Abstract
A general model of neural development is derived to fit 18 mammalian species, including humans, macaques, several rodent species, and six metatherian (marsupial) mammals. The goal of this work is to describe heterochronic changes in brain evolution within its basic developmental allometry, and provide an empirical basis to recognize equivalent maturational states across animals. The empirical data generating the model comprises 271 developmental events, including measures of initial neurogenesis, axon extension, establishment, and refinement of connectivity, as well as later events such as myelin formation, growth of brain volume, and early behavioral milestones, to the third year of human postnatal life. The progress of neural events across species is sufficiently predictable that a single model can be used to predict the timing of all events in all species, with a correlation of modeled values to empirical data of 0.9929. Each species' rate of progress through the event scale, described by a regression equation predicting duration of development in days, is highly correlated with adult brain size. Neural heterochrony can be seen in selective delay of retinogenesis in the cat, associated with greater numbers of rods in its retina, and delay of corticogenesis in all species but rodents and the rabbit, associated with relatively larger cortices in species with delay. Unexpectedly, precocial mammals (those unusually mature at birth) delay the onset of first neurogenesis but then progress rapidly through remaining developmental events.
Introduction
Current studies of the mechanisms by which neurons are specified and connected up are rapidly delivering a catalog of fundamental developmental operations (Chalupa and Williams, 2008). Work in any single species, however, cannot specify how maturation of the entire nervous system is coordinated. “Heterochrony,” the idea that the evolution of species can in part be understood by changes in the duration of developmental epochs, is a basic part of the vocabulary of developmental biology (Gould, 1977). For example, the idea that humans have an unusually extended childhood allowing prodigious abilities in cultural learning is an argument for heterochronic change in human brain plasticity compared with other primates (Hawkes, 2006; Locke and Bogin, 2006). To understand when an event is “unusual” in its duration or timing, however, requires a large comparative database.
Using animal models for medical research also requires establishing equivalent developmental times from “model” to humans, though many current laboratory practices are more traditional than empirical (Clancy et al., 2007). Many model species, for example, the zebrafish, mouse, and rat, were chosen specifically for rapid development, and the consequences of this choice for applicability to human development is unknown. An enduring problem for comparative developmental research is the altricial (early) versus precocial (late) timing of birth with respect to brain maturation, especially when using altricial rodents to model humans, whose altricial versus precocial status is ambiguous (Robson et al., 2006).
Allometric conservation has been the main conclusion of a small literature comparing brain development across mammals. Passingham (1985) noted a close correlation of developmental duration to adult brain volume. Using the method of referring all neurodevelopmental events to a single “anchor event,” eye opening, Dreher and Robinson (1988) found high predictability in visual system embryogenesis across species. We confirmed high predictability in early brain development, but also identified developmental adjustments related to speciation. For example, cortex enlargement in primates (Clancy et al., 2001) and nocturnal adaptation of the retina in the owl monkey (Dyer et al., 2009) are both associated with selective delays in neurogenesis.
Here we introduce the first major restructuring of our model of neurodevelopment using a new iterative optimization procedure. The first model was limited to the early perinatal period, and included the basic features of neurodevelopment: neurogenesis, process extension, synaptogenesis, synapse elimination, and neuron death (Clancy et al., 2000, 2001). Here, we double the species included from 9–18, triple the neurodevelopmental events modeled (95–271) and include early brain growth, myelination, electrophysiology, critical periods, and first behavioral events. The original analysis extended only to 7 months postconception in humans, while the present model extends to ∼3 years postnatal, a period of critical importance for those investigating early stress and epigenetic modification of brain development. Our overall goal is to determine how neurodevelopment is accommodated to species with different developmental rates and life history strategies, to better understand brain organization in an “evo–devo” context. We describe conservation of developmental allometry over this wide range of species in the ordering of basic developmental events, but show heterochronic changes in the order of neurogenesis associated with species-typical changes in the cortex and retina, and with precocial versus altricial life histories. Preliminary reports of these findings have been made previously (Workman et al., 2011, 2012).
Materials and Methods
Characterization of the data
Species used to construct the comparative mammalian model.
The model is designed to predict the dates of 271 neurodevelopmental events (Table 1) for 18 mammalian species (Table 2), a potential 4878 unique event-species combinations, using data collected by many investigators for 1010 of these combinations (21% of the total). For inclusion in the model, each event had to have been observed in at least 2 of our 18 species, as well as satisfy other methodological considerations discussed below. The choice of species and events was determined by availability in the research literature (including humans), and thus includes several “traditional” model species (mouse, Mus musculus; rat, Rattus norvegicus; ferret, Mustela putorius; rabbit, Oryctolagus cuniculus; cat, Felis cattus; macaque, Macaca mulatta). The presence of several eutherian species in the research literature is in part related to their unusually long or short gestations (golden hamster, Mesocricetus auratus; spiny mouse, Acomys cahirinus; guinea pig, Cavia porcellus; gerbil, Meriones unguiculatus). Six metatherian (marsupial) mammals (short-tailed possum, Monodelphis domestica; brush-tailed possum, Trichosaurus vulpecula; Northern quoll, Dasyurus hallucatus; the quokka, Setonix brachyurus; the dunnart, Sminthopsis crassicaudata; and the wallaby, Macropus eugenii) are included. In addition, we gathered data on sheep (Ovis aries) from slide libraries from the Cornell School of Veterinary Medicine Embryological Collection. This set of animals supported five supraordinate taxonomic categories, not equivalent in level or number of species: “glires,” all six rodents and the rabbit; “carnivores,” the ferret and cat; “primates,” monkey and humans; “marsupials,” all six metatherian mammals; and just one “ungulate,” the sheep.
Table 1.
List of all events with corresponding event scores, sorted by location classification
| Location | Event score | Location | Event score | Location | Event score |
|---|---|---|---|---|---|
| Brain stem | Limbic system (continued) | Cortex (continued) | |||
| Cranial motor nuclei–peak | 0.038 | Presubiculum–peak | 0.261 | Neurogenesis cortical layer II/III–peak (VC) | 0.395 |
| Oculomotor nucleus cell generation–onset | 0.041 | Dentate gyrus–peak | 0.264 | Cortical axons reach dorsal lateral geniculate nucleus | 0.396 |
| Trigeminal mesencephalon nucleus–onset | 0.041 | Anterior commissure appears | 0.267 | Primary auditory cortex neurogenesis–offset | 0.420 |
| Trigeminal mesencephalon nucleus–offset | 0.041 | Ventromedial hypothalamus neurogenesis offset | 0.269 | Corticospinal tract decussates at the pyramids | 0.428 |
| Mesencephalic trigeminal cell generation–offset | 0.042 | Mitral cells–offset | 0.274 | Corticospinal tract reaches cervical spinal cord | 0.439 |
| Trigeminal motor nucleus–peak | 0.051 | Suprachiasmatic nucleus neurogenesis offset | 0.283 | Initial differentiation of layer V (S1) | 0.444 |
| Medial longitudinal fasciculus appears | 0.062 | CA1, CA2–peak | 0.285 | Cortical axons innervate dorsal lateral geniculate nucleus | 0.465 |
| Postproliferative zone in the medulla appears | 0.073 | Nucleus accumbens—peak | 0.294 | Neurogenesis cortical layer II/III–end (VC) | 0.490 |
| Trigeminal motor nucleus–offset | 0.078 | Tufted cells–peak | 0.327 | Range of rapid synaptogenesis (VC)–start | 0.501 |
| Superior colliculus generation–onset | 0.079 | Hippocampal commissure appears | 0.337 | Onset of barrels (S1) | 0.504 |
| Superficial superior colliculus laminae–start | 0.095 | Septal nuclei–offset | 0.339 | Corticospinal tract reaches dorsal horn of cervical spinal cord | 0.514 |
| Postproliferative zone appears in the tegmentum | 0.112 | Olfactory tubercle generation–offset | 0.362 | Onset of decrease in fractional anisotropy | 0.515 |
| Posterior commissure appears | 0.112 | Entorhinal cortex neurogenesis offset | 0.375 | Onset of sublayers in layer V (S1) | 0.517 |
| Lateral cuneate nucleus–onset | 0.119 | Hippocampal CA3 neurogenesis offset | 0.380 | Onset of trilaminar plate (S1) | 0.517 |
| Lateral and medial superior olivary nucleus–onset | 0.119 | Hippocampal CA1 neurogenesis offset | 0.387 | Adult-like cortical innervation of dorsal lateral geniculate nucleus | 0.531 |
| Cranial sensory nuclei–peak | 0.120 | Subicular cortex neurogenesis offset | 0.389 | Onset of barrel field septa (S1) | 0.555 |
| Postproliferative zone appears in the pretectum | 0.125 | Granule cells in the dentate gyrus–peak | 0.583 | Visual cortical axons in SC | 0.570 |
| Medial cuneate nucleus–peak | 0.135 | Olfactory tract myelination onset | 0.634 | Corticospinal projections reach lumbar levels | 0.583 |
| Reticular nuclei—peak | 0.155 | Stria medullaris myelination onset | 0.653 | First cortical visually evoked potentials | 0.590 |
| Postproliferative zone appears in the superior colliculus | 0.157 | Fornix myelination onset | 0.675 | Corticospinal tract reaches the sacral region | 0.623 |
| Inferior colliculus generation–onset | 0.171 | Anterior commissure myelination onset | 0.677 | Middle cerebellar peduncle myelination onset | 0.652 |
| Lateral cuneate nucleus–peak | 0.186 | Mammillothalamic tract myelination onset | 0.689 | External capsule myelination onset | 0.666 |
| Substantia nigra–peak | 0.187 | Hippocampus myelination onset | 0.699 | Cingulum myelination onset | 0.687 |
