Abstract
A significant fraction of eukaryotic genomes comprises repetitive sequences, including rRNA genes, centromeres, telomeres, and retrotransposons. Repetitive elements are hotspots for recombination and represent a serious challenge for genome integrity. Maintaining these repeated elements in a compact heterochromatic structure suppresses recombination and unwanted mutagenic transposition, and is therefore indispensable for genomic stability. Paradoxically, repetitive elements are not transcriptionally inert, but produce RNA that has important functions in regulating and reinforcing the heterochromatic state. Here, we review the role of non-coding RNA (ncRNA) in recruiting chromatin-modifying enzymes to repetitive genomic loci to establish a repressive chromatin structure that safeguards chromosome integrity and genome stability.
Keywords: non-coding RNA, heterochromatin, repetitive elements, rDNA, centromeres, telomeres, retrotransposons
Introduction
A large portion of the genome in metazoans and plants is transcribed into RNA, the vast majority of it being non-coding RNA (ncRNA) that functions as structural, catalytic, or regulatory RNAs, rather than encoding proteins. Non-coding RNA has been thought of as transcriptional noise or as relics of a primordial “RNA world” that has largely been replaced by more efficient proteins. Now, though, it seems that ncRNAs are numerous and highly adapted in their functions in higher organisms, mediating processes that require highly specific nucleic acid recognition without complex catalysis, such as regulation of gene expression, guiding RNA modifications or promoting posttranscriptional events. Their mechanisms of action and biological roles are extremely diverse, indicating that, so far, we have only had a glimpse of this new class of regulatory factors. Among the most fascinating discoveries emerging from this exciting field of research is that long ncRNAs (lncRNAs) impact on chromatin structure and epigenetic control of transcription. A classical example is the XIST RNA, a 17–20 kb ncRNA with a key role in dosage compensation and X-chromosome inactivation in human and mouse.1 The list of mammalian lncRNAs is constantly growing and functional analyses have advanced our understanding of their roles in regulation of gene expression.2 For example, several lncRNAs, including Airn,3 Kcnq1ot1,4 and Nespas5 silence imprinted gene clusters. HOTAIR lncRNA derives from the HOXC locus and binds to the Polycomb-repressive complex 2 (PRC2) to repress the HoxD cluster in trans,6,7 linc-HOXA1 represses Hoxa1 by recruiting the transcriptional cofactor PURB8 and Braveheart (Bvht) ncRNA interacts with Suz12 to dictate cell fate decisions.9 Other lncRNAs, such as Evf-2, SRA, or HSR1, modulate the activity of protein-binding partners.10-12 Many of these ncRNAs have been found to cluster near the 5′- or 3′- terminal parts of genes or represent antisense RNAs that overlap coding regions. Base complementarity allows RNA to be exquisitely sequence-specific, and therefore RNA is particularly well suited for the job of specific recognition of other RNAs and distinct genomic loci. Though the function of most of the newly identified ncRNAs is yet to be elucidated, it has become apparent that ncRNA has an impact on most, if not all, chromatin-mediated processes, hence the increased interest in this research area.
Eukaryotic genomes contain a considerable fraction of constitutive heterochromatin, commonly found in blocks of repetitive DNA sequences. Repetitive elements, including rRNA genes, centromeres, telomeres, as well as short and long interspersed transposable elements, account for 30–50% of mammalian genomes. As repetitive sequences present a serious challenge for genomic stability, multicellular organisms have developed mechanisms that suppress homologous recombination and protect the structural integrity of these repeats by maintaining them in a compact, transcriptionally refractory state. For this, distinct proteins need to be guided to such sequences to establish a heterochromatic structure. Genome-wide sequencing approaches have revealed an increasing set of ncRNAs that are linked to transcriptional silencing and chromosomal integrity, indicating that epigenetic regulation represents an intimate and balanced interplay of both fields, RNA and chromatin research (for review see refs. 13–16). Transcripts originating from silent rRNA genes, telomeres, centromeres or retroelements have been implicated in heterochromatin formation and cellular homeostasis, indicating that transcription in heterochromatin is a conserved trait in the evolution of eukaryotes.17-19 Although it is not yet known how cells solve the apparent paradox of “noisy silence,” there is increasing evidence that the local plasticity of heterochromatin,20 the action of anti-silencing factors21 and the accessibility of heterochromatin to the transcription machinery during S-phase22 contribute to the expression of heterochromatic regions. This review summarizes our current knowledge of how chromatin modifying enzymes, in particular the chromatin remodeling complex NoRC, act in concert with RNA-mediated mechanisms to establish a repressive chromatin structure at repetitive elements, a mechanism that is essential for chromosome structure and genome integrity.
