Abstract
The emergence of long non-coding RNAs (lncRNAs) has shaken up our conception of gene expression regulation, as lncRNAs take prominent positions as components of cellular networks. Several cellular processes involve lncRNAs, and a significant number of them have been shown to function in cooperation with chromatin modifying enzymes to promote epigenetic activation or silencing of gene expression. Different model mechanisms have been proposed to explain how lncRNAs achieve regulation of gene expression by interacting with the epigenetic machinery. Here we describe these models in light of the current knowledge of lncRNAs, such as Xist and HOTAIR, and discuss recent literature on the role of the three-dimensional structure of the genome in the mechanism of action of lncRNAs and chromatin modifiers.
Keywords: chromatin, epigenetics, gene expression, histone modification, long noncoding RNA
Introduction
The technological advances applied to functional genomics during the last decade have depicted a new scenario in the field of RNA biology. To date, approximately 35% of the human genes identified by the ENCODE project (about 57,000; GENCODE version 17), encode for proteins.1,2 The vast majority of the remaining genes (about 65%), are transcribed into RNA but do not encode proteins, and are generally known as “non-coding” RNAs (ncRNAs). Non-coding RNAs comprise several classes of RNAs, classified in different groups in accordance to their length, function, localization, orientation or other criteria (these classifications are continuously being adjusted as new data are acquired). Long non-coding RNAs (lncRNAs) refer to non-protein coding transcripts that are longer than 200 nucleotides (though this limit is merely arbitrary). Subcategories of lncRNAs also exist and, in this case, the classification varies depending on the criterion applied: long intergenic non-coding RNAs (lincRNAs) contain the same features of mRNAs (a 5′ 7-methylguanosine cap and a 3′ poly(A) tail),3 are mainly transcribed by RNA polymerase II4 and do not overlap with protein coding genes; antisense lncRNAs (AS-lncRNAs) are transcribed in the opposite direction of protein-coding genes; intronic lncRNAs are transcribed from an intron of a protein-coding gene and; enhancer RNAs (eRNAs) are transcribed from enhancer elements.
The number of genes encoding for lncRNAs currently identified is approximately 13 000 (GENCODE version 17), representing more than the 20% of the human genome. To date, only a small number of these have been studied in detail, providing pioneer indications of their functions and mechanisms of action in regulating cellular processes. Considering the vast number of lncRNAs identified genome-wide and their low sequence conservation across species, it had originally been speculated that most of the transcripts identified as lncRNAs are actually non-functional RNAs and were generally referred to as “transcriptional noise.” However, an increasing number of lncRNAs are being reported to be functional, although many of them with strict spatial-temporal functions. The debate on the functionality of lncRNAs remains open (see refs.5 and 6) and, even though it is likely that some transcripts do not have any biological function, it is clear that those RNAs reported so far have been ascribed relevant roles in specific biological processes.
lncRNAs have been reported to play roles in many different cellular processes (e.g., cell growth and apoptosis,7,8 development,9,10 and cell pluripotency and differentiation4,11,12) possibly through multiple and diverse mechanisms. Moreover, these studies indicate that a significant number of lncRNAs function in cooperation with chromatin modifiers, implicating lncRNAs in the epigenetic regulation of gene expression.
In this review, we will focus on mammalian lncRNAs involved in the regulation of cellular processes by epigenetic mechanisms, describing in more detail some of the lncRNAs known to interact with chromatin modifiers. By reviewing the literature, we will discuss the different suggested mechanisms by which lncRNAs fine-tune gene expression.
