization, then Sanger-sequenced 16S rRNA library | 1 | 6–8 wk old + 2 mo | Female | BALB/c (Germany) | Uninfected | 60–90% Lactobacillus. 10–40% - other. | ||
| 1 | 6–8 wk old + 2 mo infection | Female | BALB/c (Germany) | HP P76 infected | Firmicutes, Proteobacteria, Bacteroidetes | Levels shift from uninfected | HP increases Firmicutes (Clostridium, Eubacterium, Ruminococcus and Streptococcus), Bacteroidetes (Bacteroides/Prevotella), Proteobacteria (Escherichia). Decreases Lactobaciillus. Effect independent of pH/pathology | ||
| Tan et al. 2007 | Culture and T-RFLP (two sets) | 9–10 mice/group | 6–8 wk old + 0.5, 3 or 6 mo | Female | C57BL/6 (Australia) | Uninfected | Firmicutes (Lactobacillus) = 99.9% | ||
| 9–10 mice/group | 6–8 wk old + 0.5, 3 or 6 mo infection | Female | C57BL/6 (Australia) | HP SS1 infected | No change from uninfected | Cultured Proteobacteria (E. coli, Moraxella osloensis, Actinobacillus muris, Pasteurella spp.) and Firmicutes(Enterococcus faecalis, Lactobacillus spp) | |||
| Lofgren et al. 2011 | 454 pyrosequencing of 16S rDNA amplicons | 2 | 6 wk old + 15 wk | Male | INS-GAS FVB/N (USA) | Uninfected | Firmicutes (55%), Bacteroidetes (25%), Cyanobacteria (15%), Proteobacteria and other (5%) | No H. pylori sequences detected. Observed 175 OTUs | |
| 3 | 6 wk old + 15 wk infection | Male | INS-GAS FVB/N (USA) | HP SS1 infected | Firmicutes (90%), Proteobacteria, Cyanobacteria, Bacteroidetes, and Tenericutes (10%) | Firmicutes (Lactobacillus) dominant | Low H. pylori reads. Increases OTUs in stomach, cecum and colon. Higher pathology. Observed 235 OTUs | ||
| Rolig et al. 2013 | PhyloChip | 5 | 7 wk old + 4 wk | Female | C57BL/6 (Taconic Farms/USA) | Uninfected | Firmicutes (73.4%), Bacteroidetes (11.8%) Verrucomicrobia (8.5%), Proteobacteria (3.2%), Actinobacteria (1.9%) (% of species) | Firmicutes (class Clostridia) 44% | |
| 4 | 7 wk old + 4 wk infection | Female | C57BL/6 (Taconic Farms/USA) | HP SS1 infected | Increases Firmicutes (class Clostridia), Proteobacteria (Helicobacter), Verrumicrobia. Decreased Firmicutes (class Bacilli), Bacteroidetes, Proteobacteria | ||||
| Lertpiriyapong et al. 2013 | qPCR for 3 ASF species | 27 | 7–11 wk old + 7 mo | 13 males, 9 females | INS-GAS FVB/N (USA) | Uninfected | Firmicutes, Bacteroidetes | 3 restricted ASF species | |
| 22 | 7–11 wk old + 7 mo infection | 15 males, 12 females | INS-GAS FVB/N (USA) | HP SS1 infected | Firmicutes, Bacteroidetes, Proteobacteria | 3 ASF species + H. pylori | Increased Firmicutes (Lactobacillus) and decreased Firmicutes (Clostridium) and Bacteroidetes (Bacteroides). H. pylori = 0.55% of bacteria in all mice. Increased pathology ~increased bacterial load |
N/A, Not available
Studies have begun to highlight the diversity in the gastric microbiota of mice and their interactions with H. pylori and its associated immunopathology. The difficulty in establishing infection in mice101,103 and the low levels of H. pylori in the mouse stomach72,77 reflect that the bacteria are not autochthonous to the mouse. Kabir et al. studied the effect of the gastric microbiota on H. pylori infection and found that multiple strains of H. pylori could colonize germ-free (GF) BALB/c mice, but failed to colonize specific-pathogen free (SPF) BALB/c mice when the predominant gastric bacteria were Lactobacillus spp103 Coinfection of H. pylori and L. salivarius of GF mice demonstrated that L. salivarius alone prevented H. pylori colonization of the mouse stomach.103 This result is similar to the clearance of H. felis from SPF C57BL/6 mice, where competition from Lactobacillus spp invading the gastric niche was postulated to have contributed to the eradication of H. felis.109
When infection is achieved, H. pylori represent 10–30% of the microbiota in the absence of significant pathology and < 5% of the bacteria in stomachs with significant disease.72,77 The decreasing levels highlights an inverse correlation between H. pylori levels and the degree of gastric pathology that is commonly observed in mice.110 In spite of a relatively small contribution in numbers, H. pylori exerts strong effects on the microbiota and overall health of the murine stomach. Using mouse-adapted H. pylori SS1, Tan et al. were able to infect C57BL/6 with a Lactobacillus-dominated gastric microbiota (> 99%).104 While the gastritis observed was mild, H. pylori were detected for the duration of the study and there was an increased gastric pH to 5, by 6 mo of infection. In spite of conditions associated with bacterial overgrowth, H. pylori infection did not cause shifts in bacterial composition.104 Rolig et al. also found that uninfected mice and mice with short-term H. pylori infection (4 wks) had little effect on the observed phyla (Firmicutes, Bacteroidetes, Verrucomicrobia, Proteobacteria, and Actinobacteria).76 As seen in the Maldonado-Contreras et al. study,82 analysis of the PhyloChip data detected specific taxa that varied with H. pylori infection. H. pylori caused increases in Firmicutes (class Clostridia), Proteobacteria (Helicobacter hepaticus) and Verrumicrobia, and was associated with decreases in Firmicutes (class Bacilli), Bacteroidetes and Proteobacteria.76 More dramatic effects were observed using H. pylori P76 in BALB/c mice with a more diverse gastric microbiota.71 H. pylori infected the stomach and increased the colonization of the stomach by lower bowel bacteria (Firmicutes (genera Clostridia, Eubacterium, Ruminococcus and Streptococcus), Bacteroidetes (genera Bacteroides/Prevotella), and Proteobacteria (genus Escherichia)), while a dramatic loss of Lactobacillus spp was observed (from > 60% in uninfected mice to 10–30% in infected mice). The shifts in the gastric microbiota were independent of significant changes in pH or pathology, implying that H. pylori infection may mediate initial alterations in the microbiota in a relatively healthy mouse stomach.71 However, vaccination against H. pylori caused a 100-fold reduction in H. pylori levels and abrogated shifts in the stomach microbiome.71
While Aebischer et al.71 and Rolig et al.76 assessed microbiota changes following acute infections with H. pylori, other models are needed to evaluate the overall effects of long-term H. pylori infection and its associated histopathological changes on the composition of the stomach microbiota. Of the models reviewed, the hypergastrinemic INS-GAS mice present the most rapid progression to atrophy and gastrointestinal intraepithelial neoplasia (GIN) in response to H. pylori,65,72,111,112 and presents an opportunity to observe changes in the microbiota associated with disease progression. Lofgren et al. demonstrated the importance of the microbiota in H. pylori-induced disease, as GF INS-GAS mice mono-associated with H. pylori had a delayed progression to neoplasia compared with SPF mice.77 In uninfected SPF mice, the phyla Firmicutes and Bacteroidetes accounted for > 80% of the represented bacteria with a large contribution of Cyanobacteria. H. pylori infection increased the percentage of Firmicutes to > 90% with a large proportion of Lactobacillus spp, which greatly reduced the contribution of Bacteroidetes (Fig. 5). Nevertheless, H. pylori infection actually increased the number of OTUs detected (235 in infected vs. 175 in uninfected),77 similar to increases in diversity observed in H. pylori infected humans.10 However, H. pylori infection induced no changes in the relative abundance of phyla in the cecum or the colon when compared with uninfected controls.77 Using a reductionist model consisting of 3 autochthonous murine bacteria, it was demonstrated that a simple gastric microbiota alone accelerated the progression to GIN compared with a GF mouse.72 Inclusion of H. pylori further accelerated the development of GIN in the INS-GAS mice with a restricted Altered Schaedler's Flora (rASF). H. pylori-infected SPF INS-GAS mice and H. pylori-infected rASF mice developed GIN at similar rates, indicating that the 3 species (ASF356 (Clostridium spp), ASF361 (Lactobacillus murinus) and ASF519 (Bacteroides spp) were able to recapitulate the effects of a complex microbiota.72 Including H. pylori, it was possible to track the concentrations of these 4 species using qPCR to gain insights into the colonization dynamics that might be associated with GIN development. The baseline composition in uninfected rASF was similar between male and female INS-GAS mice with Bacteroides species being dominant, followed by smaller, similar percentages of Lactobacillus and Clostridium species. As noted by Lofgren et al.,77 H. pylori infection increased the percentage of Lactobacillus spp (from ~15% to 65% in males and from 25% to 95% in females). Consequently, decreases were observed in Clostridium (males 25% to 12% and females from 25% to < 5%), and Bacteroides (males 60% to 20% and females 50% to < 5%).72 To test the value of the reductionist model, the authors used qPCR to track ASF species within the complex gastric microbiota in SPF males and females. In SPF INS-GAS mice, H. pylori infection decreased the levels of Bacteroides (ASF519) and Clostridium (ASF356), while increased levels were seen in two Lactobacillus spp (ASF360 and ASF361) and Eubacterium plexicaudatum (ASF492).72
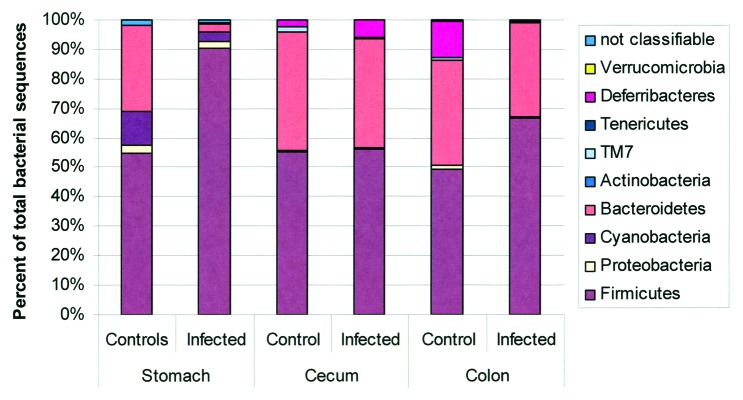
Figure 5. Microbiota composition in stomach, cecum, and colon of H pylori-infected male INS-GAS mice (n = 3, 15 weeks postinfection) vs. uninfected controls (n = 2). Note the significant increase in the relative abundance of Firmicutes and decrease of Bacteroidetes in the stomachs of H pylori–infected INS-GAS mice (p < 0.05), whereas no significant changes were observed in the colon and ceca of H. pylori-infected mice. Reproduced with permission from Lofgren et al. 2011.77
Changes in Lactobacillus levels after H. pylori infection seem contradictory. Decreasing Lactobacillus levels in BALB/c and C57BL/6 mice acutely infected with H. pylori, but minimal gastric pathology71,76 contrast to the increasing Lactobacillus levels noted in INS-GAS FVB/n mice after long-term H. pylori infection associated with gastritis and atrophy.72,77 While husbandry conditions and strain background are important factors to consider, the fluctuation in lactobacilli may reflect temporal dynamics dependent on pathology. Increased pathology with increasing gastric atrophy after long-term infection has been associated with higher gastric, bacterial levels.72 Therefore, severe gastric lesions may facilitate colonization by lower bowel bacteria.