| Sensory nucleus of the trigeminal nerve–offset | 0.199 | Fasciculus retroflexus myelination onset | 0.700 | Internal capsule myelination onset | 0.692 |
| Lateral and medial superior olivary nucleus–offset | 0.199 | Stria terminalis myelination onset | 0.703 | Corpus callosum body myelination onset | 0.722 |
| Superior colliculus–peak | 0.202 | Splenium myelination onset | 0.732 | ||
| Inferior colliculus–peak | 0.203 | Thalamus | Plasticity/OD critical period–start | 0.770 | |
| Raphe nucleus neurogenesis–offset | 0.206 | Medial geniculate nucleus onset | 0.113 | Range of rapid synaptogenesis (VC)–end | 0.849 |
| Medial cuneate nucleus–offset | 0.249 | Dorsal lateral geniculate nucleus–start | 0.127 | Visual cortex peak synaptic density (area 17) | 0.869 |
| Pontine nuclei–peak | 0.255 | Postproliferative zone appears in thalamus | 0.144 | Prefrontal cortex peak synaptic density | 0.886 |
| Lateral cuneate nucleus–offset | 0.262 | Medial geniculate nucleus–peak | 0.176 | Plasticity/OD critical period–end | 0.901 |
| Superficial superior colliculus laminae–offset | 0.279 | Ventral lateral geniculate nucleus–peak | 0.177 | Corpus callosum body myelination end | 0.971 |
| Superior colliculus generation–offset | 0.308 | Dorsal lateral geniculate nucleus–peak | 0.183 | Middle cerebellar peduncle myelination end | 1.000 |
| Dopaminergic axons from midbrain reach subplate | 0.352 | Ventroposterior and ventrobasal nuclei - peak | 0.191 | ||
| Inferior colliculus generation–offset | 0.381 | Ventrobasal neurogenesis - onset | 0.194 | Whole brain | |
| Dopaminergic axons from midbrain reach cortical plate | 0.395 | Medial geniculate nucleus offset | 0.211 | 20% maximum brain weight (day) | 0.586 |
| Dopaminergic axons from midbrain in cerebral cortex | 0.499 | Dorsal lateral geniculate nucleus–end | 0.228 | 30% maximum brain weight (day) | 0.635 |
| Superficial superior colliculus–start of lamination | 0.504 | Anteroventral, anteromedial, anterodorsal nuclei–peak | 0.238 | 40% maximum brain weight (day) | 0.682 |
| Inferior cerebellar peduncle myelination onset | 0.543 | Lateral geniculate nucleus axons in subplate | 0.305 | 50% maximum brain weight (day) | 0.729 |
| Superior colliculus segregation | 0.548 | ventrobasal neurogenesis–offset | 0.341 | 60% maximum brain weight (day) | 0.788 |
| Ipsi/contra segregation in superior colliculus | 0.561 | Pulvinar projections in interior zone of prestriate isocortex | 0.375 | 70% maximum brain weight (day) | 0.858 |
| Medial lemniscus myelination onset | 0.564 | Pulvinar projections in subplate of prestriate isocortex | 0.471 | 80% maximum brain weight (day) | 0.949 |
| Posterior commissure myelination onset | 0.591 | Pulvinar projections in cortical plate of prestriate isocortex | 0.515 | ||
| Onset of synapse elimination in neurons of the soleus | 0.629 | Lateral geniculate nucleus axons in cortical layer IV | 0.545 | Whole organism | |
| Brachium inferior colliculus myelination onset | 0.664 | Ipsi/contra segregation in lateral geniculate nucleus | 0.561 | Surface righting onset | 0.573 |
| Optic radiation myelination onset | 0.652 | Rooting reflex offset | 0.623 | ||
| Cerebellum | Auditory radiation myelination onset | 0.668 | Auditory startle reaction | 0.648 | |
| Inferior olive generation–onset | 0.058 | Lateral geniculate nucleus myelination onset | 0.680 | Preyer reflex | 0.681 |
| Inferior olivary nucleus–peak | 0.092 | Vibrissa placing adult like pattern | 0.695 | ||
| Purkinje cell generation–onset | 0.110 | Air righting reflex | 0.730 | ||
| Cerebellum (continued) | Striatum | Whole organism (continued) | |||
| Postproliferative zone appears in the cerebellum | 0.125 | Ganglionic eminence postproliferative zone appears | 0.088 | Semi-adult-like sleep cycle | 0.745 |
| Inferior olive generation–offset | 0.137 | Caudatoputamen generation–onset | 0.131 | Visual placing is mature | 0.768 |
| Purkinje cells–peak | 0.142 | Globus pallidus–peak | 0.140 | Postconception walking onset | 0.794 |
| Deep cerebellar nuclei–peak | 0.151 | Claustrum–peak | 0.147 | ||
| Anteroventral cochlear nucleus–peak | 0.179 | Caudatoputamen–peak | 0.186 | Sensory periphery | |
| Purkinje cell generation offset | 0.200 | Claustrum offset | 0.308 | Trigeminal ganglion cell generation–onset | 0.000 |
| Cochlear nuclei–peak | 0.220 | Caudatoputamen–offset | 0.519 | Trigeminal ganglion cell generation–offset | 0.195 |
| Superior cerebellar peduncle myelination onset | 0.564 | Lateral lemniscus myelination onset | 0.572 | Ears open | 0.668 |
| Lenticular fasciculus myelination onset | 0.602 | ||||
| Limbic system | Striatum myelination onset | 0.715 | Retina | ||
| Locus ceruleus–onset | 0.041 | Photoreceptor generation–onset | 0.054 | ||
| Locus ceruleus–peak | 0.059 | Cortex | Horizontal cell generation–onset | 0.059 | |
| Magnocellular basal forebrain–peak | 0.071 | Subplate—peak | 0.071 | Retinal ganglion cell generation–start | 0.091 |
| Mitral cells onset | 0.085 | Subplate–onset of neurogenesis | 0.072 | Retinal horizontal cells–peak | 0.153 |
| Olfactory tubercle generation–onset | 0.093 | Neurogenesis cortical layer VI–start (VC) | 0.117 | Amacrine generation onset | 0.158 |
| White matter appears in the hypothalamus | 0.097 | Cortical plate first observed/visible | 0.123 | axons in optic stalk | 0.163 |
| Preoptic nucleus generation–onset | 0.102 | Primary auditory cortex neurogenesis–onset | 0.136 | Retinal ganglion cells–peak | 0.195 |
| Septal nuclei–onset | 0.110 | Cortical subventricular zone–onset | 0.164 | Optic axons at chiasm of optic tract | 0.198 |
| Locus ceruleus–offset | 0.113 | Primary somatosensory cortex–layer VI offset | 0.194 | Rapid axon generation in optic nerve–start | 0.217 |
| Entorhinal cortex neurogenesis onset | 0.119 | Neurogenesis cortical layer V–start (VC) | 0.203 | Cones–peak | 0.264 |
| Raphe complex–peak | 0.120 | Neurogenesis cortical layer VI–peak (VC) | 0.204 | Optic axons invade visual centers | 0.270 |
| Medial forebrain bundle appears | 0.125 | Neocortical layer I emerges | 0.207 | Rod generation–onset | 0.282 |
| Hippocampal CA3 neurogenesis onset | 0.139 | GABA cells in subplate–start | 0.213 | Muller cell generation–onset | 0.282 |
| Subicular cortex neurogenesis onset | 0.139 | External capsule appears | 0.215 | Retinal amacrine cells–peak | 0.298 |
| Granule cell layer fascia dentata neurogenesis onset | 0.140 | Primary somatosensory cortex–layer V offset | 0.220 | Bipolar cell generation–start | 0.300 |
| Hippocampal CA1 neurogenesis onset | 0.143 | Internal capsule appears | 0.236 | Optic axons reach dorsal lateral geniculate nucleus and superior colliculus | 0.319 |
| Granule cells in the dentate gyrus–onset | 0.150 | Lhx6 first in cortex in GABAergic cells | 0.238 | Retinal ganglion cell generation–end | 0.333 |
| Postproliferative zone appears in the medial pallium | 0.151 | GABA cells in subplate–end | 0.249 | Horizontal cell generation–offset | 0.371 |
| Mammillothalamic tract appears | 0.153 | Primary somatosensory cortex–layer 2/3 onset | 0.255 | Optic nerve axon number–peak | 0.392 |
| Preoptic nucleus–peak | 0.157 | Neurogenesis cortical lamina VI–end (VC) | 0.258 | Onset of retinal waves stage II | 0.401 |
| Fasciculus retroflexus appears | 0.162 | Neurogenesis cortical lamina IV–start (VC) | 0.267 | Onset of retinal waves stage I | 0.425 |
| Stria medullaris thalami appears | 0.166 | Neurogenesis cortical layer V–peak (VC) | 0.268 | Rods–peak | 0.464 |
| Supraoptic nucleus of hypothalamus offset | 0.169 | Primary somatosensory cortex–layer IV offset | 0.283 | Retinal bipolar cells—peak | 0.478 |
| Suprachiasmatic nucleus–peak | 0.170 | GABA-ir cells in lower intermediate zone | 0.286 | End of retinal waves stage I | 0.520 |
| Amygdala–peak | 0.177 | GABA positive cells appear in lower intermediate zone | 0.287 | Bipolar cells generation–offset | 0.541 |
| Mitral cells–peak | 0.182 | Peak subventricular zone expansion in the developing isocortex | 0.289 | Amacrine cell generation–offset | 0.552 |
| Endopiriform neurogenesis offset | 0.193 | Subplate and intermediate zone apoptosis onset | 0.302 | Rod generation—offset | 0.561 |
| Anterior olfactory nucleus–peak | 0.194 | Neurogenesis cortical layer V–end (VC) | 0.305 | Onset of retinal waves stage III | 0.573 |
| Nucleus of lateral olfactory tract–peak | 0.198 | Neurogenesis cortical layer IV–peak (VC) | 0.320 | Retinal waves | 0.575 |
| Septal nuclei–peak | 0.215 | Cortical plate apoptosis onset | 0.321 | End of retinal waves stage II | 0.578 |
| Entorhinal cortex–peak | 0.230 | Neurogenesis cortical layer II/III–start (VC) | 0.342 | Rapid axon loss in optic nerve ends | 0.583 |
| Olfactory tubercle generation–peak | 0.238 | Corpus callosum appears | 0.359 | Muller cell generation–offset | 0.597 |
| Fornix appears | 0.241 | Neurogenesis cortical layer IV–end (VC) | 0.362 | Optic tract myelination onset | 0.597 |
| Stria terminalis appears | 0.243 | Layer IV genesis–start | 0.366 | Eye opening | 0.663 |
| Subiculum–peak | 0.248 | Primary somatosensory cortex–layer 2/3 offset | 0.368 | First electroretinogram | 0.665 |
| Parasubiculum–peak | 0.252 | Cortical axons reach thalamus | 0.371 | End of retinal waves stage III | 0.672 |
| Optic tract myelination end | 0.847 |
Within location subcategories, events are listed from lowest to highest event score.
Table 2.
List of species used in our analysis, sorted by species slope
| Species | Constant | Slope | Precocial score | Adult brain weight (g) | Adult male body weight (g) | Gestational length (days) |
|---|---|---|---|---|---|---|
| Spiny mouse | 2.82 | 0.622 | 1.356 | 0.7 | 42 | 39 |
| Guinea pig | 2.904 | 1.573 | 0.841 | 4 | 640 | 68.5 |
| Hamster | 2.189 | 1.644 | 0.336 | 1.12 | 108 | 15.5 |
| Rat | 2.31 | 1.705 | 0.445 | 2 | 207 | 21 |
| Gerbil | 2.532 | 1.715 | 0.401 | 1.02 | 70 | 25 |
| Mouse | 2.145 | 1.894 | 0.408 | 0.45 | 18 | 18.5 |
| Rabbit | 2.382 | 1.959 | 0.537 | 9.6 | 2170 | 31 |
| Ferret | 2.706 | 2.174 | 0.463 | 7.1 | 1900 | 41 |
| Cat | 2.784 | 2.28 | 0.61 | 28.4 | 4000 | 65 |
| Macaque | 3.27 | 2.413 | 0.761 | 93.8 | 5340 | 165 |
| Short-tailed opossum | 2.261 | 2.508 | 0.717 | 8 | 105 | 13.5 |
| Dunnart | 2.576 | 2.508 | 0.612 | 0.3 | 32 | 13.5 |
| Wallaby | 3.141 | 2.508 | 0.949 | 20 | 115 | 29.3 |
| Quoll | 2.759 | 2.508 | 0.65 | 3.38 | 650 | 18 |
| Brush-tailed opossum | 2.793 | 2.508 | 0.871 | 9.4 | 3150 | 17.5 |
| Quokka | 3.03 | 2.508 | 0.884 | 13.9 | 2700 | 27 |
| Sheep | 2.909 | 2.533 | 0.822 | 160 | 45000 | 147 |
| Human | 3.167 | 3.72 | 0.654 | 1350 | 70000 | 270 |
Included data are species constant, precocial score, adult brain and body weight, and gestational length for each species.
Criteria for inclusion and definition of measurements.
