Abstract
Background
The relationship between Fas -1377 G/A polymorphism and cancer susceptibility has been implicated in accumulating data. However, the data presented inconsistent results. This study was devised to investigate the association of Fas -1377 G/A polymorphism and cancer susceptibility in a large number of participants.
Methods
The databases of PubMed, Embase, and Web of Science were searched and a total of 27 case-control studies including 13,355 cases and 16,078 controls were included in this meta-analysis. Pooled odds ratios (ORs) with 95% confidence intervals (CIs) were calculated using the fixed-effects model. Statistical analyses were performed by using Stata software.
Results
The results suggested that Fas -1377 G/A polymorphism was overall associated with cancer susceptibility (additive model: OR, 1.16, 95%CI = 1.06–1.27, P heterogeneity = 0.381; recessive model: OR, 1.19, 95%CI = 1.10–1.29, P heterogeneity = 0.137). In the subgroup analysis by cancer type, significantly increased risk was observed in breast cancer (additive model: OR, 1.24, 95%CI = 1.04–1.58, P heterogeneity = 0.614; recessive model: OR, 1.24, 95%CI = 1.02–1.51, P heterogeneity = 0.349) and lung cancer (recessive model: OR, 1.25, 95%CI = 1.04–1.49, P heterogeneity = 0.090). Similarly, elevated cancer risk associated with Fas -1377 G/A polymorphism was revealed in Asians.
Conclusions
The combined results suggest that Fas -1377 G/A polymorphism might modulate cancer susceptibility in an Asian-specific manner.
Introduction
Cancer arises as a result of complex interactions between genetic and environmental factors and has become a major public health problem all over the world [1]–[5]. In recent years, many individual studies have set out to determine whether there is an association between genetic polymorphisms and cancer susceptibility, such as Fas -1377 G/A polymorphism and cancer susceptibility. However, these studies showed conflicting results that failed to provide compelling evidence for cancer susceptibility [6]–[9].
Apoptosis is a process of programmed cell death regulated by genes. Inappropriate regulation of apoptosis could lead to a broad range of human disorders including cancer [10]–[13]. Fas is a member of the tumor necrosis factor receptor superfamily and regulates apoptotic activities in activated lymphocytes [14]. Located on chromosome 10q24.1, Fas is highly polymorphic [15]. A functional polymorphism with a G to A substitution at -1377 position within the Fas gene has been extensively explored in the field of cancer. But there is no decisive conclusion of the role of this polymorphism in cancer development [6], [7]. In addition, several studies have been subsequently published since a previous meta-analysis was reported in 2009 [47]. In view of this, we decided to carry out a meta-analysis including 27 eligible studies published to date to systematically and comprehensively estimate the association between Fas -1377 G/A polymorphism and susceptibility to cancer.
Materials and Methods
Literature Search Strategy
The databases of PubMed, Embase, and Web of Science were searched (the last search was updated in May 2013) to identify all relevant publications on the association between Fas -1377 G/A polymorphism and cancer risk. The following search terms and their synonyms were used: “Fas”, “1377 G/A” or “CD95” or “rs2234767”, “polymorphism” or “variation”, and “cancer”. We also manually searched the reference lists of all eligible studies and review articles to obtain additional usable data that can be included in the current meta-analysis.
Inclusion Criteria and Exclusion Criteria
We selected eligible studies according to the following criteria: (1) the study must have a case-control design; (2) the association between Fas -1377 G/A polymorphisms and cancer risk must be examined; (3) adequate genotyping data must be contained such that odds ratios (ORs) with 95% confidence intervals (CIs) could be calculated; (4) the study had to be published in English and use human subjects. Exclusion criteria were: (1) insufficient information on the distribution of Fas -1377 genotypes; (2) case-only studies; (3) duplicated publications. If a study was subsequently updated, we selected the study with the largest sample size. Two investigators independently reviewed all studies to examine whether they fulfilled the inclusion criteria.
Data Extraction
Two independent investigators (Peiliang Geng and Jianjun Li) extracted the original data according to the inclusion criteria and exclusion criteria to ensure the accuracy of the retrieved information. The data extracted from each eligible study included the first author's name, year of publication, cancer type, ethnicity, source of controls, method adopted for genotyping, number of cases and controls and genotype frequencies. Disputes were settled by consulting the third person (Houjie Liang).