Another topic of interest is the role of antibiotics on H. pylori-induced disease and its effect on other gastric microbiota. Rolig et al. demonstrated that antibiotic treatment of mice altered the levels of 4400 OTUs in the stomachs of treated mice compared with untreated mice. These changes in the gastric microbiota reduced the severity of gastritis following subsequent H. pylori infection.76 Similarly, antibiotic therapy combined with Sulindac significantly delayed the normal progression of gastric pathology observed in uninfected INS-GAS mice, which like Lofgren et al., demonstrates that other microorganisms, whether in the stomach or elsewhere, may contribute to the severity of gastritis.77,113
Mongolian gerbils
The Mongolian gerbil is a useful model of H. pylori pathogenesis. Favorable attributes of the model include a low intragastric pH,105 and a male-predominant predisposition to increased susceptibility to progressive H. pylori gastritis.66,78,114-119 Limitations of the model include lack of immunologic reagents and lack of availability of inbred strains. In spite of its importance in H. pylori research, there have been no NGS studies that directly assess the abundance and structure of the gastric microbiota in gerbils. The phyla commonly observed in current, published studies are Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes, with the first two being present in all studies (Table 4).78,117-119 The gastric microbiota of uninfected Mongolian gerbils are dominated by the genus Lactobacillus similar to findings in mice.117-119 Lactobacilli are prevalent in gerbil stomachs like the mouse, due to their colonization of the non-glandular forestomach.108
Table 4. Gastric microbiota studies in Mongolian gerbils.
| Study | Method | # of Samples | Age | Sex | HP status | Phyla | Genus information | HP effect |
|---|---|---|---|---|---|---|---|---|
| Sun et al. 2003 | TTGE/NGS-sequenced 16S rDNA library | 5 | 5 wk old + 12 wk | Male | Uninfected | Firmicutes, Proteobacteria | Firmicutes (Lactobacillus) (82.4%), and Proteobacteria (Pseudomonas), Proteobacteria (Acinetobacter), Firmicutes (Bacillus, Enterococcus, Paenibacillus) (17.6%) | No info on levels, just identification |
| 5 | 5 wk old + 12 wk infection | Male | HP ATCC 43504 | Firmicutes, Proteobacteria, Actinobacteria | Firmicutes (Lactobacillus) (51.7%), Proteobacteria (Helicobacter pylori) (37.1%) and Proteobacteria (Pseudomonas, Acinetobacter), Actinobacteria (Corynebacterium), Firmicutes (Enterococcus, and Staphylococcus) (11.2%) | Severe chronic active gastritis | ||
| Zaman et al. 2010 | Culture + API | 1 | 5 wk old + 8 wk infection | Female | H pylori TK1042 (low detection) | Firmicutes, Proteobacteria, Actinobacteria | Firmicutes (Lactobacillus), Proteobacteria (Escherichia, Kluyvera), Actinobacteria (Bifidobacterium, Actinomyces) | Based on culture of single gerbil. Altered obligate anaerobe composition. |
| 1 | 5 wk old + 8 wk infection | Female | H pylori TK1402 (high detection) | Firmicutes, Proteobacteria, Actinobacteria | Firmicutes (Eubacterium), Proteobacteria (Escherichia, Kluyvera), Actinobacteria (Bifidobacterium, Actinomyces) | |||
| Yin et al. 2011 | Culture | 8 | 6–8 wk old + 12 wk | Male/ Female | Uninfected | Firmicutes, Actinobacteria | Firmicutes(Lactobacillus), Actinobacteria (Bifidobacterium) | |
| 3 | 6–8 wk old + 12 wk infection | Male/ Female | HP J99 | Firmicutes, Actinobacteria, Bacteroidetes | Observed new species and shifts in microbiota | Observed Bacteroidetes (Bacteroides) and Firmicutes (Enterococcus). Trend of increasing Actinobacteria (Bifidobacterium). Increased gastritis | ||
| Osaki et al. 2012 | qPCR | 5 | 8 wk old + 1 y | Male | Uninfected | Firmicutes, Actinobacteria | Actinobacteria (Atopobium spp, Bifidobacterium spp), Firmicutes (Clostridium coccoides, C. leptum, Enterococcus spp and Lactobacillus spp) | |
| 5 | 8 wk old + 1 y infection | Male | HP positive | Firmicutes, Actinobacteria | Actinobacteria (Atopobium spp, Bifidobacterium spp), Firmicutes (Clostridium coccoides, C. leptum, Enterococcus spp and Lactobacillus spp) | vs. uninfected - lower Clostridium coccoides | ||
| 6 | 8 wk old + 1 y infection | Male | HP negative | Firmicutes, Actinobacteria, Bacteroidetes | Actinobacteria (Atopobium, Bifidobacterium), Firmicutes (Clostridium, Enterococcus, Lactobacillus, Eubacterium) and Bacteroidetes(Prevotella) | vs. uninfected -lower Clostridium and Bifidobacterium. Higher Atopobium |
N/A, Not available
Three studies examined the effects of short, 8–12 wk H. pylori infections on the gastric microbiota, and observed few changes in the gastric phyla but documented changes in less abundant gastric genera.78,118,119 In gerbils with H. pylori infection, two groups saw decreases in lactobacilli, but both had small numbers and depended on culture for identification.78,119 The most comprehensive study utilized TTGE and NGS to sequence a 16S rRNA clone library, and found a decrease in Lactobacillus diversity with H. pylori infection and noted changes in some of the least abundant species.118 Osaki et al. used qPCR to monitor 15 species of interest after 1 y of CagA+ H. pylori TK1402 infection.117 In their control gerbils, Lactobacillus was the most abundant genus, followed closely by Enterococcus, and finally equal levels of Atopobium spp and Clostridium spp The H. pylori infection efficacy was 45%. H. pylori positive gerbils had lower levels of Clostridium coccoides compared with controls. H. pylori-infected but negative animals, had lower levels of Clostridium coccoides and C. leptum, as well as Bifidobacterium spp The H. pylori negative group had increased levels of Actinobacteria:Atopobium and were the only group with detectable levels of the Firmicutes species Eubacterium cylindroides and the Bacteroidetes genus Prevotella.117 It is possible that the gerbils that cleared H. pylori had increased gastric damage that led to altering the gastric niche.