In prior work, to assemble the most comprehensive information possible, we included any developmental observation in the citable literature (Clancy et al., 2001). In the present work, now more confident of sample adequacy, admission to the database and standardization procedures have become overall more conservative, though the developmental literature itself has persistent peculiarities. For every category of “process” now included, we made every effort to include any observation reported in the literature for each species. Observations based on an N of 1 are acceptable, as this number characterizes virtually all of the primate literature and a good part of the remainder. Measurements of body weight, brain weight, and gender of the individual in which an observation was recorded are rarely made and were not required for including an observation; when such values are used in the analyses presented here, they are taken from separate, general measurements of each species. Here we describe procedures relevant to the entire dataset and procedures for new classes of events entered; procedures for quantifying basic neuroembryological events can be found in prior publications as described previously (Clancy et al., 2000, 2001, 2007).
Postconceptional day.
Each event is designated by the day that it occurs in a particular species, and all days are measured from conception. The first postconception (PC) day is designated by convention “day 0.” PC day virtually never refers to direct demonstration of conception, but a species-appropriate estimate, for example, day of mating in most rodents, or ∼2 weeks after the last menstrual period in humans. Occasionally, staging by non-neural morphology is used in some studies. When a study cited only a postnatal day, we determined whether or not the 24 h period following birth was designated as postnatal day 0 or postnatal day 1 and adjusted the stated timing to reflect the number of days that had passed from conception. Because the ages of sheep from the Cornell School of Veterinary Medicine Embryological Collection were unknown, we converted crown rump lengths of the embryonic sheep to estimated ages in PC days. We used previously published data that quantifies the relationship between crown-rump length and PC days in sheep, focusing on rapid transformations in brain morphology (Harris, 1937; Bryden et al., 1972).
Sampling frequency and sampling delay.
The frequency with which an event is measured depends on the duration of its species' neurodevelopmental period, with rodents measured in hourly, or more typically half-day intervals, animals of intermediate brain sizes in days, and primates in intervals of days to weeks. Though we have not attempted to estimate the error of each observation in absolute days, at minimum, the density of observations had to support the fundamental claim; for example, if a “peak” of some process was claimed, observations before and after that date had to be present. We will describe the error of model values, which correspond to sampling intervals, and which should be considered in interpreting results. Sampling delay is an intrinsic problem with these data as well, as events can only be observed after they have happened. Thus, the model will always predict events to occur somewhat after their actual occurrence, this interval lengthening in the species with less frequent sampling. Overall, however, the large majority of these data are derived from studies of neurodevelopment documenting multiple developmental assessments, demonstrations of the primary data, and onset-to-offset graphs of developmental progress of the observed events, as from the laboratories of Sidman, Altman and Bayer, and Rakic.
Onsets, offsets, and peaks.
Many neurodevelopmental events are naturally described by onsets and offsets and often peaks, such as neurogenesis, development of axonal tracts, appearance of slow-wave potentials, and so forth. Often, only onsets and offsets of neurogenesis were explicitly provided in the literature, though a midpoint or peak could be derived from provided histograms. In any case where quantification of a whole process was provided, “onset” was counted as the day on which 5% of the peak was reached, and “offset” at 95%, which avoided variability resulting from the long tails of the occurrence of the “very first” or “very last” cases in phenomena like neurogenesis or axon loss. Because of the absence of demonstration of peaks or offsets in enough species, postnatal neurogenesis in the dentate gyrus and olfactory bulb could not be included.
New events: myelination, overall brain growth, and behavioral events.
Onset of myelination is defined by the first observation of a complete wrap of single Schwann cell lamellae around an axon in the brain area or tract being considered, while termination of myelination is defined as 95% of the total early brain myelin volume or axon count in a particular area.
Brain growth was measured at the point of occurrence of 10% intervals of adult brain weight or 95% of maximum weight recorded in the developmental period. The days on which each decile of growth was reached were recorded, eliminating the 10, 90, and 100% points to eliminate introducing the excessive variability of distribution tails. This left seven deciles (20–80%) of brain growth that are included in the dataset.
Behavioral and locomotor events were added and were constrained in number by the necessity of their presence across such a wide range of species. They thus focus on basic developmental features like startle, righting, rooting, and early locomotion, as well as the emergence of organized sleep patterns and other early electrophysiological events. A number of events had to be excluded when investigators named “day of birth” as the onset day, when in utero behaviors had not been measured.
Categorization of events into “location” and “process.”
In the prior model, the interactions of certain categorizations of events and species were tested ad hoc, hypotheses generated from the literature on brain allometry (Finlay and Darlington, 1995; Clancy et al., 2001). In the present analysis, every event was given an obligatory, mutually exclusive brain location and process. The ultimate intent of these classifications, past their immediate use in this paper, was to identify where evolutionary selection, genetic, epigenetic, or disease processes might affect brain regions or functions selectively. For example, all the events occurring in the isocortex might become delayed with respect to the rest of the brain by a disease process, or synaptogenesis might be delayed everywhere by a genetic defect, or production of a larger brain part might be produced by delay of neurogenesis in that species. We will discuss several such effects, but have structured this aspect of data classification to allow further exploration in future studies. Of course, this classification does not capture all possible distinctions of neurodevelopmental events, with functional systems (e.g., “visual”; “motor”) a further possibility, for example.
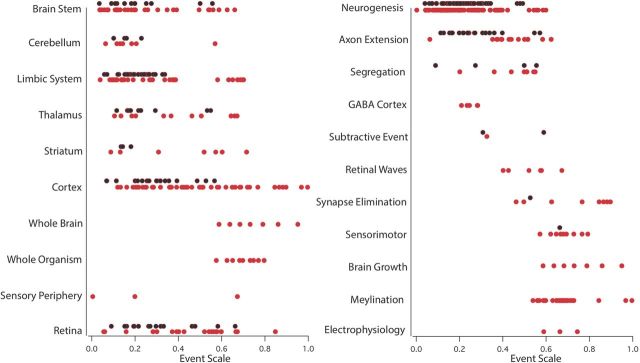
A list of Locations and Processes appears in Table 3, with a brief description of each. Their distribution and frequency in the dataset is shown in Figure 1 with respect to the “event scale,” the cross-species ordering of developmental events derived by the model. As described in Table 3, locations are gross divisions of the brain, with occasional deviations from standard medical neuroanatomical nomenclature motivated by considerations of prosomeric location (Puelles and Rubenstein, 2003). The location “Limbic” is broad, and includes the olfactory bulb; entorhinal, subicular, and olfactory cortices; hippocampus; amygdala; septum; and other basal forebrain nuclei and tracts, classified together as they have been found to covary allometrically (Reep et al., 2007). Tracts are assigned to the location of their neurons of origin, if they are singular in origin. Processes are general neuroembryological phenomena, such as “neurogenesis” or “synapse subtraction” with the atypical instances of “GABA cortex” and “retinal waves” added as the result of specific research initiatives (Finlay and Clancy, 2008; Clancy et al., 2009).
Table 3.
List of location and process classifications with brief descriptions of each. All locations are determined by the starting area of the phenomenon that is observed
| Description | |
|---|---|
| Location | |
| Brain stem | Encompasses the spinal cord, medulla, and mesencephalon |
| Cerebellum | The presumptive cerebellum |
| Limbic system | Amygdala, hippocampus, septum, olfactory bulbs, olfactory, subicular, and entorhinal cortex |
| Thalamus | All thalamic nuclei |
| Striatum | Globus pallidus, caudate nucleus |
| Cortex | Isocortex |
| Whole brain | Gross brain growth |
| Whole organism | Behavioral events |
| Sensory periphery | Peripheral nervous system |
| Retina | Retina, retinal ganglion cells |
| Process | Description |
| Neurogenesis | Cell cycle exit for any group of cells |
| Axon extension | When axons arrive at targets, form synapses, axon growth |
| Segregation | Differentiation of a population of cells |
| GABA cortex | GABAergic cells that have origins in the striatum or isocortex |
| Subtractive event | Cell death |
| Retinal waves | Patterns of developing retinal activity |
| Synapse Elimination | Decrease in synapse numbers |
| Sensorimotor | Behavioral or reflexive event |
| Brain growth | Gross brain development |
| Myelination | First wrap of a glial cell around a neuron |
| Electrophysiology | Onset of electrical activity |
Figure 1.
The distribution of the neural location (left) and general class of developmental process (right) of events used to develop the model, plotted against the event scale. The event scale, which is described in more detail in Figure 3, is a ranking of all these events on a scale from 0 to 1, and principally represents their order of occurrence across all of the species. New events added in this data analysis compared with the original model (Clancy et al., 2001) are in red and show not only the addition of late behavioral, physiological, and overall growth information to the model but also addition to the database throughout; events common to both in black.
The model
Differences from earlier models.
In our initial work, a general linear model (GLM) was used to fit developmental events related to neurogenesis for 51 brain structures in seven mammals, including one metatherian mammal (Finlay and Darlington, 1995). This model was progressively extended to include multiple neural maturational events in addition to neurogenesis, and more species, including five additional marsupial mammals and human neurodevelopmental events. Several versions of the GLM were explored, including principal component analysis (Darlington et al., 1999; Clancy et al., 2000, 2001). The most current version also serves as a Web-based utility for “translating time” between experimental animals for investigators (http://translatingtime.org).
The basic structure of all of the previous models was constant: one scale is derived from log-transformed data, to linearly rank species with respect to duration of neural development (the “species score”) and another to order developmental events (“structure score” or “event score”). The present model was fitted to the data with the “quasi-Newton” form of optimization. This method can fit models with mathematical forms that GLMs cannot handle, such as the new model. Such procedures attempt to find the values of model parameters, which minimize some value the user specifies, in this case, the sum of squared errors (SSE). The specific tool we used was the qnewton command in the Gauss statistical package (http://www.aptech.com).
When we define Y as ln (PC date), the purpose of the model is to estimate Y, the logged day of occurrence for each of 271 events in neurological development for all 18 species, with minimum expected squared error. As in regression and ANOVA models, we can break each prediction into a linear or “main” component and an interaction component, which sum to the total. In this case, the interaction component actually consists of two separate interaction terms to be described later.
In the main component, the model constructs an event scale whose scores range from 0 to 1. Each of the 271 events gets a score on this scale. The later an event occurs in development across all species, the higher its score. Simultaneously, the event scale is treated as the independent variable in 13 simple regressions, one for each eutherian species and one for all six marsupials. The marsupials had, on average, less than one-fourth the observations per species that the eutherians had, which is too small a number to support reliable estimation of six separate marsupial slopes. However, each marsupial species did receive its own additive constant. Thus the model's parameters include 18 additive constants and 13 slopes, plus a coefficient for each of the two interaction terms. Since we fixed the event scores of the earliest and latest events at 0 and 1, respectively, only the other 269 event scores remain as parameters to be estimated. Thus the total number of parameters in the model is 18 + 13 + 269 + 2 or 302.
This optimization procedure allows the event scale and the regression equations for all 18 species to be produced simultaneously. While the 18 regressions are being fitted, the event scale itself is being constructed in such a way as to yield a minimum SSE over all 18 species. The optimization software uses a trial and error process to derive simultaneously the values of all the 302 parameters that minimize SSE.