Statistical Analysis
Crude ORs with 95% CIs were calculated to evaluate the strength of the association between Fas -1377 G/A polymorphism and cancer risk. The pooled ORs were performed for additive model, dominant model and recessive model. Subgroup analysis by cancer type, ethnicity and source of control were also conducted to further assess if the Fas -1377 polymorphism was associated with cancer susceptibility in each subgroup. Heterogeneity assumption was evaluated by the chi-square based Q-test and I2 statistics [16], [17], P>0.05 for the Q test or I2<50% suggested a lack of heterogeneity. In this situation, the OR of each study was calculated by the fixed-effects model (the Mantel-Haenszel method) [18]. If P<0.05 or I2>50%, the random-effects model (the DerSimonian and Laird method) was used [19]. Sensitivity analysis was performed by removing one study at a time to ensure that our findings were not driven by any single study. The evaluation of potential publication bias was performed using the Begg's funnel plots and Egger's test [20]. Hardy-Weinberg equilibrium (HWE) of the control groups was tested by the χ2 test for goodness of fitness. All statistical analyses were performed by STATA version 12.0 (Stata Corporation, College Station, TX, USA). A level of P<0.05 was accepted as statistically significant.
Results
Study Characteristics
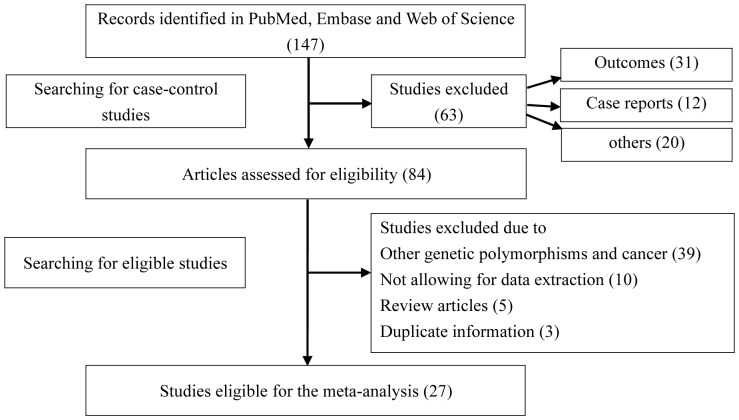
We initially identified 147 potentially relevant studies, of which 27 met the pre-described inclusion criteria and were included in the meta-analysis of the association between Fas -1377G/A polymorphism and cancer risk (Figure 1). Characteristics of all eligible case-control studies for the relationship of Fas -1377G/A polymorphism with cancer risk are summarized in Table 1. Of the twenty-seven studies included, an array of cancers including AML [6], [7], breast cancer [21]–[25], cervical cancer [26]–[28], lung cancer [8], [9], [29], [30], gastric cancer [31], [32], melanoma [33], [34], oral cancer [35], [36], and several other cancers [37]–[43] were involved. The subgroup analysis was carried out by cancer type, ethnicity and source of control, respectively. Genotype frequencies were available in all of the 27 studies.
Figure 1. Flow diagram of study identification.
Table 1. Main characteristics of the 27 eligible studies.