Highlighted differences between rodent and human gastric microbiota
The two major differences between the rodent and human gastric microbiota are 1) the prevalence and abundance of H. pylori in the human stomach, and 2) the effect of the non-glandular stomach on the bacterial species composition in the rodent stomach. The phyla observed in H. pylori positive humans in order of abundance are Proteobacteria, Firmicutes and Actinobacteria. Proteobacteria are otherwise not the main phyla in any reviewed study, whether in rodents or H. pylori-uninfected humans. In the absence of H. pylori, the most abundant phyla in humans are Firmicutes, Bacteroidetes and Actinobacteria, which arguably resembles the structure observed in the normal mouse (Firmicutes, Bacteroidetes, and varying contributions by Cyanobacteria, Verrumicrobia, Proteobacteria, and Actinobacteria). However, in humans, the main contributors to these phyla are the genera Streptococcus and Prevotella, which are not abundant in either mice or gerbils. Instead, Lactobacillus colonize the squamous epithelium (with subsequent spillover effect and presence in samples of glandular stomach) and can often outcompete all other species.
Host and Environmental Factors that Promote H. pylori Pathogenesis and Influence the Gastric Microbiota
Having discussed the important role of pH in maintaining stomach function and the studies evaluating changes in the gastric microbiota in the context of H. pylori pathogenesis, this section will evaluate other factors that affect H. pylori pathogenesis and have the potential to perturb the microbiota.
Cytokine gene polymorphisms
Cytokine gene polymorphisms in IL-1β and IL-8 have been associated with increases in gastric cancer risk. IL-1β is a proinflammatory cytokine that can inhibit acid secretion in the stomach. H. pylori–infected individuals with polymorphisms in the IL-1β gene or the IL-1 receptor antagonist have an increased risk of developing gastric atrophy and gastric cancer.120,121 In both mice and humans, overexpression of IL-1β has been linked to increased risk of gastric cancer.120,122 Another proinflammatory cytokine, IL-8, is involved in the recruitment and activation of neutrophils. A polymorphism in the IL-8 promoter region that increases IL-8 levels, is associated with increased gastric cancer risk,123,124 and stomach cancers with high levels of IL-8 levels have a poor prognosis.125 Recent clinical and epidemiological studies link increased mRNA and serum levels of CXCL1, another cytokine in the CXC chemokine family to which IL-8 belongs, to gastric cancer.126-128 In mouse models of gastric cancer, CXCL1 and transgenic IL-8 are upregulated by Helicobacter infection and increased CXCL1 expression correlates with dysplasia scores.112,129,130 It is not well understood if changes in systemic inflammatory status are produced by changes in microbiota composition or if they assist in shaping the composition of the microbiome in diverse sites of the body. However, recent studies have shown a correlation between detrimental changes in the fecal microbiota composition and increases in proinflammatory cytokines that lead to disease. Biagi et al. correlated the proliferation of Proteobacteria and reduction in Firmicutes and Bacteroidetes with increases in IL-6 and IL-8.131 The symbiont Bacteroides fragilis expressing polysaccharide A can suppress proinflammatory IL-17 production induced by Helicobacter hepaticus, a bacterium with pathogenic potential.132 The loss of a member of the Firmicutes, Faecalibacterium prausnitzii, is linked with higher risk of recurrence of Crohn Disease (CD) in humans.133 Experiments in vivo demonstrated that F. prausnitzii is protective in a chemically induced colitis model due to its antiinflammatory effects which block NF-kappaB activation and IL-8 production.133 Similarly, intestinal commensals, specifically segmented filamentous bacteria (SFB), have been implicated with the regulation of gut immune maturation and the production of IL-17.134
Age
The fecal microbiota experiences dramatic changes from birth to death. From birth to the first three years of life, a less complex microbiota increases in diversity and stabilizes, culminating in a stable and species-rich state throughout adulthood that ultimately declines in symbionts in old age.131,135-138 These observations were robust and independent of geography in three populations worldwide.135 As noted previously, the incidence of intestinal-type gastric adenocarcinomas associated with H. pylori increases with age, with a peak incidence in the eighth decade of life.139 The association of disease with increasing age fits into the Correa model of H. pylori-induced gastric pathogenesis and changes in acid secretion that are common to aging, as described above. The cumulative lifetime exposure to RONS, proinflammatory cytokines and tissue damage promoted by H. pylori are coupled in old age with a decreasing capacity to deal with antigens (immunosenescence) and reduced ability to control inflammatory responses (inflammaging). Immunosenescence, the overall decline of immunity associated with age, is demonstrated in impaired antigen presentation, reduced cytotoxic function, accumulation of effector T cells, decreased output of naive T cells and reduced B cell production in the elderly.140 Inflammaging is a process illustrative of H. pylori infection, in which chronic inflammation over time can overwhelm the body's repair capacity and promotes host damage and disease.141 The aging of the immune system increases the susceptibility to pathologies associated with inflammation, such as cardiovascular disease, autoreactivity and microbial infections.140 These processes have been shown to affect the microbiome of the elderly, for example changes in the composition of Firmicutes (leading to inflammation), and increases in the proportion of Bacteroidetes.131,136 Decreases or shifts in beneficial Clostridium clusters XIVa and IV, which include several butyrate producers, have also been observed in many elderly individuals.131,136,142,143 Greater proportions of Enterobacteriaceae were found in elderly volunteers independent of geographic location in Europe, but the importance of this finding remains unresolved.144
Gender
Irrespective of location and ethnicity, males are twice as likely as females to develop gastric cancer.139 The pattern of the M/F incidence of gastric cancer is a global phenomenon, equally seen in populations with high and low risk for gastric cancer. This remains one of the unresolved epidemiological questions given that the sexual dimorphism has not been explained by putative risk factors such as smoking, alcohol and obesity.145 Importantly, epidemiological evidence points to the protective role of female hormones146 and this variable is now being studied using in vivo models.111,112 However, the differences in gastric microbiota between males and females is not well established. Data from the Human Microbiome project found a low degree of correlation between the microbiome and gender,2 and a study of European, American and Japanese subjects identified three distinct phylogenetic classifications of gut microbiota (enterotypes) in all their subjects but found no strong correlation to gender.147 However, examining functional biomarkers, five functional modules differentiated males and females, indicating that biomarkers derived from metagenomics may be more informative than phylogenetic biomarkers.147 In contrast a European study noted strong gender effects with males having higher levels of the Bacteroides-Prevotella group than females.144 Two recent studies have also demonstrated that sex exerts an effect in murine models.72,148 Non-obese diabetic (NOD) mice develop type 1 diabetes with higher incidence in females. However, development of type 1 diabetes is prevented in GF female NOD mice and GF female NOD mice that receive a microbiota transplant from a male NOD mouse.148 In the INS-GAS model of gastric cancer, male mice develop gastric cancer while female mice develop cancer at a much lower frequency.111 The GI microbiota contributes to pathogenesis as GF INS-GAS mice have a delayed onset of cancer.77 A restricted microbiota consisting of H. pylori and three species of the 8 species of Altered Schaedler's Flora was able to promote gastric cancer in male mice, which exhibited a higher bacterial colonization levels and different distribution of bacteria compared with female mice, highlighting that differences in microbiota between males and females can affect H. pylori-associated pathogenesis.72
Ethnicity/geography