We considered dozens of possible interaction terms involving location, process, and species or taxon, but we kept just two of them in the model: (non-glires*cortex*neurogenesis) and (cat*retina*neurogenesis). Our criterion for retaining a trial interaction term was considerably stricter than mere statistical significance, since that concept refers only to the 1010 cells in our 271 × 18 matrix, which contain actual data. We found that many tested interaction terms were statistically significant by usual criteria, but increased the SEs of the estimates computed for many of the thousands of cells without observations. We chose to delete those terms, leaving only the two interactions just described.
SEs and confidence limits.
We computed a standard error of prediction (SEPRED) for each of the 4878 cells in our matrix. Standard formulas were not applicable, so we developed our own method to do this. That method recognizes that we have data in just 1010 of the 4878 cells. First we computed the model's prediction for each of those 1010 cells. Then we added to each of these values a random normal deviate with mean 0 and SD σ, chosen in a way to be described, thus creating pseudo y-values. We then used the qnewton software to fit the model to these pseudo-values, and used that fitted model to estimate Y values for all 4878 cells of the matrix. We repeated this process 1000 times, thus generating 1000 y-estimates for each cell. The SD of those 1000 values was taken to be the SEPRED value for each cell. We tested this whole process in an ordinary regression model whose SEPRED values could be calculated by well known formulas, and found that it reproduced those values very well.
We chose σ as follows. As the SEPRED values computed by this method would be proportional to the value of σ we used, it was important to choose σ carefully. In addition, the value of SSE computed for the pseudo-values by the qnewton method would be proportional to σ2. We wanted to use a value of σ that would, on the average, produce a value of SSE equal to that we had observed in the real data. So we first estimated a value of σ, then ran 100 or more trials, observed how the mean SSE of these trials differed from the SSE in the real data, and adjusted σ accordingly. After a few repetitions of this process, we found we could match the real-data SSE within 1%. We then ran the 1000 trials mentioned above.
As discussed later, some SEPRED values are many times as large as others. SEPRED values are largest for cells with few species observed for that event, few events observed for that or similar species, and no observation in that individual cell. The ranges of mean SEPRED values ordered by Species and Process may be found in Table 4.
Table 4.
Table of average SEPRED values by process and species
| Process | Rat | Mouse | Human | Gerbil | Short-tailed opossum | Dunnart | Macaque | Wallaby | Quoll | Brush-tailed opossum | Quokka | Hamster | Spiny mouse | Guinea pig | Rabbit | Sheep | Ferret | Cat |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neurogenesis | 0.06 | 0.07 | 0.14 | 0.11 | 0.10 | 0.11 | 0.09 | 0.09 | 0.10 | 0.09 | 0.10 | 0.06 | 0.04 | 0.10 | 0.08 | 0.10 | 0.08 | 0.08 |
| Axon extension | 0.05 | 0.06 | 0.11 | 0.09 | 0.09 | 0.09 | 0.07 | 0.08 | 0.08 | 0.08 | 0.08 | 0.05 | 0.04 | 0.08 | 0.06 | 0.08 | 0.07 | 0.07 |
| Segregation | 0.05 | 0.06 | 0.11 | 0.08 | 0.09 | 0.09 | 0.07 | 0.08 | 0.08 | 0.08 | 0.08 | 0.05 | 0.06 | 0.06 | 0.06 | 0.08 | 0.07 | 0.07 |
| GABA cortex | 0.06 | 0.07 | 0.14 | 0.11 | 0.10 | 0.11 | 0.09 | 0.10 | 0.10 | 0.10 | 0.10 | 0.06 | 0.03 | 0.09 | 0.08 | 0.10 | 0.08 | 0.08 |
| Subtractive event | 0.05 | 0.06 | 0.11 | 0.08 | 0.08 | 0.09 | 0.07 | 0.08 | 0.08 | 0.08 | 0.08 | 0.05 | 0.05 | 0.07 | 0.06 | 0.08 | 0.07 | 0.07 |
| Retinal waves | 0.06 | 0.06 | 0.12 | 0.08 | 0.09 | 0.10 | 0.08 | 0.09 | 0.09 | 0.09 | 0.09 | 0.06 | 0.07 | 0.06 | 0.07 | 0.09 | 0.08 | 0.08 |
| Synapse Elimination | 0.05 | 0.06 | 0.11 | 0.07 | 0.10 | 0.10 | 0.08 | 0.09 | 0.10 | 0.09 | 0.10 | 0.07 | 0.10 | 0.07 | 0.07 | 0.09 | 0.08 | 0.07 |
| Sensorimotor | 0.06 | 0.06 | 0.12 | 0.07 | 0.10 | 0.10 | 0.08 | 0.09 | 0.10 | 0.09 | 0.10 | 0.07 | 0.10 | 0.06 | 0.08 | 0.09 | 0.08 | 0.08 |
| Brain growth | 0.04 | 0.05 | 0.09 | 0.06 | 0.08 | 0.09 | 0.06 | 0.08 | 0.08 | 0.08 | 0.08 | 0.06 | 0.11 | 0.05 | 0.07 | 0.07 | 0.07 | 0.06 |
| Myelination | 0.05 | 0.06 | 0.10 | 0.06 | 0.09 | 0.10 | 0.07 | 0.09 | 0.09 | 0.09 | 0.09 | 0.06 | 0.10 | 0.06 | 0.07 | 0.08 | 0.07 | 0.07 |
| Electrophysiology | 0.05 | 0.06 | 0.11 | 0.06 | 0.09 | 0.10 | 0.07 | 0.09 | 0.09 | 0.09 | 0.09 | 0.06 | 0.09 | 0.06 | 0.07 | 0.08 | 0.07 | 0.07 |
“Leave one out” analysis.
We also performed a leave one out (LOO) analysis, which is designed to counter the suggestion that good observed fit between model and data could be caused by overfitting a model to random fluctuations in the data. Overfitting becomes a serious problem as the number of model parameters increases, and the number of parameters in our model (302) is high. However, theoretical considerations (Darlington, 1990) suggest that more important than the number of parameters is the difference between the numbers of observations and parameters. For us that number is 1010 − 302 or 708, which is very high by standard criteria. However, an LOO analysis can provide an even more convincing demonstration of the point. In this analysis, each observation is removed from the dataset one at a time, and the model is fitted to the remaining 1009 observations. Then that model is used to predict y for just the one omitted observation. Finally, the estimates of y computed in this way are correlated with the actual values of y. SEPRED values provide an even stricter measure of model stability than LOO and we consider both measures in evaluating the model. The LOO correlation, the statistical check for “overfitting”, was 0.9823.
Exclusion of outliers for statistical considerations.
A small number of events were excluded because they were identified as outliers in the SSE analysis: it proved that if only one or two events had been measured exclusively in species with similar neurodevelopmental durations (in this case, several rodents), these close values did not constrain slope fitting adequately, producing biologically implausible sequences.
Results
Basic characteristics of the database and model
Characterization of the database
The corpus of empirical data includes observations of 271 developmental events in 18 mammals, including six metatherian mammals. The five species with the most measured events were the rat (221), mouse (176), macaque (118), cat (86), and human (75); these five accounted for just over two-thirds of the 1010 observations. The numbers of events measured in 2, 3, 4, 5, or 6 species were, respectively, 80, 80, 40, 33, and 15, while 22 other events were each measured in 7–11 species. That left only eye opening, which was measured in 16 of the 18 species. Overall, the goal of the database was simply to be comprehensive and include every morphological or neurodevelopmental event given a date by any investigator: no class of event was chosen for its particular evolutionary interest or presumed ability to discriminate phylogenetic groups. Continuous processes, such as overall brain growth, were “digitized” into the minimum divisions that appeared to capture curve shapes.
In addition to the species level of analysis, five taxonomic groups were considered: glires, including the six rodents and the rabbit; six marsupials; two carnivores; two primates; and one ungulate. Important to keep in mind for the present analysis (and virtually any developmental work) is that species with adequate developmental data have a persistent confound, which is that the largest brains are primate brains, the human and macaque, and the smallest a mixture of rodents and marsupials. The earliest event in the database was “onset of trigeminal ganglion cell generation,” which we estimate to occur at 8.5 d in the mouse and 23.7 d in the human. The latest was “end of middle cerebellar myelination,” which we estimate to occur at 56.8 d in the mouse and 979 d in the human. The onset of the time period populated by data is defined to be initial neurogenesis, the first differentiation of identifiable neurons, excluding the earliest events in embryogenesis such as species differences in implantation after fertilization, diapause in metatherian mammals, and the initial organization of the body plan and neural proliferative zones. Note that the very earliest observed events were not necessarily observed in every species: the rodent samples contain observations of the generation of cranial nerve motor neurons, while the earliest observations in macaques are in retinogenesis, and in humans, first axon extension. The estimated first event converged on approximately the ninth PC day in rodents, and the 30th and 35th PC day for macaques and humans.
The 271 developmental events remain dominated by onset, peak, and offset of neurogenesis across brain locations, with ∼55% of that type, 10% events in axonogenesis, 15% myelination and overall brain growth, 10% synaptogenesis and synapse refinement, and 10% structure-level electrical activity and early behaviors (Fig. 1). It is important to note, however, that the difference in the proportions of kinds of processes in no way precludes observations of interactions between “minority” processes and species differences.
The statistical model
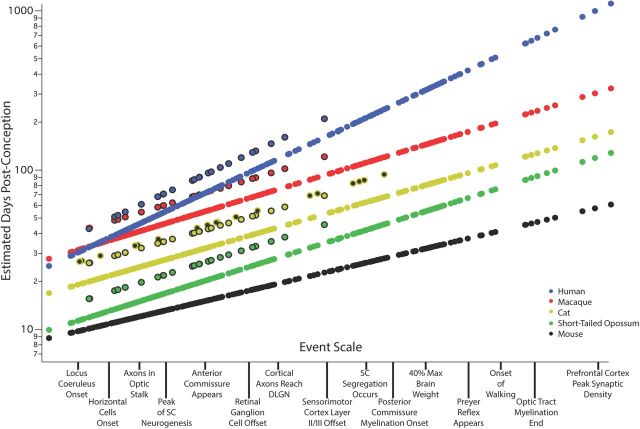
Shown in Figure 2 are the predicted values for PC day for each measured event of the model for five representative mammals spanning close to the duration range of the model: the mouse, short-tailed possum, cat, macaque, and human. The regression for each species has the form:Y = onset + slope*eventscale + (interaction term) with event scale values for every developmental event given in Table 1. The y-axis represents the predicted day of each event in each species, ranging from 8.5 to 979 PC days. The two aforementioned interaction terms (non-glires*cortex*neurogenesis) and (cat*retina*neurogenesis) can be seen as the offset lines for human, macaque, cat, and short-tailed possum, where in all cases the interaction delays the event in the relevant species compared with base values. After the general features of the model have been discussed, we will discuss the import of each modeled parameter in turn.
Figure 2.
Predicted developmental schedules for human (blue circle), macaque (red circle), cat (yellow circle), short-tailed opossum (green circle), and mouse (black circle), selected from the 18 species to illustrate the full range of developmental durations. In this graph the event scale is the x-axis, to which we have added a subset of the 271 events that were observed. This scale ranges from 0 to 1, but in this case, event scale numerical values are replaced by these example events. As will be described in Figure 3, the event scale is a common ordering of developmental events across all species. The y-axis is the estimated date of occurrence of each event in each species from conception (log scale). To determine when a particular event would be predicted to occur in any species from this graph, using the name of the event on the event scale, find where it intersects the regression line for that particular species. The y-axis value will be the predicted PC day for that event/species combination. In future graphical representations of the event scale, the event scale value for any named event can be found in Table 1. Also represented on this graph are interaction terms for corticogenesis and retinogenesis, with interaction terms always associated with individual species. The parallel lines for a subset of events in four of the species (black bordered circles for human, macaque, cat, and possum) represent delays in cortical neurogenesis with respect to their time of occurrence in the rodent and rabbit. In the cat, a second parallel line can be seen representing the delay of retinal neurogenesis (yellow circle with a black dot).
