Abstract
Long non-coding RNAs (lncRNAs) are increasingly being recognized as epigenetic regulators of gene transcription. The diversity and complexity of lncRNA genes means that they exert their regulatory effects by a variety of mechanisms. Although there is still much to be learned about the mechanism of lncRNA function, general principles are starting to emerge. In particular, the application of high throughput (deep) sequencing methodologies has greatly advanced our understanding of lncRNA gene function. lncRNAs function as adaptors that link specific chromatin loci with chromatin-remodeling complexes and transcription factors. lncRNAs can act in cis or trans to guide epigenetic-modifier complexes to distinct genomic sites, or act as scaffolds which recruit multiple proteins simultaneously, thereby coordinating their activities. In this review we discuss the genomic organization of lncRNAs, the importance of RNA secondary structure to lncRNA functionality, the multitude of ways in which they interact with the genome, and what evolutionary conservation tells us about their function.
Keywords: RNA, RNA scaffolds, RNA structure, RNAi, TGA, TGS, chromatin, epigenetics, lncRNA, ncRNA
Introduction
Epigenetics (meaning epi: “on top of,” genetics: “the study of genes and heredity”) is the study of mitotically and meiotically heritable traits that are not encoded in the primary DNA sequence itself.1,2 Covalent modification of genomic DNA nucleobases (e.g., cytosine methylation) and posttranslational modification of DNA-associated proteins (e.g., differential methylation and acetylation of N-terminal histone tails) regulate the accessibility of the genome to the transcriptional machinery.3 The patterns of DNA methylation and histone tail modifications are inherited following somatic cell division and, in some cases, also in the germ line.4 As a result, epigenetic modifications greatly expand the information content of the genome.5 Chromatin modifying proteins are the readers and writers of the histone code.6 Dynamic regulation of chromatin structure necessitates differential activity of chromatin modifying proteins. However, many chromatin modifying proteins are expressed ubiquitously,7,8 which suggests that changes in their expression are unlikely to be responsible for the establishment of differential epigenetic states in the majority of cases (but rather that an additional biochemical component imparts specificity for distinct genomic loci). Additionally, several proteins (e.g., GAPDH) have recently been found to moonlight as RNA binding proteins, suggesting that protein-RNA interactions in the cell may be much more common than was once thought.9-11 It is becoming increasingly apparent that RNA is intimately involved in the epigenetic control of gene expression, lineage commitment, and the generation of higher order complexity by conferring sequence specificity on chromatin modifying activities.8,12,13
Long Non-Coding RNAs
Long non-coding RNAs (lncRNAs) are a heterogeneous group of RNA transcripts which lack long open reading frames (and therefore exhibit low protein-coding potential) and are biochemically similar to mRNAs.14 lncRNAs are generally produced by RNA polymerase II (RNAP II) transcription, are often spliced and polyadenylated, and typically have a methylguanosine cap at their 5′ termini (although there are numerous exceptions to these general features15,16). lncRNAs are so named in order to differentiate them from the small RNAs (i.e., microRNAs, small interfering RNAs, and PIWI-interacting RNAs) and have been somewhat arbitrarily defined as being >200 nucleotides for historical reasons associated with RNA extraction methodologies.17 lncRNAs exhibit distinct spatial, temporal, cell-type specific, and sub-cellular specific expression patterns, suggesting that their transcription is tightly regulated.13,18 Many lncRNAs are rapidly turned over.19 While some lncRNAs are expressed at less than 1 copy per cell, others are more abundant than housekeeping mRNAs such as GAPDH (e.g., MALAT120). A sub-class of lncRNAs is the long intergenic non-coding RNAs (lincRNAs), which are encoded in the genomic space between protein-coding genes with no overlapping sequence. lincRNAs are not necessarily biochemically different from other lncRNAs and differ only in their genomic organization. Given that lincRNAs do not overlap with protein-coding loci, their functions can unequivocally be attributed to the lincRNA transcript itself, rather than as an indirect effect on a proximal protein-coding gene.