The incidence of gastric cancer varies in different parts of the world with the highest incidence rates documented in Eastern Asia, Eastern Europe, and South America, while North America and Africa show the lowest recorded rates.37 In the United States between 2003–2007, gastric cancer mortality rates followed ethnic divisions as mortality rates were highest among African-Americans, followed by Asian/Pacific Islanders, Native Americans, Hispanics, and Caucasians.149 The gastric cancer data are reflective of the prevalence of H. pylori which exhibits geographical variation with 80–90% prevalence in various developing countries and less than 40% prevalence in industrialized countries.22 However, racial differences alone do not explain gastric cancer rates, as migrants from high-risk regions have a decreased gastric cancer risk when they relocate to a lower risk area.150
Similarly, many studies evaluating the fecal microbiota between different geographic locations or different ethnic groups have found large variation in specific bacterial groups (but not at the phyla level) between these populations.2,135,144,151 There has been considerable interest in the variation between Prevotella spp and Bacteroides spp as they exhibit considerable variation between populations.135,144,151 However, others have classified fecal microbiomes into three enterotypes with little correlation between gut microbiomes and nationality.147 Further studies are necessary to separate the effects of ethnicity/geography from diet (and other variables) as they may be proxies for each other, as diet has been previously shown to play an important role in defining the composition of the microbiome.152
Diet
In the Correa model, it is postulated that diet plays a role in the progression of gastric cancer initiated by H. pylori. Excessive salt and nitrates may promote inflammation, while deficiencies in ascorbic acid or low intake of fresh fruits and vegetables, may decrease the stomach's ability to deal with inflammation.45,153 While vitamin C supplementation has shown no protective effects,154 high incidences of gastric cancer have been associated to countries with high salt intake.155 Animal models assessing the role of high salt in H. pylori gastric pathology have yielded mixed results.115,156 However, a recent study has demonstrated that salt upregulates the expression of H. pylori CagA.157 As the effects of salt may not be independent of CagA, studies assessing the effect of salt using the mouse-adapted H. pylori strain SS1, which lacks a functional CagA,102 may have to be interpreted with this variable in mind. Indeed, in a subsequent study, high salt diets promoted gastric adenocarcinomas in Mongolian gerbils infected with H. pylori strains with a functional CagA, but not with H. pylori lacking CagA.114 Iron is another important dietary factor, as iron deficiency has been associated with the presence of H. pylori158,159 and hypochlorhydria.49,56 Animal models have demonstrated that Helicobacter infection results in iron deficiency,158 while reduced dietary iron coupled with H. pylori infection promoted gastric cancer.116 Interestingly, reduced serum ferritin levels in patients were associated with H. pylori isolates that induced more robust proinflammatory responses in vitro.116 The contributions of diet to H. pylori pathogenesis are reviewed in greater detail by Peek and Cover.160
Diet is also believed to strongly influence the microbiota.143,152 In an elderly population, the quality and diversity of the diet correlated with microbial diversity, as well as changes in frailty, inflammation and altered abundances of short chain fatty acids (SCFAs) producing bacteria.143 The oldest and frailest subjects had the fewest copies of genes involved in SCFA production in their fecal metagenomes, and in general had increased Bacteroides spp, Parabacteroides spp or Alistipes spp coupled with a loss of Prevotella spp143 Diets rich in carbohydrates and polysaccharides result in increases in Prevotella while diets rich in protein and animal fat promote Bacteroides,151,152 and these genera seem to mutually exclude each other and define stable enterotypes.135,147 Although, Prevotella spp are associated with “healthier” diets and lifestyles, it is worth noting that the family Prevotellaceae has also been associated with the development of inflammatory bowel disease and periodontal disease.13
Animal studies have also demonstrated the strong effects of diet on the microbiome. Obese mice have a greater capacity to harvest energy from their diet compared with lean mice, and transplantation of the “obese” and “lean” microbiota to GF mice led to higher levels of body fat in the first group.161 As in H. pylori pathogenesis, iron plays an important role in the health of the lower bowel. Depletion of luminal iron in a mouse model of Crohn Disease-like ileitis increased the numbers of Bifidobacterium, Succinivibrio, Clostridium and Turicibacter, while numbers of potentially pathogenic Desulfovibrio spp were decreased.162 In a chemically induced model of colitis, decreases in Firmicutes and increases in Bacteroidetes and the family Enterobacteriaceae were noted, but these effects were prevented with ferric iron supplementation.163
Other infections
Infections, both clinical and subclinical, have a great impact on H. pylori pathogenesis, as well as the microbiota. Infections by parasites and other non-gastric helicobacters have been shown to modulate H. pylori pathogenesis. Coinfection of mice with the nematode Heligmosomoides polygyrus and H. felis induced an antiinflammatory TH2 response, and concomitant reduction in proinflammatory TH1 immune responses normally induced by H. felis.164 The development of gastric atrophy was also significantly reduced.164 The opposite effect was observed by modulating the host's response using a parasite that induces a strong TH1 response, such as Toxoplasma gondii. Coinfection with T. gondii exacerbated H. felis infection leading to increased morbidity.165 Similarly, intestinal helminths had a higher prevalence in humans residing in low gastric cancer risk areas in Colombia.166 In a Chinese population, individuals coinfected with H. pylori and Schistosoma japonicum were protected from atrophy.167 Bacteria that colonize the lower bowel in mice were able to both enhance and minimize H. pylori pathogenesis by modulating the TH1, TH17 and TREG responses.168,169 These effects were highly species-specific, as coinfections with either H. bilis or H. muridarum reduced H. pylori-induced gastric pathology and induced a TH2 immune response, while coinfection with H. hepaticus enhanced the proinflammatory TH17 response.168,169 Similarly, parasite infections and Helicobacter infections have been shown to shift the composition of intestinal bacteria, and increase the diversity of the gastric microbiota.71,170-173 H. polygyrus infection significantly elevated the numbers of lactobacilli in the ileum of infected mice,173 which is of interest as Lactobacillus spp have been reported to attenuate H. pylori gastritis in mice.174 We have conducted studies that show that Helicobacter spp infection in mice directly affects the levels of bacteria in mice with restricted microbiota,171,172 but further studies are necessary to evaluate changes in complex microbiota that may further modulate the immunomodulatory effects of subclinical Helicobacter spp infections. The idea that infection of other compartments of the GI tract may directly affect gastric pathogenesis raises the question of whether direct influence of the gastric microbiota is necessary to influence disease outcome. Conversely, H. pylori infection of the stomach has been implicated with decreased incidence of inflammatory bowel disease (IBD) and esophageal cancer,87,175 implying that infections of the stomach can alter the severity of disease of both the upper and lower GI tract.
Concluding Remarks
The role of the bacteria in the development of gastritis, ulcers and cancer has generated considerable debate. The origins of these hypotheses were the observation that bacterial overgrowth occurred in hypochlorhydric stomachs. H. pylori infection is a major cause of hypochlorhydria and has a major role in the progression from gastritis to atrophy and finally to gastric cancer. H. pylori and the associated changes in the stomach alter the ecological niche inhabited by the gastric microbiota. However, the gastric microbiota also competes, as observed in rodents and older people, with H. pylori for a gastric niche, and may play an important role in the progression of disease. More studies involving the microbiota-host-environment interactions are needed to fully understand the role of gastric bacteria in human health and disease.
Acknowledgments
This work was supported by National Institutes of Health grants P01CA028842 (JGF), P01CA026731 (JGF), and P30ES002109 (JGF).
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/26205
References
- 1.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 2.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill M. Normal and pathological microbial flora of the upper gastrointestinal tract. Scand J Gastroenterol Suppl. 1985;111:1–6. doi: 10.3109/00365528509093747. [DOI] [PubMed] [Google Scholar]