Evaluating the statistical significance and biological meaning of model components
The event scale
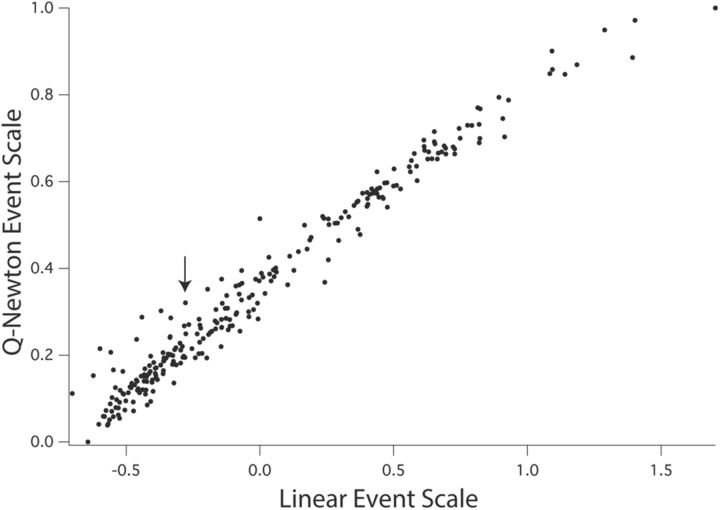
Across all 1010 observations, the correlation between Y values and their values estimated from the model was 0.9929. To understand the significance of this very high correlation, we will examine in turn both the statistical description and a biological significance translation of the model components. In Figure 3, we first examine the event scale derived by the quasi-Newton optimization technique to the ordering of the same events produced by the GLM. Before the introduction of interaction terms, for the event scale, the GLM will (unsurprisingly) produce a linear transformation of the values entered, preserving their ordinal relationship. In the qnewton model, Y = ln(PC date), within any glires species this scale correlates perfectly across events with the model's estimates of Y values. For non-glires species the correlations are perfect only within sets of events affected by the same interaction terms. There are three such sets for the cat, and two for every other non-glires species. The scale runs from 0 to 1. In Figure 3, deviations observed between the quasi-Newton and the GLM event scale could be an interesting clue to the heterochronic and other evolutionary shifts in developmental organization we are exploring. The correlation between the two, across the 271 events, is 0.9850, so the two scales are essentially very similar. A slight curvilinearity, most noticeable at the upper right of the graph in Figure 3, can be seen (test for significance of curvilinearity, t = −4.8268, p = 0.000002). The qnewton model may be producing a better fit across species by estimating the later events to occur earlier in time than the linear ranking produces suggests, possibly by mitigating the “leverage” of the very delayed primate times for these points. Comparing across the two scales, 5.84% of all event pairs are reversed in their ordinal position. This effect is most notable for a series of elevated points on the quasi-Newton event scale in the leftmost portion of the graph, near the origin (Fig. 3, elevated points left of arrow). The first six involve early axon and tract formation, five of those six in the forebrain, including the first appearance of the posterior commissure, mammillothalamic tract, external capsule, stria medularis thalami, cortical layer 1, and internal capsule. The remainder of the elevated points involve the generation of subplate and GABAergic pathways.
Figure 3.
The relation of the event scale used in this analysis to the ordering of events that was used in the prior model. Plotted on the x-axis are the event scores determined by GLM as in Clancy et al. (2001), estimating values for all species-event combinations by determining their overall ranking, and fitting those scores on a scale from 0 to 1.5. The event scale of the present analysis plotted on the y-axis was derived by the quasi-Newton optimization method described in the text. The two values derived for each event are very highly correlated, and also conform to the necessary biological ordering of events (e.g., cell birth before cell death). Each of these two scales was designed to correlate perfectly, within its own dataset, with the estimated Y values for any rodent and rabbit species. This shows that the quasi-Newton optimization method essentially ranks the order of occurrence of events in development, but it differs in two important respects. First, it shows a statistically significant nonlinearity, most obvious in the higher values on each scale. Second, some events fall noticeably above the main regression line, the largest cluster are those to the left of the arrow on the figure, and may represent events in early axonogenesis and GABA-neuron generation, which are either very substantially shifted in their order between species or nonhomologous events.
Both of these subprocesses have potential developmental interest for species differences. The first set of six axon-extension events are part of the initial “scaffolding” of major axon tracts in the brain (Easter et al., 1993), though the data are not dense enough at this point to determine phylogenetic differences. The generation of GABAergic cells has been suggested in several cases to involve species-specific innovations (Kostovic and Rakic, 1980; Chalfin et al., 2007; Clancy et al., 2009; Le Magueresse and Monyer, 2013) In these cases, some of the bouts of generation of GABAergic cells for the cortex in rodents and primates may not be homologous events and should not have been so classed, which will require much closer study. Overall, however, the close correlation of these two event scales indicates that they produce very similar sequences of neurodevelopmental events, with the two exceptions of first axonal projections and generation of GABAergic populations.
General aspects of accuracy of model estimates and accuracy by species
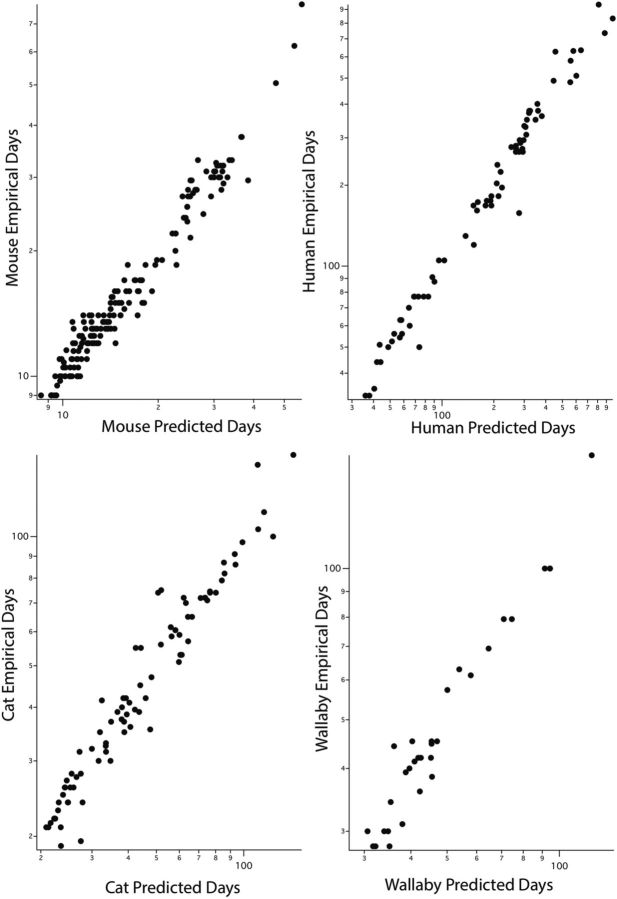
To better view the fit of data to model, the modeled versus empirical values are shown in four representative species (Fig. 4). Recall that all of the later model events are new to this analysis (Fig. 1), and the fit of data to model does not differ across the range of the event scale. The model SSE was 10.5402, so the root mean squared difference between any Y and its estimated value was √10.5402/1010 or 0.1022. If we had applied the GLM to the new 1010-case dataset and used the new interaction terms, those values would have been far higher at 23.6180 and 0.1529. Within species, by far the smallest correlation between Y and estimated Y was for the spiny mouse, at 0.7008. Values for the guinea pig and quokka were 0.9135 and 0.9384, respectively. The remaining 15 ranged from 0.9528 to 0.9885. The value for humans was 0.9878.
Figure 4.
Fit of empirical data to model values in four representative species. The x-axis is the predicted date of occurrence of all the events for which there were also empirical data points; the y-axis is the empirical value of the measured event in that species. As described in several technical aspects in the texts, the fit of predicted to actual days is very high.
The model's 4878 SEPRED values, which measure the accuracy of the model's estimates, ranged widely, and we will discuss the overall pattern of variation (Table 4) and the characteristics of the highest values. The smallest 4778 of these values (98% of the total) ranged from 0.0250 to 0.1425 with a median of 0.0813. These 4778 values had a wide range but a reasonably symmetric distribution, with the median very close to the midrange. The next 98 larger values ranged up to 0.1915, and the next two (the two highest values) were 0.1984 and 0.2501. Thus most but not all of the model's date estimates seem reasonably reliable. Unfortunately, 76 of the 100 largest SEPRED values were for humans. Even so, 195 human SEPRED values are below that range, so that many human values are predicted with good precision (Table 4). Unremarkably, estimates were most reliable when many species were observed for the event in question, including species similar to the one in question. The largest single SEPRED value was for event 147 (olfactory tubercle neurogenesis offset) in humans; that event had been observed only in rats and spiny mice.
The interaction terms
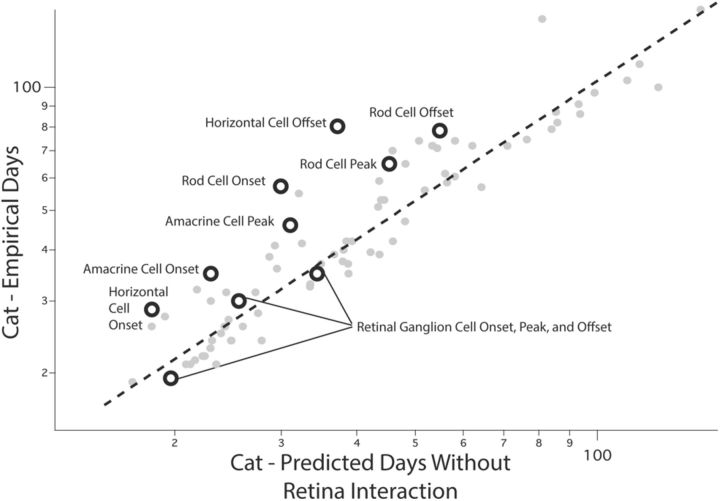
First we consider the interaction term “cat × retina × neurogenesis.” Compared with all other species, neurogenesis is selectively delayed in the cat retina. On close inspection (Fig. 5), this effect is not seen in every retinal cell class: points above the line are measures of neurogenesis in rods and horizontal and amacrine cells, but not retinal ganglion cells. The cells with delayed onset of terminal divisions are either rods or cells with direct connections to rods (bipolar cell generation has not been measured in the cat). This result is directly comparable to empirical observations we have made in the nocturnal owl monkey(Aotus azarae) retina where rod and bipolar cell numbers greatly increase, compared with much smaller changes in cone and retinal ganglion cell numbers, all compared with a closely related diurnal monkey, the capuchin monkey (Cebus apella) (Dyer et al., 2009). While the cat is not the only nocturnal mammal in this dataset, its adaptation to nocturnality resembles that seen in primates (Ross and Kirk, 2007) contrasted with the nocturnal ferret, rodents, and metatherian mammals described here.
Figure 5.