lncRNA genes have been identified through a number of complementary techniques. The application of ChIP-seq (chromatin immunoprecipitation and high throughput sequencing) has identified so called ‘K4-K36 domains’8,21. H3K4me3 is a chromatin mark of actively transcribed promoters, whereas H3K36me3 is a mark of transcriptional elongation.22 As a result, so called K4-K36 domains that do not correspond with protein-coding genes can be used to identify lncRNA genes. Similarly, enrichment of H3K3me1, H3K27ac, and p300 binding have been identified as a chromatin signature of enhancer sequences, many of which produce non-coding transcripts.23,24 High-throughput sequencing of RNA (RNA-seq) can be used to identify lncRNA transcripts at single nucleotide resolution in a strand-specific manner. Comparison of RNA-seq and ChIP-seq data sets thereby provides complementary and independent evidence of lncRNA transcription (Fig. 1A). Similarly, the application of target capture methodologies (in which cDNAs complementary to specific loci are hybridized to a microarray, eluted, and then analyzed by high-throughput sequencing [RNA CaptureSeq]) has revealed a plethora of previously unknown transcripts with well-defined intron-exon organization originating from gene deserts.25 Ribosome profiling has demonstrated that while lncRNA transcripts are generally devoid of association with ribosomes consistent with their lack of protein-coding potential, some may encode short peptide sequences.26-28
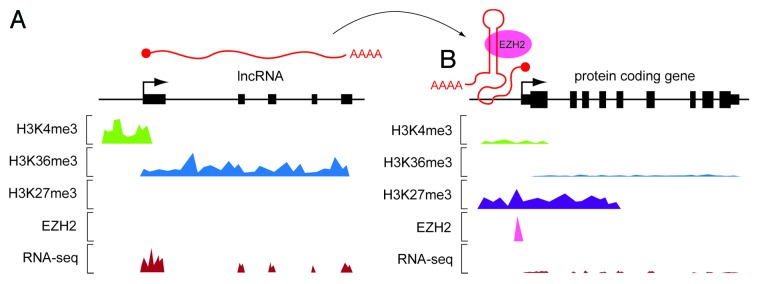
Figure 1. Application of high throughput sequencing in the identification and function of lncRNAs. Cartoon schematic of typical ChIP-seq and (poly A+) RNA-seq data tracks. (A) A hypothetical lncRNA gene is identified by the presence of a ‘K4-K36 domain’ and evidence of transcription from RNA-seq data. (B) The lncRNA transcript adopts a secondary structure that binds a chromatin-modifying protein (e.g., EZH2) and guides it to a protein-coding gene target locus. H3K4me3 and H3K36me3 ChIP-seq signals at the target locus are reduced indicating reduced transcriptional activity. The histone K27 trimethylase EZH2, and the silent state chromatin mark H3K27me3 are enriched at the target promoter indicating transcriptional silencing which corresponds with reduced RNA-seq signals from the target protein-coding gene.
Epigenetic Regulation by lncRNAs
lncRNAs have been implicated in a wide range of epigenetic processes including X-chromosome inactivation,29,30 mono-allelic expression of imprinted loci,31-33 determination of cellular differentiation and maintenance of cell identity.12,34-36 RNAi knockdown screens in embryonic stem cells revealed that many lncRNAs play crucial roles in maintaining pluripotency and lineage commitment.12 One of the best-studied lncRNAs is HOTAIR, which regulates the HOXC locus in trans through the recruitment of PRC2 (Polycomb Repressive Complex 2, an H3K27 trimethylase), leading to epigenetic silencing.37 Conversely, the RepA region of the lncRNA Xist recruits PRC2 in cis to initiate the process of X-chromosome inactivation.38 Similarly, other lncRNAs have also been shown to recruit epigenetic silencing complexes to specific loci including Air which complexes with PRC2, and Kcnq10t1 which binds to PRC2 and EHMT2 (formerly G9a, an H3K9 methylase).39,40 lncRNAs have also been shown to associate with epigenetic activating complexes (e.g., WDR5/MLL).8,13,41
A plethora of other examples of epigenetic regulation by lncRNAs have also been described. For example, bi-directional transcription regulates the CDKN1A (p21) epigenetic expression,42 and the tumor suppressor CDKN2B (p15) is epigenetically regulated by an overlapping antisense transcript.43 A natural antisense transcript has been shown to direct EZH2 (a polycomb component) to the BDNF locus,44 whereas a promoter-associated RNA recruits the DNA methyltransferase DMNT3B to the rRNA promoter.45,46 Pseudogenes can also be sources of lncRNAs, some of which exert epigenetic effects on specific target genes.47,48 For example, an antisense transcript derived from a pseudogene of PTEN recruits DMNT3A and EZH2 to the PTEN promoter leading to epigenetic silencing.47
