Abstract
Epigenetic regulation of gene expression is an increasingly well-understood concept that explains much of the contribution of an organism’s environment and experience to its biology. However, discussion persists as to which mechanisms can be classified as epigenetic. Ongoing research continues to uncover novel pathways, including the important role of non-protein coding RNA transcripts in epigenetic gene regulation. We know that the majority of human and other mammalian transcripts are not translated but that many of these are nonetheless functional. These non-coding RNAs (ncRNAs) can be short (< 200 nt) or long (< 200 nt) and are further classified by genomic origin and mechanism of action. We discuss examples of ncRNAs that interact with histone modifying complexes or DNA methyltransferases to regulate gene expression, others that are targets of these epigenetic mechanisms, and propose a model in which such transcripts feed back into an epigenetic regulatory network.
Keywords: chromatin, epigenetics, gene expression, histone, lncRNA, microRNA, noncoding RNA, piRNA, polycomb
Introduction
The most widely accepted definition of epigenetics includes stably inherited modulations in the expression of genes that do not involve changes in their DNA sequence. Developments in the field of molecular biology have uncovered a new layer of complexity lying beyond classical epigenetic mechanisms, such as methylation of DNA or posttranslational modification of histones. In this new view, gene expression can vary due to the function of RNA molecules themselves as well as their interactions with DNA and/or proteins. Specifically, increasing emphasis is being placed on the ability of non-coding RNA (ncRNA) transcripts to modulate gene expression and, thus, on their role as epigenetic modifiers. In this review, we focus on direct interactions between ncRNA and both traditional and emerging epigenetic processes, as well as instances in which targeting of ncRNAs has important downstream consequences for gene expression.
ncRNA Classification
Recent efforts by several consortia have made it clear that the vast majority of the human genome is transcribed, but that only around 2% of these transcripts are translated into protein. Many of the remaining transcripts—which do not code for protein—were at first regarded as “junk” or experimental artifact, but mounting evidence demonstrates that these RNA molecules can indeed be functional. Furthermore, the significance of this transcription to mammalian biology is supported by the strong correlation between organismal complexity and size of the non-coding, rather than protein-coding genome.1 ncRNAs less than 200 nucleotides in length are generally classified as short, while all larger transcripts are regarded as long ncRNA (lncRNA). There are several subtypes of long and short ncRNA species, many of which are involved in regulation of gene expression, and these can be further grouped according to their genomic origins and biogenic processes.
lncRNAs are highly diverse in structure and function; therefore, sequence length and lack of protein-coding capacity are perhaps their only defining characteristics. Nevertheless, several patterns have emerged which are of interest in the context of epigenomic regulation. One important consideration is that, while sequence conservation of lncRNAs is reportedly poor, transcripts with corresponding positions and directions in reference to protein coding genes are more common, indicating that their functions may well be conserved.2 lncRNAs are frequently processed similarly to mRNA and can be found in nuclear or cytosolic fractions.3,4 They are often transcribed from protein coding loci, but several lncRNA subclasses with differing origins have been described (Table 1). Some belong to introns,5 are composed of transposable genomic elements6 or form a sense-antisense pair called a “natural antisense transcript (NAT).”7 The existence of long ncRNAs has been acknowledged for several decades; however, the description of “long intergenic ncRNA (lincRNA)” was relatively recent. While the terms are occasionally used interchangeably in the literature, it bears noting that “lincRNA” refers only to the subset of lncRNAs that arises from intergenic regions.8,9 Interestingly, there is transcription at activity dependent neuronal enhancer domains10 and lncRNAs with enhancer-like properties have also been reported.11 While functional roles of lncRNAs are varied, they can be grouped into one or a combination of several patterns.12 For example, some transcripts are produced in reaction to perturbations in the cellular environment while others serve as intermediaries, facilitating or blocking known processes (including epigenetic mechanisms, discussed below) involving nucleic acids and/or proteins.
Table 1. Classification of lncRNAs.
| lncRNA subtype | Origin | Function |
|---|---|---|
| Natural Antisense Transcript (NAT) | cis-NAT: sequence complementarity to a coding RNA at the same locus | Regulates expression of sense partner transcript |
| trans-NAT: sequence complementarity to a coding RNA at a distal genomic locus | Likely regulates expression of sense partner transcript, may have other functions | |
| Long Intergenic Noncoding RNA (lincRNA) | Encoded between known protein coding genes or within introns. macroRNA or vlincRNA = very long intergenic RNA | Functions not fully characterized, likely include regulation of transcription, RNA stability, recruitment of protein complexes and other subcellular elements |
| Sense Overlapping | Transcribed from same strand of DNA as another transcript | |
| Sense Intronic | Originate from introns of coding genes; do not overlap with exons | |
| Processed Transcript | RNA transcript that is spliced and/or polyadenylated |
LncRNAs, which are all non-coding transcripts longer than 200 nucleotides, can vary greatly in their genomic origin, relationship to coding genes, and level of processing. While definitions continue to evolve, we present several of the terms currently in use.