Plotted are all of the empirically measured developmental events (y-axis) in the cat, versus predicted developmental time for each event (x-axis), before the fitting of a retinal neurogenesis interaction term. All of the events in retinal neurogenesis are highlighted by black circles and all other events are in gray. The retinal neurogenesis events all occur later than would be predicted by the general model for the cat. Consistent with the hypothesis that this developmental delay has occurred for the purpose of increasing the retinal cell populations associated with nocturnality, within the retinal neurogenesis group only retinal ganglion cells are delayed. Retinal ganglion cells are the only measured cell class in the cat retina that are neither rods nor have a connection with rods.
Next we consider the interaction term “non-glires × cortex × neurogenesis.” This interaction term is pervasive, and effects can be seen in the relevant species of Figure 2, the human, macaque, cat, and short-tailed possum. In all non-glires species, the onset of cortical neurogenesis is selectively delayed with respect to the glires (rodents and rabbits), an effect we have described previously but which now has an additional 57 data points to support the conclusion (Finlay and Darlington, 1995; Clancy et al., 2001). This developmental change corresponds to greater neuron numbers and volumes in cortex in those species where corticogenesis is relatively delayed. Our prior model characterized this change as a delay of corticogenesis in primates, but the addition of new species now makes it ambiguous whether the change should be best characterized as a primate “delay” or an advance of corticogenesis in glires.
The non-glires × cortex × neurogenesis interaction is a developmental aspect of one of the most robust features of nervous system variation across all vertebrates, the relative expansion of the olfactory–limbic system versus the isocortex (or the remaining forebrain in non-mammals) (Jerison, 1973; Reep et al., 2007; Yopak et al., 2010). Comparing the glires to the four other radiations, we can ask which condition appears ancestral, and which is “derived.” Is it that the rodents and the rabbit have been selected for a small isocortex, or have all four other radiations been independently selected for a larger isocortex? We note that non-glires was chosen rather than glires in defining the interaction term merely to make its coefficient positive, not for any biologically motivated consideration. In Figure 6 we sketch the phylogenetic distribution of the incidence of increased duration of corticogenesis, amplified by corresponding information about cortical allometry for the taxa without developmental data. The irregular distribution of the developmental schedules associated with a relatively large cortex does not clearly support either a relatively large or small cortex as the basal pattern for mammals, but a feature appearing and disappearing frequently, concordant with the allometric variation in the same feature (Reep et al., 2007; Yopak et al., 2010). As emphasized in Materials and Methods, however, this database has too few species as yet to support any more formal phylogenetic analysis.
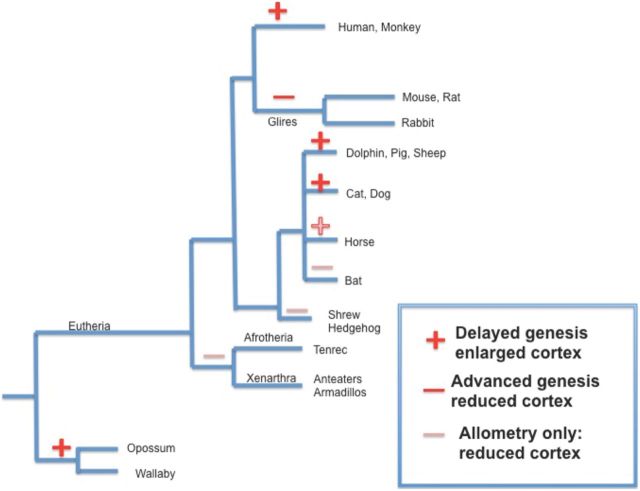
Figure 6.
Phylogenetic tree plotting the positions of relative delay in cortical neurogenesis, to determine whether delay in onset of cortical neurogenesis, associated with a taxonomic grade shift up in neocortical volume, had occurred at single or multiple times in mammalian phylogeny. Bold pluses and minuses mark branches with delays described in this dataset, and which also have species with increased adult cortical volume, while the unmarked ones represent branches where only relative cortical volume differences in adults are known (Reep et al., 2007). Increases in relative cortical volume appear to have occurred at least three times in mammals, in Metatheria (various marsupials), in the branch giving rise to carnivores, various ungulates, and dolphins, and in primates. It cannot be determined from these data as yet whether a large cortex is the “basal” state, and reduced cortical volume has been selected for in glires, bats, shrews, and Afrotheria, or the reverse. Phylogeny from Song et al. (2012).
Two replications of prior results with the extended dataset: “lateral equals late,” and overall slow development in marsupials
Brain segmentation and duration of neurogenesis
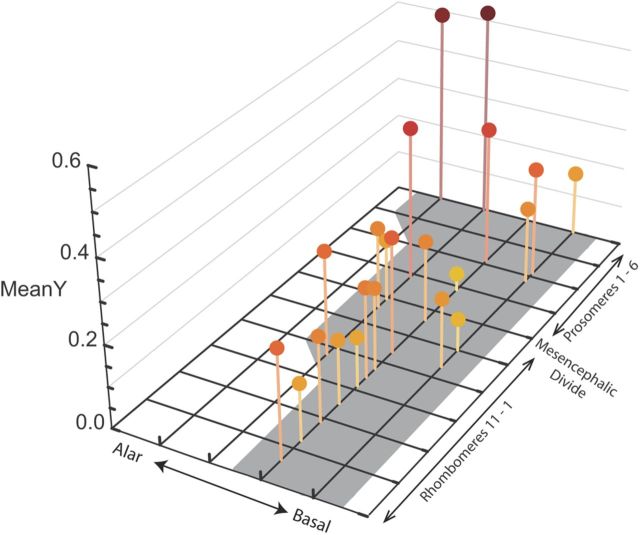
Two prior observations from the earlier limited dataset (Finlay et al., 1998, Darlington et al., 1999) required verification in the extended dataset, which we report briefly. The segmental structure of the CNS, the rhombomeres of the brainstem (Lumsden, 1990), and the prosomeres of the forebrain (Puelles, 1995) can be recognized via the patterned expression of early organizing genes in those structures. We found a statistical relationship of lateral or “alar” location in the neural tube and duration of neuron proliferation (Finlay et al., 1998). In Figure 7 we update this spatial representation of developmental duration to better reflect the current understanding of segmental organization of the brain (Puelles, and Rubenstein, 2003; Lumsden, 2004; Garcia-Lopez et al., 2009) and to include the expanded database. Note that the present data do not contain values for adult neurogenesis in the olfactory bulb and dentate gyrus, which would be located on the most lateral edge of the prosomeres. Confirming the prior result, late preservation of a productive stem cell pool is more likely in the anterior and lateral regions.
Figure 7.
The maximum duration of neurogenesis across species, plotted on the basic segmental divisions of the embryonic vertebrate nervous system. Only one side of the neural plate is plotted, with the midline to the right, and more alar or lateral locations to the left. Rhombomeres 1–11, which give rise to the medulla, pons, and cerebellum, and the prosomeres, which give rise to the diencephalon and telencephalon, are indicated. The neural plate representation compresses the segmental divisions described previously (Garcia-Lopez et al. (2009) into four medial to lateral divisions. The latest date of neurogenesis observed in each division is plotted on the z-axis, as given by the mean of the species' event scores found there. Extended duration of neurogenesis is more likely in more alar (lateral) and more rostral divisions.
Life history variations in metatherian mammals
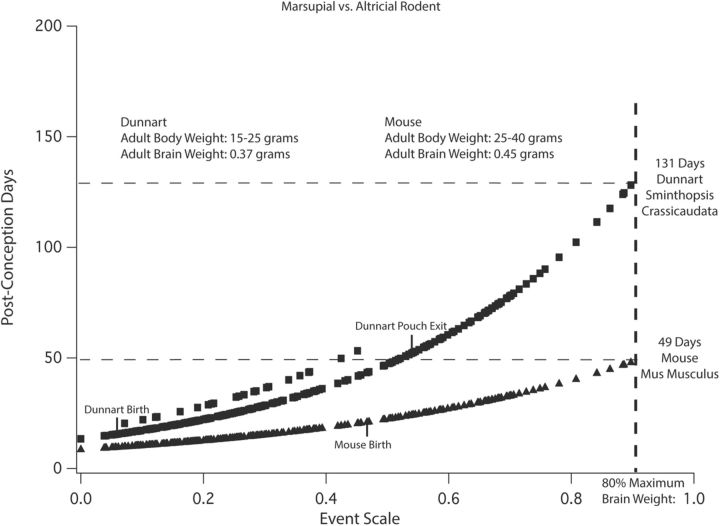
The metatherians include three from the Order Polyprodonta, carnivorous marsupials including the fat-tailed dunnart (the “marsupial mouse”), the gray short-tailed opossum, and the South American opossum. The rest are from the Order Diprodonta, herbivorous marsupials including the brush-tailed possum, the quokka, and the tammar wallaby. The principal feature distinguishing these mammals from the placental mammals is their distinctly slower rate of development (Darlington et al., 1999), and the new analysis replicates this result. Figure 8 contrasts the modeled values of the relative rate of progress along the event scale for the dunnart and the mouse, which have very similar brain sizes (Table 2). The dunnart requires more than twice as long as the mouse, 131 d from conception versus 49 d, to reach 80% of adult brain size, and earlier events are comparably slowed.
Figure 8.
Comparison of the modeled relative rates of brain development for a metatherian (marsupial) and a eutherian (placental) mammal of similar adult brain sizes. The marsupial species is the Polyprodonta dunnart, the marsupial mouse, and the placental mammal is a rodent, the common mouse M. musculus. The x-axis is the event scale, and the y-axis shows the predicted PC day of the occurrence of each event in both species. In this graph, the y-axis is a linear, not a log scale, so that the differences in duration can be better appreciated in this case where brain sizes are comparable. The elevated points near the dunnart line represent the effect of the non-glires interaction term delaying cortical neurogenesis with respect to the mouse. The dunnart takes nearly twice as long to reach 80% of its adult brain size compared with the laboratory mouse, with later developmental events disproportionately protracted compared with early events.
Substantial differences in the neurodevelopment of precocial and altricial mammals
Birth
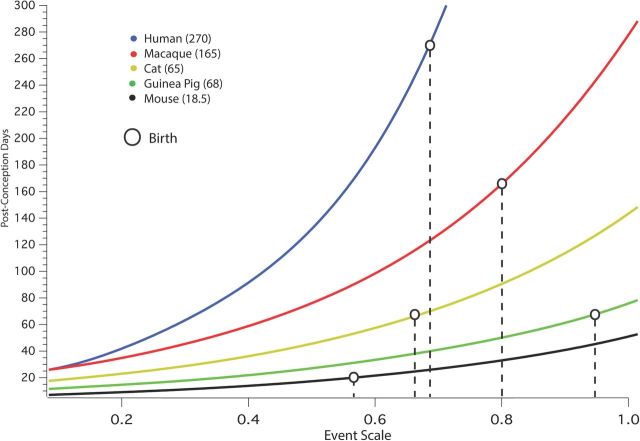
We did not include birth as one of our 271 “neural events,” and its position ranges widely relative to neural events (Fig. 9). Other partially “somatic” events that are included in the dataset, for example, eye opening or early behaviors, are in contrast highly predicted by neural events. In Figure 9, the position of birth with respect to each species' progress on the event scale is placed for five placental mammals chosen to represent close to the full range of the dataset. The events underway in each species near birth may be seen by comparing the value of birth on the event scale compared with neural events in that species (Table 1); for example, at birth in the mouse, corticogenesis is underway, while corticogenesis has been complete in humans for ∼90 d at the time of birth. The expected contrast of the position of birth with respect to neural milestones can be confirmed for the precocial guinea pig versus the altricial mouse. The births of the cat and macaque are intermediate between those two, and human birth shows evidence of “secondary altriciality,” birth moved earlier with respect to neural events than the macaque.
