Abstract
Chromatin states, quite different from changes in DNA sequence, can impact fundamental cellular processes such as determination of cell identity and development of disease. However, how chromatin states are established and regulated remain to be fully elucidated. In several lower eukaryotes, the small RNA machinery comprised of small RNA and its partners, the Argonaute proteins, is known to play important roles in the establishment of heterochromatin and silencing of repetitive sequences. In mammalian cells, however, the nuclear function of the small RNA machinery is largely unknown. Emerging evidence suggests that components of the small RNA pathway interact with chromatin to regulate nuclear events, including gene transcription and alternative splicing. In addition, these endogenous mechanisms are being exploited to target specific genomic loci for manipulation of gene expression and splicing events. In this review, I summarize current understanding of chromatin remodeling by small RNAs in mammalian cells and highlight recent efforts to map genome-wide interactions between RNAi-related factors and chromatin.
Keywords: small RNA, Argonaute, epigenetics, chromatin, RNA activation, miRNA, transcription
Introduction
The mammalian genome is pervasively transcribed; however, only a tiny portion of the transcription output codes for proteins—the transcripts that do not encode proteins are known as non-coding RNAs (ncRNAs), which are loosely classified based on size into long and short ncRNAs. Long ncRNAs (lncRNAs) are 200 nucleotides (nt) to a few thousand nt long and are estimated to be present in >8000 loci in the human genome.1 A string of recent studies has functionally characterized a number of lncRNAs and unveiled their important roles in regulating transcription and epigenetics of protein-coding genes.1-3 LncRNA-mediated epigenetic regulation depends mainly on lncRNA interactions with proteins or genomic DNA via RNA secondary structures, although some lncRNAs rely on Watson-Crick base pairing for functional activity.4
On the other hand, short ncRNAs are more prevalent in number, several classes of which have been functionally well defined, including short interfering RNAs (siRNAs), microRNAs (miRNAs), and piwi-interacting RNAs (piRNAs). Whereas piRNAs are known to function in the nucleus to maintain genomic integrity in the germline by means of epigenetic mechanisms, siRNAs and miRNAs mainly mediate posttranscriptional gene silencing through degradation of mRNA and/or inhibition of mRNA translation in the cytoplasm,5 and thereby regulate diverse biological processes such as cell growth, the cell cycle, apoptosis, and differentiation.6-8 The functions of small RNAs rely on a highly evolutionarily conserved family of RNA-binding proteins known as Argonaute (Ago).9 Human cells have a total of eight Argonaute proteins that are divided into two subfamilies: the PIWI and the EIF2C/Ago subfamilies.10 The latter subfamily has four members, Ago1 through Ago4, of which, Ago1 and Ago2 are the central protein components of the small RNA-induced silencing complex (RISC).11
Epigenetic modulation of gene expression by ncRNAs has long been recognized in animals and is often required for achieving equal expression of X-linked genes between the two sexes and for parental origin-specific monoallelic gene expression (reviewed in ref. 12). Acting through various mechanisms, including DNA methylation and histone modification, the end effect of such modulation is often transcriptional gene silencing (TGS), with one known exception, in which roX (RNA on the X chromosome) RNAs direct epigenetic activation of X-linked genes in male Drosophila.13 In plants, short double-stranded RNAs (dsRNAs) processed from long precursor RNAs are known to be able to induce de novo methylation of homologous DNA sequences, a phenomenon known as RNA-directed DNA methylation (RdDM).14 Similar epigenetic silencing mechanisms exist in other eukaryotes, including Schizosaccharomyces pombe, Drosophila melanogaster, and Caenorhabditis elegans, and are referred to as RNA-induced transcriptional silencing (RITS).15-17 Both RdDM and RITS invariably require the small RNA machinery consisting of short dsRNAs and Ago proteins.15-17 In mammals, however, the nuclear function of small RNAs and Ago proteins remains largely unknown. Despite recent examples in which small RNAs function in the nucleus to modulate gene expression, including silencing and activation at the epigenetic level and in an Ago-dependent manner,18-22 it is unclear how widespread such regulation is and what functional implications it has in normal cellular processes and diseases. In this review, I summarize current understanding of chromatin remodeling by small RNAs in mammalian cells and highlight recent high-throughput efforts in mapping genome-wide interactions between RNAi-related factors and chromatin.