lncRNAs in X-Chromosome Inactivation: Decoys and Guides for Chromatin Modifiers
The first described epigenetic mechanism involving a lncRNA was mammalian X-chromosome inactivation (XCI), the process leading to gene silencing of one of the two female X-chromosomes in order to ensure equal X-linked gene expression in both sexes. The master regulator of X inactivation in mammals is the lncRNA Xist (X-inactive specific transcript), a product of the Xist gene, which is exclusively transcribed from the inactive X (Xi) chromosome.13 Xist is known to initially associate with the X-chromosome at a discrete number of focal loci, and subsequently spread to promote chromosome-wide heterochromatization (see refs.14-18). Since its discovery, the mechanism of X inactivation has been the subject of intensive study and nowadays represents a model mechanism for the understanding of lncRNA function. Several other lncRNAs have been identified that play essential roles in the process, such as the lncRNA Tsix, transcribed from the active X (Xa) chromosome in the antisense direction from Xist, and known to negatively regulate the expression of Xist19 in cis, and Jpx, which activates Xist by removing the repressive RNA-binding protein CTCF from the Xist promoter.20,21 Moreover, Tsix expression was shown to be sustained by another lncRNA, Xite, which has been shown to influence Tsix promoter activity. However, it remains unclear whether the genetic control of Tsix locus by Xite depends only on its RNA, since truncated Xite RNAs were shown to function as well as the full-length Xite lncRNA in regulating Tsix expression.22 As briefly described above, the mechanism of X inactivation is indeed an elegant interplay between several partners, finely regulating correct gene silencing. Much more is known about Xist and X chromosome inactivation, and several aspects of this intriguing cellular mechanism still remain unsolved (comprehensive reviews can be found elsewhere).19,23,24 However, what is relevant to the scope of this review is the notion that X inactivation was the first mechanism of gene silencing identified to depend on lncRNA and Polycomb group (PcG) proteins,25 elucidated by the finding that Xist was able to interact with the Polycomb repressive complex 2 (PRC2),26,27 opening the way to an increasing number of studies on epigenetic regulation of gene expression mediated by lncRNAs.
PcG proteins were originally identified in Drosophila melanogaster as essential components for the proper development of the organism that function by silencing the expression of homeotic genes. PcGs were later shown to have homologs in many other species, including mouse and human. PcG proteins assemble into enzymatically active in multisubunit complexes, of which the best characterized are the polycomb repressive complex 1 (PRC1) and PRC2. PRC1 is known to catalyze lysine 119 monoubiquitylation of histone H2A (H2AUb1), whereas PRC2 is able to dimethylate and trimethylate lysine 27 of histone H3 (H3K27me2/3), both known as repressive chromatin marks. PRC2, which is required for the initial targeting of genomic regions to be silenced, contains three core subunits, although other proteins, transiently interacting with the core, have been described.28 The three core subunits of PRC2 are the histone methyltransferase (HMT) catalytic subunit EZH2, the zinc finger protein Suz12, and EED (embryonic ectoderm development), the subunit involved in substrate recognition.29 PRC1, on the other hand, is required for stabilizing the gene silencing initiated by PRC2, and is a much more heterogeneous complex with several different forms identified in mammals, characterized by different subunit paralogues, composition and function.29
As mentioned before, Xist associates with PRC2 and their interaction leads to H3K27me2/3 of the Xi. Their binding has been shown to rely on a repeat motif contained in the 5′ portion of Xist RNA26 and, interestingly, a second non-coding transcript containing the same repeat, known as RepeatA (RepA), encoded within Xist first exon was also identified and found to independently bind PRC2 in vitro.26 Therefore, a simplified model sees RepA and Tsix, also shown to bind PRC2,26 detaining the chromatin complex until Jpx transcription and loss of Tsix expression activate Xist production, which is in turn able to interact with PRC2 and spread along the future Xi resulting in the H3K27 methylation of the entire chromosome.16 This model allows us to introduce two mechanisms of action through which lncRNAs have been suggested to work: (1) as “decoys” (RepA and Tsix function as decoys for PRC2) and (2) as “guides” (Xist is a guide for the complex to localize to the correct regions for silencing) (Fig. 1A and B).
