Abstract
Pregnancy outcome is inextricably linked to placental development, which is strictly controlled temporally and spatially through mechanisms that are only partially understood. However, increasing evidence suggests non-coding RNAs (ncRNAs) direct and regulate a considerable number of biological processes and therefore may constitute a previously hidden layer of regulatory information in the placenta. Many ncRNAs, including both microRNAs and long non-coding transcripts, show almost exclusive or predominant expression in the placenta compared with other somatic tissues and display altered expression patterns in placentas from complicated pregnancies. In this review, we explore the results of recent genome-scale and single gene expression studies using human placental tissue, but include studies in the mouse where human data are lacking. Our review focuses on the ncRNAs epigenetically regulated through genomic imprinting or X-chromosome inactivation and includes recent evidence surrounding the H19 lincRNA, the imprinted C19MC cluster microRNAs, and X-linked miRNAs associated with pregnancy complications.
Keywords: non-coding RNA, placenta, microRNA, preeclampsia, epigenetics, pregnancy
Introduction
The best-known function of the placenta is to mediate fetal-maternal exchange throughout pregnancy, but it also plays a major role in directing maternal adaptation to pregnancy by secreting a variety of steroid and peptide hormones that modulate maternal physiology without which pregnancy could not be sustained. The placenta is a unique organ in several respects. First, although the placenta is a shared organ between mother and fetus, it is an extra-embryonic tissue and is therefore primarily regulated by the fetal genome. Second, the placenta completely separates from mother and fetus after birth, making it the only truly transient organ. For this reason, the placenta may not be under the same lifetime epigenetic constraints as other somatic tissues. Placental development in humans begins shortly after an embryo implants into the lining of the uterus, where it begins a strikingly invasive process that remodels the uterine spiral arterioles to sequester a maternal blood supply to facilitate efficient feto-maternal exchange. This invasive process, which has many similarities with cancer metastasis,1 appears to be strictly controlled both spatially and temporally in humans through mechanisms that are only partially understood.2,3 However, emerging evidence, particularly from high-throughput gene expression technologies, suggests non-coding RNA molecules (ncRNAs) direct and regulate a considerable number of biological processes and cellular functions. Therefore, ncRNAs may constitute a previously hidden layer of regulatory information in the placenta. In this review, we focus on the imprinted and X-linked ncRNAs, which are typically expressed from only one allele. We explore the regulation of these ncRNAs in the context of human placental development. Examining particular influential genomic regions, a key focus of this review will be the role that ncRNA expression in the placenta plays in pregnancy complications, such as preeclampsia, that are attributed to abnormal placental development. Although this review is focused on human placental development, studies in the mouse are also discussed where human data are lacking.
The Placenta is Key to Fetal and Maternal Health
The placenta is part of the conceptus and therefore is genetically identical to the fetus. Its development is initiated at implantation about 5–6 d after conception and follows a dynamic and constantly changing trajectory providing gaseous and nutrient exchange functions between the maternal and fetal circulations to support fetal growth.4 Impaired placental (trophoblast) invasion has been implicated in several complications of pregnancy such as preeclampsia and intrauterine growth restriction (IUGR)5 and pre-term labor.6,7 For example, in preeclampsia, invasion of the spiral arterioles and the maternal decidual stroma is shallow, resulting in poor maternal blood flow to the placenta.5,8,9 Despite huge research efforts, our understanding of the highly complex molecular regulation of both normal and abnormal placentation is still inadequate. However, ncRNAs are emerging as key regulators of development10,11 and therefore provide new avenues of inquiry relating to placental differentiation and function. If so, the perturbed regulation of ncRNAs in the placenta may result in one or more of a continuum of pregnancy complications that compromise the health of both mother and infant.
Classification and Detection of Non-Coding RNA
There are many different classes of ncRNAs, as these molecules vary greatly with regards to sequence length and complexity, splicing isoforms, polyadenylation, regulation, and biological function. The most well-characterized class of ncRNAs are the infrastructural RNAs (rRNAs, tRNAs, snRNAs, snoRNAs), which constitute many integral cellular components and are involved in processes such as translation, transcript splicing and higher level regulatory processes, including DNA methylation.10 Other ncRNAs are typically classed based on their sequence length, with RNAs shorter than ~200 nucleotides termed short non-coding RNAs (sncRNAs), and those greater than ~200 nucleotides are termed long non-coding RNAs (lncRNAs).11 The sncRNAs include the microRNAs (miRNAs), piwi-interacting RNAs (piRNAs), and the small interfering RNAs (siRNAs). These short RNAs, particularly the miRNAs, have received the most attention to date, and currently dominate the ncRNA literature. However, there has been a steady accumulation of evidence indicating that lncRNA transcripts, as a class, have a diverse repertoire of biological functions11 and constitute a significant proportion of total cellular RNA.12
Although the central dogma of biology has previously allowed little scope for the regulatory capabilities of ncRNA (for a review see ref. 13), the ability to detect and measure ncRNAs has also hindered progress toward appreciating the gamut of their functional abilities. Detecting ncRNAs in any tissue has posed challenges for several reasons. First, distinguishing if a transcript has protein-coding ability can be difficult as ncRNA transcripts can originate from intronic and untranslated regions of coding transcripts, or can be alternative splice variations that abolish a transcript’s coding potential.14,15 Second, lncRNAs can be transcribed from DNA that spans intergenic and coding regions resulting in transcripts that host protein-coding DNA sequence. Third, many ncRNAs do not end with a poly-A signal,12 which is characteristic of protein coding genes. This difference has profound implications regarding ncRNA detection as many cDNA, SAGE, microarray, and RNA-Seq methods rely on poly-A labeling, enrichment or priming. For these reasons among several others (see ref. 16), ncRNA transcripts can be difficult to discover and measure, which has subsequently hampered our ability to annotate and functionally classify ncRNAs in health and disease.
The Roles of Non-Coding RNAs in Genomic Imprinting in the Placenta
Imprinted genes are known to have significant effects on placental development and are implicated in many placental pathologies.17–19 Imprinted genes are expressed in a parent-of-origin-dependent manner, with the imprinted alleles being epigenetically silenced.20,21 Genomic imprinting is typically observed in clusters of ~2–12 genes, with most of these clusters having at least one lncRNA gene.22 The epigenetic regulation of imprinting can involve DNA methylation imprints, repressive histone modifications, and complex enhancer competition scenarios involving cis-acting lncRNA transcripts.22–25 Ablation of lncRNAs within imprinting clusters typically results in the loss of imprinting,22 demonstrating that lncRNAs can act as cis regulators of autosomal gene expression.