Non-Coding RNAs Regulate the Epigenetic State of rRNA Genes
Mammalian genomes contain several hundred copies of rRNA genes (rDNA) that are tandemly arrayed in clusters, known as nucleolus organizer regions (NORs). The overall rDNA transcriptional activity of a given cell depends on the demand for protein synthesis, and hence on metabolic activity. However, even in cycling cells that exhibit high rRNA synthetic activity, only a fraction of rDNA copies is transcribed, whereas other repeats are silenced in a cell and/or tissue-specific manner. In mammals, several epigenetic features distinguish transcriptionally active from silent rRNA genes. Generally, an “open” chromatin structure that is characterized by DNA hypomethylation, acetylation of histone H4 (H4ac) and dimethylation of histone H3 at lysine 4 (H3K4me2) marks transcriptionally active gene promoters, whereas silent rDNA repeats are characterized by promoter hypermethylation, histone H4 hypoacetylation, and methylation of H3K9, H3K27, and H4K20.23 Recently, the existence of a third, intermediate chromatin configuration has been identified that marks rRNA genes that exhibit an open chromatin structure, but are not transcribed.24 Like at active rRNA genes, the promoter of such transcriptional permissive, “poised” genes is unmethylated, is associated with components of the preinitiation complex, but the promoter-bound nucleosome carries both euchromatic (H3K4me3) and heterochromatic (H3K27me3) histone modifications. Active and silent rDNA repeats are not only characterized by specific epigenetic marks but also by distinct nucleosome positions. At active genes, the promoter-bound nucleosome covers nucleotides (nt) from ‒157 to ‒2, and this specific nucleosomal architecture places the core promoter and the upstream control element into close proximity, allowing cooperative binding of transcription initiation factors. At silent rRNA genes, the nucleosome is positioned 24 nt further downstream, placing the nucleosome into a translational position that is unfavorable for transcription complex assembly.25 Taken together, three structurally and functionally distinct chromatin states of rRNA genes exist, i.e., active, silent, and poised ones.
The silent state of rRNA genes is established by NoRC (nucleolar remodeling complex), an ATP-dependent chromatin remodeling machine consisting of the ATPase SNF2h and the nucleolar protein TIP5.26 NoRC interacts with DNA methyltransferases, histone deacetylases and histone methyltransferases, thereby triggering promoter methylation, heterochromatin formation and transcriptional silencing.23,27,28 Thus, NoRC coordinates the activities of macromolecular complexes that modify histones, methylate DNA and establish a “closed” heterochromatic state. Recent studies have demonstrated that poly(ADP-ribose)-polymerase-1 (PARP-1) plays a role in the inheritance of the silent state of rDNA. PARP-1 binds to silent rRNA genes in mid‒late S-phase, associates with TIP5 and PARylates nucleolar histones.29 Thus, PARP-1 may act as a co-repressor, cooperating with NoRC to facilitate the inheritance of heterochromatic histone marks through cell cycle progression.
Studies in several organisms have shown that the intergenic spacer contains duplicated sequences that resemble the core element of the major gene promoter and direct RNA Polymerase I (RNAP I) transcription in vitro and in vivo.30-32 In mouse, transcripts from a RNAP I promoter located ~2 kilobases upstream of the pre-rRNA transcription start site maintains the balance between transcriptionally active and silent rRNA genes. These spacer transcripts are processed into a heterogeneous population of 150–250 nt RNAs, dubbed pRNA (promoter-associated RNA) as their sequence overlaps the rDNA promoter.28 pRNA can interact with TIP5, the large subunit of NoRC, through a phylogenetically conserved hairpin structure. This interaction between pRNA and TIP5 is required to recruit NoRC to nucleoli. RNase footprinting and protease sensitivity experiments revealed that TIP5 binds to pRNA in an induced fit mechanism, leading to structural changes of TIP5 that might facilitate the interaction with co-repressors, thus promoting heterochromatin formation and rDNA silencing (Fig. 1).33 Notably, depletion of pRNA caused translocation of NoRC from nucleoli to the nucleoplasm, whereas refeeding with ectopic pRNA restored nucleolar localization, underscoring the essential role of pRNA in guiding NoRC to rDNA and establishing the repressive chromatin structure. A recent study has shown that pRNA is synthesized from a sub-fraction of hypomethylated rRNA genes during mid S-phase, acting in trans to inherit DNA methylation and transcriptional repression of late-replicating silent rDNA copies.34 Although it remains obscure how pRNA discriminates between silent and active rDNA repeats, these results indicate that individual rDNA repeats are functionally different and this variability has distinct functional consequences.
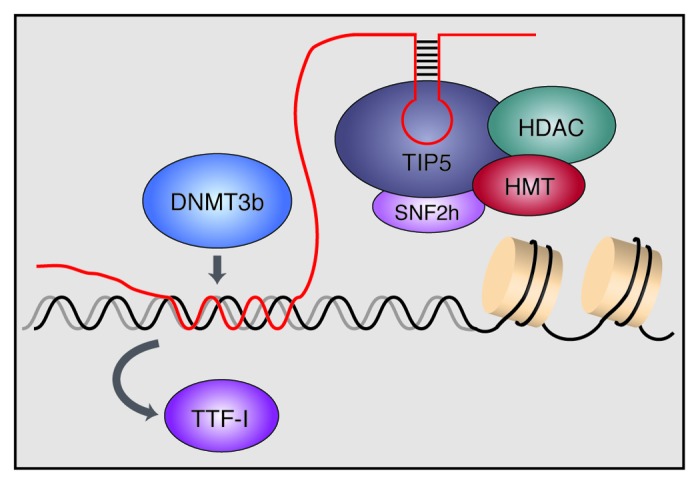
Figure 1. Model illustrating the role of pRNA in recruiting chromatin modifying enzymes to rDNA. Transcripts that match the rDNA promoter, dubbed pRNA (promoter-associated RNA), form a specific secondary structure that is recognized by TIP5, the large subunit of the chromatin remodeling complex NoRC. NoRC is associated with histone deacetylases (HDACs) and histone methyltransferases (HMTs) that establish heterochromatic features at the rDNA promoter. In addition, pRNA directly interacts with DNA, forming a DNA:DNA:RNA triple helix with the binding site of the transcription factor TTF-I, leading to displacement of TTF-I. The triple helical structure is recognized by the DNA methyltransferase DNMT3b, which methylates the rDNA promoter, leading to transcriptional silencing.