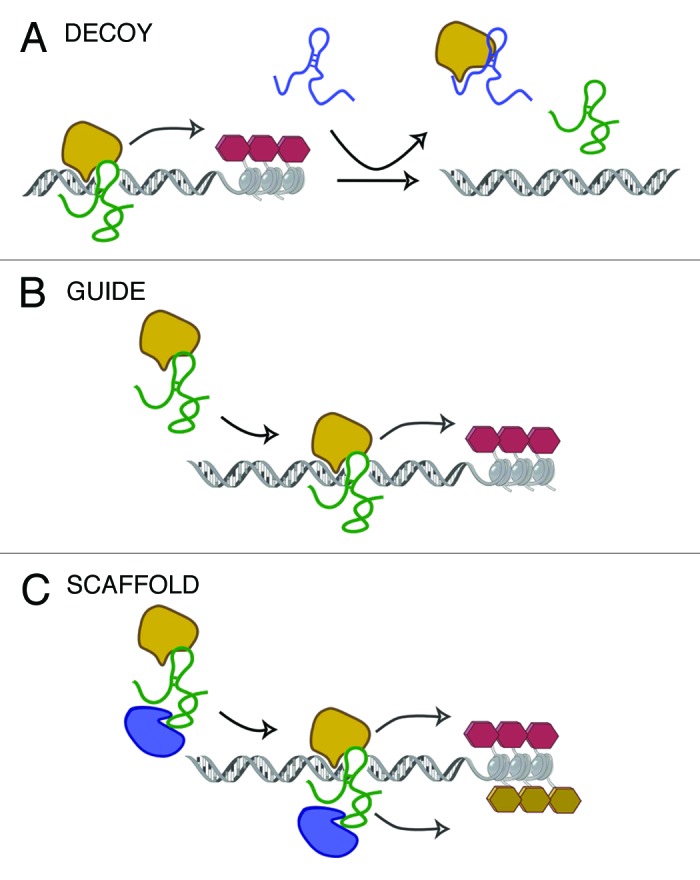
Figure 1. Proposed models for lncRNAs-mediated chromatin modification. (A) lncRNAs may function as “decoys,” sequestering chromatin-modifying enzymes away from other interacting partners or (B) as “guides” for chromatin modifiers by driving them to the correct locations in the genome. (C) lncRNAs could alternatively act as “scaffolds” to tether more than one protein complex and mediate different chromatin modifications.
The direct interaction between Xist and PRC2 and their role in XCI remains a source of debate due to disagreeing results or experimental limitations in determining the specific binding of Xist to PRC2.30 Certainly, the crosstalk complexity between all the partners involved in the establishment of XCI allows us to foresee a mechanism of action for Xist and the other lncRNAs involved that requires additional proteic factors apart from PRC2. However, it cannot be excluded that in in the presence of novel interacting partners the “decoy” and “guide” models would remain valid, and that the interplay between lncRNAs and different interacting proteins would be responsible for the regulation of gene expression.
HOTAIR and Kcnq1ot1 lncRNAs: Scaffolds for the Binding of Chromatin Complexes
PRC2 probably represents the most studied chromatin complex associated with lncRNAs. Many other lncRNAs, some of which are listed in Table 1, have been reported to interact with PRC2 and to promote gene silencing. Among them, the lincRNA HOTAIR (HOX Antisense Intergenic RNA), which is encoded antisense to the HOXC locus, one of the four chromosomal loci (HOXA to D) containing the clustered HOX genes.31,32 Briefly, HOTAIR regulates the transcriptional silencing of genes of the HOXD locus and other gene loci by binding PRC2 to its 5′ end and driving it to the specific locus where methylation and epigenetic silencing of gene expression is achieved.31 As opposed to Xist, HOTAIR has been shown to act in trans by regulating expression of genes in distant chromosomal locations.31 Moreover, HOTAIR interacts with another chromatin complex, the lysine specific demethylase 1 (LSD1)-CoREST complex, which mediates the removal of mono- and di-methylation of H3K4 in nucleosomes, a histone mark associated with gene activation.33 It seems therefore that HOTAIR is able to operate not only as a guide lncRNA, but also as a “scaffold,” due to its ability to bind and bring together two complexes that cooperate in establishing the repressive chromatin state (Fig. 1C).
Table 1.