Imprinting is largely, although not exclusively, observed in eutherian mammals and is thought to have arisen with viviparity and the evolutionary emergence of the placenta.26,27 The prevailing evolutionary hypothesis of imprinting suggests that paternally-expressed genes have been selected to maximize fetal resource acquisition from the mother, while maternally-expressed genes have been selected to balance resources allocated to current and future offspring.27 Since imprinted genes are suggested to facilitate a tug-of-war between maternal and paternal genomes, this hypothesis predicts that imprinted genes are heavily involved in fetal and placental growth and development throughout pregnancy.21,27,28 Not surprisingly, more imprinted genes are expressed in the placenta than in any other tissue, with several being placenta specific.29
Although the exact mechanisms regulating imprinted regions remain unclear, the maintenance of imprints appears to differ between embryonic and extra-embryonic tissues.29 This suggests that extra-embryonic cell lineages, many of which make up the placenta, may employ regulatory mechanisms involving ncRNAs that are not observed in embryonic cell lineages. Despite the fact that much of our understanding of placental imprinting comes from studies in mice,29 the evidence from human research to date suggests that many human placental abnormalities and pregnancy complications are associated with altered imprinting involving ncRNAs.
The Imprinted H19 Long Non-Coding RNA and miR-675
H19 was one of the first lncRNAs to be discovered and is considered a key regulatory molecule in placental development. H19 lies within a large imprinted domain (> 1 MB), and is predominantly expressed from the maternal chromosome. H19 placental expression is largely monoallelic30 and is one of the most highly expressed genes in the human placenta.31 However, the functional roles of H19 are only now beginning to emerge.
H19, and the adjacent and reciprocally-imprinted IGF2 gene, make up one of the most widely studied imprinted genomic regions in humans. Both H19 and IGF2 share many cis-regulatory elements, with the prevailing regulatory model of this locus indicating a complex interaction of DNA methylation, CTCF binding and enhancer competition scenarios mainly elucidated through targeted deletion and transgenic techniques in murine models.32
Somewhat consistent with observations in humans, studies in mice have further demonstrated that altered imprinting of H19 is associated with placental and fetal growth abnormalities.32–34 For example, (epi)mutations in the H19–IGF2 region are associated with Silver-Russell and Beckwith-Wiedemann syndromes, which manifest phenotypically in utero as severe growth-restriction and overgrowth, respectively.35 Furthermore, altered epigenetic regulation of the H19-IGF2 region in human placentas has been associated with the pregnancy complication preeclampsia, which is attributed to abnormal placental development early in gestation.36,37 Biallelic expression of H19 has been observed at higher rates during the first trimester of pregnancy compared with term,36,38,39 with the early first trimester placenta showing patterns of imprinting plasticity.30 Together, these studies suggest H19 plays an important regulatory role in early placental development.
Recent work suggests that H19 is a trans regulator of an imprinted gene network for growth and development40 involving miRNAs hosted within the H19 transcript,32,41,42 which may account for some of H19’s bioactivity. Most recently, Keniry et al. have described H19 as a developmental reservoir of miR-675 in the mouse.43 This study shows the miR-675 microRNA is processed from the first exon of H19 in a developmental stage specific manner in the placenta. They also showed that levels of miR-675 increased with gestation acting as a placental growth suppressor.43 Although overall H19 expression remained unchanged throughout gestation, the RNA-binding protein Elavl1 (also known as HuR) appeared to bind to the H19 transcript preventing excision of miR-675 early in gestation.43 Elavl1 abundance decreased as gestation progressed, enabling miR-675 to be processed and to act as a placental growth suppressor.43 Although this study has increased our knowledge of H19 function in the placenta, it may not accurately portray the situation in humans for several reasons. First, the human and mouse H19 transcripts show notable sequence divergence. Second, a microarray analysis by Sitras et al. found no significant difference in ELAVL1 expression between first trimester and third trimester human placentas,44 (Fig. 1A) which is contrary to the observation in mice. However, as suggested by Keniry et al., the excision of miR-675 may also be regulated by additional RNA binding proteins.43 To examine this possibility, we performed an in silico analysis of RNA binding protein domains within the human and mouse H19 transcripts. We note that the ELAVL1 binding sites that flank the miR-675 locus in mouse are not present in the human transcript (Fig. 1A). However, we observed that binding domains flanking miR-675 existed for the RNA binding proteins NONO and YBX1 in the human H19 transcript, and these proteins show a significant decrease in expression as gestation progresses (Fig. 1B). These differences between mouse and human indicate further work is required to elucidate the true extent of H19 and miR-675 regulation and functionality in the human placenta. This would require miR-675 expression across human gestation to be evaluated, followed by a careful analysis of how miR-675 excision is repressed in humans. These experiments using human derived samples will be a fundamental step toward determining why H19 is implicated in human pregnancy complications attributed to abnormal placental development.
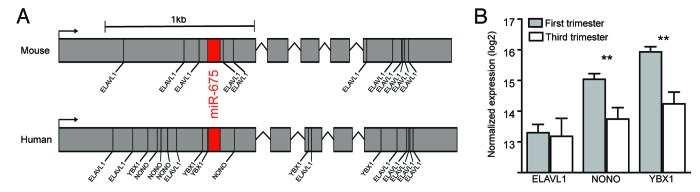
Figure 1. The H19 lncRNA transcript and RNA binding proteins in human and mouse. (A) Schematic representation of human and mouse H19 transcripts indicating locations of RNA binding protein motifs. The ELAVL1 binding motifs that surround the miR-675 locus in the mouse transcript are not present in the human transcript, however binding motifs for the NONO and YBX1 proteins are present in the human transcript. (B) Expression of genes that encode the RNA binding proteins ELAVL1, NONO, and YBX1 in human first and third trimester placentas. Expression of NONO, and YBX1 show a significant decrease in expression as gestation progresses, with ELAVL1 showing no difference across gestation. Data for RNA binding protein expression differences between first and third trimester were reported in ref.44 and the figures were generated using normalized array data obtained from the NCBI Gene Expression Omnibus (accession GSE28551). The RNA binding protein recognition sequences were predicted using the RNA binding protein database.107 Human and mouse H19 transcript sequences obtained from UCSC reference genomes hg19 and mm10 respectively.
The Imprinted C19MC miRNA Cluster
An intriguing observation of placental-expressed miRNAs arises from the largest known miRNA cluster discovered to date; C19MC. This cluster, located at human chromosome 19 (19q13.42), features ~46 miRNA genes transcribed from a ~100 kb region. C19MC is imprinted, with only the paternally inherited chromosome being expressed45,46 predominantly, if not exclusively, in the placenta.47 Furthermore, C19MC is unique to the primate lineage, excluding model organism research to determine the functions of miRNAs in this cluster.47
Transcription of C19MC miRNAs can be activated in some cells by treatment with DNA methylation inhibitors indicating that the region is under DNA methylation-dependent epigenetic control.45,48,49 Further evidence also suggests that the C19MC miRNAs are excised from a much larger lncRNA, which is transcribed from an RNAP II promoter within a CpG-rich region.45,47 C19MC miRNAs are also expressed in exosomes released from primary human trophoblast cells and are detectable in the serum of pregnant women,50 highlighting their potential as fetal-maternal signaling molecules that may modulate maternal adaptation to pregnancy. Although the precise functional mechanisms of C19MC miRNAs remain largely unknown, the abundance of C19MC transcripts in the placenta, their imprinted regulation, and detection in the maternal circulation, suggest a significant role in placental biology.