While these studies strongly suggested a key role for lncRNAs as guides for epigenetic modifier complexes in a number of specific cases, the generality of this phenomenon has also been tested. In a landmark study, Khalil et al. performed RNA-immunoprecipitation followed by microarray analysis (RIP-chip) using antibodies against components of the chromatin-remodeling complexes PRC2, CoREST (a repressor of neuronal genes), and SMCX (a H3K3me3 demethylase). This study revealed that ~40% of known lncRNAs associated with one or more of these epigenetic modifier complexes.8 Interestingly, lncRNAs associated with CoREST were typically not found to associate with PRC2, suggesting that each complex binds to a distinct subset of lncRNA transcripts.8 In the same study, depletion of some of these lncRNAs by RNAi resulted in activation of known polycomb-regulated genes. Protein-coding genes proximal to the lncRNA gene were not significantly affected, implying that these transcripts regulate their target in trans.8 Similarly, Xhao et al. performed RIP-seq with antibodies against the PRC2 component Ezh2 in order to identify polycomb-associated lncRNAs in an unbiased manner.49 This analysis estimated that over 9000 transcripts (including many novel antisense RNAs and other un-annotated transcripts) are bound by PRC2.
Taken together, these studies suggest that a major function of lncRNAs is to direct chromatin modifying activities or transcription factors to specific genomic loci. This function establishes differential chromatin states, induces large-scale changes in gene expression, and determines cell fate (Fig. 1B). Although not the focus of this review, it is important to also mention that lncRNAs are also involved in non-epigenetic, posttranscriptional gene regulation.50-53
RNA Structure and lncRNA Function
Decades of research has been mainly concerned with the role of RNA as a transient intermediary which facilitates the flow of information from the genome to the proteome. However, it is now recognized that the structural plasticity of RNA enables it to perform organizational, catalytic and regulatory functions. Long RNA transcripts are flexible and may adopt one or more complex and dynamic secondary structures. RNA structures can act as riboswitches, regulatory motifs that act to sense the concentration of specific small molecules (e.g., cellular metabolites and metal ions, etc.).54 This property of long RNA transcripts may mean that lncRNAs have the potential to modulate their structure (and therefore their binding partners) in response to environmental conditions.55
RNA structure can be predicted using in silico folding programs (generally based on minimum free energy models)56,57 or determined experimentally through the use chemical agents or enzymes which selectively cleave or modify RNA bases (i.e., RNA footprinting).58 To this end, an RNA transcript is transcribed and folded in vitro, and then subjected to digestion with either RNase VI (which selectively cleaves dsRNA) or RNase A (which selectively cleaves ssRNA). The resulting fragments are separated by electrophoresis and RNA secondary structure inferred. In the case of the related technique, SHAPE (Selective 2-Hydroxyl Acylation analyzed by Primer Extension), flexible ribonucleobases are selectively acetylated. This chemical modification blocks the procession of reverse transcriptase during subsequent cDNA synthesis. Flexible regions of the transcript of interest are inferred by the cloning and sequencing of the resultant cDNA fragments.
A limitation of these techniques is that they generally require RNA to be folded in vitro before analysis and so the biological relevance of the information gained may be limited. Similarly, structural motifs that are dependent on binding of a protein or nucleic acid co-factor will be undetectable by these methods. To circumvent this problem, in vivo methods have also been developed.59,60 For example, dimethyl sulfide (DMS) can be used to selectively modify solvent-accessible RNA bases in vivo. While this method takes into account higher order RNA structure and intermolecular interactions, it is potentially confounded by the heterogeneity of structures that can be adopted by an RNA molecule in the cell.