Depletion of pRNA decreased DNA methylation and enhanced rDNA transcription, whereas transfection of cells with synthetic pRNA triggered de novo methylation of the rDNA promoter and repressed transcription. Importantly, the 5′-end of pRNA can induce promoter hypermethylation and rDNA silencing independently of the NoRC complex. This is achieved by Hoogsten base pairing between pRNA and the rDNA promoter, forming a triple-stranded DNA-RNA structure at the binding site of the transcription factor TTF-I.35 The triple-helical DNA-RNA structure displaces TTF-I from its cognate site T0, initiating transcriptional repression. The triplex can then be specifically recognized by the de novo DNA methyltransferase DNMT3b, leading to methylation of a critical CpG residue in the promoter, which impairs binding of the transcription factor UBF and prevents transcription complex assembly.36 These results uncover a novel mechanism of epigenetic regulation, whereby ncRNAs bound to regulatory gene sequences by Hoogsten base pairing act as a “local address” to direct specific proteins to regulatory genomic loci or to replace them from their target sequence.
As epigenetic states are reversible, they can be modified by environmental factors that impinge on chromatin structure and transcriptional regulation. However, the pathways that alter chromatin structure in response to environmental or developmental cues remain largely unknown. Given that non-coding RNAs can act as a molecular trafficking system that guides chromatin modifiers to distinct genomic sites to establish specific epigenetic landscapes, it is plausible that ncRNA may mediate short-term changes in chromatin structure in response to external challenges. In accord with this view, strand-specific PCR-walking across rDNA revealed low levels of antisense transcripts that are synthesized by RNA polymerase II.37 These antisense transcripts, termed PAPAS (promoter and pre-rRNA antisense), comprise a heterogeneous population of RNA covering sequences of pre-rRNA and the rDNA promoter. Notably, the synthesis and/or stability of PAPAS vary among different cell lines and depend on the physiological state of the cells. PAPAS levels anticorrelate with the level of pre-rRNA, i.e., PAPAS is upregulated in quiescent cells where rDNA transcription is low, whereas PAPAS levels are low in cancer cells with high rRNA synthetic activity. The inverse correlation between the levels of PAPAS and 45S pre-rRNA suggests a role of these antisense transcripts in repression of RNAP I transcription upon growth arrest. In fact, chromatin immunoprecipitation (ChIP) assays comparing growing and quiescent NIH3T3 cells revealed that elevated levels of PAPAS correlate with enhanced trimethylation of histone H4 at lysine 20 (H4K20me3). This link between inhibition of rDNA transcription and upregulation of both PAPAS and H4K20me3 in response to growth factor deprivation suggested that PAPAS guides Suv4–20h2, the histone methyltransferase that mediates H4K20 trimethylation, to rDNA. In support of such a targeting mechanism, in vitro and in vivo RNA-protein interaction assays revealed that PAPAS binds with high affinity to Suv4–20h2. Once targeted to the rDNA promoter, Suv4-20h2 mediates trimethylation of H4K20 and compaction of chromatin, creating a chromatin environment that impairs transcription, thus enforcing signal-dependent transcriptional shutdown in response to growth arrest (Bierhoff et al., unpublished data). These results reveal a compelling mechanism of epigenetic regulation, establishing a repressive chromatin structure in response to external signals, thus ensuring shutdown of RNAP I transcription when cells become quiescent (Fig. 2). Such structural changes make heterochromatin intrinsically less vulnerable to DNA damage, thereby increasing chromosomal stability. Given that increased homologous recombination among the rDNA repeats correlates with tumorigenesis and developmental disorders,38,39 impaired heterochromatin formation by dysregulation of either pRNA or PAPAS will increase the risk of rDNA instability, nucleolar disintegration and cellular senescence.
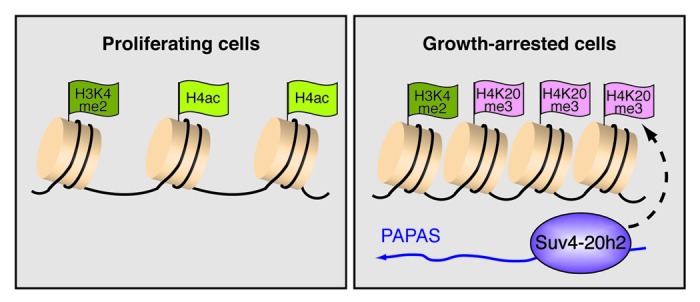
Figure 2. PAPAS induces trimethylation of H4K20 and chromatin compaction in growth-arrested cells. The scheme depicts epigenetic features of the rDNA promoter in proliferating cells and quiescent cells. In growth-arrested cells, increased levels of antisense transcripts, termed PAPAS (promoter and pre-rRNA antisense), recruit the H4K20 methyltransferase Suv4–20h2 to the rDNA promoter, thereby triggering H4K20 trimethylation and chromatin compaction. Increased levels of H4K20me3 are accompanied by decreased histone H4 acetylation, whereas H3K4 methylation is not affected.