Figure 9.
The position of birth for six placental mammals relative to the event scale (x-axis); the age of each mammal in PC days can be read for birth (or any event scale value) on the y-axis. The five placental mammals are chosen to represent close to the full range of the dataset and include one highly precocial mammal, the guinea pig. For an example, in the mouse at birth cortical neurogenesis is still underway and synaptogenesis in the forebrain is only beginning, while in the guinea pig at birth, cortical neurogenesis, cortical cell migration, and basic axonogenesis is entirely complete, and the point of peak synaptic density has passed (human (blue dot), macaque (red dot), cat (yellow cat), guinea pig (green dot), and mouse (black dot).
Slope and onset of progression through neural events
Considering again only placental mammals, one major difference that can be seen is the time required to reach specified neural milestones (Fig. 2, log scale). For example, it takes the mouse ∼12 d to reach 40% of adult brain size, while it takes humans ∼120 d to reach the same milestone.
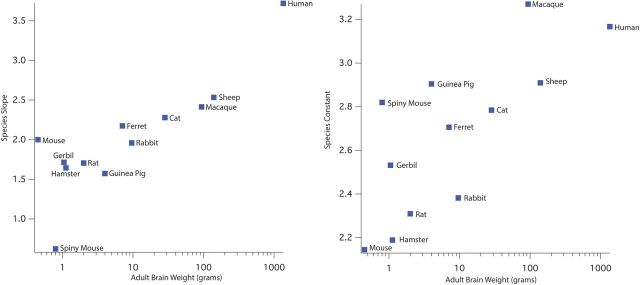
To determine whether there is a systematic relationship between these developmental parameters and adult species characteristics, we compared the relationship (for the 12 eutherian species) between our “onset” and “slope” scales to four other species characteristics: logged brain size; brain size adjusted for body size; gestational length; and a measure of maturity at birth, defined as log (date of weaning) minus log (gestation). For the latter measure of maturity, we assume that the longer weaning comes after birth, the more altricial the animal. Each of the two scales (onset and slope) was predicted by stepwise backward regression from the four species characteristics. Each stepwise process left only one species characteristic as important. Adult brain weight (ln) correlates 0.8932 with slope (Fig. 10, left) but correlates poorly with gestational length (Fig. 10, right). In Figure 10, right, a noticeable ranking of species with precociality on the y-axis can be observed. Gestational length, however, correlates 0.9285 with the species intercept, which finds onset of neurogenesis (not graphed). As slope is also highly (but not perfectly) correlated with the total duration of neurodevelopment, from first neurogenesis to 80% of adult brain weight, this observation echoes Passingham's (1985) observation of the log-linear relationship of the brain size at birth and duration of gestation across a variety of placental mammals.
Figure 10.
The relationship of adult brain weight (x-axis, log scale), modeled slope (left), and intercept (right) values for all eutherian mammals. The slope of the modeled regression line (left) can be used as a measure of the number of days, and thus the total developmental duration required to reach the last maturational point measured is closely related to brain weight. The intercept (right), which gives the PC age when the first neurodevelopmental events occur, distinguishes precocial from altricial animals, and also relates to brain size. Of the rodents, for example, the precocial spiny mouse and guinea pig have the highest intercepts, while the altricial mouse and hamster have the lowest.
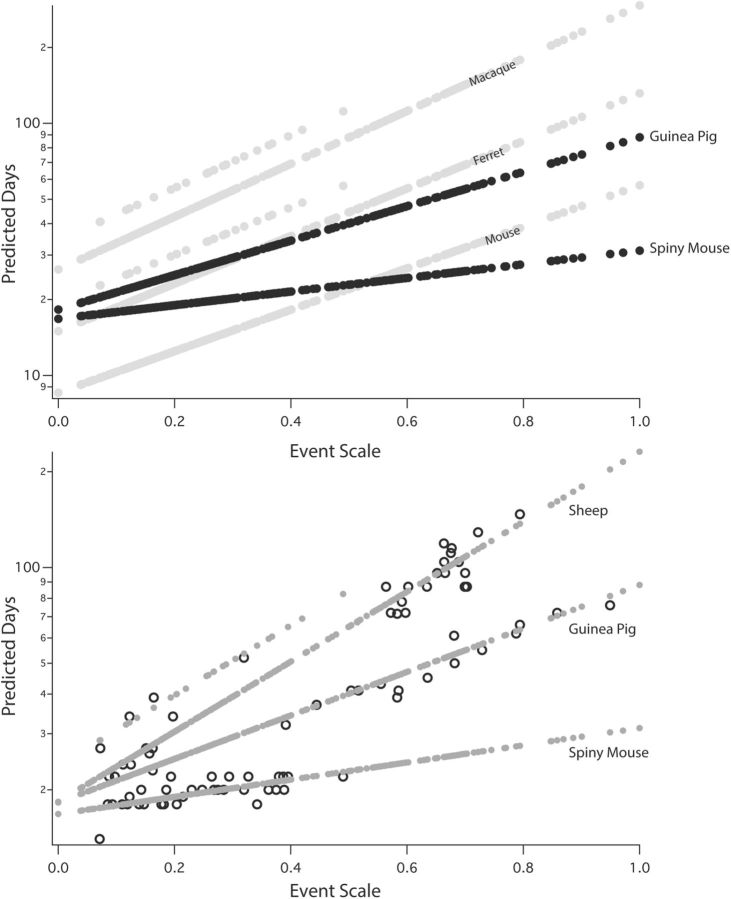
Comparing the spiny mouse to the mouse, for example, the first neurodevelopmental events occur quite late, beginning at around PC day 16, compared with approximately PC day 9 in the mouse, and thereafter, the progress through stages of development occurs extremely rapidly until its late birth at PC Day 39 (Fig. 11, top). This observation is very surprising, as it would seem that a species with a premium on advanced maturation at birth should not “waste” a full week in the generation of its progenitor pool, which is unlikely to be different in number of progenitors in the laboratory mouse and the spiny mouse. A similar distinction can be seen for the guinea pig versus the ferret. As we were concerned that some unexpected underpopulation of the dataset might be responsible for this, we plotted the empirical values versus modeled values for three representative precocial animals: the sheep, guinea pig, and spiny mouse (Fig. 11, bottom). We confirmed that there was both adequate data and good fit of data to model for both early neuroembryological events and later events close to birth, particularly so for the sheep.
Figure 11.
Top, Modeled regression slopes for two highly precocial rodents, the spiny mouse and guinea pig, compared with the altricial mouse (M. musculus) whose brain weight is similar to the spiny mouse, and the relatively altricial ferret, whose brain weight is close to the guinea pig; the regression slope for the macaque is also plotted. x-axis is the event scale, and the y-axis is predicted PC day (logged). Both precocial cases show the delayed onset of in utero neurodevelopment and very rapid progress through developmental stages characteristic of the precocial group. Elevated points associated with the ferret and macaque regression lines are the non-glires*corticogenesis interaction term. Bottom, Empirical data density and distribution, and fit of data to model, for the three most precocial species, the spiny mouse, guinea pig, and sheep. All modeled points are represented in gray, while the points for which there are empirical measurements are black circles.
Calculating an index of precociality
Omitting the few events involved in an interaction, the predicted time of any event can be given as y = onset + slope*eventscore, where the onset and slope values are specific to the species, and the eventscore is the event's value on the event scale. Here, gestation is gest = log(days of gestation). If we define gest = y = onset+ slope*eventscore and solve for eventscore, we get eventscore = (gest − onset)/slope.
This gives the eventscore of events, which we estimate will occur at the same time as birth. For our 271 events, eventscores run from 0 to 1. So the more developed a species is at birth (i.e., the more precocial it is), the higher that eventscore.
The Precocial Score column of Table 2 lists the eventscores calculated in this manner. The precocial index could be used as an empirically defined definition of precociality for eutherians, at least relative to neural development. This index could be useful to researchers concerned about using altricial mammals, like rats and mice, to model events in humans to guide the choice of alternate models, or account for scaling differences. Table 2 also shows precocial scores calculated for marsupials, but using pouch-exit instead of uterus-exit as comparable to birth in placental mammals (Darlington at al., 1999) and the common slope for all marsupials described earlier (2.5083). The marsupial values fall in the same general range as the non-glires eutherians, though higher than all the glires except the guinea pig and spiny mouse.
Discussion
We describe a model of extended neural development for a large number of mammalian species, incorporating all of the basic neuroembryological events such as neurogenesis, axon extension, synaptogenesis, and axonal sorting, but also brain growth, postnatal myelination, and early behavioral events. Neural maturation can be described by a single model predicting the timing of all events in all examined species, with a correlation of modeled values to empirical data of 0.9929. Considering the very wide range of species and developmental durations encompassed by the model, the data transformations of the model producing this close fit are minimal: one essentially linear event scale fits all species and a separate regression equation for each species characterizing days required for each one to progress through the event scale. The duration of neurodevelopment in days, as indicated by the slope of each species' regression line, is highly correlated with eventual brain size. Prior observations of the relationship of prosomeric location to neurogenesis (lateral = late) and slow maturation in marsupials were replicated in this tripled dataset. Unexpectedly, precocial mammals delay the onset of neurodevelopment in utero, when it would seem more likely that they would begin neural development early to reach their highly mature state at birth. Finally, selective delay of isocortical genesis in all species but glires (i.e., rodents, rabbit) was observed, casting doubt on the idea that a relatively enlarged cortex is a derived characteristic of primates alone.
Considerations in the choice of model
The representation of development that this model produces differs from our prior models (Finlay and Darlington, 1995; Clancy et al., 2001, 2007) in that each species' rate of development, the number of days required to move through fixed development, varies independently for each species (but with metatherians kept as a single, slower developing group). In the prior model, all eutherian species were forced to have a single slope, which fit adequately because the data range was truncated before the point of human birth (compare the data ranges before this truncation in Figs. 2, 9). This point occurs before the relative differences in the slopes in the largest brained species begin to show substantial effects. The prior model seriously underestimates the time of occurrence of late events in large-brained species. Considering the evolutionary significance of this description, it suggests that selection for any trait associated with large brain size could also be selection for a particular developmental duration and rate, and the reverse (Charvet and Finlay, 2012).
Because of the possibility of overfitting, we were very conservative in the interaction terms we accepted. In that way, we have almost certainly understated the occurrences of shifts in the timing of neural processes that are one likely source of “grade shifts” between taxa observed in the literature on vertebrate brain allometry (de Winter and Oxnard, 2001; Iwaniuk et al., 2004; Charvet et al., 2010; Kingsbury et al., 2012). However, the organization of this database is designed to be hospitable to evo–devo hypothesis testing, where suggestive trends observed in the existing data can be followed by predictions of the relative timing in new species for neural events not yet measured, or predictions can be made for categories not yet explored here. Our prediction and test of later retinogenesis in the cat, derived from our observations in the nocturnal owl monkey (Dyer et al., 2009), are of that kind.