- 6.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–5. doi: 10.1016/S0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 7.Warren JR, Marshall BJ. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273–5. [PubMed] [Google Scholar]
- 8.Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, Lau JY, Sung JJ, Ding C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–7. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theisen J, Nehra D, Citron D, Johansson J, Hagen JA, Crookes PF, DeMeester SR, Bremner CG, DeMeester TR, Peters JH. Suppression of gastric acid secretion in patients with gastroesophageal reflux disease results in gastric bacterial overgrowth and deconjugation of bile acids. J Gastrointest Surg. 2000;4:50–4. doi: 10.1016/S1091-255X(00)80032-3. [DOI] [PubMed] [Google Scholar]
- 12.Simon GL, Gorbach SL. Intestinal flora in health and disease. Gastroenterology. 1984;86:174–93. [PubMed] [Google Scholar]
- 13.Korecka A, Arulampalam V. The gut microbiome: scourge, sentinel or spectator? J Oral Microbiol 2012; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manson JM, Rauch M, Gilmore MS. The commensal microbiology of the gastrointestinal tract. Adv Exp Med Biol. 2008;635:15–28. doi: 10.1007/978-0-387-09550-9_2. [DOI] [PubMed] [Google Scholar]
- 16.Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest. 2007;117:60–9. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schubert ML, Peura DA. Control of gastric acid secretion in health and disease. Gastroenterology. 2008;134:1842–60. doi: 10.1053/j.gastro.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Martinsen TC, Bergh K, Waldum HL. Gastric juice: a barrier against infectious diseases. Basic Clin Pharmacol Toxicol. 2005;96:94–102. doi: 10.1111/j.1742-7843.2005.pto960202.x. [DOI] [PubMed] [Google Scholar]
- 19.Bhaskar KR, Garik P, Turner BS, Bradley JD, Bansil R, Stanley HE, LaMont JT. Viscous fingering of HCl through gastric mucin. Nature. 1992;360:458–61. doi: 10.1038/360458a0. [DOI] [PubMed] [Google Scholar]
- 20.Corfield AP, Carroll D, Myerscough N, Probert CS. Mucins in the gastrointestinal tract in health and disease. Front Biosci. 2001;6:D1321–57. doi: 10.2741/Corfield. [DOI] [PubMed] [Google Scholar]
- 21.Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:G922–9. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 22.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–90. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11:385–99. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauerfeind P, Garner R, Dunn BE, Mobley HL. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. The HP0165-HP0166 two-component system (ArsRS) regulates acid-induced expression of HP1186 alpha-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J Bacteriol. 2007;189:2426–34. doi: 10.1128/JB.01492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun. 2007;75:3747–57. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol. 2006;188:2656–65. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navabi N, Johansson ME, Raghavan S, Lindén SK. Helicobacter pylori infection impairs the mucin production rate and turnover in the murine gastric mucosa. Infect Immun. 2013;81:829–37. doi: 10.1128/IAI.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz JM, Durham CG, Ho SB, Lorenz RG. Gastric mucus alterations associated with murine Helicobacter infection. J Histochem Cytochem. 2009;57:457–67. doi: 10.1369/jhc.2009.952473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiedemann T, Hofbaur S, Tegtmeyer N, Huber S, Sewald N, Wessler S, Backert S, Rieder G. Helicobacter pylori CagL dependent induction of gastrin expression via a novel αvβ5-integrin-integrin linked kinase signalling complex. Gut. 2012;61:986–96. doi: 10.1136/gutjnl-2011-300525. [DOI] [PubMed] [Google Scholar]
- 31.Saha A, Backert S, Hammond CE, Gooz M, Smolka AJ. Helicobacter pylori CagL activates ADAM17 to induce repression of the gastric H, K-ATPase alpha subunit. Gastroenterology. 2010;139:239–48. doi: 10.1053/j.gastro.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Keeffe J, Moran AP. Conventional, regulatory, and unconventional T cells in the immunologic response to Helicobacter pylori. Helicobacter. 2008;13:1–19. doi: 10.1111/j.1523-5378.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, Liu XF, Zhuang Y, Zhang JY, Liu T, Yin Z, Wu C, Mao XH, Jia KR, Wang FJ, et al. Helicobacter pylori-induced Th17 responses modulate Th1 cell responses, benefit bacterial growth, and contribute to pathology in mice. J Immunol. 2010;184:5121–9. doi: 10.4049/jimmunol.0901115. [DOI] [PubMed] [Google Scholar]
- 34.Hitzler I, Sayi A, Kohler E, Engler DB, Koch KN, Hardt WD, Müller A. Caspase-1 has both proinflammatory and regulatory properties in Helicobacter infections, which are differentially mediated by its substrates IL-1β and IL-18. J Immunol. 2012;188:3594–602. doi: 10.4049/jimmunol.1103212. [DOI] [PubMed] [Google Scholar]
- 35.Oertli M, Sundquist M, Hitzler I, Engler DB, Arnold IC, Reuter S, Maxeiner J, Hansson M, Taube C, Quiding-Järbrink M, et al. DC-derived IL-18 drives Treg differentiation, murine Helicobacter pylori-specific immune tolerance, and asthma protection. J Clin Invest. 2012;122:1082–96. doi: 10.1172/JCI61029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 37.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 38.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 39.Chan AO, Wong BC, Lam SK. Gastric cancer: past, present and future. Can J Gastroenterol. 2001;15:469–74. doi: 10.1155/2001/850308. [DOI] [PubMed] [Google Scholar]
- 40.Blaser MJ. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999;179:1523–30. doi: 10.1086/314785. [DOI] [PubMed] [Google Scholar]
- 41.Kumar V, Abbas AK, Fausto N, Aster J. Robbins and Cotran Pathologic Basis of Disease. Philadelphia, PA: Saunders Elsevier, 2010. [Google Scholar]
- 42.Lauren P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. An Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 43.Cuello C, López J, Correa P, Murray J, Zarama G, Gordillo G. Histopathology of gastric dysplasias: correlations with gastric juice chemistry. Am J Surg Pathol. 1979;3:491–500. doi: 10.1097/00000478-197912000-00002. [DOI] [PubMed] [Google Scholar]
- 44.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–5. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 45.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–40. [PubMed] [Google Scholar]
- 46.Genta RM. Helicobacter pylori, inflammation, mucosal damage, and apoptosis: pathogenesis and definition of gastric atrophy. Gastroenterology. 1997;113(Suppl):S51–5. doi: 10.1016/S0016-5085(97)80012-1. [DOI] [PubMed] [Google Scholar]
- 47.Li Q, Karam SM, Gordon JI. Diphtheria toxin-mediated ablation of parietal cells in the stomach of transgenic mice. J Biol Chem. 1996;271:3671–6. doi: 10.1074/jbc.271.7.3671. [DOI] [PubMed] [Google Scholar]
- 48.van den Brink GR, Hardwick JC, Tytgat GN, Brink MA, Ten Kate FJ, Van Deventer SJ, Peppelenbosch MP. Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology. 2001;121:317–28. doi: 10.1053/gast.2001.26261. [DOI] [PubMed] [Google Scholar]
- 49.Stockbruegger RW. Bacterial overgrowth as a consequence of reduced gastric acidity. Scand J Gastroenterol Suppl. 1985;111:7–16. doi: 10.3109/00365528509093749. [DOI] [PubMed] [Google Scholar]
- 50.Svendsen JH, Dahl C, Svendsen LB, Christiansen PM. Gastric cancer risk in achlorhydric patients. A long-term follow-up study. Scand J Gastroenterol. 1986;21:16–20. doi: 10.3109/00365528609034615. [DOI] [PubMed] [Google Scholar]
- 51.Ahn JS, Eom CS, Jeon CY, Park SM. Acid suppressive drugs and gastric cancer: a meta-analysis of observational studies. World J Gastroenterol. 2013;19:2560–8. doi: 10.3748/wjg.v19.i16.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tannenbaum SR, Wishnok JS, Leaf CD. Inhibition of nitrosamine formation by ascorbic acid. Am J Clin Nutr. 1991;53(Suppl):247S–50S. doi: 10.1093/ajcn/53.1.247S. [DOI] [PubMed] [Google Scholar]
- 53.Gramlich G, Zhang J, Nau WM. Increased antioxidant reactivity of vitamin C at low pH in model membranes. J Am Chem Soc. 2002;124:11252–3. doi: 10.1021/ja026927b. [DOI] [PubMed] [Google Scholar]
- 54.Williams C, McColl KE. Review article: proton pump inhibitors and bacterial overgrowth. Aliment Pharmacol Ther. 2006;23:3–10. doi: 10.1111/j.1365-2036.2006.02707.x. [DOI] [PubMed] [Google Scholar]
- 55.Yeomans ND, Brimblecombe RW, Elder J, Heatley RV, Misiewicz JJ, Northfield TC, Pottage A. Effects of acid suppression on microbial flora of upper gut. Dig Dis Sci. 1995;40(Suppl):81S–95S. doi: 10.1007/BF02214873. [DOI] [PubMed] [Google Scholar]
- 56.Sanduleanu S, Jonkers D, de Bruïne A, Hameeteman W, Stockbrügger RW. Changes in gastric mucosa and luminal environment during acid-suppressive therapy: a review in depth. Dig Liver Dis. 2001;33:707–19. doi: 10.1016/S1590-8658(01)80050-5. [DOI] [PubMed] [Google Scholar]