In contrast to lncRNAs, short ncRNAs have been extensively classified based on their genomic origins and precise mechanisms of action. Many of these subgroups have important consequences for gene expression, affecting transcription itself as well as downstream processes. Perhaps the best studied of these are the microRNAs (miRNAs), which are 20–23 nucleotides (nts) in length and usually recognize target mRNAs by complementarity to a 2–7 nt long seed region in the 3′-UTR,13,14 but may also target the 5′-UTR.15 This binding activates the miRNA-induced silencing complex (miRISC) through which miRNAs act as posttranscriptional repressors of gene expression. This process includes the endoribonuclease Dicer, which cleaves double stranded RNA (dsRNA), and is involved in the formation of another emerging class of short ncRNAs, the endogenous siRNAs (endo-siRNAs). These transcripts are not yet well understood, but it has been reported that unlike miRNAs, endo-siRNAs require extensive sequence complementarity to induce gene repression.16 To produce these small endo-siRNAs, Dicer cleaves long double-stranded ncRNAs formed from natural sense-antisense transcript pairs (cis-nat siRNAs) at a single locus or from gene-pseudogene pairs (trans-nat siRNAs), which may include transcripts from distinct chromosomes,17 or from hairpin loops formed at inverted repeat-containing sequences.18 Another category of endogenous short RNAs, the PIWI-interacting RNAs (piRNAs), are 26–30 nt, single-stranded and Dicer independent. piRNA precursor transcripts undergo endonucleolytic cleavage in the cytoplasm, are loaded into the Aubergine (Aub) or Piwi chaperone complex for 3′ trimming and methylation, completing the primary biogenesis pathway. When loaded into Aub, piRNAs can then be amplified via secondary biogenesis, a similar process that first involves recognition and loading of a cognate transcript onto the Piwi protein, Argonaute 3 (Ago3).19 piRNAs can silence transcription of target RNAs (frequently transposable elements recognized as “non-self”) through perfect or mismatched base pairing, leading to trimethylation of histone 3 lysine 9 (H3K9me3, a marker of inactive chromatin) by one of several histone methyltransferases.19
Many of the gene regulatory functions performed by the ncRNAs described above are dynamic, sensitive to environmental changes and therefore may fit an expanded view of epigenetics as discussed in the following section. We limit our focus on these long and short ncRNA species; other publications provide excellent overviews of known classes of functional ncRNAs.20,21 Additionally, the overall transcriptional landscape of human cells has been extensively described by the ENCODE and FANTOM projects.22,23
Defining Epigenetics
As mentioned above, epigenetics is widely known as the regulation of gene expression without changes to DNA sequence. Currently, there is discussion in the field as to how stable and heritable a mechanism must be in order to be classified as “epigenetic” and, whether it is acceptable to include not only those marks retained across generations of organisms, but also those passed on during cell divisions, along with other categories of self-perpetuating marks.24 Patterns of DNA methylation, a classical epigenetic mark linked to gene repression, have indeed been shown in both plants and mammals to be stably transmissible to offspring.25,26 On the other hand, DNA methylation status is not transmitted with perfect fidelity, causing differences in pattern even among clonal cell populations.27,28 Furthermore, although histone protein modifications are considered traditional epigenetic mechanisms, they are dynamically edited by demethylases (discussed below) and their distribution can turn over within hours in response to environmental stimuli.29-31 Consequently, others have argued for an expansion of our working definition of epigenetics that includes alterations to chromatin which reflect dynamic cellular environments and activity states.32,33 Nonetheless, the biology of stable maintenance of chromatin marks is an important component of epigenetic phenomena that is yet to be worked out, and regulation by ncRNA is likely part of the picture.
It is known that during fertilization and implantation of embryos, DNA demethylation and subsequent re-methylation occurs34 and that DNA methyltransferases (DNMTs) 1–3 propagate CpG methylation after S-phase across mitoses by associating with hemimethylated DNA (reviewed in ref. 35). Transmission of histone modifications could involve Polycomb proteins (discussed below) and has been alternately hypothesized to follow a “semi-conservative” model (similar to DNA replication)36 or a random pattern, in either case most likely relying on a “reader” molecule to recognize and spread the parental methylation to newly synthesized histones. 37 As we will illustrate with specific examples below, ncRNA species are involved in gene silencing and activation by influencing the mechanisms mentioned above. The role of ncRNA in epigenetic inheritance appears therefore to be in recruitment and guidance of these processes at the appropriate moment in cell cycling or throughout the development and lifespan of an organism.