| lncRNA | Chromatin modifier | Technique used (and references) |
|---|---|---|
| Xist | PRC2 | Co-localization by immunostaining (antibodies α-EZH2, and α-EED),34 RIP-qPCR (antibodies α-EZH2, and α-Suz12),26 RIP-Chip27 |
| Kcnq1ot1 | G9a and PRC2, Dnmt1 | RIP-qPCR (antibodies α-G9a, α-EZH2, and α-Suz12),35 RIP-qPCR (antibody α-Dnmt1)36 |
| Air | G9a | RIP-qPCR (antibody α-G9a)37 |
| HOTAIR | PRC2 and LSD1-CoREST | RIP-qPCR (antibodies α-LSD1 and α-EZH2),31,33 in vitro transcribed biotinylated RNA pull-down with cell extracts or purified PRC2 and LSD1 complexes,31,33 RIP-Chip27 |
| ANRIL | PRC1 (CBX7) and PRC2 | RIP-qPCR (CBX7), RNA EMSA, biotinylated RNA pull-down,38 RIP-qPCR (antibody α-Suz12)39 |
| pRNA | Dnmt3b | Southwestern blot and pull-down experiments40 |
| Mistral | MLL-Trx (MLL1) | RIP-qPCR (MLL1), Recombinant MLL1 - in vitro binding assays41 |
| HOTTIP | MLL-Trx (WDR5) | RIP-qPCR (antibody α-WDR5), Recombinant GST-WDR5 - in vitro transcribed RNA interaction42 |
| DBE-T | MLL-Trx (Ash1L) | RIP-qPCR (antibody α-Ash1L), Recombinant GST-Ash1L - in vitro transcribed RNA interaction43 |
| PINC | PRC2 | RIP-qPCR (antibodies α-EZH2, α-Suz12 and α-RbAp46)44 |
| PTENpg1 asRNA α | Dnmt3a | RIP-qPCR (antibody α-Dnmt3a)45 |
| Fendrr | PRC2 and MLL-Trx (WDR5) | RIP-qPCR (antibodies α-EZH2, α-Suz12, and α-WDR5)9 |
| Braveheart | PRC2 | RIP-qPCR (antibody α-Suz12), Biotinylated RNA pull-down11 |
| NeST | MLL-Trx (WDR5) | RIP-qPCR (FLAG-WDR5 transfection and IP with antibody α-FLAG)46 |
| UBC1 | PRC2 | RIP-qPCR (antibodies α-EZH2, and α-Suz12)47 |
| Pint | PRC2 | RIP-qPCR (antibody α-Suz12), in vitro transcribed biotinylated RNA pull-down with cell extracts or purified PRC248 |
The ability to tether more than one protein complex has been shown for several other lncRNAs, some listed in Table 1. This list is likely to expand as genome-wide studies have reported a large number of transcripts able to interact with different chromatin complexes.4,27,33 Moreover, RNA molecules should not be thought as linear entities able to interact with proteins only by sequence-specific elements but, on the contrary, lncRNAs are likely to make use of their secondary structures to interact with their protein partners as well as with the DNA.49
Kcnq1ot1 represents another example of a lncRNA able to bind different chromatin modifiers. It is a 91 kb-long lncRNA that is transcribed in antisense orientation from intron 10 of the murine Kcnq1 gene.35 Briefly, this lncRNA is expressed only from the paternal allele and is responsible for the silencing of a cluster of ten genes spread over a 1 Mb region around its location.35Kcnq1ot1 was originally found to correlate with repressive histone modifications, such as H3K9me3 and H3K27me3, in the Kcnq1 domain,50 and was later identified to interact with two chromatin modifiers responsible for these histone modifications, the HMT G9a, which mono- and di-methylates H3K9, and PRC2.35 As opposed to HOTAIR, no evidence for coexisting interaction of the two enzymes has been reported for Kcnq1ot1, suggesting that the lncRNA may work as a scaffold by bringing, simultaneously or independently, different complexes in proximity to the genes to be silenced. Moreover, the interaction between Kcnq1ot1 and these enzymatic complexes has been shown to occur in placenta but not in liver, and the presence or absence of interaction correlates with lineage-specific differences observed for these chromatin modifications in the Kcnq1 locus.35
Interestingly, the genes regulated in cis by Kcnq1ot1, which are imprinted both in the embryo and in the placenta of the paternal chromosome, are located closer to the promoter, while the genes repressed only in the placenta are those located more distally from the promoter.36 Since the interaction between Kcnq1ot1 and G9a or PRC2 was found to occur only in the placenta,35 it remained unclear how the lncRNA could mediate the silencing of genes in the embryo. A more recent publication has identified the DNA methyltransferase Dnmt1 as a novel interacting partner of Kcnq1ot1 in the embryo, suggesting that the RNA might mediate the silencing of imprinted genes in the embryo by promoting specific DNA methylation.36 It seems therefore that Kcnq1ot1 is able to mediate lineage-specific imprinting through different mechanisms, being able to recruit chromatin modifiers for the silencing of placental-specific genes or by promoting allele-specific methylation of DNA for the imprinting of genes in the embryo.
The Role of Chromatin in the Coordinated Function of lncRNAs and Chromatin Modifiers
Only recently chromatin has started being considered as an active player in the mechanism by which lncRNAs and chromatin modifiers achieve regulation of gene expression. Therefore, it seems that, although the “decoy,” “guide,” and “scaffold” models described above may remain valid overall, the mechanism of action of lncRNAs still needs to be fully uncovered and, presumably, will be more complicated than this.