The Small RNA Machinery in the Nucleus
Small RNAs are pervasively present in the nucleus and have been found to be an integral component of chromatin.23,24 Many studies have also demonstrated the nuclear localization of Ago proteins and small RNA species, such as miRNA, by biochemical, immunological and deep sequencing analyses.25-29 These observations raise the interesting possibility that the small RNA machinery may interact with chromatin to affect nuclear processes regulated by epigenetic programs in mammalian cells.
Initial evidence for a nuclear function of small RNAs came from studies that showed that short dsRNAs and miRNAs can target Ago proteins to specific chromosomal loci to regulate gene transcriptional activity either negatively22,30,31 or positively.19,26,30,32-35 Additional roles of nuclear small RNAs and Agos in the DNA-damage response and alternative splicing have recently been reported.36,37
Small RNA and DNA Methylation
DNA methylation is one of the most important epigenetic mechanisms and regulates diverse biological processes. Despite intensive studies on this topic, fundamental questions that remain to be answered are how sequence-specific DNA methylation is encoded and regulated. Obviously, ncRNAs are attractive candidates for initiating sequence-specific methylation given the established example (i.e., RdDM) in plants. There are, however, inconsistent reports regarding whether small RNAs can mediate DNA methylation in mammalian cells.18,38-40 In 2004, Morris et al. observed, by restriction enzyme-based methylation analysis, that targeting the promoter of elongation factor 1 α (EF1A) using synthetic dsRNAs could induce DNA methylation and TGS.18 Later, Suzuki et al. used an RNA duplex to target the conserved tandem NF-κB binding motifs in the LTR of HIV-1 virus and found that such targeting leads to suppression of productive HIV-1 infection, which is associated with de novo CpG methylation within the target region and reduced HIV-1 transcription.41 However, when the duplex RNA was introduced as an shRNA, it failed to induce DNA methylation, instead it caused an increase in H3K27me3.42 Very recently, Park et al. observed that an expressed shRNA targeting a segment of the hepatitis B virus (HBV) genome resulted in increased (31.3% vs. 12.8% in controls) methylation at CpG sites near the target, as assayed in virus-transfected HepAD38 hepatoma cells.43 By high-resolution bisulfite genomic sequencing mapping of CpG methylation, Ting et al.38 examined the E-cadherin promoter in cancer cells transfected with synthetic dsRNAs designed to target a region within a CpG island surrounding the E-cadherin promoter, and observed that the siRNA efficiently induced TGS, which was associated with increased H3K9me2 but not DNA methylation.38 Similarly, Corey targeted three TATA-less and one TATA-containing promoters using dsRNAs and observed TGS that was neither a result of increased promoter DNA methylation, as analyzed by methylation-specific PCR and bisulfite genomic sequencing, nor affected by inhibition of DNA methyltransferase (DNMT) activity using 5-aza-2'-deoxycytidine or DNMT siRNAs.39
Despite the conflicting reports, perhaps an endogenous mechanism similar to plant RdDM still exists in mammalian cells. An optimal strategy other than using 21-nt duplex RNA optimized for posttranscriptional RNAi has yet to be developed to exploit this mechanism. In this regard, piRNAs, the cousins of miRNAs, are known to induce de novo DNA methylation in mouse germ cells.44,45 For example, the de novo methylation of the imprinted Rasgrf1 locus in male mouse germ cells requires trans-acting piRNAs.45 In human colon cancer HCT116 cells hypomorphic for Dicer (with the helicase domain in exon 5 deleted), the expression of a number of genes could be reactivated to a level comparable to that in parental HCT116 cells by treatment with demethylating agents with concurrent loss of promoter hypermethylation. The loss of methylation was not due to changes in the expression or activities of DNMTs.46 This study suggests that Dicer-processed small RNAs could still play a role in the establishment or maintenance of DNA methylation at certain genomic loci. By deep sequencing of small RNAs, a recent study identified a new class of small RNAs originating from LINE-1 elements, which the authors termed endo-siRNAs. Overexpression of endo-siRNAs in cancer cells can silence LINE-1 expression through increased DNA methylation of the LINE-1 5′-UTR promoter in a manner dependent on Dicer and Ago2.47
Small RNAs and Histone Modifications
Various histone modifications have been shown to be associated with genomic loci targeted by artificially designed dsRNAs or by naturally occurring small RNAs that act in cis or in trans.