It is in this context that we incorporate ncRNA regulation of gene expression into a larger epigenetic network. It should be noted that transcription factors creating lasting effects through positive feedback loops could also be included, but are beyond the scope of this review (see ref. 24 for more information). First, genomic loci producing non-coding transcripts are subject to epigenetic regulation similar to that of protein-coding genes.38-42 Therefore, as with coding genes, environmental influences can lead to changes in their expression and to functional disturbances. We can include in this category the transcription of lncRNAs designated as “signals,”12 as these are examples of activity-dependent, sequence-independent changes in gene expression. In turn, both short and long ncRNAs modulate gene expression through mechanisms that will be discussed in detail below. As both targets and enactors of expression changes, ncRNAs are important messengers of gene regulation. In the following sections, we outline mechanisms of ncRNA-dependent regulation of gene expression, many of which involve traditional epigenetics.
lncRNA-Mediated Gene Silencing Through Polycomb Mechanisms
The Polycomb group (PcG) of proteins was identified in D. melanogaster for their ability to cause heritable gene silencing, in particular through mediation of X inactivation.43,44 Polycomb complexes occurring in different species are composed of analogous subunits with specialized roles; these include proteins with putative RNA binding domains (RBDs) or catalytic domains with histone-modifying activity (e.g., SET) as in mammalian EZH2 and CLF in Arabidopsis thaliana.45-49 Other subunits, including the chromodomains found in core Polycomb Repressive Complex 1 (PRC1) proteins, have been shown to bind modified histones such as trimethylated histone 3 lysine 27 (H3K27me3) thereby targeting the locus for further modification and gene repression.50,51
As was first shown for ANRIL, the antisense lncRNA located in the p14/p15/INK4 locus, these transcripts can cause gene repression via PcG recruitment and formation of heterochromatin.52-54 Another well-studied example of a ncRNA involved in gene silencing through Polycomb recruitment is that which takes place during X–chromosome inactivation (XCI), which ensures equal X-linked gene dosage in female mammals. XIST, a conserved 17kb lncRNA, is widely known to be essential to this process and to be inhibited by its antisense partner TSIX (also non-coding).55-58 A short repeat RNA (RepA) found within XIST recruits Polycomb repressive complex 2 (PRC2) to the locus, leading to H3K27 trimethylation, silencing and spreading of XCI throughout the X chromosome.59-62 It has been shown that PRC1 is also recruited to the X-inactivation center,63,64 likely due to the deposition of H3K27me3 marks as described above, as well as through an H3K27me3-independent pathway.65
In contrast to the widespread X-chromosome gene silencing resulting from XCI, other lncRNAs invoke PcG proteins for more targeted repression. On a somewhat smaller scale, imprinting of a region on human chromosome 11p15.5 (involved in Beckwith-Wiedemann syndrome) appears to be controlled at least in part by the paternally-expressed antisense ncRNA KvDMR1/LIT1/KCNQ1OT1.66-68 This 91 kb-long transcript mediates bidirectional silencing by interacting with chromatin in a tissue lineage-specific manner and recruits histone methyltransferase members of the PRC2 complex, as well as the enzyme G9a, resulting in an increase in repressive histone modifications in the Kcnq1 domain.69 Even more targeted still is the repression of the negative regulator of flowering (at the flowering control (FLC) locus) in A. thaliana by the COLDAIR RNA. In this organism, vernalization causes transcription from within the FLC intron 1 of COLDAIR, which appears to recruit PRC2 through CLF binding.70 Another targeted mechanism of PcG recruitment is that of the NAT to brain-derived neurotrophic factor (BDNF), BDNF-AS. Chromatin immunoprecipitation of mammalian cells treated with siRNAs against BDNF-AS demonstrated reduced promoter occupancy by the PRC2 component EZH2, as well as a decrease in H3K27me3 at that locus.71
It is clear from the above that lncRNAs play an important role in targeting PcG proteins to coding regions; however, they may be significant even as targets of Polycomb complexes. For example, the enhancer region of mouse Neurog1, a proneural basic helix-loop-helix protein important for neuronal fate specification and differentiation, is home to a 1.4-kb sense-oriented ncRNA called utNgn1. This transcript was shown be a target of PcG silencing during later (gliogenic) stages of development. Both Neurog1 and utNgn1 are more highly expressed during earlier developmental stages, and the non-coding transcript was shown to positively regulate expression of Neurog1. In later development, utNgn1 becomes a target of Ring1B (a component of PRC1), and suppression of Neurog1 is achieved indirectly by PcG regulation of the enhancer-derived RNA utNgn1.72 This illustrates a potential functional paradigm for ncRNAs as intermediaries, in which they not only serve to recruit epigenetic (and other) enzymes to a particular locus, but are targeted themselves in order to modify their ability to regulate their partner genes.