Recently, these techniques have also been combined with next generation sequencing technologies. Parallel Analysis of RNA Structure (PARS) and FragSeq involve the deep sequencing of cDNA libraries generated from nuclease digested RNA samples,61,62 whereas SHAPE-seq is a high throughput version of the SHAPE methodology.63 The application of genome-wide sequencing techniques to the biochemical analysis of RNA structure determination will likely provide valuable insights into lncRNA biology and function. For example, the basis for targeting of specific genomic loci by lncRNAs is currently poorly understood. A complete catalog of structural features within non-coding transcripts will likely reveal the mechanism(s) by which lncRNAs interact with genome, and thereby accelerate the study of lncRNA gene function. It may even facilitate the design of synthetic lncRNA genes tailored to epigenetically regulate target genes of interest.
Interactions of lncRNAs with Chromatin
It has long been known that RNA comprises a substantial component of chromatin,64 that higher-order chromatin structure is dependent on RNA-chromatin interactions,65 and that non-coding RNAs are intimately involved in epigenetic processes.66 The functionality of lncRNAs is a result of their ability to act as adaptors that mediate interactions between chromatin, proteins, and other RNAs. The primary nucleotide sequence provides a basis for lncRNA targeting to a specific nucleic acid by complementary Watson-Crick base-paring at single-stranded DNA regions (e.g., at R-loops) (Fig. 2A).67 lncRNAs can also form triplex structures through Hoogsteen/Reverse-Hoogsteen base pairing (Fig. 2B).45,68 For example, a triplex-forming ncRNA binds to the promoter of rRNA genes in order to recruit the DNA methyltransferase DNMT3B and induce epigenetic silencing.45,46 Nascent lncRNAs can also be tethered to the locus from which they are transcribed through association with RNAP II (Fig. 2C).69,70 As a result, lncRNA transcripts can act as unique molecular signatures for their specific allele of origin.70 These cis-acting tethered lncRNAs are platforms for interactions with other ncRNAs or ribonucleoprotein complexes. For example, miRNAs can guide Argonaute complexes to tethered promoter associated RNAs in order to induce transcriptional gene silencing or activation.71-77
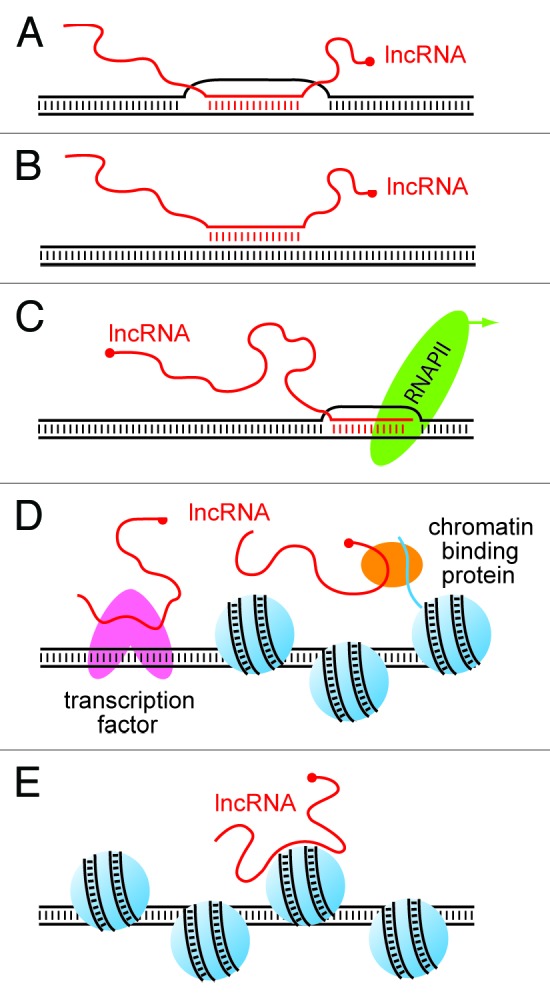
Figure 2. Interactions of lncRNAs with the genome. lncRNAs interact with genomic DNA and chromatin in a variety of ways. (A) lncRNAs can bind to regions of single stranded DNA to form a DNA-RNA heteroduplex by Watson-Crick base pairing. (B) lncRNAs can also form DNA-DNA-RNA triplexes by Hoogsteen or reverse Hoogsteen base pairing. (C) lncRNAs can be tethered to a chromatin through association with RNAP II and thereby act as an allele-specific signatures for specific locus. (D) lncRNAs can be indirectly bound to chromatin through binding to chromatin-associated proteins or transcription factors. (E) lncRNAs can form structures which directly sense chromatin structure.