Studies of pregnancy complications attributed to abnormal placental development, in particular those focusing on placental gene expression in preeclampsia where a transcriptome-wide method (microarrays or high-throughput RNA sequencing) is employed, have shown differential regulation in the placental expression of some miRNAs in the C19MC family (Table 1).51–54
Table 1. Imprinted or X-linked miRNA differentially expressed in preeclampsia (PE) and pre-term birth (PTB) with validated targets and potential mechanisms.
| Pregnancy complication |
miRNA (cytoband) |
Expression in complication vs. control | Experimentally validated target genes109,110 detectable in the human placenta31 | Potential roles and contributing mechanisms |
|---|---|---|---|---|
| PE | miR-20b53,54,102 (Xq26.2) |
Increased | ARID4B, BAMBI, CDKN1A, CRIM1, ESR1, HIF1A, HIPK3, MYLIP, PPARG, STAT3, VEGFA | Impairing placental function through suppression of genes (CRIM1, HIF1A, VEGFA) that have a role in maintaining endothelial cell function and angiogenesis104,111–113 Repression of genes (CDKN1A, HIPK3, STAT3) involved in apoptosis and trophoblast invasion.114–116 |
| PE | miR-22254,103 (Xq11.3) |
Increased | BBC3, BCL2L11, CDKN1B, CDKN1C, CORO1A, ESR1, FOS, FOXO3, ICAM1, MMP1, PPP2R2A, PTEN, SOD2, SSSCA1, STAT5A, TCEAL1, TNFSF10, TP53 | Downregulation of genes (BBC3, BCL2, CORO1A, FOS, FOXO3, TNFSN10, TP53) that can promote apoptosis.117–122 Downregulation of genes (ICAM1) involved in endothelial cell function.123 Downregulation of genes (CDKN1B, CDKN1C) that regulate cell cycle progression and trophoblast differentiation.124 |
| PE | miR-22352,53 (Xq12) |
Decreased | CHUK, Il6, IRS1, LMO2, NFIX, RHOB, STMN1 | Upregulation of a gene (RHOB) involved in apoptosis signaling.125 Upregulation of a gene (IL6) involved with immune response and inflammation.126 |
| PE | miR-519b53,54 (19q13.42) |
Increased | CDKN1A, ELAVL1 | Alteration of apoptosis signals (CDKN1A).114 Downregulation of ELAVL1, potentially altering miR-675 excision from the H19 lincRNA.43 |
| PE | miR-519e52,53 (19q13.42) |
Increased | CDKN1A | Alteration of apoptosis signals (CDKN1A).114 |
| PE | miR-520 g53,54 (19q13.42) |
Increased | VEGFA | Impaired endothelial cell function and angiogenesis (VEGFA).104 |
| PE | miR-52453,102 (19q13.42) |
Increased | - | - |
| PE/PTB | miR-517a52,65 (19q13.42) |
Increased | - | Regulation of apoptosis127 |
| PE/PTB | miR-518b52,53,65,128 (19q13.42) |
Increased | - | |
| PE/PTB | miR-520h53,128 (19q13.42) |
Increased | ABCG2, CDKN1A, ID1, ID3, SMAD6, VEGFA | Downregulation of a gene (ABCG2) involved in protecting fetal exposure to xenobiotics ingested by the mother.129 Alteration of apoptosis signals (CDKN1A).114 Impaired endothelial cell function and angiogenesis (VEGFA)104 |
| PE/PTB | miR-526b53,65 (19q13.42) |
Increased | - | - |
Together, these studies have identified 21 miRNAs with increased placental expression in preeclampsia and/or pre-term birth when compared with normal pregnancies, with eight of these miRNAs showing increased expression in at least two studies (Table 1). Although empirical evidence is currently lacking for the targets of many C19MC miRNAs, miR-520g, and miR-520h have been shown experimentally to repress expression of VEGF, an angiogenic gene implicated in preeclampsia.55 Furthermore, expression of the VEGF receptor gene, FLT1, has also shown consistently higher expression in placentas from preeclamptic pregnancies.56–62 Additionally, the cell cycle inhibitor and apoptosis associated CDKN1A (p21) gene is a validated target of several C19MC miRNAs differentially expressed in preeclampsia (Table 1), which further implicates these miRNA genes given the links preeclampsia has with perturbed apoptosis.63
These results are of particular interest for several reasons, particularly with respect to preeclampsia. First, as the C19MC region is imprinted, increased miRNA expression may result from loss of imprinting, and the loss of imprinting of other placental ncRNAs such as H19 has known associations with preeclampsia.36 Second, C19MC allelic repression is regulated by DNA methylation imprints,45 and alteration to DNA methylation in the placenta is also associated with preeclampsia.64 Third, at least two miRNAs in the C19MC cluster target the VEGFA gene that is closely networked to the FLT1 gene. Fourthly, at least three C19MC miRNAs have been shown to target the ELAVL1 gene, which may be involved in regulating miR-675 excision from H19 transcript (see discussion of ref. 43 above).
The differential expression of several C19MC miRNAs in the placenta is also associated with preterm labor.65 Preterm birth and subsequent preterm delivery allows the investigation of placental gene expression at a much earlier time-point than normal labored deliveries. As such, the changes in miRNA expression may simply reflect the changing patterns of C19MC miRNAs as gestation progresses, and not be indicative of pathology. However, expression of C19MC miRNAs in cells derived from matched placentas sampled during both first trimester and term is comparable,46 suggesting the aberrant C19MC regulation in preterm birth is not due to developmental stage differences. Together, these findings warrant further inquiry into the biological role of the C19MC miRNAs, particularly in identifying their regulatory potential, as this increased understanding could reveal novel therapeutic targets.
X-chromosome ncRNAs in Placental Development and Pregnancy Complications
During intrauterine development, there are distinct sex differences in fetal growth trajectories and hence in birth weight,66–68 with a sex bias in the prevalence of preterm birth,69,70 pregnancy complications such as preeclampsia71,72 and perinatal death.73 As fetal growth and development are highly dependent on the exchange efficiency and capacity of the placenta, sex-specific differences in normal and pathological fetal development are most likely due to sex differences in placental function. Recent work has shown that there is a distinct male sex bias in the prevalence of placental dysfunction,72 which supports the findings of previous studies74–78 showing sex biases in a spectrum of pregnancy conditions and fetal health associated with abnormal placentation. However, the underlying mechanisms that predispose one sex over the other to deviate from a normal course of fetal development remain unknown.