Most of the aforementioned lncRNAs interacting with Polycomb complexes are examples of transcripts operating in cis, meaning they target the same genomic region from which they are transcribed. Other lncRNAs have been described that work in trans, using PcG mechanisms to modify expression of distant loci. HOTAIR, a 2.2-kb trans-active lncRNA transcribed from chromosome 12, represses a 40-kb stretch of homeobox genes on human chromosome 2 by recruiting PRC2 and LSD1; this causes H3K27 trimethylation and loss of the activating mark H3 lysine 4 trimethylation (H3K4me3).73-75 Recently, overexpression of a trans-acting lincRNA, termed linc-UBC1, was reported in a sample of bladder cancer tissues. This transcript associates with at least two PRC2 subunits—EZH2 and SUZ12—and may be involved in repression of several known targets of that complex.76 Furthermore lncRNA-H19, originally described as a key player in genomic imprinting,77-79 was shown to promote metastasis (also in bladder cancer) by derepressing the Wnt pathway in trans through a PRC2-dependent mechanism.80
Despite the significant evidence that PRC2 and other Polycomb proteins are recruited by lncRNAs and function largely in gene repression, association of PRC2 with active chromatin (as defined by the presence of activating histone marks and RNA polymerase II occupancy) has been reported. Specifically, the mammalian PRC2 subunit EZH1 may bind active domains and promote transcriptional elongation during myocyte development, as demonstrated with ChIP-seq and siRNA knockdown of Ezh1.81 More broadly, a recent study found that human PRC2 promiscuously binds RNA both in vitro and in vivo. This includes RNA from other species—even one that does not possess PcG proteins, such as E. coli.82 Furthermore, the authors report that while EZH2 binds both active and repressed chromatin, transcription of the active subset of genes is not affected by loss of SUZ12, the subunit responsible for PRC2 inhibition. Therefore, binding of active chromatin likely represents a distinct function, adding to the complexity surrounding the interaction between PRC2 and RNA species.
lncRNAs and Other Epigenetic Complexes
We have discussed above many of the known interactions of lncRNAs with PcG protein complexes; however, this class of transcripts has also been shown to regulate gene expression through interactions with several other epigenetic enzymes. In fact, Fendrr, a lncRNA involved in cardiac development, is linked to down or upregulation of individual genes by specifically binding and/or modulating the promoter occupancy of both PRC2 and the H3K4 methyltransferase TrxG/MLL.83 In contrast to lncRNAs that function exclusively through PRC2 mechanisms, other transcripts act by increasing expression of target genes. HOTTIP is expressed primarily in distal cells (e.g., foreskin and foot fibroblasts) and has been shown to be necessary for activation of HOXA genes through its binding and recruitment of the MLL1 subunit WDR5 to the locus.84
Beyond regulation of gene expression through histone modifications, lncRNAs have also been implicated in gene silencing through DNA methylation. One example is Kcnq1ot1, the mouse homolog of the imprinting control RNA described above. In addition to interacting with PRC2 and G9a, it appears that through a critical 890-bp region, this transcript recruits the DNA methyltransferase Dnmt1 to CpG islands of imprinted genes.85 Another interesting case is the natural antisense transcript (NAT) AS1DHRS4. lncRNAs in this class frequently regulate their sense partners in cis86; this NAT does so as well by maintaining epigenetic silencing through G9a and EZH2. However, AS1DHRS4 may also induce repression in trans, by controlling localization of several DNMTs to the promoter regions of two nearby transcripts, DHRS4L1 and DHRS4L2.87
Most of the epigenetic actions of ncRNAs discussed thus far have involved the addition of a particular mark; however, the dynamic landscape of histone and DNA modifications suggests a mechanism for their removal as well. While histone deacetylation and dephosphorylation and DNA demethylation have been recognized for some time,88,89 the idea of histone demethylation was controversial until the discovery of the nuclear amine oxidase homolog LSD1 in 2004.90 As mentioned above, this enzyme has since been shown to cause gene repression when anchored to target genes by the lncRNA HOTAIR by removing the activating mark H3K4me375; however, other demethylase species may potentially activate expression by targeting other methylated histones. Recently, the locus-specific H3K4me3 demethylase Fbxl10 was shown to associate with several gene promoters, including that of Xist, through its PHD domain.91 Flowering of A. thaliana also appears to be controlled in part by FLD, a homolog of LSD1, which may require 3′ processing of an antisense RNA in order to be active.92-93 In the spore-forming yeast Saccharomyces cerevisiae, global H3K4 demethylation has been shown to repress sense and antisense non-coding transcription surrounding the transcription start sites of coding genes.94 The downstream consequences of this silencing have yet to be worked out; nevertheless, given our current knowledge of ncRNA action, this observation suggests that in addition to direct cooperation of lncRNAs with demethylases, targeting of these transcripts by epigenetic mechanisms may serve as an indirect mechanism of effecting changes in gene expression.