Centromere Specification and Function Requires ncRNAs
Centromeres are specialized chromosomal structures that govern kinetochore assembly and ensure equal chromosome segregation during mitosis. They are composed of short tandem repeats forming large arrays up to several megabases.40,41 Like rRNA genes, centromeres are organized in tandemly repeated clusters and exhibit a repressive heterochromatic structure. Maintaining a heterochromatic structure at centromeres is important for kinetochore function and chromosome integrity. Murine centromeres comprise arrays of minor satellite sequences that are embedded in a larger pericentromeric region, composed of major satellite repeats.42 Human centromeres comprise arrays of 171-bp α-satellite repeats that are surrounded by arrays of longer repetitive elements, including satellites II and III (Sat II and III) that are interspersed with unique sequence elements.43 Centric and pericentric regions differ in chromatin structure, forming structurally distinct domains, both of which are required for kinetochore assembly and proper chromosome segregation. Centromere identity is epigenetically determined by a unique chromatin signature in the core region, containing the centromere-specific histone H3 variant CENP-A and a nucleoprotein complex containing other centromeric proteins.44,45 Chromatin containing CENP-A is periodically interrupted by nucleosomes containing H3K4me2, whereas the flanking pericentromeric heterochromatin is characterized by heterochromatic histone modifications, including di- and trimethylated H3K9 and H4K20 as well as associated HP1 proteins.46
Surprisingly, centromeres have been discovered to localize close to nucleoli and several centromeric proteins contain a nucleolar targeting domain. This raises the possibility that the nucleolus plays a role in assembly of the kinetochore, providing a hub for heterochromatin formation and ncRNA-dependent gene repression.47,48 In fact, heterochromatin formation at centric and pericentric repeats appears to be mediated by similar mechanisms used to silence rDNA, NoRC playing a central role in this process. Analysis of TIP5 localization by ChIP assays and immunostaining revealed that a significant fraction of NoRC was associated with pericentromeric chromatin. Depletion of TIP5 reduced H3K9me3 and H4K20me3 at major satellite repeats, whereas overexpression of TIP5 increased the level of heterochromatic histone marks.49,50 Consistent with NoRC-dependent heterochromatin formation being critical for proper centromere function, abnormally large spindles appeared in TIP5-deficient cells, thus emphasizing the relevance of NoRC for centromere structure and function.50 Together, these results demonstrate that NoRC function is not restricted to rDNA silencing but also serves an essential function in heterochromatin formation at centromeres.
Recent studies have demonstrated that centromeres are transcriptionally active, and satellite DNA transcription is linked to centromere maintenance. Transcripts originating from the centromeric core region have been detected in several organisms, including vertebrates, invertebrates, plants and yeast. In most species, satellite repeats are differentially expressed in a development- or tissue-specific manner, and a balanced, low-level synthesis of satellite transcripts safeguards the integrity of heterochromatin at centromeres.47,51-55 Transcripts from minor and α-satellite repeats are required for loading CEN proteins to centromeres, including INCENP, Survivin and aurora B kinase (Fig. 3).47,56,57 Moreover, single stranded RNA has been shown to bind to CENP-C and enhance binding of CENP-C to DNA.58 Demethylation of H3K4me2 by the lysine demethylase LSD1 induced loss of centromeric transcription and mislocalization of the CENP-A chaperone HJURP.59 On the other hand, acetylation of H3K9 and overexpression of satellite RNA resulted in centromere inactivation and failure to recruit CENP-A,60 highlighting the role of RNA in protein composition and epigenetic properties of centric chromatin.
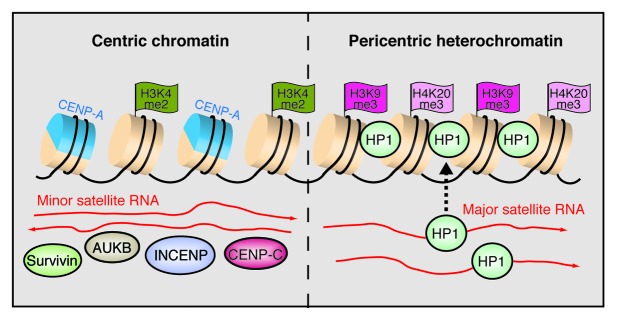
Figure 3. RNA shapes the epigenetic landscape at centromeres. The model shows the chromatin organization of minor and major satellite repeats at murine centromeres. Minor satellites in the centric region are characterized by alternating nucleosomes that harbor either the histone H3 variant CENP-A or H3K4me2. Transcripts in both orientations facilitate the association with centric proteins including CENP-C, INCENP, Survivin and aurora B kinase (AUKB). At pericentric heterochromatin, major satellite transcripts mediate the deposition of HP1α on nucleosomes marked by H3K9me3 and H4K20me3.