Exogenous small RNA-mediated regulation of gene transcription/epigenetics and mRNA splicing
Manipulating specific gene transcription or splicing at will has remained challenging. Inspired by the successful use of 21-nt siRNAs in mammalian cells to exploit the RNAi mechanism48 originally discovered in C. elegans,49 several laboratories have attempted to use the same duplex design strategy to target genomic loci, especially gene promoters, to manipulate gene transcription but with a variable degree of success. Although the detailed molecular mechanism remains elusive in these cases, epigenetic changes are evident at the target sites and often lead to an effect on gene expression that lasts longer than those observed for cytoplasmic RNAi.21,46,50-54
Small RNA-induced TGS
In the aforementioned study by Ting et al., targeting the CpG island region of the E-cadherin promoter using dsRNAs was able to induce efficient TGS in human colon and breast cancer cell lines with increased H3K9me2 at the target promoter.38 This silencing effect lasted for at least 8 days after dsRNA transfection. 38 In another study, dsRNA-guided TGS of the CCR5 and RASSF1A genes involved the co-localization of Ago1, TRBP2, and EZH2 to the dsRNA targets on promoters, leading to an increase in H3K9me2.22 Similarly, targeting the promoter of the ubiquitin C (UbC) gene by dsRNAs induced TGS via increased levels of H3K9me2 and H3K27me2.54 In addition, the establishment of these repressive epigenetic marks requires Ago1 and histone deacetylase 1 (HDAC1).54 Furthermore, small RNA-induced TGS has been exploited to suppress replication of HIV-1 via heterochromatin formation by targeting the HIV-1 5′LTR region using dsRNAs or shRNAs.27,42,51,55,56
Small RNA-induced transcriptional gene activation (RNAa)
Surprisingly, targeting selected genomic regions including the promoter and 3′ terminus of genes can lead to an increase in gene expression at the transcriptional/epigenetic level,21,32,57 a phenomenon known as RNAa.32,50 In our initial work reporting RNAa,32 the original goal was to test whether dsRNAs could be used to induce DNA methylation. We initially designed two dsRNAs for the E-cadherin promoter by following the rational siRNA design rules which disfavor targets with a high GC content.58 As a result, the targets of the two dsRNAs fell outside the CpG island in the E-cadherin promoter, which is frequently hypermethylated in cancer cells leading to epigenetic silencing of the E-cadherin gene.59 When the dsRNAs were introduced into prostate cancer cells, they both induced, rather than silenced, the expression of E-cadherin.32 However, in HeLa cells, in which E-cadherin expression is normally silenced via promoter hypermethylation,32 dsRNAs alone were unable to reactivate E-cadherin expression unless the cells were co-treated with the demethylating agent 5-aza-2'-deoxycytidine, suggesting that epigenetic states may affect accessibility of promoters to RNAa.32 Although, in our study, we did not observe the expected change in DNA methylation at the target sites, loss of H3K9me2 and H3K9me3 was noted.32
In several other studies reporting the RNAa phenomenon, similar epigenetic modifications were found involving either the loss of suppressive epigenetic marks such as H3K9me2, H3K9me3, H3K9 acetylation (H3K9ac), and H3K4ac or the gain of active marks such as H3K4me2 and H3K4me3.21,32,60 Despite these observations, questions remain as to whether the epigenetic changes are the causes or consequences of increased gene expression upon promoter targeting. Further studies are needed to identify the protein partners responsible for chromatin remodeling and the epigenetic marks characteristic of RNAa.
Small RNA-mediated alternative splicing
Targeting pre-mRNA splicing sites using dsRNAs has been utilized to affect splicing events by means of chromatin remodeling. Allo et al. found that targeting intronic or exonic sequences close to an alternative exon using dsRNAs can affect the splicing of the exon. This process depends on Ago1 and Ago2, and is associated with an increase in H3K9me2 and H3K27me3.61 However, another similar study found that dsRNA-directed alternative splicing of the SMN2 and dystrophin genes mainly requires Ago2, and does not involve the two types of chromatin changes (H3K9me2 and H3K27me3),62 which is different from the report by Allo et al.61 An interesting observation common to both studies is that intronic dsRNAs do not elicit cleavage of the target mRNA as they were predicted via the RNAi pathway,61,62 which may suggest that the targeting process is coupled to transcription.