lncRNAs can also associate with chromatin indirectly through interactions with chromatin modifying enzymes or other DNA binding proteins (e.g., transcription factors). In these cases, the proteins act as binding adaptors between chromatin and the lnRNA (Fig. 2D). For example, the lncRNA Xist interacts with the X chromosome via an interaction with YY1.78 Interestingly, many transcription factors also bind to RNA. Transcription factors typically recognize short nucleic acid motifs which occur with high frequency in the genome. It is therefore likely that lncRNA binding partners may act as additional determinants of target site recognition. For example, the lncRNA Evf-2 binds to the transcription factor Dlx-2 in order to enhance its role as a transcriptional activator.79 lncRNAs can also fold into secondary motifs that can directly bind to specific chromatin surface features (Fig. 2E).69
A key concept when considering the functionality of lncRNAs is modularity.80 lncRNAs may consist of multiple modules, each with the ability to bind to distinct protein or nucleic acid partners.81 As such lncRNAs can act as scaffolds that coordinate the activities of multiple epigenetic modifier proteins. For example, the lncRNA HOTAIR acts as scaffold which binds to PRC2 at its 5′ end82 and the LSD1/CoREST/REST complex at its 3′ end.83 These two protein complexes act to trimethylate H3K27 and to demethylate H3K4 respectively, which together facilitate transcriptional silencing of HOTAIR target loci.
Higher order chromatin structure and sub-nuclear organization are also subject to regulation by lncRNAs. The lncRNA TUG1 binds methylated polycomb 2 protein 2 (Pc2, a component of the PRC1 complex), which directs the accompanying genomic DNA to polycomb bodies and leads to epigenetic silencing. Conversely, unmethylated Pc2 binds to a different lncRNA, MALAT1, which localizes its associated chromatin to interchromatin granules (ICGs) which are associated with active transcription.84 Similarly, the lncRNA SRA (Steroid Receptor RNA Activator) associates with CTCF (CCCTC-binding factor) via an interaction with the DEAD-box RNA helicase p68 (DDX5) and is essential for CTCF activity.85 CTCF defines insulator regions and regulates chromatin macro-structure by facilitating gene loop formation.86 Chromosome three-dimensional structure is also important in the case of HOTTIP, a lncRNA transcribed in the antisense orientation from the 5′ end of the HOXA locus, which coordinates the differential activation of distal HOX genes via recruitment of WDR5-MLL complexes.41 Despite regulating the expression of distant genes, HOTTIP acts as a cis-regulator as it is physically transcribed in the vicinity of its target genes on account of gene looping.
Recently, two landmark studies have utilized RNA pulldown strategies in order to probe genomic lncRNA binding sites on a global scale. These techniques have been termed Chromatin Isolation by RNA Purification (ChIRP-seq)87 and Capture Hybridization Analysis of RNA Targets (CHART-seq).88 In both ChIRP-seq and CHART-seq, chemical crosslinking is used to bind chromatin protein, RNA, and DNA as in a conventional ChIP assay. Samples are then probed using biotinylated/desthiobiotinylated DNA oligonucleotides complementary to a specific lncRNA of interest. Associated gDNA is eluted or removed by RNase H digestion, and analyzed by deep sequencing.