RNA is also a structural component of pericentric heterochromatin, RNA-protein interactions being required for the localization of HP1α and H3K9me3 at centromeres.61 Moreover, a recent analysis of HP1α binding to RNA revealed that the SUMO-modified form of HP1α specifically binds to major satellite transcripts, promoting loading of HP1α to pericentric heterochromatin.62 Given that too much euchromatin or heterochromatin disrupts centromere function,63 the transcriptional properties and chromatin composition at centromeres needs to be intricately regulated. The transcription factors Pax3 and Pax9 have been shown to play a pivotal role in keeping pericentric transcription in check.64 In mouse cells, Pax3 and Pax9 associate with conserved binding motifs in major satellite repeats and repress transcription from both DNA strands. Simultaneous depletion of both factors causes an over 1000-fold upregulation of transcription, leading to loss of H3K9me3 and H4K20me3 from pericentric heterochromatin and severe mitotic defects. Pericentric heterochromatin has been shown to acquire euchromatic features upon exposure to stress, facilitating transcription of RNAs containing a subset of Sat III repeats in response to heat shock, DNA damage or osmotic stress.65,66 The Sat III transcripts nucleate the formation of nuclear stress bodies (nSBs) in which RNAP II together with transcription and splicing factors are sequestered.66 Thus, upregulation of Sat III transcription and formation of nSBs is presumably a mechanism cells use to repress transcription and RNA processing under stress conditions.
Finally, digital gene expression analysis revealed a tremendous generation of bidirectional ncRNAs from major satellite repeats in cancer cells, suggesting that global alteration in pericentric RNAs may affect differentiation programs implicated in cancer.67 Indeed, both DNA methylation and H3K9 trimethylation are critical for the maintenance of satellite repression and dysregulation of these epigenetic marks is linked to carcinogenesis. For example, abrogation of H3K9 trimethylation by knockout of Suv39h1/2 leads to decreased genome stability and increased the susceptibility to develop B-cell lymphomas,68 reinforcing the role of heterochromatin in safeguarding genome integrity. It remains to be determined whether the act of transcription per se, or the transcripts themselves, play a direct part in this process.
TERRA-Mediated Heterochromatin Formation Facilitates Telomere Homeostasis
Telomeres are heterochromatic nucleoprotein structures located at the ends of eukaryotic chromosomes, comprising hexameric 5′-TTAGGG-3′ repeats in vertebrates that are flanked by repeat-rich, gene-poor subtelomeric regions. Telomeres are bound by specialized proteins, known as Shelterins, that are essential for telomere structure and function.69 Sequestration of telomeres into nucleoprotein caps shields chromosome ends from exposure to DNA damage and prevents telomere fusions, thus protecting chromosome integrity. However, recent work has revealed that packaging into a compact higher-order chromatin structure is important for telomere maintenance. The heterochromatic structure of telomeric and subtelomeric chromatin is conserved in higher eukaryotes. Both telomeric and subtelomeric repeats are marked by heterochromatic histone modifications, e.g., H3K9me3, H4K20me3, and hypoacetylated histones H3 and H4.70,71 Loss of telomeric and subtelomeric heterochromatin leads to chromatin decondensation, telomere shortening and increased frequency of telomere recombination and genomic instability, i.e., features that are associated with cellular senescence and transformation.70,71 Considering the longstanding idea that telomeres are transcriptionally silent genomic regions, it came as a surprise that telomeres are actively transcribed into a ncRNA, termed TERRA (telomeric repeat-containing RNA). TERRA transcription originates from a promoter-like CpG island within subtelomeres, proceeding toward chromosome ends.72 TERRA transcripts comprise UUAGGG repeats, are polyadenylated and range in size from about 100 bases to even more than 9 kilobases. They form discrete nuclear foci that co-localize with telomeric repeats in interphase and mitotic cells, indicating that TERRA is an integral part of telomeric heterochromatin.73,74 TERRA is abundant in adult mouse tissues but hardly detectable in mouse embryos and in human cancer,74 indicating that the synthesis and/or stability of TERRA is downregulated in highly proliferative cells.
Hypomethylation of subtelomeres in human cells by depletion of the DNA methyltransferases DNMT1 and DNMT3b leads to elevated TERRA levels and elongated telomeres,72 suggesting a link between TERRA levels and telomere length. Likewise, treatment with trichostatin A, an inhibitor of histone deacetylases, was accompanied by upregulation of TERRA,73 reinforcing the link between TERRA synthesis, epigenetic regulation and telomere length control. In support of TERRA affecting telomere structure, siRNA-mediated knockdown of TERRA decreased H3K9me3 at telomeric chromatin and induced a massive DNA damage response at telomeres.75 These observations demonstrate that TERRA impacts on the establishment and/or maintenance of the repressive chromatin structure at chromosome ends, a process that is perturbed in telomere-related diseases, such as cancer and premature aging. In accord with TERRA protecting chromosome ends from DNA damage by promoting heterochromatin formation, TERRA accumulates during conditions of proliferative stress, forming nuclear aggregates that are required for chromosome end capping and prevention of DNA damage.75,76
Regarding the mechanism underlying heterochromatin formation at telomeric repeats, we have recently shown that NoRC binds to a significant fraction of telomeres, recruiting histone modifying enzymes, such as Suv4-20h2 and SIRT6, to establish heterochromatic features at telomeres.50 Overexpression of TIP5 increased the levels of H3K9me3 and K4K20me3 and decreased acetylation of histone H4 at telomeres and subtelomeres, whereas knockdown of TIP5 had opposite effects. Notably, overexpression or depletion of TIP5 led to corresponding changes of TERRA levels, indicating that NoRC cooperates with TERRA to preserve the structural and functional integrity of telomeres. Indeed, TERRA was shown to be associated with TIP5, supporting that TERRA serves a guiding function in NoRC-dependent heterochromatin formation at telomeres (Fig. 4). Noteworthy, depletion of TIP5 increased the association of telomeres with promyelocytic leukemia (PML) nuclear bodies and increased homologous recombination, emphasizing the impact of NoRC on telomere and chromosome stability.