Since the majority of genes are autosomal, many sexually dimorphic traits are driven by the sex-biased expression of autosomal genes.79 For decades, much of the scientific literature has solely attributed the influence of sex hormones to sexual dimorphism, yet increasing evidence suggests sex chromosome genes are also implicated in the regulation of autosomal gene expression (for a review see ref. 79). While many sex-specific gene expression differences have been appreciated for some time, their phenotypic and clinical implications, particularly in the placenta and in pregnancy complications, remain relatively unexplored.80–83
X-linked miRNAs as Potential Drivers of Sex Differences in Placental Gene Expression
When comparing the X chromosome to the 22 autosomes, the human X chromosome (with 140 annotated miRNAs in miRBase84) appears to be enriched for miRNA genes when considering its size and genomic content. Only chromosome 1, which features 11 more miRNA genes than the X chromosome, is richer in miRNA content and is ~100 MB larger in size. In contrast, the Y chromosome has only two annotated miRNA genes in a pseudo-autosomal region, which undergoes recombination with the X chromosome.
The observation that X chromosomes have high miRNA gene content highlights the potential of X-linked miRNAs to contribute to sex-biased autosomal gene expression. As X-linked miRNAs can potentially target multiple autosomal genes, sex-biased expression of X-linked miRNAs could trigger cascade-like effects, potentially driving sex-biased expression of many autosomal genes.85 Additionally, ~35 X-linked miRNAs are located within introns of protein-coding genes which are likely to share transcriptional elements with their host genes, potentially resulting in co-regulation. For example, the X-linked gene CHM, which hosts miR-361, is known to escape X chromosome inactivation (XCI), which could therefore lead to sex-biased expression of miR-361, and through autosomal gene targeting result in sex biased expression of autosomal genes.
Previous Studies Provide Few Clues to which miRNAs Escape XCI
Although X-linked miRNAs are potential drivers of sex differences in placental development and function, there is little evidence to suggest which, if any, of these miRNAs actually escape XCI, leading to biallelic (and potentially increased) expression in females. The inactivation of X chromosomes in human extra-embryonic tissues has been a topic of continual research over the past three decades, although the status and extent of XCI in the human placenta remains controversial. Most early studies focused on allele-specific expression from the G6PD (Xq28) locus to determine if the X chromosome was randomly inactivated or skewed toward paternal or maternal XCI, and showed that XCI varies notably across samples, with patterns of random and skewed X-inactivation.86–90 In later attempts to clarify the status of XCI in the human placenta, research shifted to different loci, again showing mixed results of skewed and random XCI.91–96 Taken together, these results suggest a high degree of XCI heterogeneity in the extra-embryonic tissues of female fetuses. Although only assessing genes on the q-arm of the X chromosome, these studies suggest that when XCI deviates from random, it is the paternal X chromosome that is most often inactivated.
What has become increasingly apparent over the past decade, however, is that multiple X chromosome regions escape inactivation, and that these extend beyond chromosomal regions with Y chromosome homologs.80,97 As the results of any XCI assay are dependent on the loci under investigation, it now appears spurious to infer the regulation of a whole chromosome (or region) based on the assessment of one or two X chromosome loci. In an attempt to widen the scope of our understanding of XCI in the human placenta, recent work has demonstrated allele-specific expression profiles of 22 genes spread across the X chromosome.98 The results of this most comprehensive placental study to date suggest that XCI in the human placenta is random, with localized mosaic patterns of maternal and paternal XCI.98 However, given the number of samples and the methodological limitations,99–101 placental XCI studies still lack the depth of XCI research conducted using other human tissues. Subsequently, we have very little indication of what ncRNAs, particularly miRNAs, escape XCI and potentially contribute to sex-biased placental gene expression in normal and complicated pregnancies.
X-linked miRNAs Associated with Preeclampsia
Recent studies have shown many X chromosome miRNAs that occur in clusters are differentially regulated in placentas from preeclamptic pregnancies (Fig. 2). When summarizing the results of these studies, increased expression of miR-20b53,54,102 and miR-22254,103 and decreased expression of miR-22352,53 is supported by two or more studies. Of particular interest, these miRNA have been shown experimentally to target multiple genes involved in processes such as apoptosis, angiogenesis and immune response (Table 1), all of which are implicated in the pathogenesis of preeclampsia.63,104,105
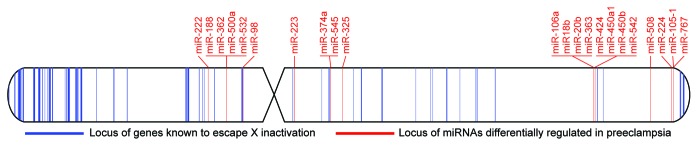
Figure 2. Idiogram representation of the human X chromosome showing the X-linked miRNAs associated with preeclampsia (red) occur in clusters in close proximity of genes that escape X inactivation (blue). The data for miRNAs showing altered expression in preeclampsia were derived from refs.51–54,103,108 and the data for genes that escape inactivation were adapted from ref.80 with genomic coordinates converted to hg19 coordinates using the UCSC liftOver tool.
There is also evidence implicating the miRNA cluster at Xq26.3, which flanks the placenta-specific PLAG1 gene. This cluster contains six miRNAs, four of which (miR-424, miR-542, miR-450a-1, and miR-450b) have been shown to be differentially regulated in placentas from preeclamptic pregnancies.51–54 Curiously, at the individual miRNA level, miR-424 is upregulated in preeclampsia, while miR-542 and miR-450b are downregulated, and for miR-450a-1 the data are conflicting.52,54 miR-424 is an interesting case since it overlaps the transcription start site of MGC16121, a lincRNA that is virtually unstudied. Additionally, miR-424 is a hypoxia-induced regulator of HIF1A and involved in angiogenesis,106 highlighting its potential role in preeclampsia.
Although many of these miRNAs are in close proximity to genes shown to escape XCI (Fig. 2), sample sex information would be required to determine if these X-linked miRNA expression differences are indeed sex related and resulting from differential X chromosome epigenetic regulation. However tenuous the link between XCI and expression differences in preeclampsia, the preliminary evidence discussed here justifies further investigation. Future work focused on delineating the boundaries of XCI in the human placenta and validating the targets of miRNAs that escape XCI may provide clues to the mechanisms giving rise to sex-biases in placental development and the downstream implications for adverse pregnancy outcomes such as preeclampsia.