Epigenetic Mechanisms of Short ncRNAs
Many short ncRNAs control gene expression by affecting transcription or mRNA stability, but here we consider the evidence that these transcripts influence some familiar epigenetic mechanisms. First, transcription of 50–200 nt short RNAs was described from H3K27me3-rich PRC2 target genes undergoing cell-type specific silencing. Indeed, these short RNAs interact with PRC2 through stem-loop structures, stabilizing the complex near the site of transcription and leading to chromatin repression in cis.95 We have already presented one such example – the short repeat RNA RepA, which is key for binding of Xist and PRC2.62
Furthermore, miRNAs are known to regulate expression of several components of Polycomb complexes, thereby affecting their downstream actions. At least two miRNAs that are downregulated by oncogenes or other players involved in tumorigenesis appear to target the histone methyltransferase EZH2, resulting in EZH2 overexpression.96-99 Several other miRNAs act as tumor suppressors by inhibiting the stem cell factor Bmi1, which is also a component of PRC1.100-102 Another, miR-214, reduces Ezh2 expression and promotes skeletal muscle differentiation.103 The RNAi pathway also influences posttranscriptional chromatin modifications104 and is crucial for formation of heterochromatin and its propagation across cell divisions105; this suggests a mechanism for endo-siRNA-induced gene expression changes. Indeed, H3K9 methylation of heterochromatin in Drosophila is dependent on cytoplasmic localization of transposable element-derived endo-siRNAs.106
Histone deacetylases (HDACs), like PcG proteins, are involved in gene silencing but do so by removing activating marks, rather than through deposition of repressive modifications. Furthermore, some HDACs have been implicated in disease—for example, HDAC1 overexpression has been observed in certain types of cancer, and (at least in prostate cancer cells), may normally be targeted for repression by miR-449a.107 However, in hepatocellular carcinoma (HCC) cells overexpressing HDACs 1–3, decreased miR-449a signaling was reported and was linked to decreased proliferation and tumor formation.108 These contrasting findings may reflect differences between tissue types, or represent a more complex feedback system that remains to be clarified.
miRNAs have also been shown to downregulate DNA methylation, most frequently resulting in increased gene expression. In lung cancer and leukemia cells, the miR-29 family has been shown to target DNMT1 and 3, and restored expression of these miRNAs is associated with decreased tumorigenicity in vitro and in vivo.109,110 Interestingly, the DNMT3b3 splice variant of DNMT3b lacks the conserved site targeted by miR-148. This renders it resistant to repression by the miRNA, and means that downregulation of other DNMT3b variants results in increased relative abundance of DNMT3b3 due to miR-148.111 miR-132, which is dysregulated in schizophrenia and is involved in antiviral innate immunity and circadian timing, has been to shown regulate gene expression by directly inhibiting DNMT3A, the transcription factor GATA2, EP300 (a transcriptional co-activator), as well as several members of histone modifying complexes in vitro and in vivo.112-114 Endo-siRNAs may also be involved in de novo DNA methylation, as seen with the A. thaliana gene FWA. In this case the process appears to occur in two steps, including initial recruitment of siRNAs to pre-methylated FWA and then either cis- or trans-directed DNA methylation of transgenic copies introduced by transformation with Agrobacterium.115,116 Thus, we see that there are many interactions between short ncRNAs and traditional epigenetic pathways, but modulation of these transcripts themselves can also have important consequences.
Short ncRNAs as Epigenetic Intermediaries
As mentioned in the above examples, in addition to influencing the expression and function of epigenetic enzymes, many ncRNAs are targets of these same processes. Unlike other epigenetically regulated genes, however, this often has the effect of propagating further expression changes downstream. This concept is well illustrated by both normal biology and perturbations occurring in disease. For example, hypermethylation and decreased expression of one group of miRNAs was linked to human breast cancer117,118 and that of another group to the metastatic process.119 Also, several miRNAs (including miR-128a) are more highly expressed in adult compared with pediatric cerebellum, and are downregulated in medulloblastoma, a cerebellar tumor. Normally associated with neuronal differentiation, miR-128a appears to inhibit Bmi-1, a member of the epigenetic polycomb repressor (PRC1) complex, promoting buildup of reactive oxygen species and arresting proliferation of medulloblastoma cells.120 Thus, the normal developmental program involves dynamic expression of this ncRNA, while reversion to an early developmental state is associated with cancer. Loss of posttranscriptional repression of the single miRNA target results in increased action by the PRC1 complex, likely causing widespread changes in chromatin state.
miRNAs are not the only short RNAs that may be involved in disease pathogenesis through epigenetic pathways. Endo-siRNA function was originally described in oocytes, but there is now evidence that these RNAs may also cause environmentally sensitive gene expression changes in somatic tissues. In Caenorhabditis elegans, orthologs of the acute lymphoblastic leukemia (ALL-1)-fused gene (AF10) and Retinoblastoma (lin-35) appear to control gene expression through endo-siRNAs.121 A study of A. thaliana revealed that a bacterial pathogen found in its environment, Pseudomonas syringae, upregulates the endogenous cis-nat siRNA nat-siRNAATGB2, resulting in suppression of PPRL expression.122 Human HCC tissue was shown to have significantly downregulated expression of a set of endo-siRNA precursors and upregulation of their cognate genes and others. Normally, the ψPPM1K-derived endo-siRNA decreases HCC cell proliferation by directly suppressing NEK8.123 As HCC (among other cancers) is commonly associated to diet and other environmental stimuli, future studies would do well to establish a direct link between these factors and ncRNA tumorigenesis.