Endogenous small RNA-mediated epigenetic gene regulation
The observations that genomic loci can be targeted using artificially designed small RNAs imply that epigenetic regulation by naturally occurring small RNAs may also operate under physiological conditions. Given the similarities between artificially designed dsRNAs and naturally occurring miRNAs with regard to their size, structure and dependence on Ago proteins, miRNAs would be the best candidates for targeting Ago proteins and chromatin modifiers to specific genomic loci. In fact, computational methods predict that miRNAs have as many targets in gene promoters as in 3′UTR regions.22,50,63 However, these targets are generally poorly conserved due to weak conservation of promoter sequences per se across species and only a few have been experimentally validated.
miRNA-induced RNAa
The first example came from the study of miR-373 in human cells.33 By scanning the promoter of the E-cadherin gene for miRNA targets, Place et al. identified a highly complementary target for miR-373. Transfection of either a mature miR-373 mimic or its precursor induces E-cadherin expression at the transcriptional level. To identify genes that are potentially activated by miRNAs in a physiological context, Huang et al. found that expression of a group of cell-cycle-related genes depends on the synthesis of miRNA in both human and mouse cells. Among these genes is cyclin B1 (Ccnb1), a critical gene that promotes mitotic cell-cycle entry. The expression of Ccnb1 decreases when Drosha, Dicer or Ago1 is depleted by RNAi, and increases upon forced expression of Ago1, suggesting that Ccnb1 is a potential miRNA-induced gene.26,64 To determine which miRNA(s) stimulates Ccnb1 expression, bioinformatics analysis of the mouse Ccnb1 promoter identified highly complementary targets for several miRNAs, including miR-744 and miR-1186. Overexpression of these miRNAs by either transfecting their mimics or cloning their precursors into a lentiviral vector induces increased Ccnb1 transcription with a concurrent increase in RNAP II recruitment and H3K4me3 mark at the target promoter. On the other hand, knockdown of endogenous miR-744 causes the opposite change in Ccnb1 expression. This study provides the first example that endogenous miRNAs could function through epigenetic mechanisms to sustain gene expression.
This additional layer of gene regulation by miRNAs may have profound implications in diseases such as cancer. In fact, overexpression of miR-744 confers on cells a proliferative advantage via stimulating Ccnb1, leading to a shortened cell-doubling time. Long-term overexpression of miR-744 in mouse prostate cancer cells can further induce chromosomal instability and inhibit in vivo tumor formation.26
Very recently, another endogenous miRNA, miR-589, was shown to target the promoter of the COX-2 gene to induce its expression at the transcriptional level in human cancer cells.65 Blockage of endogenous miR-589 using an anti-miR causes downregulation of COX-2, whereas introducing miR-589 into cells has the opposite effect on COX-2 expression.65 Mechanistically, miR-589 uses sense promoter RNAs derived from the COX-2 promoter as the binding targets and the binding requires the small RNA pathway components Ago2 and GW182. A perfect complementary dsRNA targeting a nearby region in the COX-2 promoter can also activate COX-2 expression, which is associated with an increase in H3K4me3 and H4ac.65
miRNA-induced TGS
miRNAs have also been shown to epigenetically silence target genes. miR-320, transcribed from the promoter of POLR3D, transcriptionally silences the POLR3D gene in cis by directing the association of Ago1, EZH2, a polycomb group (PcG) component, and H3K27me3 with the POLR3D promoter.66 Epigenetic regulation by miRNAs in trans has also been reported.31,67,68 Younger et al.67 showed that several miRNAs with predicted targets in the promoter of the progesterone receptor (PR) gene can silence PR expression by inducing H3K9me and the recruitment of Ago2, but not Ago1, to target sites. In addition, certain miRNAs can regulate the same target genes both posttranscriptionally and epigenetically. For example, miR-223 is known to downregulate nuclear factor I-A (NFI-A) by pairing with its target in the 3′UTR of NF1-A, leading to granulopoiesis in hematopoietic stem cells.69 In a study by Zardo et al., miR-223 was found to also target PcG proteins to a target site on the NF1-A promoter to induce a silent chromatin state characterized by a decrease in H3K4me3 and an increase in H3K27me3 and the recruitment of PcG member YY1 and Suz12 during retinoic acid (RA)-induced granulopoiesis.31
The Epigenetic Landscape of Ago-Chromatin Interactions
The small RNA pathway is commonly regarded as a genome defense mechanism to suppress genome parasites by initiating heterochromatin formation and is thus supposed to operate mainly on repetitive sequences, as found in fission yeast.70,71 Surprisingly, genome-wide mapping of Drosophila Ago2 (dAgo2) binding sites on chromosomes in two recent studies revealed that dAgo2 preferentially binds to euchromatin, as evidenced by chromatin-immunoprecipitation (ChIP)-sequencing (ChIP-seq) and immunofluorescence staining of polytene chromosomes. dAgo2 binding occurs almost exclusively to actively transcribed promoters72,73 and depends on additional components of the small RNA machinery, including small RNAs and Dicer.73
In human cells, biochemical analyses provide convincing evidence that nuclear Ago1 and Ago2 are in a complex with chromatin29,37 and additional factors involved in chromatin modification and alternative splicing such as the H3K9me3-binding chromobox protein HP1γ, and the H3K9-specific lysine methyltransferase Suv39h1.37 These observations strongly suggest that the small RNA machinery may modulate broad chromatin domains in the genome.