| Authors | Year | Source of control | Ethnicity | Cancer type | Genotyping method | Case | Control | HWE | ||||||||||
| Sample size | GG | GA | AA | G | A | Sample size | GG | GA | AA | G | A | |||||||
| Sibley | 2003 | Population | European | AML | PCR–RFLP | 471 | 319 | 136 | 16 | 774 | 168 | 931 | 726 | 186 | 19 | 1638 | 224 | 0.087 |
| Sun | 2004 | Population | Asian | Esophageal | PCR–RFLP | 588 | 250 | 234 | 104 | 734 | 442 | 648 | 273 | 306 | 69 | 852 | 444 | 0.218 |
| Kripple | 2004 | Population | European | Breast | TaqMan | 499 | 371 | 120 | 8 | 862 | 136 | 497 | 401 | 92 | 4 | 894 | 100 | 0.610 |
| Lai | 2005 | Hospital | Asian | Cervical | TaqMan | 318 | 127 | 138 | 53 | 392 | 244 | 318 | 99 | 165 | 54 | 3633 | 273 | 0.293 |
| Sun | 2005 | Population | Asian | Cervical | PCR–RFLP | 314 | 144 | 144 | 26 | 432 | 196 | 615 | 282 | 277 | 56 | 841 | 389 | 0.304 |
| Zhang | 2005 | Population | Asian | Lung | PCR–RFLP | 1000 | 413 | 433 | 154 | 1259 | 741 | 1270 | 539 | 601 | 130 | 1679 | 861 | 0.046 |
| Li | 2006 | Hospital | Asian | Bladder | PCR–RFLP | 216 | 66 | 104 | 46 | 236 | 196 | 252 | 81 | 124 | 47 | 286 | 218 | 0.970 |
| Park | 2006 | Hospital | Asian | Lung | PCR–RFLP | 582 | 187 | 300 | 95 | 674 | 490 | 582 | 172 | 313 | 97 | 657 | 507 | 0.024 |
| Li | 2006 | Hospital | European | Melanoma | PCR–RFLP | 602 | 486 | 107 | 9 | 1079 | 125 | 603 | 459 | 134 | 10 | 1052 | 154 | 0.951 |
| Zhang | 2006 | Hospital | European | SCCHN | PCR–RFLP | 721 | 562 | 142 | 17 | 1266 | 176 | 1234 | 957 | 264 | 13 | 2178 | 290 | 0.268 |
| Gormas | 2007 | Population | European | Lung | PCR | 94 | 21 | 73 | 0 | 115 | 73 | 50 | 13 | 37 | 0 | 63 | 37 | >0.05 |
| Zhang | 2007 | Population | Asian | Breast | PCR–RFLP | 840 | 293 | 418 | 129 | 1004 | 676 | 839 | 345 | 382 | 112 | 1072 | 606 | 0.700 |
| Crew | 2007 | Population | European | Breast | TaqMan | 1057 | 809 | 225 | 23 | 1843 | 271 | 1106 | 847 | 234 | 25 | 1928 | 284 | 0.069 |
| Koshkina | 2007 | Hospital | European | Osteosarcoma | PCR–RFLP | 123 | 99 | 22 | 2 | 220 | 26 | 510 | 400 | 100 | 10 | 900 | 120 | 0.210 |
| Zhang | 2007 | Population | European | Melanoma | PCR–RFLP | 229 | 183 | 41 | 5 | 407 | 51 | 351 | 269 | 70 | 12 | 608 | 94 | 0.009 |
| Ter-Minassi | 2008 | Hospital | European | Lung | TaqMan | 2174 | 1645 | 492 | 37 | 3782 | 566 | 1497 | 1138 | 336 | 23 | 2612 | 382 | 0.751 |
| Kang | 2008 | Population | Asian | Cervical | PCR–RFLP | 154 | 54 | 69 | 31 | 177 | 131 | 168 | 56 | 82 | 20 | 194 | 142 | 0.998 |
| Yang | 2008 | Population | Asian | Pancreatic | PCR–RFLP | 397 | 186 | 169 | 42 | 541 | 253 | 907 | 420 | 376 | 111 | 1216 | 598 | 0.062 |
| Zhou | 2009 | Population | Asian | Gastric | PCR–RFLP | 262 | 124 | 117 | 21 | 365 | 159 | 524 | 225 | 251 | 48 | 701 | 347 | 0.062 |
| Cao | 2010 | Population | Asian | Nasopharyngeal | PCR–RFLP | 576 | 141 | 264 | 171 | 546 | 606 | 608 | 172 | 303 | 133 | 647 | 569 | 0.984 |
| Kim | 2010 | Population | Asian | AML | PCR | 592 | 195 | 303 | 94 | 693 | 491 | 858 | 286 | 427 | 145 | 999 | 717 | 0.501 |
| Wang | 2010 | Population | Asian | Oral | PCR–RFLP | 431 | 146 | 208 | 77 | 500 | 362 | 333 | 115 | 165 | 53 | 395 | 271 | 0.628 |
| Zhu | 2010 | Hospital | Asian | Renal | PCR–RFLP | 353 | 124 | 173 | 56 | 421 | 285 | 365 | 161 | 161 | 43 | 483 | 247 | 0.777 |
| Kupcinskas | 2011 | Hospital | European | Gastric | TaqMan | 114 | 95 | 18 | 1 | 208 | 20 | 238 | 197 | 40 | 1 | 434 | 42 | 0.492 |
| Wang | 2012 | Hospital | Asian | Breast | PCR–RFLP | 375 | 138 | 171 | 66 | 447 | 303 | 496 | 197 | 246 | 53 | 640 | 352 | 0.064 |
| Hashemi | 2013 | Population | Asian | Breast | PCR | 134 | 20 | 106 | 8 | 146 | 122 | 152 | 26 | 115 | 11 | 167 | 137 | >0.05 |
| Karimi | 2013 | Population | Asian | Oral | PCR–RFLP | 139 | 88 | 42 | 9 | 218 | 60 | 126 | 84 | 30 | 12 | 198 | 54 | 0.001 |
PCR: polymerase chain reaction; PCR-RFLP: PCR-restriction fragment length polymorphism; TaqMan: TaqManSNP; AML: acute myeloid leukemia; SCCHN: squamous cell carcinoma of the head and neck; HWE: Hardy-Weinberg equilibrium.