Conclusions
Non-coding RNAs are increasingly implicated in many developmental and pathological processes; placental development is no exception. Although much research points toward the central role of ncRNAs in placental development and function, large gaps in our knowledge remain. In particular, the ncRNAs under epigenetic regulatory control through mechanisms such as XCI and genomic imprinting appear to be influential players. However, many questions remain regarding the functional actions of these transcripts and whether their change in expression in associated pregnancy complications is a cause or consequence. In either case, increasing our understanding of the epigenetically regulated ncRNAs in normal placental development is essential if these perplexing molecules are ever to be used as diagnostic or predictive biomarkers.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
SB is supported by a Healthy Development Adelaide and Channel 7 Children’s Research Foundation PhD Scholarship and an Australian Postgraduate Award. TB-M is supported by the Cancer Council SA and SAHMRI Beat Cancer Project (TBM APP1030945). CTR is supported by a National Health and Medical Research Council (NHMRC) Senior Research Fellowship APP1020749 (http://www.nhmrc.gov.au). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Glossary
Abbreviations:
- Non-coding RNA (ncRNA)
micro RNA (miRNA), long non-coding RNA (lincRNA)
Footnotes
Previously published online: www.landesbioscience.com/journals/epigenetics/article/26197
References
- 1.Murray MJ, Lessey BA. Embryo implantation and tumor metastasis: common pathways of invasion and angiogenesis. Semin Reprod Endocrinol. 1999;17:275–90. doi: 10.1055/s-2007-1016235. [DOI] [PubMed] [Google Scholar]
- 2.Graham CH, Lala PK. Mechanisms of placental invasion of the uterus and their control. Biochem Cell Biol. 1992;70:867–74. doi: 10.1139/o92-135. [DOI] [PubMed] [Google Scholar]
- 3.Lyall F. The human placental bed revisited. Placenta. 2002;23:555–62. doi: 10.1053/plac.2002.0850. [DOI] [PubMed] [Google Scholar]
- 4.Roberts CT. IFPA Award in Placentology Lecture: Complicated interactions between genes and the environment in placentation, pregnancy outcome and long term health. Placenta. 2010;31(Suppl):S47–53. doi: 10.1016/j.placenta.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Khong TY, De Wolf F, Robertson WB, Brosens I. Inadequate maternal vascular response to placentation in pregnancies complicated by pre-eclampsia and by small-for-gestational age infants. Br J Obstet Gynaecol. 1986;93:1049–59. doi: 10.1111/j.1471-0528.1986.tb07830.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim YM, Bujold E, Chaiworapongsa T, Gomez R, Yoon BH, Thaler HT, Rotmensch S, Romero R. Failure of physiologic transformation of the spiral arteries in patients with preterm labor and intact membranes. Am J Obstet Gynecol. 2003;189:1063–9. doi: 10.1067/S0002-9378(03)00838-X. [DOI] [PubMed] [Google Scholar]
- 7.Kim YM, Chaiworapongsa T, Gomez R, Bujold E, Yoon BH, Rotmensch S, Thaler HT, Romero R. Failure of physiologic transformation of the spiral arteries in the placental bed in preterm premature rupture of membranes. Am J Obstet Gynecol. 2002;187:1137–42. doi: 10.1067/mob.2002.127720. [DOI] [PubMed] [Google Scholar]
- 8.Rinkenberger JL, Cross JC, Werb Z. Molecular genetics of implantation in the mouse. Dev Genet. 1997;21:6–20. doi: 10.1002/(SICI)1520-6408(1997)21:1<6::AID-DVG2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Y, Genbacev O, Damsky CH, Fisher SJ. Oxygen regulates human cytotrophoblast differentiation and invasion: implications for endovascular invasion in normal pregnancy and in pre-eclampsia. J Reprod Immunol. 1998;39:197–213. doi: 10.1016/S0165-0378(98)00022-9. [DOI] [PubMed] [Google Scholar]
- 10.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 11.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 12.Kapranov P, St Laurent G, Raz T, Ozsolak F, Reynolds CP, Sorensen PH, Reaman G, Milos P, Arceci RJ, Thompson JF, et al. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–9. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 14.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankish A, Mudge JM, Thomas M, Harrow J. The importance of identifying alternative splicing in vertebrate genome annotation. Database (Oxford) 2012;2012:bas014. doi: 10.1093/database/bas014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowden AL, Coan PM, Angiolini E, Burton GJ, Constancia M. Imprinted genes and the epigenetic regulation of placental phenotype. Prog Biophys Mol Biol. 2011;106:281–8. doi: 10.1016/j.pbiomolbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bressan FF, De Bem TH, Perecin F, Lopes FL, Ambrosio CE, Meirelles FV, Miglino MA. Unearthing the roles of imprinted genes in the placenta. Placenta. 2009;30:823–34. doi: 10.1016/j.placenta.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–9. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 21.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 22.Koerner MV, Pauler FM, Huang R, Barlow DP. The function of non-coding RNAs in genomic imprinting. Development. 2009;136:1771–83. doi: 10.1242/dev.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Constância M, Pickard B, Kelsey G, Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- 24.Kacem S, Feil R. Chromatin mechanisms in genomic imprinting. Mamm Genome. 2009;20:544–56. doi: 10.1007/s00335-009-9223-4. [DOI] [PubMed] [Google Scholar]
- 25.Koerner MV, Barlow DP. Genomic imprinting-an epigenetic gene-regulatory model. Curr Opin Genet Dev. 2010;20:164–70. doi: 10.1016/j.gde.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci U S A. 2006;103:6623–8. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haig D. Altercation of generations: genetic conflicts of pregnancy. Am J Reprod Immunol. 1996;35:226–32. doi: 10.1111/j.1600-0897.1996.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 28.Haig D. Genetic conflicts in human pregnancy. Q Rev Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- 29.Wagschal A, Feil R. Genomic imprinting in the placenta. Cytogenet Genome Res. 2006;113:90–8. doi: 10.1159/000090819. [DOI] [PubMed] [Google Scholar]
- 30.Buckberry S, Bianco-Miotto T, Hiendleder S, Roberts CT. Quantitative allele-specific expression and DNA methylation analysis of H19, IGF2 and IGF2R in the human placenta across gestation reveals H19 imprinting plasticity. PLoS One. 2012;7:e51210. doi: 10.1371/journal.pone.0051210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Zhao K, Jiang P, Lu ZX, Wang J, Murray JC, Xing Y. Transcriptome landscape of the human placenta. BMC Genomics. 2012;13:115. doi: 10.1186/1471-2164-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabory A, Ripoche MA, Yoshimizu T, Dandolo L. The H19 gene: regulation and function of a non-coding RNA. Cytogenet Genome Res. 2006;113:188–93. doi: 10.1159/000090831. [DOI] [PubMed] [Google Scholar]