Using ChIRP-seq, Chu et al. showed that the lncRNA HOTAIR binds to the genome at numerous locations, many of which overlap with binding sites for the polycomb components EZH2 and SUZ12,87 consistent with its known activity as a trans acting lncRNA guide for PRC2.37,82 Depletion of EZH2 by RNAi did not affect HOTAIR binding suggesting that the PRC2 binding and chromatin interaction modules of HOTAIR are functionally independent.87 In contrast with histone modifications, and similar to transcription factors, lncRNA binding is focal (typically corresponding to sharp peaks spanning <500 base pairs of gDNA), suggesting that binding to chromatin occurs at highly specific loci. Similarly, using CHART-seq, Simon et al. analyzed the genomic binding sites of the ncRNA roX2 in Drosophila S2 cells.88 The highest roX2 occupancy sites were located on the X-chromosome and overlapped with known binding sites of MSL (Male Specific Lethal) complex, consistent with the function of roX2 as a regulator of X-chromosome dosage compensation.88 Both ChIRP-seq and CHART-seq have enormous potential in unraveling the function of lncRNAs. However, it should be noted that neither technique provides direct evidence for the basis for interaction between lncRNA and chromatin. Importantly, both studies found that GA-rich homopurine motifs were enriched at lncRNA target sites. Given that the formation of DNA-DNA-RNA triplexes requires a stretch of purines, this finding suggests that triplex formation may be the primary means by which lncRNAs like HOTAIR and roX2 interact with the genome. Indeed lncRNA-gDNA triplex interactions have been described in other contexts.45,68 The results of these studies are also consistent with indirect chromatin association via an RNA binding protein intermediate. This alternative seems unlikely, as it would necessitate an increase in complexity not afforded by the limited genomic repertoire of DNA/RNA binding proteins. An advantage of the ChIRP/CHART-seq methods is that, by capturing the RNA component, it is also possible to analyze the co-precipitated protein by mass spectrometry. As a result, future studies may be able to differentiate between the direct RNA-gDNA and indirect protein intermediate models of lncRNA-chromatin binding.
Evolution of lncRNA Genes
There is some disagreement over the degree of conservation of lncRNA genes, although it is widely recognized that they are less well conserved than protein-coding genes. Wang et al. suggest that the majority of murine lncRNAs are evolving neutrally.89 Conversely, Ponjavic et al. provide evidence of purifying selection in lncRNA promoters, primary sequence, and splice sites.90 Similarly, analysis of lncRNA genes has demonstrated that exonic regions tend to have lower base substitution rates than intronic regions, which is suggestive of evolutionary conservation.21,91 In a separate study, Pang et al. showed that, while conservation between human and mouse lncRNAs was low, when the lncRNA sequences were analyzed as 50 nt segments the levels of conservation were much higher. Consequently, lncRNAs may consist of conserved functional modules residing within long stretches of sequence which is under little evolutionary constraint.92 These studies suggest that lncRNAs are under different evolutionary pressures than protein-coding genes or other types of ncRNAs such as tRNAs, rRNAs or microRNAs. This is exemplified by the classic lncRNAs: Xist and Air, which have well established functional roles (in X-chromosome inactivation and imprinting of the Igfr2 locus) but are poorly conserved between human and mouse.93,94 Furthermore, the relative lack of conservation is not necessarily evidence for lack of functionality (or for biological irrelevance) as some lncRNA genes may represent relatively recent evolutionary innovations, be lineage specific, or have temporally restricted functions. It has been proposed that differential lncRNA expression may go some way toward explaining the vast morphological differences between organisms with similar protein-coding repertoires (and by extension, between individuals within the same species).95
The activity of lncRNAs is dependent on the formation of RNA structure motifs that act as binding domains for proteins or chromatin structures. As a result, there may be selection pressure acting to conserve these structural features which would not necessarily be apparent at the primary sequence level.96 To this end, the use of programs such as FOLDALIGN,97 which can compare sequences based on predicted local RNA structures, has identified thousands of loci in the human and mouse genomes with conserved secondary RNA structures but very poor homology at the primary nucleotide sequence.98 It is important to also consider that some lncRNAs may not produce functional transcripts per se, but rather their transcription in itself may be sufficient to regulate the transcription of neighboring genes in cis.99-101 In this case their ‘functionality’ may be maintained in the absence of sequence conservation.