The development of techniques to study three-dimensional structure of the genome, such as chromosome conformation capture51-55 and techniques to enrich endogenous lncRNAs along with their protein partners and DNA targets, such as capture hybridization analysis of RNA targets (CHART)56 or chromatin isolation by RNA purification (ChIRP),57 are allowing a better understanding of the role of chromatin in the mechanism of action of lncRNAs. eRNAs, for instance, have been suggested to mediate chromatin looping between the enhancer region and the promoter of the genes they regulate, although working as a scaffold for transcriptional activators rather than for chromatin modifiers.58 A representative example of the tripartite interplay between chromatin, lncRNAs and chromatin modifiers is given by HOTAIR. Genome-wide mapping of its occupancy sites has shown a significant pattern of co-occupancy with the binding sites of the PRC2 subunits EZH2 and Suz12 as well as H3K27me3, suggesting that localization of PRC2 to target sites depends on the interaction between HOTAIR and chromatin.57 Moreover, HOTAIR occupancy was shown to be preserved upon EZH2 depletion,57 supporting a mechanistic model that proposes that HOTAIR binds to its target chromatin regions independently of PRC2. The evidence seems therefore to sustain a model in which the lncRNA makes contact with the chromatin and, once located in the correct place, promotes the recruitment of the chromatin-modifying complex.
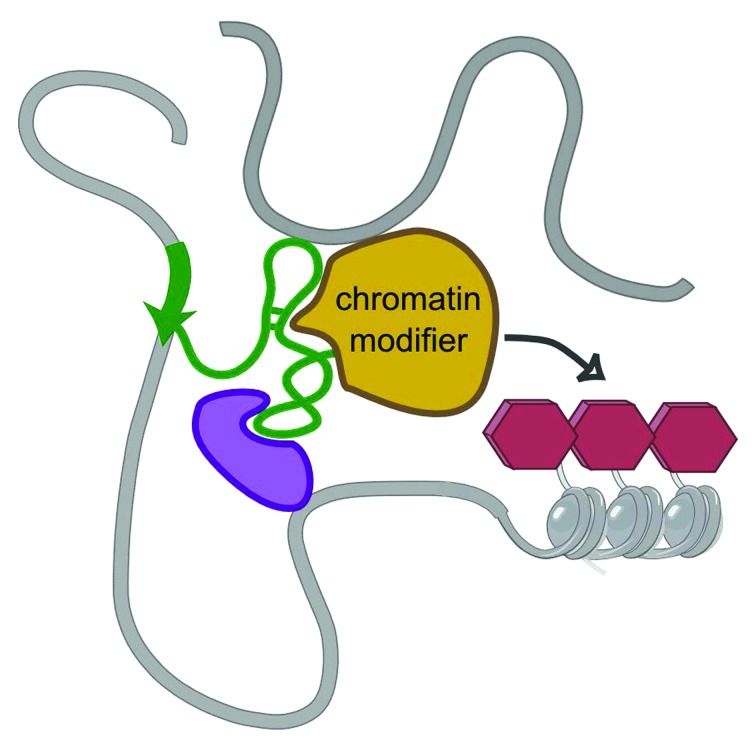
To support the notion that the three-dimensional structure of the genome contributes to the function of lncRNAs, a recently published study uses XCI and Xist lncRNA as a model to provide new insights into the mechanism of lncRNA localization to DNA and silencing of gene expression.17 By using a methodology to capture RNA interactions with chromatin, named RNA antisense purification (RAP), combined with genome-wide chromosome conformation capture, Engreitz and coworkers analyzed Xist localization across the X chromosome at different time points of Xist induction and therefore XCI establishment.17 The obtained results seem to suggest that Xist spreading across the X chromosome is initiated by its transfer to specific distal sites from its transcription locus and that such initial contacts depend on spatial proximity rather than sequence specificity.17 Interestingly, initial localization of Xist was observed on the periphery of regions enriched in active genes both when using either wild-type Xist or a Xist RNA lacking the A-repeat domain, the domain required for the binding of PRC2.17 However, later access of Xist to these active regions was found to be dependent on the presence of the A-repeat domain, suggesting a model in which chromatin-bound Xist (in proximity of active genes) recruits PRC2, which, in turn, drives gene silencing by promoting heterochromatization.17
In conclusion, the results obtained for Xist, as well as those described above for HOTAIR, seem to point to a mechanism of action for these two lncRNAs that relies upon the interaction between the lncRNA and chromatin to occur in the first place (Fig. 2). Given the lack of obvious RNA-DNA sequence complementarity, the mechanism by which lncRNA-chromatin interactions are established in the first place remains as yet unsolved. Some studies have suggested a direct interaction between DNA and lncRNA through the formation of triple helix structures.9,40 However, the mediation of additional protein factors in the chromatin-lncRNA complexes cannot be excluded.