Meta-analysis
Major results of the meta-analysis are presented in Table 2. No significant between-study heterogeneity was detected across studies and thus we selected the fix-effects model to summarize the ORs. Overall, we found a significant association between Fas -1377G/A polymorphism and cancer risk under the additive model (OR, 1.16, 95%CI = 1.06–1.27, P heterogeneity = 0.381), but the association was more pronounced under the recessive model (OR, 1.19, 95%CI = 1.10–1.29, P heterogeneity = 0.137) (Figure 2, 3). In the subgroup analysis by cancer type, significantly increased risk was observed in breast cancer (additive model: OR, 1.24, 95%CI = 1.04–1.58, P heterogeneity = 0.614; recessive model: OR, 1.24, 95%CI = 1.02–1.51, P heterogeneity = 0.349) and lung cancer (recessive model: OR, 1.25, 95%CI = 1.04–1.49, P heterogeneity = 0.090).
Table 2. Main results of the pooled data in the meta-analysis.
| Additive model | Dominant model | Recessive model | |||||||
| Subtypes | OR (95% CI) | Heterogeneity | OR (95% CI) | Heterogeneity | OR (95% CI) | Heterogeneity | |||
| Ph | I2 (%) | Ph | I2 (%) | Ph | I2 (%) | ||||
| Cancer type | |||||||||
| AML | 1.07 (0.82, 1.14) | 0.080 | 67.4 | 1.14 (0.99, 1.30) | 0.011 | 84.6 | 1.02 (0.79, 1.32) | 0.125 | 57.6 |
| Breast | 1.28 (1.04, 1.58) | 0.614 | 0 | 1.08 (0.99, 1.19) | 0.602 | 0 | 1.24 (1.02, 1.51) | 0.349 | 10.0 |
| Cervical | 0.96 (0.72, 1.29) | 0.432 | 0 | 0.96 (0.83, 1.12) | 0.590 | 0 | 1.08 (0.82, 1.42) | 0.245 | 28.9 |
| Lung | 1.19 (0.98, 1.43) | 0.163 | 45.0 | 1.01 (0.92, 1.10) | 0.960 | 0 | 1.25 (1.04, 1.49) | 0.090 | 58.4 |
| Melanoma | 0.74 (0.37, 1.47) | 0.659 | 0 | 0.82 (0.66, 1.03) | 0.795 | 0 | 0.77 (0.39, 1.53) | 0.627 | 0 |
| Gastric | 0.85 (0.50, 1.46) | 0.526 | 0 | 0.93 (0.74, 1.17) | 0.885 | 0 | 0.90 (0.53, 1.52) | 0.547 | 0 |
| Oral | 1.03 (0.71, 1.48) | 0.444 | 0 | 1.03 (0.84, 1.26) | 0.749 | 0 | 1.04 (0.74, 1.47) | 0.313 | 1.9 |
| Other | 1.26 (1.08, 1.47) | 0.301 | 16.9 | 1.02 (0.94, 1.11) | 0.940 | 0 | 1.31 (1.13, 1.52) | 0.139 | 38.0 |
| Ethnicity | |||||||||
| European | 1.23 (0.94, 1.60) | 0.419 | 1.8 | 1.04 (0.96, 1.13) | 0.069 | 43.4 | 1.22 (0.93, 1.58) | 0.519 | 0 |
| Asian | 1.15 (1.05, 1.26) | 0.318 | 11.6 | 1.02 (0.97, 1.07) | 0.994 | 0 | 1.19 (1.09, 1.30) | 0.060 | 37.4 |
| Source of control | |||||||||
| Population | 1.16 (1.05, 1.29) | 0.383 | 6.1 | 1.05 (0.99, 1.10) | 0.587 | 0 | 1.19 (1.08, 1.32) | 0.073 | 36.4 |
| Hospital | 1.15 (0.97, 1.35) | 0.311 | 14.3 | 0.99 (0.92, 1.06) | 0.783 | 0 | 1.19 (1.02, 1.39) | 0.419 | 2.2 |
| Total | 1.16 (1.06, 1.27) | 0.381 | 5.7 | 1.02 (0.98, 1.07) | 0.722 | 0 | 1.19 (1.10, 1.29) | 0.137 | 23.7 |
| Total* | 1.16 (1.05, 1.28) | 0.484 | 0 | 1.03 (0.98, 1.08) | 0.583 | 0 | 1.19 (1.08, 1.30) | 0.249 | 15.8 |
AML: acute myeloid leukemia; Ph: p-value of heterogeneity test; CI: confidence interval; OR: odds ratio;
*meta-analysis results after removing the studies deviating from Hardy-Weinberg equilibrium (HWE).