- 57.Moayyedi P, Wason C, Peacock R, Walan A, Bardhan K, Axon AT, Dixon MF. Changing patterns of Helicobacter pylori gastritis in long-standing acid suppression. Helicobacter. 2000;5:206–14. doi: 10.1046/j.1523-5378.2000.00032.x. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki M, Miura S, Suematsu M, Fukumura D, Kurose I, Suzuki H, Kai A, Kudoh Y, Ohashi M, Tsuchiya M. Helicobacter pylori-associated ammonia production enhances neutrophil-dependent gastric mucosal cell injury. Am J Physiol. 1992;263:G719–25. doi: 10.1152/ajpgi.1992.263.5.G719. [DOI] [PubMed] [Google Scholar]
- 59.Koh TJ, Chen D. Gastrin as a growth factor in the gastrointestinal tract. Regul Pept. 2000;93:37–44. doi: 10.1016/S0167-0115(00)00176-2. [DOI] [PubMed] [Google Scholar]
- 60.Iijima K, Sekine H, Koike T, Imatani A, Ohara S, Shimosegawa T. Long-term effect of Helicobacter pylori eradication on the reversibility of acid secretion in profound hypochlorhydria. Aliment Pharmacol Ther. 2004;19:1181–8. doi: 10.1111/j.1365-2036.2004.01948.x. [DOI] [PubMed] [Google Scholar]
- 61.Annibale B, Aprile MR, D’ambra G, Caruana P, Bordi C, Delle Fave G. Cure of Helicobacter pylori infection in atrophic body gastritis patients does not improve mucosal atrophy but reduces hypergastrinemia and its related effects on body ECL-cell hyperplasia. Aliment Pharmacol Ther. 2000;14:625–34. doi: 10.1046/j.1365-2036.2000.00752.x. [DOI] [PubMed] [Google Scholar]
- 62.Eissele R, Brunner G, Simon B, Solcia E, Arnold R. Gastric mucosa during treatment with lansoprazole: Helicobacter pylori is a risk factor for argyrophil cell hyperplasia. Gastroenterology. 1997;112:707–17. doi: 10.1053/gast.1997.v112.pm9041231. [DOI] [PubMed] [Google Scholar]
- 63.Kuipers EJ, Lundell L, Klinkenberg-Knol EC, Havu N, Festen HP, Liedman B, Lamers CB, Jansen JB, Dalenback J, Snel P, et al. Atrophic gastritis and Helicobacter pylori infection in patients with reflux esophagitis treated with omeprazole or fundoplication. N Engl J Med. 1996;334:1018–22. doi: 10.1056/NEJM199604183341603. [DOI] [PubMed] [Google Scholar]
- 64.Takaishi S, Cui G, Frederick DM, Carlson JE, Houghton J, Varro A, Dockray GJ, Ge Z, Whary MT, Rogers AB, et al. Synergistic inhibitory effects of gastrin and histamine receptor antagonists on Helicobacter-induced gastric cancer. Gastroenterology. 2005;128:1965–83. doi: 10.1053/j.gastro.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 65.Wang TC, Dangler CA, Chen D, Goldenring JR, Koh T, Raychowdhury R, Coffey RJ, Ito S, Varro A, Dockray GJ, et al. Synergistic interaction between hypergastrinemia and Helicobacter infection in a mouse model of gastric cancer. Gastroenterology. 2000;118:36–47. doi: 10.1016/S0016-5085(00)70412-4. [DOI] [PubMed] [Google Scholar]
- 66.Hagiwara T, Mukaisho K, Nakayama T, Sugihara H, Hattori T. Long-term proton pump inhibitor administration worsens atrophic corpus gastritis and promotes adenocarcinoma development in Mongolian gerbils infected with Helicobacter pylori. Gut. 2011;60:624–30. doi: 10.1136/gut.2010.207662. [DOI] [PubMed] [Google Scholar]
- 67.Ekström AM, Held M, Hansson LE, Engstrand L, Nyrén O. Helicobacter pylori in gastric cancer established by CagA immunoblot as a marker of past infection. Gastroenterology. 2001;121:784–91. doi: 10.1053/gast.2001.27999. [DOI] [PubMed] [Google Scholar]
- 68.Cai X, Carlson J, Stoicov C, Li H, Wang TC, Houghton J. Helicobacter felis eradication restores normal architecture and inhibits gastric cancer progression in C57BL/6 mice. Gastroenterology. 2005;128:1937–52. doi: 10.1053/j.gastro.2005.02.066. [DOI] [PubMed] [Google Scholar]
- 69.El-Zimaity HM, Ota H, Graham DY, Akamatsu T, Katsuyama T. Patterns of gastric atrophy in intestinal type gastric carcinoma. Cancer. 2002;94:1428–36. doi: 10.1002/cncr.10375. [DOI] [PubMed] [Google Scholar]
- 70.Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, Welling GW. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–75. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aebischer T, Fischer A, Walduck A, Schlötelburg C, Lindig M, Schreiber S, Meyer TF, Bereswill S, Göbel UB. Vaccination prevents Helicobacter pylori-induced alterations of the gastric flora in mice. FEMS Immunol Med Microbiol. 2006;46:221–9. doi: 10.1111/rp10.1016-j.femsim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 72.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge Z, Wang TC, Fox JG. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2013 doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heilig HG, Zoetendal EG, Vaughan EE, Marteau P, Akkermans AD, de Vos WM. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol. 2002;68:114–23. doi: 10.1128/AEM.68.1.114-123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monstein HJ, Tiveljung A, Kraft CH, Borch K, Jonasson J. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J Med Microbiol. 2000;49:817–22. doi: 10.1099/0022-1317-49-9-817. [DOI] [PubMed] [Google Scholar]
- 75.Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58:509–16. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- 76.Rolig AS, Cech C, Ahler E, Carter JE, Ottemann KM. The degree of Helicobacter pylori-triggered inflammation is manipulated by preinfection host microbiota. Infect Immun. 2013;81:1382–9. doi: 10.1128/IAI.00044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, Mobley M, Potter A, Varro A, Eibach D, Suerbaum S, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–20. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zaman C, Osaki T, Hanawa T, Yonezawa H, Kurata S, Kamiya S. Analysis of the microflora in the stomach of Mongolian gerbils infected with Helicobacter pylori. J Gastroenterol Hepatol. 2010;25(Suppl 1):S11–4. doi: 10.1111/j.1440-1746.2009.06215.x. [DOI] [PubMed] [Google Scholar]
- 79.Hu Y, He LH, Xiao D, Liu GD, Gu YX, Tao XX, Zhang JZ. Bacterial flora concurrent with Helicobacter pylori in the stomach of patients with upper gastrointestinal diseases. World J Gastroenterol. 2012;18:1257–61. doi: 10.3748/wjg.v18.i11.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 2013;65:763–72. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- 81.Yang I, Nell S, Suerbaum S. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev. 2013;37:736–61. doi: 10.1111/1574-6976.12027. [DOI] [PubMed] [Google Scholar]
- 82.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–9. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, et al. Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol. 2005;3:733–9. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 84.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7:887–94. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zilberstein B, Quintanilha AG, Santos MA, Pajecki D, Moura EG, Alves PR, Maluf Filho F, de Souza JA, Gama-Rodrigues J. Digestive tract microbiota in healthy volunteers. Clinics (Sao Paulo) 2007;62:47–54. doi: 10.1590/S1807-59322007000100008. [DOI] [PubMed] [Google Scholar]
- 89.Sharma BK, Santana IA, Wood EC, Walt RP, Pereira M, Noone P, Smith PL, Walters CL, Pounder RE. Intragastric bacterial activity and nitrosation before, during, and after treatment with omeprazole. Br Med J (Clin Res Ed) 1984;289:717–9. doi: 10.1136/bmj.289.6447.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adamsson I, Nord CE, Lundquist P, Sjöstedt S, Edlund C. Comparative effects of omeprazole, amoxycillin plus metronidazole versus omeprazole, clarithromycin plus metronidazole on the oral, gastric and intestinal microflora in Helicobacter pylori-infected patients. J Antimicrob Chemother. 1999;44:629–40. doi: 10.1093/jac/44.5.629. [DOI] [PubMed] [Google Scholar]
- 91.Sjöstedt S, Heimdahl A, Kager L, Nord CE. Microbial colonization of the oropharynx, esophagus and stomach in patients with gastric diseases. Eur J Clin Microbiol. 1985;4:49–51. doi: 10.1007/BF02148660. [DOI] [PubMed] [Google Scholar]
- 92.Thorens J, Froehlich F, Schwizer W, Saraga E, Bille J, Gyr K, Duroux P, Nicolet M, Pignatelli B, Blum AL, et al. Bacterial overgrowth during treatment with omeprazole compared with cimetidine: a prospective randomised double blind study. Gut. 1996;39:54–9. doi: 10.1136/gut.39.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mowat C, Williams C, Gillen D, Hossack M, Gilmour D, Carswell A, Wirz A, Preston T, McColl KE. Omeprazole, Helicobacter pylori status, and alterations in the intragastric milieu facilitating bacterial N-nitrosation. Gastroenterology. 2000;119:339–47. doi: 10.1053/gast.2000.9367. [DOI] [PubMed] [Google Scholar]
- 94.Kato S, Fujimura S, Kimura K, Nishio T, Hamada S, Minoura T, Oda M. Non-Helicobacter bacterial flora rarely develops in the gastric mucosal layer of children. Dig Dis Sci. 2006;51:641–6. doi: 10.1007/s10620-006-3185-0. [DOI] [PubMed] [Google Scholar]
- 95.Zavros Y, Rieder G, Ferguson A, Merchant JL. Gastritis and hypergastrinemia due to Acinetobacter lwoffii in mice. Infect Immun. 2002;70:2630–9. doi: 10.1128/IAI.70.5.2630-2639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. Bacterial biogeography of the human digestive tract. Sci Rep. 2011;1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 2009;5:e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Biarc J, Nguyen IS, Pini A, Gossé F, Richert S, Thiersé D, Van Dorsselaer A, Leize-Wagner E, Raul F, Klein JP, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis) Carcinogenesis. 2004;25:1477–84. doi: 10.1093/carcin/bgh091. [DOI] [PubMed] [Google Scholar]