Another study of HCC cell lines and patient tissue uncovered a novel piRNA, piR-Hep1, that is overexpressed in tumors and may increase cell viability and migration.124 However, the majority of literature on piRNAs describes their involvement in spermatogenesis, and disruptions in piRNA processing have been reported in studies of male infertility. A genome-wide analysis of DNA methylation in testicular biopsies of infertile males revealed promoter hypermethylation and transcriptional silencing of the piRNA biogenesis genes PIWIL1/2 and TDRD1/9, resulting in decreased expression of selected piRNAs and their target genes.125 A similar observation was made in a study of Drosophila brain tumors: microarray profiling and deep sequencing showed dysregulated expression of four piRNA-biogenesis genes and 19 piRNAs, respectively.126
Concluding remarks
In the preceding sections, we considered the diverse mechanisms through which long and short ncRNAs exert changes in gene expression. Many of these transcripts are necessary for proper targeting of histone modifying complexes or play a role in DNA methylation. Perhaps a contemporary definition of epigenetics ought to also include the gene silencing or upregulation caused by non-coding RNAs, which occurs in normal biology and is subject to the influence of epigenetic complexes. This broader interpretation could then incorporate disease-causing environmental influences that are passed on across cell divisions, as seen in cancer. It would also cover critical discrepancies in gene expression, observed between cells with identical DNA sequence, during differentiation or among developing tissues. Importantly, when ncRNAs are themselves targeted for repression or overexpression, this can be an effective way of amplifying changes in the levels of downstream effectors.
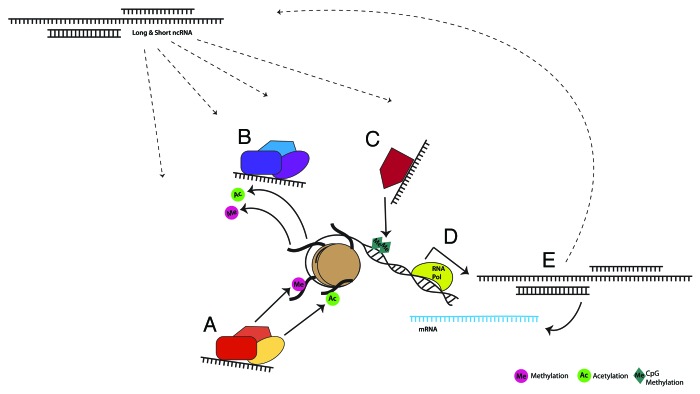
Future studies clarifying the link between environmental stimuli and changes in ncRNA function are key to a more complete understanding of the role of these transcripts. This information would contribute to the modified version of Francis Crick’s “central dogma,” in which RNA is not only a stop on the journey between DNA and protein, but provides important regulatory feedback influencing both transcription and translation. Therefore, in addition to the role of ncRNA in classical epigenetics, we propose that transcripts targeted by these mechanisms serve as epigenetic intermediaries, transmitting the message throughout a vast network of dynamic regulation of gene expression (Fig. 1).
Figure 1. Environmentally sensitive, dynamic regulation of gene expression by long and short ncRNAs occurs through direct and indirect interaction with classical epigenetic mechanisms, forming a larger epigenetic network. ncRNAs are known to bind and recruit histone modifying complexes to either add (A) or remove (B) methyl and acetyl groups. These transcripts also modulate DNA methyltransferases (C), thereby facilitating or suppressing DNA methylation. Loci that are targeted by each of these mechanisms can encode protein coding and/or noncoding RNA transcripts (D). ncRNAs in turn can affect gene expression by interacting with mRNA, or through feedback into the previously discussed pathways (E).
Disclosure of Potential Conflicts of Interest
CW is a consultant for Opko Health (formerly CuRNA) and a co-founder of Epigenetix Inc..