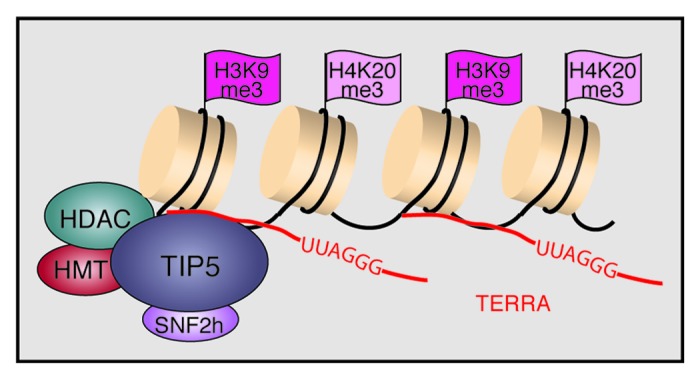
Figure 4. TERRA recruits NoRC to telomeres to establish/maintain heterochromatin at chromosome ends. TIP5 interacts with histone deacetylases (HDACs) and histone methyltransferases (HMTs), leading to deacetylation of histone H4 and trimethylation of H3K9 and H4K20.
Mastering Genomic Parasites: Non-Coding RNAs Silence Retroelements
Retrotransposons are remnants of parasitic DNA elements that have shaped host genomes, thus driving genomic evolution. Retrotransposons constitute a large part of eukaryotic genomes and pose a potential threat to genomic integrity. Although most retroelements have lost their ability for transposition due to mutations or truncations, some of them are intact and contain active promoters. Hence, retrotransposons, including the long-terminal repeat (LTR)-containing intracisternal A particle (IAP) elements or the long interspersed nuclear elements-1 (LINE-1), are multifaceted regulators of the transcriptome that may affect the expression of neighboring genes.77,78 Apparently, in a co-evolutionary “arms race” between hosts and mobile DNA elements, several mechanisms have emerged that counteract retrotransposon activity, including RNA-dependent epigenetic silencing.79 Retrotransposition has been observed in different types of human cancers, emphasizing the need for repression of retroelements in somatic cells.80 Interestingly, the epigenetic mechanisms that silence retrotransposons appear to be different in cycling and postmitotic cells. In somatic cells, retroelements are generally silenced by CpG methylation, whereas histone marks seem to play a minor role.81 Preliminary data indicate that postmitotic cells use an additional mechanism to silence retrotransposons and to protect the genome from mutagenic transposition. Similar to PAPAS-mediated recruitment of Suv4-20h2 to the rDNA promoter, transcripts from IAP and LINE-1 elements appear to trigger upregulation of H4K20 trimethylation and chromatin compaction in quiescent cells (Bierhoff et al., unpublished data). These data imply that RNA originating from retroelements serves as a signal to recruit Suv4-20h2 and establish a repressive chromatin structure at these repetitive sequences (Fig. 5A). Moreover, bidirectional transcripts from LINE-1 elements in human cells generate short interfering RNAs to trigger an RNA interference response that suppresses transposition.82
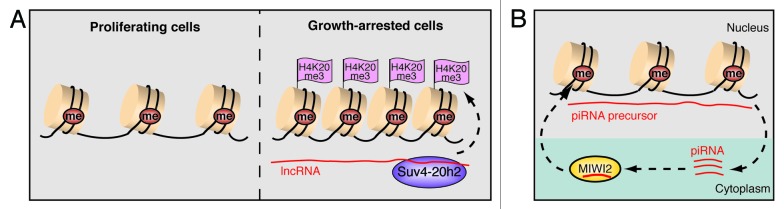
Figure 5. Non-coding RNA silences transposable elements in somatic and germ cells. (A) Somatic cells suppress transposon activity by DNA methylation and acquire H4K20 trimethylation (H3K20me3) upon growth arrest. Heterochromatin formation in quiescent cells involves upregulation of transposon-derived lncRNAs that guide Suv4–20h2 to the locus and induce H4K20me3. (B) Retrotransposons are repressed by piRNA-induced DNA methylation in murine fetal germ cells. Binding of the Piwi protein MIWI2 to mature piRNAs that originate from retrotransposon transcripts (piRNA precursors) facilitates nuclear localization and MIWI2/piRNA-dependent gene silencing.