Figure 2. Meta-analysis for the association between Fas -1377 G/A polymorphism and cancer risk by fixed-effects model (additive model; stratified by ethnicity).
Figure 3. Meta-analysis for the association between Fas -1377 G/A polymorphism and cancer risk by fixed-effects model (recessive model; stratified by ethnicity).
Subgroup analysis by ethnicity also provided evidence for an association in Asian populations (additive model: OR, 1.15, 95%CI = 1.05–1.26, P heterogeneity = 0.318; recessive model: OR, 1.19, 95%CI = 1.09–1.30, P heterogeneity = 0.060), but not in European populations. In the succeeding analysis by source of control, an elevated cancer risk was observed in both population-based and hospital-based studies (Table 2).
Sensitivity Analysis
We performed a leave-one-out sensitivity analysis by omitting one study at a time to assess the stability of the combined results. The results suggested that our findings were not substantially affected by any single study (data not shown).
Publication Bias
Begg's funnel plot and Egger's test were performed to detect publication bias. No statistically significant evidence of publication bias was revealed (Begg's test: P = 0.826; Egger's test: P = 0.721, additive model) (Figure 4).
Figure 4. Publication bias test for all included studies (additive model).
Discussion
The human Fas gene mapped on chromosome 10q24.1 consists of nine exons and eight introns [15]. -1377 G/A polymorphism, located in the promoter region of the Fas gene, has been investigated in a variety of previous studies looking at cancer risk [8], [21], [22], [26]. However, these findings remain controversial rather than conclusive. This might be attributed to the different ethnicities, distinct study design, and sample inadequacy in each of the published studies. But meta-analysis could avoid the shortcomings and convincingly estimate the genetic association through including all relevant studies.
In our meta-analysis, we observed Fas -1377G/A polymorphism was overall associated with cancer susceptibility under the additive model and the recessive model. Several published meta-analyses observed the same finding that Fas -1377 G/A polymorphism was associated with cancer risk as well as some common diseases, such as autoimmune rheumatic diseases, systemic lupus erythematosus [44]–[47]. The detection power of the four meta-analyses, however, may be limited largely because of sample insufficiency: 4 publications (996 cases and 1,160 controls) were included by Lu et al. [44], 5 (615 cases and 622 controls) by Lee et al. [45], 3 (444 cases and 442 controls) by Xiang et al. [46] and 17 (10,564 cases and 12,075 controls) by Qiu et al [47]. Our meta-analysis nevertheless summarized data from 27 studies composed of 13,355 cases and 16,078 controls. It should be noted that study size is obviously important to know the proportion of false positive findings of meta-analysis. Therefore, the relatively larger sample may assure the statistical power of our study. Deviation from HWE was observed in several studies, which may result from misclassification of genotypes, because multiple genotyping methods were used across studies. When we reanalyzed the studies without departure form HWE, the general results were not significantly altered, suggesting our findings are robust and convincing.