By conducting a ChIP-on-chip analysis using a pan-Ago antibody, Benhamed et al. examined genome-wide Ago binding in promoter regions in senescent and pre-senescent fibroblasts. They found increased Ago binding to E2F-regulated promoters of proliferation-promoting genes whose expression is transcriptionally repressed by the RB1/E2F repressor complex in senescent cells.74 By deep-sequencing small RNAs co-immunoprecipitated with Ago proteins and H3K9me2, they further showed a strong positive correlation between Ago-bound and heterochromatin-bound miRNAs, as exemplified by members of the let-7 family in senescent cells. Subsequent study focusing on Ago2 further showed that Ago2 and miRNAs are recruited to the promoter of E2F-responsive genes (i.e., CDCA8 and CDC2), leading to increased H3K27me3 at the target promoters and TGS. However, it remains to be validated whether other Ago proteins (e.g., Ago1) are also involved in this process.
Epigenetic modulation induced by the binding of Ago proteins, including Ago1 and Ago2, has also been shown to play a role in alternative splicing. In a study by Ameyar-Zazoua et al.,37 Ago1 and Ago2 were found to be recruited throughout the CD44 gene but especially at alternative splicing sites to induce H3K9me3, which subsequently causes RNAP II slowdown and facilitates recruitment of spliceosomes, leading to preferential inclusion of CD44 variant exons in the mature mRNA. This mechanism seems to be small RNA-dependent and to act on many other genes since the small RNAs recovered from Ago-chromatin complexes could be mapped to the CD44 splicing sites, and gene expression profiling showed that knockout of Ago2 or Dicer affects more than 2000 common splicing events in mouse fibroblasts.37
In a recent effort to assess how pervasively the small RNA machinery interacts with chromatin, my laboratory mapped genome-wide chromosomal binding of nuclear Ago1 and Ago2 by ChIP-seq in human cancer cells. Consistent with the binding preference observed for dAgo2 in Drosophila,72,73 human Ago1, but not Ago2, preferentially binds to euchromatic regions with the binding centered on actively transcribed promoters as marked by H3K4me3, whereas Ago1 binding in repetitive sequences is significantly depleted.29 Remarkably, Ago1-bound genes are enriched for several major oncogenic signaling pathways that promote cell growth and survival and cell-cycle progression. Loss- and gain-of-function analyses revealed that Ago1-binding events within promoters have a largely positive effect on gene transcription. Although the exact mechanism underlying Ago1 binding of chromosomal loci remains to be determined, these new findings provide the first glimpse of Ago deposition on the genomic landscape and suggest an oncogenic role for Ago1, possibly by orchestrating oncogenic transcriptional/epigenetic programs that confer to cells an advantage for growth and survival. As such, the nuclear small RNA pathway may contribute to cancer initiation and progression.