Conclusions
A major role of lncRNAs is in the regulation of epigenetic states by modulating chromatin structure and nuclear organization. lncRNAs are adaptors that mediate interactions between chromatin and proteins involved in the epigenetic regulation of transcription. While lncRNAs generally show low levels of conservation at the primary sequence level, they may contain shorter internal stretches of conserved sequence. These are likely functional motifs that adopt structures capable of binding to proteins or other nucleic acids. lncRNAs consist of multiple binding modules, thus enabling complex, coordinated recruitment of epigenetic modifying complexes or transcription factors to specific genomic loci in both cis and trans. lncRNAs therefore confer specificity upon epigenetic processes (which explains the existence of differential chromatin states despite many chromatin modifying activities being ubiquitously expressed). In this review we have discussed the importance of RNA structure on the function of lncRNAs and the plethora of ways in which they interact with the genome.
The degree to which lncRNAs are functional or merely represent ‘transcriptional noise’ is an open debate.102 Regardless, clear biological functions have now been assigned to a small, but growing, number of non-coding transcripts. lncRNAs are less amenable to studies of gene function than protein-coding genes as conventional knockout or knockdown studies may not produce immediately obvious phenotypes.103 Similarly, as many lncRNAs are not conserved between human and mouse, classical gene knockout studies are not possible. Alternatively, so-called ‘guilt by association’ methods have been utilized to infer lncRNA function by using gene set enrichment analysis (GSEA) to group lncRNA genes with protein-coding genes that exhibit similar patterns of expression and are likely involved in related cellular processes.21,104-107
Advances in high-throughput sequencing methodologies have enabled rapid progress in this field by enabling genome-wide inferences to be made concerning the generality of lncRNA function. The recent explosion of interest in non-coding RNA biology is unlikely to abate as the ‘dark matter of the genome’ is progressively illuminated by future studies. As we learn more about the interface between non-coding RNAs and epigenetics it is likely that the etiology of many complex diseases will be elucidated and novel therapeutic targets identified.108,109
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Roberts TC is supported by a MRC (UK) Centenary Early Career Award. Morris KV is supported by NIAID grants R56 AI096861-01 and P01 AI099783-01. Weinberg MS acknowledges funding from the MRC (South Africa).
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26700
References
- 1.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–3. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teif VB, Rippe K. Predicting nucleosome positions on the DNA: combining intrinsic sequence preferences and remodeler activities. Nucleic Acids Res. 2009;37:5641–55. doi: 10.1093/nar/gkp610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paro R. Propagating memory of transcriptional states. Trends Genet. 1995;11:295–7. doi: 10.1016/S0168-9525(00)89081-2. [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 6.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 8.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cieśla J. Metabolic enzymes that bind RNA: yet another level of cellular regulatory network? Acta Biochim Pol. 2006;53:11–32. [PubMed] [Google Scholar]
- 10.Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674–90. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–63. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 12.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–45. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–22. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Yin Q-F, Yang L, Zhang Y, Xiang J-F, Wu Y-W, Carmichael GG, Chen L-L. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–30. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 17.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 18.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–23. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C-L, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr., Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–8. doi: 10.1016/S0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- 30.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–28. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 31.Rougeulle C, Heard E. Antisense RNA in imprinting: spreading silence through Air. Trends Genet. 2002;18:434–7. doi: 10.1016/S0168-9525(02)02749-X. [DOI] [PubMed] [Google Scholar]
- 32.Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Göndör A, Grange T, Ohlsson R, Kanduri C. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol Cell Biol. 2004;24:7855–62. doi: 10.1128/MCB.24.18.7855-7862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 34.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501–12. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 36.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 40.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 41.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris KV, Santoso S, Turner A-M, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–9. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz K-M, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–9. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayer C, Schmitz K-M, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–61. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 47.Johnsson P, Ackley A, Vidarsdottir L, Lui W-O, Corcoran M, Grandér D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–6. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–75. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–8. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 53.Li J-T, Zhang Y, Kong L, Liu Q-R, Wei L. Trans-natural antisense transcripts including noncoding RNAs in 10 species: implications for expression regulation. Nucleic Acids Res. 2008;36:4833–44. doi: 10.1093/nar/gkn470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–92. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 55.Henkin TM. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–90. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36(Web Server issue):W70-4. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol. 2006;16:270–8. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol. 2010;20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zemora G, Waldsich C. RNA folding in living cells. RNA Biol. 2010;7:634–41. doi: 10.4161/rna.7.6.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liebeg A, Waldsich C. Probing RNA structure within living cells. Methods Enzymol. 2009;468:219–38. doi: 10.1016/S0076-6879(09)68011-3. [DOI] [PubMed] [Google Scholar]
- 61.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–7. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci U S A. 2011;108:11063–8. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 65.Maison C, Bailly D, Peters AHFM, Quivy J-P, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–34. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 66.Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol Cell Biochem. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 67.Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science. 2013;340:619–21. doi: 10.1126/science.1234848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 69.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–6. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–42. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim DH, Saetrom P, Snøve O, Jr., Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–5. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol. 2012;14:266–75. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adilakshmi T, Sudol I, Tapinos N. Combinatorial action of miRNAs regulates transcriptional and post-transcriptional gene silencing following in vivo PNS injury. PLoS One. 2012;7:e39674. doi: 10.1371/journal.pone.0039674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N, et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119:4034–46. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 76.Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–49. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Place RF, Li L-C, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–33. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–74. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 82.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–55. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–72. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 87.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK-S. Mouse transcriptome: Neutral evolution of “non-coding” complementary DNAs. Nature [Internet] 2004 [cited 2013 Jul 10]; 431. Available from: http://www.nature.com/nature/journal/v431/n7010/abs/nature03016.html [PubMed]
- 90.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–65. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 93.Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11:833–49. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oudejans CB, Westerman B, Wouters D, Gooyer S, Leegwater PA, van Wijk IJ, Sleutels F. Allelic IGF2R repression does not correlate with expression of antisense RNA in human extraembryonic tissues. Genomics. 2001;73:331–7. doi: 10.1006/geno.2001.6522. [DOI] [PubMed] [Google Scholar]
- 95.Taft RJ, Mattick JS. Increasing biological complexity is positively correlated with the relative genome-wide expansion of non-protein-coding DNA sequences. Genome Biol. 2003;5:1. doi: 10.1186/gb-2003-5-1-p1. [DOI] [Google Scholar]
- 96.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–51. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Havgaard J, Kaur S, Gorodkin J. Comparative ncRNA gene and structure prediction using Foldalign and FoldalignM. Curr Protoc Bioinformatics 2012; Chapter 12:Unit12.11. [DOI] [PubMed] [Google Scholar]
- 98.Torarinsson E, Sawera M, Havgaard JH, Fredholm M, Gorodkin J. Thousands of corresponding human and mouse genomic regions unalignable in primary sequence contain common RNA structure. Genome Res. 2006;16:885–9. doi: 10.1101/gr.5226606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmitt S, Paro R. Gene regulation: a reason for reading nonsense. Nature. 2004;429:510–1. doi: 10.1038/429510a. [DOI] [PubMed] [Google Scholar]
- 100.Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell. 2000;5:377–86. doi: 10.1016/S1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 101.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–21. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625–, discussion e1001102. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lewejohann L, Skryabin BV, Sachser N, Prehn C, Heiduschka P, Thanos S, Jordan U, Dell’Omo G, Vyssotski AL, Pleskacheva MG, et al. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav Brain Res. 2004;154:273–89. doi: 10.1016/j.bbr.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 104.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–78. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Loewer S, Cabili MN, Guttman M, Loh Y-H, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberts TC, Wood MJA. Therapeutic targeting of non-coding RNAs. Essays Biochem. 2013;54:127–45. doi: 10.1042/bse0540127. [DOI] [PubMed] [Google Scholar]
- 109.Varela MA, Roberts TC, Andaloussi SE, Wood MJ. Natural Antisense Makes Sense for Gene-specific Activation in Brain. Mol Ther Nucleic Acids. 2012;1:e24. doi: 10.1038/mtna.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]