Apart from the comparison among all subjects, we also performed stratification analysis by cancer type. We found that Fas -1377 G/A polymorphism increased the risk of some cancers, such as breast cancer and lung cancer. Our findings were consistent with those revealed in the previous studies [6], [9], [21], [26], but contradictory discoveries that there was no association between Fas -1377 G/A polymorphism and lung cancer were also suggested in two studies [7], [8]. The underlying etiology mechanisms differ substantially across cancers, and the role of Fas -1377 G/A polymorphism in various caners requires to be identified by future larger studies.
In addition, in the subgroup analysis by ethnicity, Fas -1377 G/A polymorphism was found to increase cancer risk in Asian populations under several genetic models, such as the recessive model and the additive model. However, this association was obtained in European populations. There is obvious disparity in genotype frequencies between the two ethnic groups (GA: 21.3% vs 47.7%; AA: 1.5% vs 13.2%). It is known that different genetic background donates a series of differences between ethnic groups, for instance, frequency of exposure to cancer-causing agents and diverse lifestyles, which are important components in the process of cancer progression.
In the final subgroup analysis by control source, we observed significant association in both population-based and hospital-based studies. However, investigators demonstrated a different discovery of significantly increased cancer risk associated with Fas -1377 AA genotype among studies based on population-based controls, but not among studies of hospital-based controls [47]. Control subjects in some hospital-based studies may be poorly-defined reference populations and failed to well represent the general population, leading to some biases in the analysis, but the relatively small sample may be responsible for a large part of the inconsistency.
Some limitations in our meta-analysis need to be addressed. To begin with, in the subgroup analysis by cancer type, significant association was not observed in several cancers, such as gastric cancer, melanoma cancer and oral cancer. Fas -1377 G/A polymorphism and these cancers may be positively correlated, which may be masked due to the small sample size in this study. Furthermore, there existed heterogeneity between studies. The reason might be attributable to the different genetic backgrounds of the subjects and study design in each of the included studies. Finally, this meta-analysis was carried out among Asian and European populations, thus the results can not be applicable in other ethnicities.
In summary, the meta-analysis provided evidence that Fas -1377 G/A polymorphism might be associated with an increased cancer risk. Significant association was also found in subgroup analyses by cancer type, ethnicity and source of control. In future, studies with a larger sample size and multiple ethnic groups are required to further validate the relationship between Fas -1377 G/A polymorphism and cancer susceptibility.
Funding Statement
This work was supported in part by grant number 30973430 from the National Natural Science Foundation of China (to HJ.L). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bredberg A (2011) Cancer: more of polygenic disease and less of multiple mutations? A quantitative viewpoint. Cancer 117: 440–445. [DOI] [PubMed] [Google Scholar]
- 2. Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- 3. Pharoah PD, Dunning AM, Ponder BA, Easton DF (2004) Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer 4: 850–860. [DOI] [PubMed] [Google Scholar]
- 4. Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010. CA Cancer J Clin 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 5. Siegel R, Ward E, Brawley O, Jemal A (2011) Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 61: 212–236. [DOI] [PubMed] [Google Scholar]
- 6. Sibley K, Rollinson S, Allan JM, Smith AG, Law GR, et al. (2003) Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res 63: 4327–4330. [PubMed] [Google Scholar]