Acknowledgments
We would like to thank Andrea Johnstone, PhD and Mohammad Faghihi, MD, PhD for their insightful comments and for critically reading the manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/27473
References
- 1.Liu G, Mattick JS, Taft RJ. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle. 2013;12:2061–72. doi: 10.4161/cc.25134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louro R, El-Jundi T, Nakaya HI, Reis EM, Verjovski-Almeida S. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics. 2008;92:18–25. doi: 10.1016/j.ygeno.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magistri M, Faghihi MA, St Laurent G, 3rd, Wahlestedt C. Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts. Trends Genet. 2012;28:389–96. doi: 10.1016/j.tig.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–93. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X, Duan X, Qian J, Li F. Abundant conserved microRNA target sites in the 5′-untranslated region and coding sequence. Genetica. 2009;137:159–64. doi: 10.1007/s10709-009-9378-7. [DOI] [PubMed] [Google Scholar]
- 16.Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–6. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Röther S, Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93:1905–15. doi: 10.1016/j.biochi.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 19.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–34. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 20.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–41. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sabin LR, Delás MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Mol Cell. 2013;49:783–94. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. RIKEN Genome Exploration Research Group. Genome Science Group (Genome Network Project Core Group) FANTOM Consortium Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 24.Ptashne M. Epigenetics: core misconcept. Proc Natl Acad Sci U S A. 2013;110:7101–3. doi: 10.1073/pnas.1305399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–61. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 26.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 27.Silva AJ, Ward K, White R. Mosaic methylation in clonal tissue. Dev Biol. 1993;156:391–8. doi: 10.1006/dbio.1993.1086. [DOI] [PubMed] [Google Scholar]
- 28.Laird CD, Pleasant ND, Clark AD, Sneeden JL, Hassan KM, Manley NC, Vary JC, Jr., Morgan T, Hansen RS, Stöger R. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc Natl Acad Sci U S A. 2004;101:204–9. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Thomas PM, Kelleher NL. Measurement of acetylation turnover at distinct lysines in human histones identifies long-lived acetylation sites. Nat Commun. 2013;4:2203. doi: 10.1038/ncomms3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu H, Maunakea AK, Martin MM, Huang L, Zhang Y, Ryan M, Kim R, Lin CM, Zhao K, Aladjem MI. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet. 2013;9:e1003542. doi: 10.1371/journal.pgen.1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8. doi: 10.1083/jcb.146.5.905. [DOI] [PubMed] [Google Scholar]
- 33.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Gen. 2011;13:97–109. doi: 10.1083/jcb.146.5.905. [DOI] [PubMed] [Google Scholar]
- 34.Silva AJ, White R. Inheritance of allelic blueprints for methylation patterns. Cell. 1988;54:145–52. doi: 10.1016/0092-8674(88)90546-6. [DOI] [PubMed] [Google Scholar]
- 35.Blomen VA, Boonstra J. Stable transmission of reversible modifications: maintenance of epigenetic information through the cell cycle. Cell Mol Life Sci. 2011;68:27–44. doi: 10.1007/s00018-010-0505-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakatani Y, Tagami H, Shestakova E. How is epigenetic information on chromatin inherited after DNA replication? Ernst Schering Res Found Workshop. 2006;57::89–96. doi: 10.1007/3-540-37633-x_5. [DOI] [PubMed] [Google Scholar]
- 37.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- 38.Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M, Oakes C, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9:e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Treppendahl MB, Qiu X, Søgaard A, Yang X, Nandrup-Bus C, Hother C, Andersen MK, Kjeldsen L, Möllgård L, Hellström-Lindberg E, et al. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood. 2012;119:206–16. doi: 10.1182/blood-2011-06-362541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C, Teng ZQ, McQuate AL, Jobe EM, Christ CC, von Hoyningen-Huene SJ, Reyes MD, Polich ED, Xing Y, Li Y, et al. An epigenetic feedback regulatory loop involving microRNA-195 and MBD1 governs neural stem cell differentiation. PLoS One. 2013;8:e51436. doi: 10.1371/journal.pone.0051436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dallosso AR, Hancock AL, Malik S, Salpekar A, King-Underwood L, Pritchard-Jones K, Peters J, Moorwood K, Ward A, Malik KT, et al. Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. RNA. 2007;13:2287–99. doi: 10.1261/rna.562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X, He D, Dong XD, Dong F, Wang J, Wang L, Tang J, Hu DN, Yan D, Tu L. MicroRNA-124a is epigenetically regulated and acts as a tumor suppressor by controlling multiple targets in uveal melanoma. Invest Ophthalmol Vis Sci. 2013;54:2248–56. doi: 10.1167/iovs.12-10977. [DOI] [PubMed] [Google Scholar]
- 43.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Mage J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–5. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 45.Katz A, Oliva M, Mosquna A, Hakim O, Ohad N. FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 2004;37:707–19. doi: 10.1111/j.1365-313X.2003.01996.x. [DOI] [PubMed] [Google Scholar]
- 46.Doyle MR, Amasino RM. A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 2009;151:1688–97. doi: 10.1104/pp.109.145581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denisenko O, Shnyreva M, Suzuki H, Bomsztyk K. Point mutations in the WD40 domain of Eed block its interaction with Ezh2. Mol Cell Biol. 1998;18:5634–42. doi: 10.1128/mcb.18.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 51.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–9. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 53.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- 56.Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–42. doi: 10.1016/0092-8674(92)90520-M. [DOI] [PubMed] [Google Scholar]
- 57.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 58.Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/S0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 59.Mak W, Baxter J, Silva J, Newall AE, Otte AP, Brockdorff N. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive x chromosome in trophoblast stem cells. Curr Biol. 2002;12:1016–20. doi: 10.1016/S0960-9822(02)00892-8. [DOI] [PubMed] [Google Scholar]
- 60.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 61.Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–95. doi: 10.1016/S1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- 62.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol. 2004;167:1025–35. doi: 10.1083/jcb.200409026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–22. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet. 1999;8:1209–17. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 67.Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci U S A. 1999;96:8064–9. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engemann S, Strödicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J. Sequence and functional comparison in the Beckwith-Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum Mol Genet. 2000;9:2691–706. doi: 10.1093/hmg/9.18.2691. [DOI] [PubMed] [Google Scholar]