- 33.Lau MM, Stewart CE, Liu Z, Bhatt H, Rotwein P, Stewart CL. Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 1994;8:2953–63. doi: 10.1101/gad.8.24.2953. [DOI] [PubMed] [Google Scholar]
- 34.Constância M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, Fundele R, Stewart F, Kelsey G, Fowden A, Sibley C, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–8. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 35.Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187–200. doi: 10.1042/bse0480187. [DOI] [PubMed] [Google Scholar]
- 36.Yu L, Chen M, Zhao D, Yi P, Lu L, Han J, Zheng X, Zhou Y, Li L. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta. 2009;30:443–7. doi: 10.1016/j.placenta.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Gao WL, Li D, Xiao ZX, Liao QP, Yang HX, Li YX, Ji L, Wang YL. Detection of global DNA methylation and paternally imprinted H19 gene methylation in preeclamptic placentas. Hypertens Res. 2011;34:655–61. doi: 10.1038/hr.2011.9. [DOI] [PubMed] [Google Scholar]
- 38.Pozharny Y, Lambertini L, Ma Y, Ferrara L, Litton CG, Diplas A, Jacobs AR, Chen J, Stone JL, Wetmur J, et al. Genomic loss of imprinting in first-trimester human placenta. Am J Obstet Gynecol. 2010;202:e1–8. doi: 10.1016/j.ajog.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 39.Jinno Y, Ikeda Y, Yun K, Maw M, Masuzaki H, Fukuda H, Inuzuka K, Fujishita A, Ohtani Y, Okimoto T, et al. Establishment of functional imprinting of the H19 gene in human developing placentae. Nat Genet. 1995;10:318–24. doi: 10.1038/ng0795-318. [DOI] [PubMed] [Google Scholar]
- 40.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–80. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 41.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–6. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steck E, Boeuf S, Gabler J, Werth N, Schnatzer P, Diederichs S, Richter W. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med (Berl) 2012;90:1185–95. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- 43.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–65. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sitras V, Fenton C, Paulssen R, Vårtun A, Acharya G. Differences in gene expression between first and third trimester human placenta: a microarray study. PLoS One. 2012;7:e33294. doi: 10.1371/journal.pone.0033294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefèvre A, Coullin P, Moore GE, Cavaillé J. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–82. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 46.Flor I, Neumann A, Freter C, Helmke BM, Langenbuch M, Rippe V, Bullerdiek J. Abundant expression and hemimethylation of C19MC in cell cultures from placenta-derived stromal cells. Biochem Biophys Res Commun. 2012;422:411–6. doi: 10.1016/j.bbrc.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Bortolin-Cavaillé ML, Dance M, Weber M, Cavaillé J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–73. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito Y, Suzuki H, Tsugawa H, Nakagawa I, Matsuzaki J, Kanai Y, Hibi T. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA-512-5p with downregulation of Mcl-1 in human gastric cancer cells. Oncogene. 2009;28:2738–44. doi: 10.1038/onc.2009.140. [DOI] [PubMed] [Google Scholar]
- 49.Tsai KW, Kao HW, Chen HC, Chen SJ, Lin WC. Epigenetic control of the expression of a primate-specific microRNA cluster in human cancer cells. Epigenetics. 2009;4:587–92. doi: 10.4161/epi.4.8.10230. [DOI] [PubMed] [Google Scholar]
- 50.Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, Sadovsky Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod. 2012;18:417–24. doi: 10.1093/molehr/gas013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204:e12–21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu XM, Han T, Sargent IL, Yin GW, Yao YQ. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200:e1–7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 53.Ishibashi O, Ohkuchi A, Ali MM, Kurashina R, Luo SS, Ishikawa T, Takizawa T, Hirashima C, Takahashi K, Migita M, et al. Hydroxysteroid (17-β) dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension. 2012;59:265–73. doi: 10.1161/HYPERTENSIONAHA.111.180232. [DOI] [PubMed] [Google Scholar]
- 54.Hu Y, Li P, Hao S, Liu L, Zhao J, Hou Y. Differential expression of microRNAs in the placentae of Chinese patients with severe pre-eclampsia. Clin Chem Lab Med. 2009;47:923–9. doi: 10.1515/CCLM.2009.228. [DOI] [PubMed] [Google Scholar]
- 55.Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y. The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One. 2008;3:e1719. doi: 10.1371/journal.pone.0001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enquobahrie DA, Meller M, Rice K, Psaty BM, Siscovick DS, Williams MA. Differential placental gene expression in preeclampsia. Am J Obstet Gynecol. 2008;199:566.e1-11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McCarthy C, Cotter FE, McElwaine S, Twomey A, Mooney EE, Ryan F, Vaughan J. Altered gene expression patterns in intrauterine growth restriction: potential role of hypoxia. Am J Obstet Gynecol. 2007;196:e1–6. doi: 10.1016/j.ajog.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 58.Nishizawa H, Pryor-Koishi K, Kato T, Kowa H, Kurahashi H, Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta. 2007;28:487–97. doi: 10.1016/j.placenta.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 59.Winn VD, Gormley M, Paquet AC, Kjaer-Sorensen K, Kramer A, Rumer KK, Haimov-Kochman R, Yeh RF, Overgaard MT, Varki A, et al. Severe preeclampsia-related changes in gene expression at the maternal-fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology. 2009;150:452–62. doi: 10.1210/en.2008-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jarvenpaa J, Vuoristo JT, Savolainen ER, Ukkola O, Vaskivuo T, Ryynanen M. Altered expression of angiogenesis-related placental genes in pre-eclampsia associated with intrauterine growth restriction. Gynecol Endocrinol. 2007;23:351–5. doi: 10.1080/09513590701350291. [DOI] [PubMed] [Google Scholar]