Are the observed Ago1-chromosomal interactions in human cells mediated by small RNAs such as miRNAs? The answer could be “yes,” based on the following lines of evidence found in our work.29 First, computational prediction reveals significantly enriched targets for about 50 miRNAs in Ago1-bound sequences compared with control sequences. Among these miRNAs, one-third belongs to several onco-miR clusters, including the miR-17-92 and miR-106b-25 clusters and the miR-520/373 family. Second, Ago1 physically interacts with RNAP II, and disruption of mRNA biogenesis pathways affects the association. Third, the expression of Ago1-bound genes can only be induced by forced expression of wild-type Ago1 but not by mutant Ago1 lacking the PAZ domain, suggesting that the Ago–small RNA association is partly required.75 It is also possible that other small RNA species such as nascent promoter transcripts could serve as binding scaffolds for Ago proteins. In this regard, Ago proteins have been shown to be able to load single-stranded RNAs76-78 and the location of most Ago1 peaks in our study coincides with that of promoter transcripts known as tiRNAs (transcription-initiation RNAs) or PASRs (promoter-associated sRNAs). (reviewed in ref 79)
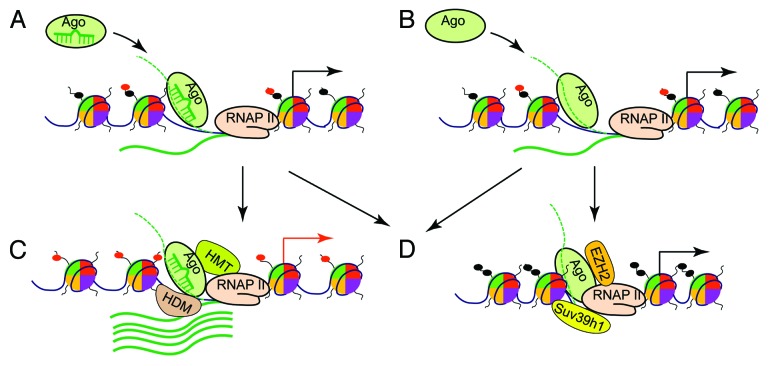
Together, two putative mechanistic models exist for the observed Ago–chromatin association: one is miRNA-guided in trans targeting of Ago proteins to chromosome sites throughout the genome, and the other is promoter RNA-mediated in cis recruitment of Ago proteins to promoter sites. In both targeting models, RNA-loaded Ago proteins would recruit additional chromatin-modifying factors to affect local chromatin structure, leading to changes in gene transcription or altered alternative splicing events (Fig. 1).
Figure 1. Models for chromatin remodeling by the small RNA machinery. Ago proteins, especially Ago1, are guided to chromatin loci at gene promoters by trans-acting miRNAs, which bind to complementary targets on nascent promoter RNAs (dotted green line) or promoter DNA (black line) (A). Alternatively, nascent promoter RNAs act in cis to recruit Ago proteins (B). In both models, the RNA-loaded Ago proteins further recruit chromatin-modifying proteins (CMPs) to remodel local chromatin structure. At certain promoters, as depicted in (C), histone methyltransferases (HMTs) or demethylases (HDMs) are recruited, leading to the removal of repressive histone modifications such as H3K9me2 and H3K9me3 or to the addition of active histone modifications such as H3K4me3 (red dots on histone tails). As a result, the affected promoters assume an open chromatin structure to allow for increased transcriptional activity. As illustrated in (D), RNA-loaded Agos can also recruit CMPs such as EZH2 and Suv39h1 to induce heterochromatin typified by the modifications of H3K9me and H3K27me (black dots on histone tails), leading to attenuated promoter activity.
Conflicting Views of the Roles of Different Ago Proteins in Nuclear Events
Among the four mammalian Ago proteins, Ago1 and Ago2 are the most thoroughly studied for their roles in RNAi and RNAi-related nuclear events. Ago2 differs from other Ago members in that it is the only Ago possessing slicer activity.80 As such, Ago1 and Ago2 are known to have a different preference in small RNA loading. It is generally believed that Ago1 preferentially loads miRNAs to mediate translational repression and mRNA destabilization whereas Ago2 prefers perfect complementary duplex RNAs leading to mRNA target cleavage.81,82 In some other studies, however, Ago1 and Ago2 were shown either to have completely overlapping functions in mediating RNAi regardless of the guide RNAs (siRNA or miRNA)9 or to have non-redundant functions, with Ago1 capable of associating with additional small RNA species other than siRNAs and miRNAs.78
When it comes to nuclear events involving Ago proteins, there are very conflicting views about which Ago plays the major role. For example, some studies found that only Ago1 is recruited to the target promoter by dsRNA to mediate TGS,22,54 whereas others found the involvement of Ago2, but not Ago1, in TGS.30,83 In dsRNA-guided alternative splicing, a similar discrepancy exists.61,62 High-throughput mapping studies also showed conflicting results with regard to the presence or absence of an Ago2–chromatin association.29,37,74
These conflicting observations could sometimes be attributed to differences in the model genes and cell types examined. Technical issues such as specificity of the antibodies used may also be a contributing factor. Nevertheless, there is evidence for intrinsic differences between nuclear Ago1 and Ago2. For example, two recent studies examining Ago subcellular localization showed that Ago1 and Ago2 have distinct nuclear compartmentalization, with Ago1 localizing in the nucleoplasm and Ago2 mainly in the inner nuclear envelope,27,29 indicative of their potential functional partitioning in the nucleus.