As genomic stability of germ cells is crucial for transmission of the genetic information to the next generation, retrotransposition in the germline would exert deleterious effects, implying the need for a specific defense mechanism. The PIWI clade of Argonaute proteins is predominantly expressed in germ cells and interacts with 24–30 nucleotides sized RNAs (piRNAs), constituting a piRNA-induced silencing complex, termed piRISC.83 piRNAs are processed from long single stranded precursor RNAs that map to intergenic repeat-rich loci.84,85 These piRNA clusters comprise various transposable elements, thereby giving rise to piRISC-mediated repression of transposons at the transcriptional and posttranscriptional level. In mouse, three PIWI-like proteins, MILI, MIWI, and MIWI2, are expressed in male germ cells. Knockout of each of these proteins causes male-specific sterility, demonstrating their essential role in spermatogenesis.86-88 During germ cell development, MILI is broadly expressed in embryonic and post-natal stages, while MIWI2 expression is restricted to the pre-pachytene stage.89 Significantly, MIWI2 is associated with piRNAs that are enriched in transposon sequences.89,90 piRNA-MIWI2 complexes are translocated into the nucleus where they induce de novo DNA methylation and epigenetic silencing of retroelements (Fig. 5B). Consistently, LINE-1 and IAP retroelements are hypomethylated in fetal germ cells lacking MILI or MIWI2.91
A recent study has shown that expression of retroelements is dynamic and stage specific, most elements becoming repressed during early mouse embryogenesis.92 Silencing of repetitive elements was caused by loss of active histone marks rather than acquisition of repressive marks. After fertilization and global DNA demethylation, LINE-1 and IAP elements become activated, being transcribed from both parental genomes. Their transcriptional activity decreases as development proceeds, finally attaining DNA methylation and H3K9 trimethylation to ensure constitutive heterochromatin formation and propagation of the silent state. Scanning the sequence of a LINE-1 element revealed a stretch of polypurine residues within the ORF1 region that has the potential to form a DNA:RNA triplex structure.92 Immunofluorescence with an antibody that recognizes triple helices showed nuclear staining of late 2-cell embryos. Staining was almost undetectable at the 8-cell stage, demonstrating that transcriptional activation of LINE-1 elements occurs within a narrow time frame during early embryogenesis. Together, these results suggest that repetitive elements can autoregulate their activity by generating small RNAs that affect the epigenetic state of their respective target sequences. Thus, mammalian cells control the activity of mobile elements by producing long and short non-coding RNAs to silence retroelements and protect the genome from mutagenic transposition.
Perspectives
Given their abundance and functional relevance, repetitive elements can be viewed as building blocks of eukaryotic genomes. As they present a serious challenge for genome stability, repetitive sequences are maintained in a compact, transcriptionally refractory state. A common emerging theme is that non-coding RNA serves a key role in the establishment and maintenance of heterochromatic features. Non-coding RNAs can carry out their functions by association with chromatin-modifying complexes, by interaction with protein(s) bound to specific genomic sites, or by direct interaction with nucleic acids, e.g., forming DNA:RNA heteroduplexes, DNA:DNA:RNA triplexes, annealing with nascent RNA chains, and likely by many other mechanisms. A major challenge is to understand the precise mechanism by which individual ncRNAs establish a repressive chromatin structure at specific genome loci. As many ncRNAs act in concert with distinct protein partners, elucidation of the full spectrum of ncRNA binding proteins using high-throughput screens, such as the interactome capture technique,93,94 will be informative. Identification of the sites of RNA-protein interactions will give further insights into the versatility of RNA recognition motifs and their involvement in diverse biological functions. This holds especially true for NoRC, which is involved in epigenetic silencing of rRNA genes, telomeres and centromeres. Whereas the function of NoRC at rDNA and telomeres is clearly RNA-dependent, its interaction with centromeric transcripts has yet to be proven. It is tempting to speculate that the interaction of NoRC with pRNA and TERRA involves a common structural motif that is shared between these two RNAs. Moreover, as in many cases the secondary structure of ncRNAs dictates their functions, it is important to understand the structural features of specific RNAs. Chemical and enzymatic probing of secondary structure will uncover general structural modules in regulatory RNA and delineate structure-function relationships. One question of particular interest is whether a given ncRNA acts in cis (on neighboring genes) or trans (on distantly located genes) to induce intricate patterns of epigenetic modifications. Further insights into the crosstalk between ncRNAs and repetitive elements will uncover the link between aberrant ncRNA expression and uncontrolled activity of these abundant genomic loci. Given that homologous recombination, retrotransposition and telomeric dysfunction have a high mutagenic potential and are dysregulated in various diseases, especially cancer, ncRNA-dependent heterochromatin formation is essential for preserving chromosome structure and genome integrity. Thus, understanding the function of repeat-derived RNAs will foster their use as diagnostic markers or even as therapeutic targets in human disease and neoplasia.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26485
References
- 1.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 2.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–3. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 4.Mancini-DiNardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–82. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson CM, Ball ST, Dawson C, Mehta S, Beechey CV, Fray M, Teboul L, Dear TN, Kelsey G, Peters J. Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet. 2011;7:e1001347. doi: 10.1371/journal.pgen.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maamar H, Cabili MN, Rinn J, Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev. 2013;27:1260–71. doi: 10.1101/gad.217018.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/S0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 12.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–60. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 13.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 14.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malecová B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther. 2010;12:214–22. [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bühler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14:1041–8. doi: 10.1038/nsmb1315. [DOI] [PubMed] [Google Scholar]
- 18.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–5. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 19.Schoeftner S, Blasco MAA. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28:2323–36. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad K, Henikoff S. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell. 2001;104:839–47. doi: 10.1016/S0092-8674(01)00281-1. [DOI] [PubMed] [Google Scholar]