- 7. Kim HJ, Jin XM, Kim HN, Lee IK, Park KS, et al. (2010) Fas and FasL polymorphisms are not associated with acute myeloid leukemia risk in Koreans. DNA Cell Biol 29: 619–624. [DOI] [PubMed] [Google Scholar]
- 8. Park SH, Choi JE, Kim EJ, Jang JS, Lee WK, et al. (2006) Polymorphisms in the FAS and FASL genes and risk of lung cancer in a Korean population. Lung Cancer 54: 303–308. [DOI] [PubMed] [Google Scholar]
- 9. Gormus U, Ergen A, Yaylim-Eraltan I, Yilmaz H, Turna A, et al. (2007) Fas-1377 A/G polymorphism in lung cancer. In Vivo 21: 663–666. [PubMed] [Google Scholar]
- 10. Thompson CB (1995) Apoptosis in the pathogenesis and treatment of disease. Science 267: 1456–1462. [DOI] [PubMed] [Google Scholar]
- 11. Raff M (1998) Cell suicide for beginners. Nature 396: 119–122. [DOI] [PubMed] [Google Scholar]
- 12. Shivapurkar N, Reddy J, Chaudhary PM, Gazdar AF, et al. (2003) Apoptosis and lung cancer: a review. J Cell Biochem 88: 885–898. [DOI] [PubMed] [Google Scholar]
- 13. Hajra KM, Liu JR (2004) Apoptosome dysfunction in human cancer. Apoptosis 9: 691–704. [DOI] [PubMed] [Google Scholar]
- 14. Li LH, Li WX, Wu O, Zhang GQ, Pan HF, et al. (2009) Fas expression on peripheral blood lymphocytes in systemic lupus erythematosus: relation to the organ damage and lymphocytes apoptosis. Mol Biol Rep 36: 2047–2052. [DOI] [PubMed] [Google Scholar]
- 15. Inazawa J, Itoh N, Abe T, Nagata S (1992) Assignment of the human Fas antigen gene (Fas) to 10q24.1. Genomics 14: 821–822. [DOI] [PubMed] [Google Scholar]
- 16. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 17. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327 p. 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 19. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 20. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crew KD, Gammon MD, Terry MB, Zhang FF, Agrawal M, et al. (2007) Genetic polymorphisms in the apoptosis-associated genes FAS and FASL and breast cancer risk. Carcinogenesis 28: 2548–2551. [DOI] [PubMed] [Google Scholar]
- 22. Koshkina NV, Kleinerman ES, Li G, Zhao CC, Wei Q, et al. (2007) Exploratory analysis of Fas gene polymorphisms in pediatric osteosarcoma patients. J Pediatr Hematol Oncol 29: 815–821. [DOI] [PubMed] [Google Scholar]
- 23. Zhang B, Sun T, Xue L, Han X, Zhang B, et al. (2007) Functional polymorphisms in FAS and FASL contribute to increased apoptosis of tumor infiltration lymphocytes and risk of breast cancer. Carcinogenesis 28: 1067–1073. [DOI] [PubMed] [Google Scholar]
- 24. Wang W, Zheng Z, Yu W, Lin H, Cui B, et al. (2012) Polymorphisms of the FAS and FASL genes and risk of breast cancer. Oncol Lett 3: 625–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hashemi M, Fazaeli A, Ghavami S, Eskandari-Nasab E, Arbabi F, et al. (2013) Functional polymorphisms of FAS and FASL gene and risk of breast cancer - pilot study of 134 cases. PLoS One 8: e53075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang S, Dong SM, Seo SS, Kim JW, Park SY (2008) FAS -1377 G/A polymorphism and the risk of lymph node metastasis in cervical cancer. Cancer Genet Cytogenet 180: 1–5. [DOI] [PubMed] [Google Scholar]
- 27. Lai HC, Lin WY, Lin YW, Chang CC, Yu MH, et al. (2005) Genetic polymorphisms of FAS and FASL (CD95/CD95L) genes in cervical carcinogenesis: An analysis of haplotype and gene-gene interaction. Gynecol Oncol 99: 113–118. [DOI] [PubMed] [Google Scholar]
- 28. Sun T, Zhou Y, Li H, Han X, Shi Y, et al. (2005) FASL -844C polymorphism is associated with increased activation-induced T cell death and risk of cervical cancer. J Exp Med 202: 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ter-Minassian M, Zhai R, Asomaning K, Su L, Zhou W, et al. (2008) Apoptosis gene polymorphisms, age, smoking and the risk of non-small cell lung cancer. Carcinogenesis 29: 2147–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X, Miao X, Sun T, Tan W, Qu S, et al. (2005) Functional polymorphisms in cell death pathway genes FAS and FASL contribute to risk of lung cancer. J Med Genet 42: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou RM, Wang N, Chen ZF, Duan YN, Sun DL, et al. (2010) Polymorphisms in promoter region of FAS and FASL gene and risk of cardia gastric adenocarcinoma. J Gastroenterol Hepatol 25: 555–561. [DOI] [PubMed] [Google Scholar]