It is possible that the general loading preference toward dsRNA and miRNA also applies to nuclear Ago1 and Ago2, which may explain the scantier nuclear localization of Ago2 in contrast to the diffuse nuclear presence of Ago1.27,29 For dsRNA-triggered nuclear mechanisms, Ago2 may be the primary loading partner for its slicer activity, which is required for the initial processing of the duplex,50,84,85 Otherwise, Ago1 may be the major RNA-binding partner of naturally occurring small RNAs in the nucleus.
Concluding Remarks
We are just beginning to realize the unexpected roles of the small RNA machinery in regulating gene transcription and many other nuclear events by interacting with chromatin to affect local epigenetic states. Future studies should focus on elucidating the underlying mechanisms, especially the exact targeting mechanism, the small RNA species and the types of proteins involved. High-throughput approaches to systemically map the interactions between the small RNA machinery and chromatin, and the associated chromatin changes will help us appreciate the scope of small RNA-mediated epigenetic regulation and its implications in cell physiology and disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from the National Institutes of Health (1R01GM090293-0109) and Department of Defense (W81XWH-08-1-0260).
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26830
References
- 1.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–6. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 8.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–17. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank-Kamenetsky M, Grefhorst A, Anderson NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R, Fan Y, et al. Therapeutic RNAi targeting PCSK9 acutely lowers plasma cholesterol in rodents and LDL cholesterol in nonhuman primates. Proc Natl Acad Sci U S A. 2008;105:11915–20. doi: 10.1073/pnas.0805434105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–23. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Franke A, Baker BS. The rox1 and rox2 RNAs are essential components of the compensasome, which mediates dosage compensation in Drosophila. Mol Cell. 1999;4:117–22. doi: 10.1016/S1097-2765(00)80193-8. [DOI] [PubMed] [Google Scholar]
- 14.Wassenegger M, Heimes S, Riedel L, Sänger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–76. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 15.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 16.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–72. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 17.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–4. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 18.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 19.Li LC. The multifaceted small RNAs. RNA Biol. 2008;5:61–4. doi: 10.4161/rna.5.2.5989. [DOI] [PubMed] [Google Scholar]
- 20.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol. 2005;1:216–22. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 21.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–73. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 22.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–7. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 23.Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodríguez-Campos A, Azorín F. RNA is an integral component of chromatin that contributes to its structural organization. PLoS One. 2007;2:e1182. doi: 10.1371/journal.pone.0001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohrt T, Mütze J, Staroske W, Weinmann L, Höck J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–49. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahlenstiel CL, Lim HG, Cooper DA, Ishida T, Kelleher AD, Suzuki K. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res. 2012;40:1579–95. doi: 10.1093/nar/gkr891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC. Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells. PLoS Genet. 2013;9:e1003821. doi: 10.1371/journal.pgen.1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–48. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N, et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119:4034–46. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 32.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–42. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem Biol. 2010;17:1344–55. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren S, Kang MR, Wang J, Huang V, Place RF, Sun Y, Li LC. Targeted induction of endogenous NKX3-1 by small activating RNA inhibits prostate tumor growth. Prostate. 2013;73:1591–601. doi: 10.1002/pros.22709. [DOI] [PubMed] [Google Scholar]
- 36.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–5. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 38.Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–10. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corey DR. Regulating mammalian transcription with RNA. Trends Biochem Sci. 2005;30:655–8. doi: 10.1016/j.tibs.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Jiang G, Zheng L, Pu J, Mei H, Zhao J, Huang K, Zeng F, Tong Q. Small RNAs targeting transcription start site induce heparanase silencing through interference with transcription initiation in human cancer cells. PLoS One. 2012;7:e31379. doi: 10.1371/journal.pone.0031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]
- 42.Yamagishi M, Ishida T, Miyake A, Cooper DA, Kelleher AD, Suzuki K, Watanabe T. Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infect. 2009;11:500–8. doi: 10.1016/j.micinf.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Park HK, Min BY, Kim NY, Jang ES, Shin CM, Park YS, Hwang JH, Jeong SH, Kim N, Lee DH, et al. Short hairpin RNA induces methylation of hepatitis B virus covalently closed circular DNA in human hepatoma cells. Biochem Biophys Res Commun. 2013;436:152–5. doi: 10.1016/j.bbrc.2013.04.108. [DOI] [PubMed] [Google Scholar]