- 61.Sitras V, Paulssen RH, Grønaas H, Leirvik J, Hanssen TA, Vårtun A, Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30:424–33. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 62.Meng T, Chen H, Sun M, Wang H, Zhao G, Wang X. Identification of differential gene expression profiles in placentas from preeclamptic pregnancies versus normal pregnancies by DNA microarrays. OMICS. 2012;16:301–11. doi: 10.1089/omi.2011.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levy R. The role of apoptosis in preeclampsia. Isr Med Assoc J. 2005;7:178–81. [PubMed] [Google Scholar]
- 64.Novakovic B, Saffery R. The ever growing complexity of placental epigenetics - role in adverse pregnancy outcomes and fetal programming. Placenta. 2012;33:959–70. doi: 10.1016/j.placenta.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Mayor-Lynn K, Toloubeydokhti T, Cruz AC, Chegini N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod Sci. 2011;18:46–56. doi: 10.1177/1933719110374115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crawford MA, Doyle W, Meadows N. Gender differences at birth and differences in fetal growth. Hum Reprod. 1987;2:517–20. doi: 10.1093/oxfordjournals.humrep.a136581. [DOI] [PubMed] [Google Scholar]
- 67.Bertino E, Coscia A, Boni L, Rossi C, Martano C, Giuliani F, Fabris C, Spada E, Zolin A, Milani S. Weight growth velocity of very low birth weight infants: role of gender, gestational age and major morbidities. Early Hum Dev. 2009;85:339–47. doi: 10.1016/j.earlhumdev.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 68.de Onis M, Siyam A, Borghi E, Onyango AW, Piwoz E, Garza C. Comparison of the World Health Organization growth velocity standards with existing US reference data. Pediatrics. 2011;128:e18–26. doi: 10.1542/peds.2010-2630. [DOI] [PubMed] [Google Scholar]
- 69.Brettell R, Yeh PS, Impey LW. Examination of the association between male gender and preterm delivery. Eur J Obstet Gynecol Reprod Biol. 2008;141:123–6. doi: 10.1016/j.ejogrb.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 70.Ingemarsson I. Gender aspects of preterm birth. BJOG. 2003;110(Suppl 20):34–8. doi: 10.1016/s1470-0328(03)00022-3. [DOI] [PubMed] [Google Scholar]
- 71.Elsmén E, Källén K, Marsál K, Hellström-Westas L. Fetal gender and gestational-age-related incidence of pre-eclampsia. Acta Obstet Gynecol Scand. 2006;85:1285–91. doi: 10.1080/00016340600578274. [DOI] [PubMed] [Google Scholar]
- 72.Murji A, Proctor LK, Paterson AD, Chitayat D, Weksberg R, Kingdom J. Male sex bias in placental dysfunction. Am J Med Genet A. 2012;158A:779–83. doi: 10.1002/ajmg.a.35250. [DOI] [PubMed] [Google Scholar]
- 73.Vatten LJ, Skjaerven R. Offspring sex and pregnancy outcome by length of gestation. Early Hum Dev. 2004;76:47–54. doi: 10.1016/j.earlhumdev.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 74.Clifton VL. Review: Sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31(Suppl):S33–9. doi: 10.1016/j.placenta.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 75.Hodyl NA, Wyper H, Osei-Kumah A, Scott N, Murphy VE, Gibson P, Smith R, Clifton VL. Sex-specific associations between cortisol and birth weight in pregnancies complicated by asthma are not due to differential glucocorticoid receptor expression. Thorax. 2010;65:677–83. doi: 10.1136/thx.2009.123091. [DOI] [PubMed] [Google Scholar]
- 76.Di Renzo GC, Rosati A, Sarti RD, Cruciani L, Cutuli AM. Does fetal sex affect pregnancy outcome? Gend Med. 2007;4:19–30. doi: 10.1016/S1550-8579(07)80004-0. [DOI] [PubMed] [Google Scholar]
- 77.Edwards A, Megens A, Peek M, Wallace EM. Sexual origins of placental dysfunction. Lancet. 2000;355:203–4. doi: 10.1016/S0140-6736(99)05061-8. [DOI] [PubMed] [Google Scholar]
- 78.Ghidini A, Salafia CM. Gender differences of placental dysfunction in severe prematurity. BJOG. 2005;112:140–4. doi: 10.1111/j.1471-0528.2004.00308.x. [DOI] [PubMed] [Google Scholar]
- 79.Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–40. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 80.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 81.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–22. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Graves JA. Review: Sex chromosome evolution and the expression of sex-specific genes in the placenta. Placenta. 2010;31(Suppl):S27–32. doi: 10.1016/j.placenta.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 83.Arnold AP, Lusis AJ. Understanding the sexome: measuring and reporting sex differences in gene systems. Endocrinology. 2012;153:2551–5. doi: 10.1210/en.2011-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33:791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- 86.Ropers HH, Wolff G, Hitzeroth HW. Preferential X inactivation in human placenta membranes: is the paternal X inactive in early embryonic development of female mammals? Hum Genet. 1978;43:265–73. doi: 10.1007/BF00278833. [DOI] [PubMed] [Google Scholar]
- 87.Migeon BR, Do TT. In search of non-random X inactivation: studies of fetal membranes heterozygous for glucose-6-phosphate dehydrogenase. Am J Hum Genet. 1979;31:581–5. [PMC free article] [PubMed] [Google Scholar]
- 88.Harrison KB, Warburton D. Preferential X-chromosome activity in human female placental tissues. Cytogenet Cell Genet. 1986;41:163–8. doi: 10.1159/000132221. [DOI] [PubMed] [Google Scholar]
- 89.Harrison KB. X-chromosome inactivation in the human cytotrophoblast. Cytogenet Cell Genet. 1989;52:37–41. doi: 10.1159/000132835. [DOI] [PubMed] [Google Scholar]
- 90.Mohandas TK, Passage MB, Williams JW, 3rd, Sparkes RS, Yen PH, Shapiro LJ. X-chromosome inactivation in cultured cells from human chorionic villi. Somat Cell Mol Genet. 1989;15:131–6. doi: 10.1007/BF01535073. [DOI] [PubMed] [Google Scholar]
- 91.Goto T, Wright E, Monk M. Paternal X-chromosome inactivation in human trophoblastic cells. Mol Hum Reprod. 1997;3:77–80. doi: 10.1093/molehr/3.1.77. [DOI] [PubMed] [Google Scholar]
- 92.Looijenga LH, Gillis AJ, Verkerk AJ, van Putten WL, Oosterhuis JW. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am J Hum Genet. 1999;64:1445–52. doi: 10.1086/302382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zeng SM, Yankowitz J. X-inactivation patterns in human embryonic and extra-embryonic tissues. Placenta. 2003;24:270–5. doi: 10.1053/plac.2002.0889. [DOI] [PubMed] [Google Scholar]
- 94.Willemsen R, Bontekoe CJ, Severijnen LA, Oostra BA. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet. 2002;110:601–5. doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- 95.Dhara SK, Benvenisty N. Gene trap as a tool for genome annotation and analysis of X chromosome inactivation in human embryonic stem cells. Nucleic Acids Res. 2004;32:3995–4002. doi: 10.1093/nar/gkh746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uehara S, Tamura M, Nata M, Ji G, Yaegashi N, Okamura K, Yajima A. X-chromosome inactivation in the human trophoblast of early pregnancy. J Hum Genet. 2000;45:119–26. doi: 10.1007/s100380050197. [DOI] [PubMed] [Google Scholar]