- 21.Zofall M, Grewal SI. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol Cell. 2006;22:681–92. doi: 10.1016/j.molcel.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 23.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32:393–6. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 24.Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci U S A. 2012;109:8161–6. doi: 10.1073/pnas.1201262109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Längst G, Grummt I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006;25:5735–41. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–40. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–61. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Guetg C, Santoro R. Noncoding RNAs link PARP1 to heterochromatin. Cell Cycle. 2012;11:2217–8. doi: 10.4161/cc.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moss T, Boseley PG, Birnstiel ML. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980;8:467–85. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cassidy BG, Yang-Yen HF, Rothblum LI. Additional RNA polymerase I initiation site within the nontranscribed spacer region of the rat rRNA gene. Mol Cell Biol. 1987;7:2388–96. doi: 10.1128/mcb.7.7.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn A, Grummt I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 1987;6:3487–92. doi: 10.1002/j.1460-2075.1987.tb02673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–80. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santoro R, Schmitz KM, Sandoval J, Grummt I. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep. 2010;11:52–8. doi: 10.1038/embor.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–9. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell. 2001;8:719–25. doi: 10.1016/S1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 37.Bierhoff H, Schmitz K, Maass F, Ye J, Grummt I. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol. 2010;75:357–64. doi: 10.1101/sqb.2010.75.060. [DOI] [PubMed] [Google Scholar]
- 38.Killen MW, Stults DM, Adachi N, Hanakahi L, Pierce AJ. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum Mol Genet. 2009;18:3417–28. doi: 10.1093/hmg/ddp282. [DOI] [PubMed] [Google Scholar]
- 39.Stults DM, Killen MW, Williamson EP, Hourigan JS, Vargas HD, Arnold SM, Moscow JA, Pierce AJ. Human rRNA gene clusters are recombinational hotspots in cancer. Cancer Res. 2009;69:9096–104. doi: 10.1158/0008-5472.CAN-09-2680. [DOI] [PubMed] [Google Scholar]
- 40.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 41.Schueler MG, Sullivan BA. Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet. 2006;7:301–13. doi: 10.1146/annurev.genom.7.080505.115613. [DOI] [PubMed] [Google Scholar]
- 42.Buscaino A, Allshire R, Pidoux A. Building centromeres: home sweet home or a nomadic existence? Curr Opin Genet Dev. 2010;20:118–26. doi: 10.1016/j.gde.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Hayden KE. Human centromere genomics: now it’s personal. Chromosome Res. 2012;20:621–33. doi: 10.1007/s10577-012-9295-y. [DOI] [PubMed] [Google Scholar]
- 44.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–46. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 46.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–60. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010;29:2135–46. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Postepska-Igielska A, Krunic D, Schmitt N, Greulich-Bode KM, Boukamp P, Grummt I. The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 2013;14:704–10. doi: 10.1038/embor.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diaz MO, Barsacchi-Pilone G, Mahon KA, Gall JG. Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell. 1981;24:649–59. doi: 10.1016/0092-8674(81)90091-X. [DOI] [PubMed] [Google Scholar]
- 52.Trapitz P, Wlaschek M, Bünemann H. Structure and function of Y chromosomal DNA. II. Analysis of lampbrush loop associated transcripts in nuclei of primary spermatocytes of Drosophila hydei by in situ hybridization using asymmetric RNA probes of four different families of repetitive DNA. Chromosoma. 1988;96:159–70. doi: 10.1007/BF00331048. [DOI] [PubMed] [Google Scholar]
- 53.Rudert F, Bronner S, Garnier JM, Dollé P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome. 1995;6:76–83. doi: 10.1007/BF00303248. [DOI] [PubMed] [Google Scholar]
- 54.Li YX, Kirby ML. Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev Dyn. 2003;228:72–81. doi: 10.1002/dvdy.10355. [DOI] [PubMed] [Google Scholar]
- 55.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferri F, Bouzinba-Segard H, Velasco G, Hubé F, Francastel C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009;37:5071–80. doi: 10.1093/nar/gkp529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, Wong LH. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc Natl Acad Sci U S A. 2012;109:1979–84. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergmann JH, Rodríguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–40. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergmann JH, Jakubsche JN, Martins NM, Kagansky A, Nakano M, Kimura H, Kelly DA, Turner BM, Masumoto H, Larionov V, et al. Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J Cell Sci. 2012;125:411–21. doi: 10.1242/jcs.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–34. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 62.Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet. 2011;43:220–7. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 63.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–22. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bulut-Karslioglu A, Perrera V, Scaranaro M, de la Rosa-Velazquez IA, van de Nobelen S, Shukeir N, Popow J, Gerle B, Opravil S, Pagani M, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol. 2012;19:1023–30. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 65.Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–51. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biamonti G, Vourc’h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–6. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–37. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 69.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 70.García-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–9. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 71.Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol. 2007;178:925–36. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, Giulotto E, Azzalin CM. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–94. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 74.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–36. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 75.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–13. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng Z, Wang Z, Xiang C, Molczan A, Baubet V, Conejo-Garcia J, Xu X, Lieberman PM, Dahmane N. Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J Cell Sci. 2012;125:4383–94. doi: 10.1242/jcs.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kazazian HH., Jr. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 78.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 79.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–85. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 80.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–71. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci. 2012;37:127–33. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 82.Yang N, Kazazian HH., Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–71. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 83.Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26:2361–73. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 85.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 86.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–30. doi: 10.1016/S1534-5807(02)00165-X. [DOI] [PubMed] [Google Scholar]
- 87.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–49. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 88.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 89.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–63. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 91.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–17. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol. 2013;20:332–8. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- 93.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 94.Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc. 2013;8:491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]