- 99.Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1:e00129–10. doi: 10.1128/mBio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A. 2004;101:4250–5. doi: 10.1073/pnas.0306398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee A, O’Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–97. doi: 10.1016/S0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 102.Crabtree JE, Ferrero RL, Kusters JG. The mouse colonizing Helicobacter pylori strain SS1 may lack a functional cag pathogenicity island. Helicobacter. 2002;7:139–40, author reply 140-1. doi: 10.1046/j.1083-4389.2002.00071.x. [DOI] [PubMed] [Google Scholar]
- 103.Kabir AM, Aiba Y, Takagi A, Kamiya S, Miwa T, Koga Y. Prevention of Helicobacter pylori infection by lactobacilli in a gnotobiotic murine model. Gut. 1997;41:49–55. doi: 10.1136/gut.41.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tan MP, Kaparakis M, Galic M, Pedersen J, Pearse M, Wijburg OL, Janssen PH, Strugnell RA. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Appl Environ Microbiol. 2007;73:1010–3. doi: 10.1128/AEM.01675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mollenhauer-Rektorschek M, Hanauer G, Sachs G, Melchers K. Expression of UreI is required for intragastric transit and colonization of gerbil gastric mucosa by Helicobacter pylori. Res Microbiol. 2002;153:659–66. doi: 10.1016/S0923-2508(02)01380-3. [DOI] [PubMed] [Google Scholar]
- 106.Cynthia M, Kahn M. The merck veterinary manual. 10th ed. Philadelphia (PA): Merck Sharp & Dohme Corp.; 2005. [Google Scholar]
- 107.Porter JR, Rettger LF. Influence of Diet on the Distribution of Bacteria in The Stomach, Small Intestine and Cecum of the White Rat. J Infect Dis. 1940;66:104–10. doi: 10.1093/infdis/66.2.104. [DOI] [Google Scholar]
- 108.Takahashi H, Nakano Y, Matsuoka T, Kumaki N, Asami Y, Koga Y. Role of indigenous lactobacilli in gastrin-mediated acid production in the mouse stomach. Appl Environ Microbiol. 2011;77:6964–71. doi: 10.1128/AEM.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schmitz JM, Durham CG, Schoeb TR, Soltau TD, Wolf KJ, Tanner SM, McCracken VJ, Lorenz RG. Helicobacter felis--associated gastric disease in microbiota-restricted mice. J Histochem Cytochem. 2011;59:826–41. doi: 10.1369/0022155411416242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sheh A, Lee CW, Masumura K, Rickman BH, Nohmi T, Wogan GN, Fox JG, Schauer DB. Mutagenic potency of Helicobacter pylori in the gastric mucosa of mice is determined by sex and duration of infection. Proc Natl Acad Sci U S A. 2010;107:15217–22. doi: 10.1073/pnas.1009017107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohtani M, García A, Rogers AB, Ge Z, Taylor NS, Xu S, Watanabe K, Marini RP, Whary MT, Wang TC, et al. Protective role of 17 beta -estradiol against the development of Helicobacter pylori-induced gastric cancer in INS-GAS mice. Carcinogenesis. 2007;28:2597–604. doi: 10.1093/carcin/bgm150. [DOI] [PubMed] [Google Scholar]
- 112.Sheh A, Ge Z, Parry NM, Muthupalani S, Rager JE, Raczynski AR, Mobley MW, McCabe AF, Fry RC, Wang TC, et al. 17β-estradiol and tamoxifen prevent gastric cancer by modulating leukocyte recruitment and oncogenic pathways in Helicobacter pylori-infected INS-GAS male mice. Cancer Prev Res (Phila) 2011;4:1426–35. doi: 10.1158/1940-6207.CAPR-11-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee CW, Rickman B, Rogers AB, Muthupalani S, Takaishi S, Yang P, Wang TC, Fox JG. Combination of sulindac and antimicrobial eradication of Helicobacter pylori prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res. 2009;69:8166–74. doi: 10.1158/0008-5472.CAN-08-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gaddy JA, Radin JN, Loh JT, Zhang F, Washington MK, Peek RM, Jr., Algood HM, Cover TL. High dietary salt intake exacerbates Helicobacter pylori-induced gastric carcinogenesis. Infect Immun. 2013;81:2258–67. doi: 10.1128/IAI.01271-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kato S, Tsukamoto T, Mizoshita T, Tanaka H, Kumagai T, Ota H, Katsuyama T, Asaka M, Tatematsu M. High salt diets dose-dependently promote gastric chemical carcinogenesis in Helicobacter pylori-infected Mongolian gerbils associated with a shift in mucin production from glandular to surface mucous cells. Int J Cancer. 2006;119:1558–66. doi: 10.1002/ijc.21810. [DOI] [PubMed] [Google Scholar]
- 116.Noto JM, Gaddy JA, Lee JY, Piazuelo MB, Friedman DB, Colvin DC, Romero-Gallo J, Suarez G, Loh J, Slaughter JC, et al. Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans. J Clin Invest. 2013;123:479–92. doi: 10.1172/JCI64373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Osaki T, Matsuki T, Asahara T, Zaman C, Hanawa T, Yonezawa H, Kurata S, Woo TD, Nomoto K, Kamiya S. Comparative analysis of gastric bacterial microbiota in Mongolian gerbils after long-term infection with Helicobacter pylori. Microb Pathog. 2012;53:12–8. doi: 10.1016/j.micpath.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 118.Sun YQ, Monstein HJ, Nilsson LE, Petersson F, Borch K. Profiling and identification of eubacteria in the stomach of Mongolian gerbils with and without Helicobacter pylori infection. Helicobacter. 2003;8:149–57. doi: 10.1046/j.1523-5378.2003.00136.x. [DOI] [PubMed] [Google Scholar]
- 119.Yin YN, Wang CL, Liu XW, Cui Y, Xie N, Yu QF, Li FJ, Lu FG. Gastric and duodenum microflora analysis after long-term Helicobacter pylori infection in Mongolian Gerbils. Helicobacter. 2011;16:389–97. doi: 10.1111/j.1523-5378.2011.00862.x. [DOI] [PubMed] [Google Scholar]
- 120.El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA, Herrera J, Lissowska J, Yuan CC, Rothman N, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 121.El-Omar EM, Rabkin CS, Gammon MD, Vaughan TL, Risch HA, Schoenberg JB, Stanford JL, Mayne ST, Goedert J, Blot WJ, et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology. 2003;124:1193–201. doi: 10.1016/S0016-5085(03)00157-4. [DOI] [PubMed] [Google Scholar]
- 122.Tu S, Bhagat G, Cui G, Takaishi S, Kurt-Jones EA, Rickman B, Betz KS, Penz-Oesterreicher M, Bjorkdahl O, Fox JG, et al. Overexpression of interleukin-1beta induces gastric inflammation and cancer and mobilizes myeloid-derived suppressor cells in mice. Cancer Cell. 2008;14:408–19. doi: 10.1016/j.ccr.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lu W, Pan K, Zhang L, Lin D, Miao X, You W. Genetic polymorphisms of interleukin (IL)-1B, IL-1RN, IL-8, IL-10 and tumor necrosis factor alpha and risk of gastric cancer in a Chinese population. Carcinogenesis. 2005;26:631–6. doi: 10.1093/carcin/bgh349. [DOI] [PubMed] [Google Scholar]
- 124.Taguchi A, Ohmiya N, Shirai K, Mabuchi N, Itoh A, Hirooka Y, Niwa Y, Goto H. Interleukin-8 promoter polymorphism increases the risk of atrophic gastritis and gastric cancer in Japan. Cancer Epidemiol Biomarkers Prev. 2005;14:2487–93. doi: 10.1158/1055-9965.EPI-05-0326. [DOI] [PubMed] [Google Scholar]
- 125.Kido S, Kitadai Y, Hattori N, Haruma K, Kido T, Ohta M, Tanaka S, Yoshihara M, Sumii K, Ohmoto Y, et al. Interleukin 8 and vascular endothelial growth factor -- prognostic factors in human gastric carcinomas? Eur J Cancer. 2001;37:1482–7. doi: 10.1016/S0959-8049(01)00147-2. [DOI] [PubMed] [Google Scholar]
- 126.Junnila S, Kokkola A, Mizuguchi T, Hirata K, Karjalainen-Lindsberg ML, Puolakkainen P, Monni O. Gene expression analysis identifies over-expression of CXCL1, SPARC, SPP1, and SULF1 in gastric cancer. Genes Chromosomes Cancer. 2010;49:28–39. doi: 10.1002/gcc.20715. [DOI] [PubMed] [Google Scholar]
- 127.Jung JJ, Noh S, Jeung HC, Jung M, Kim TS, Noh SH, Roh JK, Chung HC, Rha SY. Chemokine growth-regulated oncogene 1 as a putative biomarker for gastric cancer progression. Cancer Sci. 2010;101:2200–6. doi: 10.1111/j.1349-7006.2010.01666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Resnick MB, Sabo E, Meitner PA, Kim SS, Cho Y, Kim HK, Tavares R, Moss SF. Global analysis of the human gastric epithelial transcriptome altered by Helicobacter pylori eradication in vivo. Gut. 2006;55:1717–24. doi: 10.1136/gut.2006.095646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Okumura T, Ericksen RE, Takaishi S, Wang SS, Dubeykovskiy Z, Shibata W, Betz KS, Muthupalani S, Rogers AB, Fox JG, et al. K-ras mutation targeted to gastric tissue progenitor cells results in chronic inflammation, an altered microenvironment, and progression to intraepithelial neoplasia. Cancer Res. 2010;70:8435–45. doi: 10.1158/0008-5472.CAN-10-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Asfaha S, Dubeykovskiy AN, Tomita H, Yang X, Stokes S, Shibata W, Friedman RA, Ariyama H, Dubeykovskaya ZA, Muthupalani S, et al. Mice that express human interleukin-8 have increased mobilization of immature myeloid cells, which exacerbates inflammation and accelerates colon carcinogenesis. Gastroenterology. 2013;144:155–66. doi: 10.1053/j.gastro.2012.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, Nikkïla J, Monti D, Satokari R, Franceschi C, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 133.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–91. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis. 2002;34(Suppl 2):S12–8. doi: 10.1016/S1590-8658(02)80157-8. [DOI] [PubMed] [Google Scholar]