- 44.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–52. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ting AH, Suzuki H, Cope L, Schuebel KE, Lee BH, Toyota M, Imai K, Shinomura Y, Tokino T, Baylin SB. A requirement for DICER to maintain full promoter CpG island hypermethylation in human cancer cells. Cancer Res. 2008;68:2570–5. doi: 10.1158/0008-5472.CAN-07-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Dahlstrom JE, Lee SH, Rangasamy D. Naturally occurring endo-siRNA silences LINE-1 retrotransposons in human cells through DNA methylation. Epigenetics. 2012;7:758–71. doi: 10.4161/epi.20706. [DOI] [PubMed] [Google Scholar]
- 48.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 49.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 50.Portnoy V, Huang V, Place RF, Li LC. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA. 2011;2:748–60. doi: 10.1002/wrna.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, Rao S, Kelleher AD. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008;283:23353–63. doi: 10.1074/jbc.M709651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Place RF, Noonan EJ, Földes-Papp Z, Li LC. Defining features and exploring chemical modifications to manipulate RNAa activity. Curr Pharm Biotechnol. 2010;11:518–26. doi: 10.2174/138920110791591463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Wang J, Huang V, Place RF, Li LC. Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem J. 2012;443:821–8. doi: 10.1042/BJ20111491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–95. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Hum Gene Ther. 2012;23:473–83. doi: 10.1089/hum.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh A, Palanichamy JK, Ramalingam P, Kassab MA, Bhagat M, Andrabi R, Luthra K, Sinha S, Chattopadhyay P. Long-term suppression of HIV-1C virus production in human peripheral blood mononuclear cells by LTR heterochromatization with a short double-stranded RNA. J Antimicrob Chemother. 2013 doi: 10.1093/jac/dkt348. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 57.Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat Chem Biol. 2010;6:621–9. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–30. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 59.Li LC, Zhao H, Nakajima K, Oh BR, Ribeiro Filho LA, Carroll P, Dahiya R. Methylation of the E-cadherin gene promoter correlates with progression of prostate cancer. J Urol. 2001;166:705–9. doi: 10.1016/S0022-5347(05)66047-8. [DOI] [PubMed] [Google Scholar]
- 60.Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, Glass CK, Väisänen S, Ylä-Herttuala S. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ Res. 2009;105:604–9. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- 61.Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–24. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 62.Liu J, Hu J, Corey DR. Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucleic Acids Res. 2012;40:1240–50. doi: 10.1093/nar/gkr780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Younger ST, Pertsemlidis A, Corey DR. Predicting potential miRNA target sites within gene promoters. Bioorg Med Chem Lett. 2009;19:3791–4. doi: 10.1016/j.bmcl.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang V, Li LC. miRNA goes nuclear. RNA Biol. 2012;9:269–73. doi: 10.4161/rna.19354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt777. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim DH, Saetrom P, Snøve O, Jr., Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16230–5. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39:5682–91. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol Biol. 2009;10:12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–31. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 70.Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37:809–19. doi: 10.1038/ng1602. [DOI] [PubMed] [Google Scholar]
- 71.Zofall M, Grewal SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–96. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]
- 72.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–5. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol. 2012;14:266–75. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu S, Jin L, Huang Y, Zhang F, Kay MA. Slicing-independent RISC activation requires the argonaute PAZ domain. Curr Biol. 2012;22:1536–42. doi: 10.1016/j.cub.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okamura K, Ladewig E, Zhou L, Lai EC. Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev. 2013;27:778–92. doi: 10.1101/gad.211698.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–74. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 78.Polikepahad S, Corry DB. Profiling of T helper cell-derived small RNAs reveals unique antisense transcripts and differential association of miRNAs with argonaute proteins 1 and 2. Nucleic Acids Res. 2013;41:1164–77. doi: 10.1093/nar/gks1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–44. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 80.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 81.Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009;16:1259–66. doi: 10.1038/nsmb.1712. [DOI] [PubMed] [Google Scholar]
- 82.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–19. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 85.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–20. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]