- 32. Kupcinskas J, Wex T, Bornschein J, Selgrad M, Leja M, et al. (2011) Lack of association between gene polymorphisms of Angiotensin converting enzyme, Nod-like receptor 1, Toll-like receptor 4, FAS/FASL and the presence of Helicobacter pylori-induced premalignant gastric lesions and gastric cancer in Caucasians. BMC Med Genet 12: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li C, Larson D, Zhang Z, Liu Z, Strom SS, et al. (2006) Polymorphisms of the FAS and FAS ligand genes associated with risk of cutaneous malignant melanoma. Pharmacogenet Genomics 16: 253–263. [DOI] [PubMed] [Google Scholar]
- 34. Zhang H, Sun XF, Synnerstad I, Rosdahl I (2007) Importance of FAS-1377, FAS-670, and FASL-844 polymorphisms in tumor onset, progression, and pigment phenotypes of Swedish patients with melanoma: a case-control analysis. Cancer J 13: 233–237. [DOI] [PubMed] [Google Scholar]
- 35. Wang LH, Ting SC, Chen CH, Tsai CC, Lung O, et al. (2010) Polymorphisms in the apoptosis-associated genes FAS and FASL and risk of oral cancer and malignant potential of oral premalignant lesions in a Taiwanese population. J Oral Pathol Med 2010 39(2): 155–161. [DOI] [PubMed] [Google Scholar]
- 36. Karimi MY, Kapoor V, Sharma SC, Das SN (2013) Genetic polymorphisms in FAS (CD95) and FAS ligand (CD178) promoters and risk of tobacco-related oral carcinoma: gene-gene interactions in high-risk Indians. Cancer Invest 31: 1–6. [DOI] [PubMed] [Google Scholar]
- 37. Krippl P, Langsenlehner U, Renner W, Köppel H, Samonigg H (2004) Re: Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96: 1478–1479. [DOI] [PubMed] [Google Scholar]
- 38. Li C, Wu W, Liu J, Qian L, Li A, et al. (2006) Functional polymorphisms in the promoter regions of the FAS and FAS ligand genes and risk of bladder cancer in south China: a case-control analysis. Pharmacogenet Genomics 16: 245–251. [DOI] [PubMed] [Google Scholar]
- 39. Sun T, Miao X, Zhang X, Tan W, Xiong P, et al. (2004) Polymorphisms of death pathway genes FAS and FASL in esophageal squamous-cell carcinoma. J Natl Cancer Inst 96: 1030–1036. [DOI] [PubMed] [Google Scholar]
- 40. Yang M, Sun T, Wang L, Yu D, Zhang X, et al. (2008) Functional variants in cell death pathway genes and risk of pancreatic cancer. Clin Cancer Res 14: 3230–3236. [DOI] [PubMed] [Google Scholar]
- 41. Zhang Z, Wang LE, Sturgis EM, El-Naggar AK, Hong WK, et al. (2006) Polymorphisms of FAS and FAS ligand genes involved in the death pathway and risk and progression of squamous cell carcinoma of the head and neck. Clin Cancer Res 12: 5596–602. [DOI] [PubMed] [Google Scholar]
- 42. Zhu J, Qin C, Wang M, Yan F, Ju X, et al. (2010) Functional polymorphisms in cell death pathway genes and risk of renal cell carcinoma. Mol Carcinog 49: 810–817. [DOI] [PubMed] [Google Scholar]
- 43. Cao Y, Miao XP, Huang MY, Deng L, Lin DX, et al. (2010) Polymorphisms of death pathway genes FAS and FASL and risk of nasopharyngeal carcinoma. Mol Carcinog 49: 944–950. [DOI] [PubMed] [Google Scholar]
- 44. Lu MM, Ye QL, Feng CC, Yang J, Zhang T, et al. (2012) Association of FAS gene polymorphisms with systemic lupus erythematosus: A case-control study and meta-analysis. Exp Ther Med 4: 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee YH, Bae SC, Choi SJ, Ji JD, Song GG (2012) Associations between the FAS -670 A/G and -1,377 G/A polymorphisms and susceptibility to autoimmune rheumatic diseases: a meta-analysis. Mol Biol Rep 39: 10671–9. [DOI] [PubMed] [Google Scholar]
- 46. Xiang N, Li XM, Wang GS, Tao JH, Li XP (2013) Association of Fas gene polymorphisms with systemic lupus erythematosus: a meta-analysis. Mol Biol Rep 40: 407–415. [DOI] [PubMed] [Google Scholar]
- 47. Qiu LX, Shi J, Yuan H, Jiang X, Xue K, et al. (2009) FAS -1,377 G/A polymorphism is associated with cancer susceptibility: evidence from 10,564 cases and 12,075 controls. Hum Genet 125: 431–435. [DOI] [PubMed] [Google Scholar]