- 69.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 70.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–9. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 71.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–9. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Onoguchi M, Hirabayashi Y, Koseki H, Gotoh Y. A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc Natl Acad Sci U S A. 2012;109:16939–44. doi: 10.1073/pnas.1202956109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He W, Cai Q, Sun F, Zhong G, Wang P, Liu H, Luo J, Yu H, Huang J, Lin T. linc-UBC1 physically associates with polycomb repressive complex 2 (PRC2) and acts as a negative prognostic factor for lymph node metastasis and survival in bladder cancer. Biochim Biophys Acta. 2013;1832:1528–37. doi: 10.1016/j.bbadis.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 77.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 78.Zemel S, Bartolomei MS, Tilghman SM. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat Genet. 1992;2:61–5. doi: 10.1038/ng0992-61. [DOI] [PubMed] [Google Scholar]
- 79.Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet. 1994;7:433–9. doi: 10.1038/ng0794-433. [DOI] [PubMed] [Google Scholar]
- 80.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–21. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 81.Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell. 2012;45:255–62. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–7. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–14. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–9. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 86.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–43. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Q, Su Z, Xu X, Liu G, Song X, Wang R, Sui X, Liu T, Chang X, Huang D. AS1DHRS4, a head-to-head natural antisense transcript, silences the DHRS4 gene cluster in cis and trans. Proc Natl Acad Sci U S A. 2012;109:14110–5. doi: 10.1073/pnas.1116597109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–71. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 89.Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 90.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 91.Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J. The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology, and the metabolome of fibroblasts. J Biol Chem. 2012;287:30984–92. doi: 10.1074/jbc.M112.341040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C. The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell. 2007;28:398–407. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 93.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–7. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 94.Xu M, Soloveychik M, Ranger M, Schertzberg M, Shah Z, Raisner R, Venkatasubrahmanyan S, Tsui K, Gebbia M, Hughes T, et al. Timing of transcriptional quiescence during gametogenesis is controlled by global histone H3K4 demethylation. Dev Cell. 2012;23:1059–71. doi: 10.1016/j.devcel.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Möller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–12. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 97.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–9. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 99.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009;69:2287–95. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 100.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–95. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 101.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 102.Liu S, Tetzlaff MT, Cui R, Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am J Pathol. 2012;181:1823–35. doi: 10.1016/j.ajpath.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kloc A, Martienssen R. RNAi, heterochromatin and the cell cycle. Trends Genet. 2008;24:511–7. doi: 10.1016/j.tig.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 105.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–7. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 106.Fagegaltier D, Bougé AL, Berry B, Poisot E, Sismeiro O, Coppée JY, Théodore L, Voinnet O, Antoniewski C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci U S A. 2009;106:21258–63. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–24. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 108.Buurman R, Gürlevik E, Schäffer V, Eilers M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kühnel F, Skawran B. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012;143:811–20, e1-15. doi: 10.1053/j.gastro.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 109.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–8. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Duursma AM, Kedde M, Schrier M, le Sage C, Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–7. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–30. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lagos D, Pollara G, Henderson S, Gratrix F, Fabani M, Milne RS, Gotch F, Boshoff C. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12:513–9. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 114.Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HY. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20:731–51. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- 116.Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS Biol. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lehmann U, Hasemeier B, Römermann D, Müller M, Länger F, Kreipe H. [Epigenetic inactivation of microRNA genes in mammary carcinoma] Verh Dtsch Ges Pathol. 2007;91:214–20. [PubMed] [Google Scholar]
- 118.Hsu PY, Deatherage DE, Rodriguez BA, Liyanarachchi S, Weng YI, Zuo T, Liu J, Cheng AS, Huang TH. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009;69:5936–45. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5:e10748. doi: 10.1371/journal.pone.0010748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grishok A, Hoersch S, Sharp PA. RNA interference and retinoblastoma-related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:20386–91. doi: 10.1073/pnas.0810589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr., Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A. 2006;103:18002–7. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chan WL, Yuo CY, Yang WK, Hung SY, Chang YS, Chiu CC, Yeh KT, Huang HD, Chang JG. Transcribed pseudogene ψPPM1K generates endogenous siRNA to suppress oncogenic cell growth in hepatocellular carcinoma. Nucleic Acids Res. 2013;41:3734–47. doi: 10.1093/nar/gkt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–73. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 125.Heyn H, Ferreira HJ, Bassas L, Bonache S, Sayols S, Sandoval J, Esteller M, Larriba S. Epigenetic disruption of the PIWI pathway in human spermatogenic disorders. PLoS One. 2012;7:e47892. doi: 10.1371/journal.pone.0047892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–7. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]