- 139.Sipponen P, Correa P. Delayed rise in incidence of gastric cancer in females results in unique sex ratio (M/F) pattern: etiologic hypothesis. Gastric Cancer. 2002;5:213–9. doi: 10.1007/s101200200037. [DOI] [PubMed] [Google Scholar]
- 140.Weiskopf D, Weinberger B, Grubeck-Loebenstein B. The aging of the immune system. Transpl Int. 2009;22:1041–50. doi: 10.1111/j.1432-2277.2009.00927.x. [DOI] [PubMed] [Google Scholar]
- 141.Franceschi C, Capri M, Monti D, Giunta S, Olivieri F, Sevini F, Panourgia MP, Invidia L, Celani L, Scurti M, et al. Inflammaging and anti-inflammaging: a systemic perspective on aging and longevity emerged from studies in humans. Mech Ageing Dev. 2007;128:92–105. doi: 10.1016/j.mad.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 142.Mäkivuokko H, Tiihonen K, Tynkkynen S, Paulin L, Rautonen N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br J Nutr. 2010;103:227–34. doi: 10.1017/S0007114509991553. [DOI] [PubMed] [Google Scholar]
- 143.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–84. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 144.Mueller S, Saunier K, Hanisch C, Norin E, Alm L, Midtvedt T, Cresci A, Silvi S, Orpianesi C, Verdenelli MC, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72:1027–33. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lindblad M, Rodríguez LA, Lagergren J. Body mass, tobacco and alcohol and risk of esophageal, gastric cardia, and gastric non-cardia adenocarcinoma among men and women in a nested case-control study. Cancer Causes Control. 2005;16:285–94. doi: 10.1007/s10552-004-3485-7. [DOI] [PubMed] [Google Scholar]
- 146.Chandanos E, Lagergren J. Oestrogen and the enigmatic male predominance of gastric cancer. Eur J Cancer. 2008;44:2397–403. doi: 10.1016/j.ejca.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 147.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. MetaHIT Consortium Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–8. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 149.Altekruse SF, Kosary CL, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, et al. SEER Cancer Statistics Review, 1975-2007. Bethesda, MD: National Cancer Institute, 2010. [Google Scholar]
- 150.Parkin DM. International variation. Oncogene. 2004;23:6329–40. doi: 10.1038/sj.onc.1207726. [DOI] [PubMed] [Google Scholar]
- 151.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsugane S, Sasazuki S. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer. 2007;10:75–83. doi: 10.1007/s10120-007-0420-0. [DOI] [PubMed] [Google Scholar]
- 154.Bjelakovic G, Nikolova D, Gluud LL, Simonetti RG, Gluud C. Mortality in randomized trials of antioxidant supplements for primary and secondary prevention: systematic review and meta-analysis. JAMA. 2007;297:842–57. doi: 10.1001/jama.297.8.842. [DOI] [PubMed] [Google Scholar]
- 155.D’Elia L, Rossi G, Ippolito R, Cappuccio FP, Strazzullo P. Habitual salt intake and risk of gastric cancer: a meta-analysis of prospective studies. Clin Nutr. 2012;31:489–98. doi: 10.1016/j.clnu.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 156.Rogers AB, Taylor NS, Whary MT, Stefanich ED, Wang TC, Fox JG. Helicobacter pylori but not high salt induces gastric intraepithelial neoplasia in B6129 mice. Cancer Res. 2005;65:10709–15. doi: 10.1158/0008-5472.CAN-05-1846. [DOI] [PubMed] [Google Scholar]
- 157.Loh JT, Friedman DB, Piazuelo MB, Bravo LE, Wilson KT, Peek RM, Jr., Correa P, Cover TL. Analysis of Helicobacter pylori cagA promoter elements required for salt-induced upregulation of CagA expression. Infect Immun. 2012;80:3094–106. doi: 10.1128/IAI.00232-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thomson MJ, Pritchard DM, Boxall SA, Abuderman AA, Williams JM, Varro A, Crabtree JE. Gastric Helicobacter infection induces iron deficiency in the INS-GAS mouse. PLoS One. 2012;7:e50194. doi: 10.1371/journal.pone.0050194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Duque X, Moran S, Mera R, Medina M, Martinez H, Mendoza ME, Torres J, Correa P. Effect of eradication of Helicobacter pylori and iron supplementation on the iron status of children with iron deficiency. Arch Med Res. 2010;41:38–45. doi: 10.1016/j.arcmed.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 160.Peek RM, Cover TL. Diet, microbial virulence and Helicobacter pylori-induced gastric cancer. Gut Microbes. 2013 doi: 10.4161/gmic.26262. Forthcoming (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 162.Werner T, Wagner SJ, Martínez I, Walter J, Chang JS, Clavel T, Kisling S, Schuemann K, Haller D. Depletion of luminal iron alters the gut microbiota and prevents Crohn’s disease-like ileitis. Gut. 2011;60:325–33. doi: 10.1136/gut.2010.216929. [DOI] [PubMed] [Google Scholar]
- 163.Ettreiki C, Gadonna-Widehem P, Mangin I, Coëffier M, Delayre-Orthez C, Anton PM. Juvenile ferric iron prevents microbiota dysbiosis and colitis in adult rodents. World J Gastroenterol. 2012;18:2619–29. doi: 10.3748/wjg.v18.i21.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fox JG, Beck P, Dangler CA, Whary MT, Wang TC, Shi HN, Nagler-Anderson C. Concurrent enteric helminth infection modulates inflammation and gastric immune responses and reduces helicobacter-induced gastric atrophy. Nat Med. 2000;6:536–42. doi: 10.1038/75015. [DOI] [PubMed] [Google Scholar]
- 165.Stoicov C, Whary M, Rogers AB, Lee FS, Klucevsek K, Li H, Cai X, Saffari R, Ge Z, Khan IA, et al. Coinfection modulates inflammatory responses and clinical outcome of Helicobacter felis and Toxoplasma gondii infections. J Immunol. 2004;173:3329–36. doi: 10.4049/jimmunol.173.5.3329. [DOI] [PubMed] [Google Scholar]
- 166.Whary MT, Sundina N, Bravo LE, Correa P, Quinones F, Caro F, Fox JG. Intestinal helminthiasis in Colombian children promotes a Th2 response to Helicobacter pylori: possible implications for gastric carcinogenesis. Cancer Epidemiol Biomarkers Prev. 2005;14:1464–9. doi: 10.1158/1055-9965.EPI-05-0095. [DOI] [PubMed] [Google Scholar]
- 167.Du Y, Agnew A, Ye XP, Robinson PA, Forman D, Crabtree JE. Helicobacter pylori and Schistosoma japonicum co-infection in a Chinese population: helminth infection alters humoral responses to H. pylori and serum pepsinogen I/II ratio. Microbes Infect. 2006;8:52–60. doi: 10.1016/j.micinf.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 168.Lemke LB, Ge Z, Whary MT, Feng Y, Rogers AB, Muthupalani S, Fox JG. Concurrent Helicobacter bilis infection in C57BL/6 mice attenuates proinflammatory H. pylori-induced gastric pathology. Infect Immun. 2009;77:2147–58. doi: 10.1128/IAI.01395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ge Z, Feng Y, Muthupalani S, Eurell LL, Taylor NS, Whary MT, Fox JG. Coinfection with Enterohepatic Helicobacter species can ameliorate or promote Helicobacter pylori-induced gastric pathology in C57BL/6 mice. Infect Immun. 2011;79:3861–71. doi: 10.1128/IAI.05357-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kuehl CJ, Wood HD, Marsh TL, Schmidt TM, Young VB. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect Immun. 2005;73:6952–61. doi: 10.1128/IAI.73.10.6852-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Whary MT, Danon SJ, Feng Y, Ge Z, Sundina N, Ng V, Taylor NS, Rogers AB, Fox JG. Rapid onset of ulcerative typhlocolitis in B6.129P2-IL10tm1Cgn (IL-10-/-) mice infected with Helicobacter trogontum is associated with decreased colonization by altered Schaedler’s flora. Infect Immun. 2006;74:6615–23. doi: 10.1128/IAI.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ge Z, Feng Y, Taylor NS, Ohtani M, Polz MF, Schauer DB, Fox JG. Colonization dynamics of altered Schaedler flora is influenced by gender, aging, and Helicobacter hepaticus infection in the intestines of Swiss Webster mice. Appl Environ Microbiol. 2006;72:5100–3. doi: 10.1128/AEM.01934-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Walk ST, Blum AM, Ewing SA, Weinstock JV, Young VB. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis. 2010;16:1841–9. doi: 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sgouras DN, Panayotopoulou EG, Martinez-Gonzalez B, Petraki K, Michopoulos S, Mentis A. Lactobacillus johnsonii La1 attenuates Helicobacter pylori-associated gastritis and reduces levels of proinflammatory chemokines in C57BL/6 mice. Clin Diagn Lab Immunol. 2005;12:1378–86. doi: 10.1128/CDLI.12.12.1378-1386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Luther J, Dave M, Higgins PD, Kao JY. Association between Helicobacter pylori infection and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Inflamm Bowel Dis. 2010;16:1077–84. doi: 10.1002/ibd.21116. [DOI] [PMC free article] [PubMed] [Google Scholar]