- 97.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12:815–26. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 98.Moreira de Mello JC, de Araújo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ge B, Gurd S, Gaudin T, Dore C, Lepage P, Harmsen E, Hudson TJ, Pastinen T. Survey of allelic expression using EST mining. Genome Res. 2005;15:1584–91. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang H, Elbein SC. Detection of allelic imbalance in gene expression using pyrosequencing. Methods Mol Biol. 2007;373:157–76. doi: 10.1385/1-59745-377-3:157. [DOI] [PubMed] [Google Scholar]
- 101.Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–6. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 102.Wang W, Feng L, Zhang H, Hachy S, Satohisa S, Laurent LC, Parast M, Zheng J, Chen DB. Preeclampsia up-regulates angiogenesis-associated microRNA (i.e., miR-17, -20a, and -20b) that target ephrin-B2 and EPHB4 in human placenta. J Clin Endocrinol Metab. 2012;97:E1051–9. doi: 10.1210/jc.2011-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.PengFei L YaLi H, Sha H, Liu L, JunLi Z, YaYi H. The expression of microRNA in placenca from severe preeclampsia patients. Chinese Journal of Practical Gynecology and Obstetrics. 2009;25:911–4. [Google Scholar]
- 104.Andraweera PH, Dekker GA, Roberts CT. The vascular endothelial growth factor family in adverse pregnancy outcomes. Hum Reprod Update. 2012;18:436–57. doi: 10.1093/humupd/dms011. [DOI] [PubMed] [Google Scholar]
- 105.Borzychowski AM, Sargent IL, Redman CW. Inflammation and pre-eclampsia. Semin Fetal Neonatal Med. 2006;11:309–16. doi: 10.1016/j.siny.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 106.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, et al. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–54. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011;39(Database issue):D301–8. doi: 10.1093/nar/gkq1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lázár L, Nagy B, Molvarec A, Szarka A, Rigó J., Jr. Role of hsa-miR-325 in the etiopathology of preeclampsia. Mol Med Rep. 2012;6:597–600. doi: 10.3892/mmr.2012.954. [DOI] [PubMed] [Google Scholar]
- 109.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37(Database issue):D155–8. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database issue):D163–9. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Glienke J, Sturz A, Menrad A, Thierauch KH. CRIM1 is involved in endothelial cell capillary formation in vitro and is expressed in blood vessels in vivo. Mech Dev. 2002;119:165–75. doi: 10.1016/S0925-4773(02)00355-6. [DOI] [PubMed] [Google Scholar]
- 112.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–84. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 113.Pringle KG, Kind KL, Sferruzzi-Perri AN, Thompson JG, Roberts CT. Beyond oxygen: complex regulation and activity of hypoxia inducible factors in pregnancy. Hum Reprod Update. 2010;16:415–31. doi: 10.1093/humupd/dmp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 115.Curtin JF, Cotter TG. Live and let die: regulatory mechanisms in Fas-mediated apoptosis. Cell Signal. 2003;15:983–92. doi: 10.1016/S0898-6568(03)00093-7. [DOI] [PubMed] [Google Scholar]
- 116.Poehlmann TG, Fitzgerald JS, Meissner A, Wengenmayer T, Schleussner E, Friedrich K, Markert UR. Trophoblast invasion: tuning through LIF, signalling via Stat3. Placenta. 2005;26(Suppl A):S37–41. doi: 10.1016/j.placenta.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 117.Thomadaki H, Scorilas A. BCL2 family of apoptosis-related genes: functions and clinical implications in cancer. Crit Rev Clin Lab Sci. 2006;43:1–67. doi: 10.1080/10408360500295626. [DOI] [PubMed] [Google Scholar]
- 118.Chan KT, Creed SJ, Bear JE. Unraveling the enigma: progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 2011;21:481–8. doi: 10.1016/j.tcb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Preston GA, Lyon TT, Yin Y, Lang JE, Solomon G, Annab L, Srinivasan DG, Alcorta DA, Barrett JC. Induction of apoptosis by c-Fos protein. Mol Cell Biol. 1996;16:211–8. doi: 10.1128/mcb.16.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Urbich C, Knau A, Fichtlscherer S, Walter DH, Brühl T, Potente M, Hofmann WK, de Vos S, Zeiher AM, Dimmeler S. FOXO-dependent expression of the proapoptotic protein Bim: pivotal role for apoptosis signaling in endothelial progenitor cells. FASEB J. 2005;19:974–6. doi: 10.1096/fj.04-2727fje. [DOI] [PubMed] [Google Scholar]
- 121.Gonzalvez F, Ashkenazi A. New insights into apoptosis signaling by Apo2L/TRAIL. Oncogene. 2010;29:4752–65. doi: 10.1038/onc.2010.221. [DOI] [PubMed] [Google Scholar]
- 122.Halperin R, Peller S, Sandbank J, Bukovsky I, Schneider D. Expression of the p53 gene and apoptosis in gestational trophoblastic disease. Placenta. 2000;21:58–62. doi: 10.1053/plac.1999.0442. [DOI] [PubMed] [Google Scholar]
- 123.Dye JF, Jablenska R, Donnelly JL, Lawrence L, Leach L, Clark P, Firth JA. Phenotype of the endothelium in the human term placenta. Placenta. 2001;22:32–43. doi: 10.1053/plac.2000.0579. [DOI] [PubMed] [Google Scholar]
- 124.Ullah Z, Kohn MJ, Yagi R, Vassilev LT, DePamphilis ML. Differentiation of trophoblast stem cells into giant cells is triggered by p57/Kip2 inhibition of CDK1 activity. Genes Dev. 2008;22:3024–36. doi: 10.1101/gad.1718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Prendergast GC. Actin’ up: RhoB in cancer and apoptosis. Nat Rev Cancer. 2001;1:162–8. doi: 10.1038/35101096. [DOI] [PubMed] [Google Scholar]
- 126.Akira S, Hirano T, Taga T, Kishimoto T. Biology of multifunctional cytokines: IL 6 and related molecules (IL 1 and TNF) FASEB J. 1990;4:2860–7. [PubMed] [Google Scholar]
- 127.Yoshitomi T, Kawakami K, Enokida H, Chiyomaru T, Kagara I, Tatarano S, Yoshino H, Arimura H, Nishiyama K, Seki N, et al. Restoration of miR-517a expression induces cell apoptosis in bladder cancer cell lines. Oncol Rep. 2011;25:1661–8. doi: 10.3892/or.2011.1253. [DOI] [PubMed] [Google Scholar]
- 128.Montenegro D, Romero R, Kim SS, Tarca AL, Draghici S, Kusanovic JP, Kim JS, Lee DC, Erez O, Gotsch F, et al. Expression patterns of microRNAs in the chorioamniotic membranes: a role for microRNAs in human pregnancy and parturition. J Pathol. 2009;217:113–21. doi: 10.1002/path.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kolwankar D, Glover DD, Ware JA, Tracy TS. Expression and function of ABCB1 and ABCG2 in human placental tissue. Drug Metab Dispos. 2005;33:524–9. doi: 10.1124/dmd.104.002261. [DOI] [PubMed] [Google Scholar]