Figure 2. Model for lncRNAs-mediated chromatin modifications in a three-dimensional context. The lncRNA (green) may contact specific chromatic regions (gray) placed in proximity by the three-dimensional organization of the genome. The contact can be direct or mediated by another protein (purple). lncRNA may then recruit the chromatin modifier and promote epigenetic modifications to regulate gene expression.
The potential of combining three-dimensional structure of the genome with DNA-binding maps of lncRNAs and proteins will certainly help us uncover the complexity behind lncRNAs-mediated regulation of gene expression. Additionally, a better understanding of the biochemical and structural nature of the lncRNA-protein and lncRNA-DNA interactions is needed to define the underlying molecular mechanisms involved in the process. These studies will undoubtedly help refine the suggested model and will tell us whether these are applicable to other lncRNAs.
Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/27472
References
- 1.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M, ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 6.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–8. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loewer S, Cabili MN, Guttman M, Loh Y-H, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 14.Pollex T, Heard E. Recent advances in X-chromosome inactivation research. Curr Opin Cell Biol. 2012;24:825–32. doi: 10.1016/j.ceb.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Brockdorff N. Chromosome silencing mechanisms in X-chromosome inactivation: unknown unknowns. Development. 2011;138:5057–65. doi: 10.1242/dev.065276. [DOI] [PubMed] [Google Scholar]
- 16.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 17.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013 doi: 10.1038/nature12719. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 20.Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–51. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11:731–43. doi: 10.1016/S1097-2765(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 23.Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. 2011;130:223–36. doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senner CE, Brockdorff N. Xist gene regulation at the onset of X inactivation. Curr Opin Genet Dev. 2009;19:122–6. doi: 10.1016/j.gde.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–5. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nayak V, Xu C, Min J. Composition, recruitment and regulation of the PRC2 complex. Nucleus. 2011;2:277–82. doi: 10.4161/nucl.2.4.16266. [DOI] [PubMed] [Google Scholar]
- 29.Lanzuolo C, Orlando V. Memories from the polycomb group proteins. Annu Rev Genet. 2012;46:561–89. doi: 10.1146/annurev-genet-110711-155603. [DOI] [PubMed] [Google Scholar]
- 30.Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–42. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 35.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 36.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–9. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 37.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 38.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou M-M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment toand silencing of p15. Oncogene. 2010;30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–9. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–6. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149:819–31. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shore AN, Kabotyanski EB, Roarty K, Smith MA, Zhang Y, Creighton CJ, Dinger ME, Rosen JM. Pregnancy-induced noncoding RNA (PINC) associates with polycomb repressive complex 2 and regulates mammary epithelial differentiation. PLoS Genet. 2012;8:e1002840. doi: 10.1371/journal.pgen.1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnsson P, Ackley A, Vidarsdottir L, Lui W-O, Corcoran M, Grandér D, Morris KV. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–6. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez JA, Wapinski OL, Yang YW. Bureau J-F, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST Long ncRNA Controls Microbial Susceptibility and EpigeneticActivation of the Interferon-g Locus. Cell. 2013;152:743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He W, Cai Q, Sun F, Zhong G, Wang P, Liu H, Luo J, Yu H, Huang J, Lin T. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta. 2013;1832:1528–37. doi: 10.1016/j.bbadis.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 48.Marín-Béjar O, Marchese FP, Athie A, Sánchez Y, González J, Segura V, Huang L, Moreno I, Navarro A, Monzó M, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanduri C, Thakur N, Pandey RR. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. EMBO J. 2006;25:2096–106. doi: 10.1038/sj.emboj.7601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 52.Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–33. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 54.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belton J-M, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–76. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ørom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21:194–8. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]