Onset, duration, and rate in the development of the nervous system
Overall patterns
The most striking feature of mammalian neural development is the preservation of the order of developmental events across species in the face of the extreme range of developmental duration. The basic order of the 271 events we have analyzed is preserved over developmental periods ranging between 25 and 1000 d. Some of this order is necessary and unremarkable: for example, neuron birth must precede axon growth, which must precede behavior. The coordination of potentially separable systems, however, appears to be equally conserved with such required features. For example, large features like the order of deployment and organization of separate sensory systems, like visual and somatosensory systems, stay constant. From the neurons that comprise them to the behaviors dependent on each sensory system, none become notably offset, or heterochronic comparing the species examined here. We have no case where any sensory system has wholesale acceleration or retardation with respect to any other. Finer points of organization, like the progression of neurogenesis in the thalamus from primary sensory nuclei to association nuclei, are also preserved from the smallest brains to the largest ones, and across taxa (Finlay et al., 2001; Stevens, 2001). Some often noted phylogenetic regularities in brain scaling, like the relatively constant ratio relationship of the cerebellum and isocortex, would appear to fall naturally out of their consistent relative timing of neurogenesis of the brain's largest structures, keeping them in a reasonably constant relationship even though they share no direct synaptic or trophic contact that might serve to directly regulate their neuronal numbers (Clark et al., 2001; Herculano-Houzel, 2010).
Changes in duration
In altricial mammals, the duration of development correlates closely with the eventual size of the brain. The transformation of development seen in animals with small brains compared with larger brains is nonlinear: progress through later events is systematically slowed (Fig. 2). For example, it takes the rat ∼19 d to complete corticogenesis and a human ∼180, while to reach 80% of adult brain weight then requires an additional 50 d in the rat and 750 d in humans. Failure to properly calibrate for the exponential relationship of late developmental durations to brain size may potentially account for the widespread assumption in paleontology that humans have been selected for “extended development” decoupled from brain size (Locke and Bogin, 2006). Just as our disproportionately large isocortex is entirely predictable for a primate of our brain size, the duration of our development may be simultaneously “disproportionate” and entirely predictable, representing a necessary developmental linkage. This developmental linkage may supply an additional piece of the puzzle of unusually late skill competence in humans (Schuppli et al., 2012).
Changes in rate
The overall slowed developmental rates of metatherian mammals compared with eutherian mammals (Fig. 8), and the different rates of development producing equivalent amounts of neural mass in precocial and altricial species (Fig. 10), demonstrate conclusively that there is no unique amount of time associated with production of a unit amount of brain tissue, even though there may be predictable relationships of the two in subgroups like “altricial eutherian mammals.” Thus, the rate of neural development has been and could be a focus for selection pressure. Given the overwhelming evidence for relative size of both body and brain as a major focus of selection in vertebrate evolution (Eisenberg, 1981; Smaers et al., 2012), the necessity of understanding those features of the genome that control the duration or rate of somatic versus neural development becomes all the more important.
Interactions and grade shifts
We show direct evidence for heterochrony for the relative size of the cortex and for retinal adaptations, and a comparative approach allows a better classification of within-brain shifts in developmental timing. For example, it is often implicit that the enlarged primate isocortex and its associated selective delay in isocortical neurogenesis are derived features in primates and that special selection has occurred to enable it (Dunbar and Shultz, 2007). However, there are multiple grade shifts in both forebrain volume across vertebrates and in relative cortex volume within mammalian taxa, not simply a singular shift in primates (Finlay and Darlington, 1995; Yopak et al., 2010). The smaller isocortex of rodents and rabbits could just as well be the derived condition—cortical volume may have been selected against in those lineages to devote, perhaps, more neural tissue to the mesencephalon, or olfactory processing in the forebrain. Studies comparing gene expression across these taxonomic boundaries should consider this possibility explicitly and not assume primates are the derived pattern.
Neurogenesis onset and precociality
A major surprise of this model was the systematic delay in the onset of first neurogenesis in precocial mammals compared with altricial ones followed by an extremely rapid progression (as measured in days) through developmental milestones up to their late birth. At about the point of eye opening, maturational points appear to be approximately equivalent, but before that point, altricial species animals have spent more time in the period of neurogenesis, axon extension, and synaptogenesis, while the precocial animal species have spent more relative time in the generation of its precursor pool. The spiny mouse, compared with the mouse, is the most extreme example of this discrepancy, but other precocial–altricial pairs show the same pattern of compression versus extension. The empirical values of multiple early developmental events support the model, making it unlikely that later maturational events have somehow forced bad values on early events in precocial mammals. For example, the mitral cells of the olfactory bulb are produced at 10.5 d in the laboratory mouse, but at 18 d in the spiny mouse. Why compress the events organizing the nervous system into a shortened period if there is a premium on completed brain function at birth for a precocial mammal? Perhaps the constraints that early birth puts on the relative functional development of non-neural organ systems (respiration, immunity, heart, and circulatory systems) dictates a different deployment of developmental resources toward critical organ systems development early on, which this dataset is blind to because of its focus on neural characteristics.
A different and also inexplicable delay in completion of the precursor pool can be seen in altricial animals. Comparing mouse to macaque, the first neurons are produced in mice at ∼9 d PC, while comparable neurons are not produced until about the 28th day in the macaque. While a first guess for this might be that these 20 d reflect the time required to produce the larger primordia required in monkeys, in fact, preliminary data from our laboratory focusing on rodents and carnivores suggest that the sizes of the isocortical primordia are only approximately twice as large at those equivalent developmental stages, with both tissue volumes quite small. As it seems unlikely any direct physical constraint prevents the monkey from supporting minimally larger precursor regions, a deeper answer should be sought in the mutual regulation of brain with other somatic organ systems or control of cytogenesis over the lifetime of the animal.
Biomedical research and the “animal model”
Provision of accurate information about the equivalence of developmental time periods for animal models of human brain disorders, and to provide a broader reference frame to understand “abnormality” is a central motivation for this work. Little empirically based vocabulary exists to describe normality and abnormality of neurodevelopmental timing. Though the words “retardation” or “developmental delay” are sometimes used to describe developmental disorders, or “precocity” for unusual intelligence, these terms have a paradoxical relationship to cross-species comparisons in that it is the largest, most complex brains that have the most “delayed” development. As reference corpora of brain development of atypically and typically developing children using various imaging methods are rapidly being established, it will be possible to address abnormalities in the rate and order of development with respect to the stability of neurodevelopment across species and describe them appropriately (Brooksbank et al., 1989; Kesslak et al., 1994; Caviness et al., 1999; Shaw et al., 2006; Raznahan et al., 2012). The extensive literature on the evolution of vertebrate, mammalian, and hominin life histories and how they are adapted to resources and reproductive tactics, describes the advance, delay, extension, and compression of a wide number of such events as reproductive onset and cessation, interbirth interval, and duration of childhood (Hawkes, 2006; Flatt and Heyland, 2011). Comparative neuroembryologists, anthropologists, and clinicians are now holding separate parts of a larger developmental puzzle, which might now be merged.
Footnotes
This work is supported by National Science Foundation IBN 0849612 and NSF 0849627, National Institutes of Health (NIH) Grant #P20 RR-16460 from the IDeA Networks of Biomedical Research Excellence Program of the National Center for Research Resources, NIH CHHD Grant #U10 HD-500009, and an NIH CHHD 1F32HD067011-01A1 fellowship to C.J.C. We thank Douglas Elrod for his help in the implementation of the quasi-Newton optimization techniques; Diarmuid Cahalane for help in graphics; and Christopher D. Gaffney, Nikki Kimura, B.A. Choi, Terri Teague-Ross, Julie Staudinger, and Amanda James for their work in development of the database. We thank Dr. Drew Noden for access and help with the Cornell School of Veterinary Medicine Embryological Collection.
The authors declare no competing financial interests.
References
- Brooksbank BW, Walker D, Balázs R, Jørgensen OS. Neuronal maturation in the foetal brain in Down's syndrome. Early Hum Dev. 1989;18:237–246. doi: 10.1016/0378-3782(89)90019-4. [DOI] [PubMed] [Google Scholar]
- Bryden MM, Evans HE, Binns W. Embryology of the sheep. I. Extraembryonic membranes and the development of body form. J Morphol. 1972;138:169–185. doi: 10.1002/jmor.1051380204. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Lange NT, Makris N, Herbert MR, Kennedy DN. MRI-based brain volumetrics: emergence of a developmental brain science. Brain Dev. 1999;21:289–295. doi: 10.1016/S0387-7604(99)00022-4. [DOI] [PubMed] [Google Scholar]
- Chalfin BP, Cheung DT, Muniz JA, de Lima Silveira LC, Finlay BL. Scaling of neuron number and volume of the pulvinar complex in New World primates: comparisons with humans, other primates and mammals. J Comp Neurol. 2007;504:265–274. doi: 10.1002/cne.21406. [DOI] [PubMed] [Google Scholar]
- Chalupa LM, Williams RW. Eye, retina and visual system of the mouse. Cambridge, Massachusetts: MIT; 2008. [Google Scholar]
- Charvet CJ, Finlay BL. Embracing covariation in brain evolution: large brains, extended development and flexible primate social systems. Prog Brain Res. 2012;195:71–87. doi: 10.1016/B978-0-444-53860-4.00004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Sandoval AL, Striedter GF. Phylogenetic origins of early alterations in brain region proportions. Brain Behav Evol. 2010;75:104–110. doi: 10.1159/000300573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. The course of human events: predicting the timing of primate neural development. Dev Sci. 2000;3:57–66. doi: 10.1111/1467-7687.00100. [DOI] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. doi: 10.1016/S0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- Clancy B, Kersh B, Hyde J, Anand KJ, Darlington RB, Finlay BL. Web-based method for translating neurodevelopment from laboratory species to humans. Neuroinformatics. 2007;5:79–94. doi: 10.1385/ni:5:1:79. [DOI] [PubMed] [Google Scholar]
- Clancy B, Teague-Ross TJ, Nagarajan R. Cross-species analyses of the cortical GABAergic and subplate neural populations. Front Neuroanat. 2009;3:20. doi: 10.3389/neuro.05.020.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DA, Mitra PP, Wang SS. Scalable architecture in mammalian brains. Nature. 2001;411:189–193. doi: 10.1038/35075564. [DOI] [PubMed] [Google Scholar]
- Darlington RB. Regression and linear models. New York: McGraw-Hill; 1990. [Google Scholar]
- Darlington RB, Dunlop SA, Finlay BL. Neural development in metatherian and eutherian mammals: variation and constraint. J Comp Neurol. 1999;411:359–368. doi: 10.1002/(SICI)1096-9861(19990830)411:3<359::AID-CNE1>3.3.CO%3B2-A. [DOI] [PubMed] [Google Scholar]
- de Winter W, Oxnard CE. Evolutionary radiations and convergences in the structural organization of mammalian brains. Nature. 2001;409:710–714. doi: 10.1038/35055547. [DOI] [PubMed] [Google Scholar]
- Dreher B, Robinson SR. Development of the retinofugal pathway in birds and mammals: evidence for a common timetable. Brain Behav Evol. 1988;31:369–390. doi: 10.1159/000116602. [DOI] [PubMed] [Google Scholar]
- Dunbar RI, Shultz S. Evolution in the social brain. Science. 2007;317:1344–1347. doi: 10.1126/science.1145463. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Martins R, da Silva Filho M, Muniz JA, Silveira LC, Cepko CL, Finlay BL. Developmental sources of conservation and variation in the evolution of the primate eye. Proc Natl Acad Sci U S A. 2009;106:8963–8968. doi: 10.1073/pnas.0901484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter SS, Jr, Ross LS, Frankfurter A. Initial tract formation in the mouse brain. J Neurosci. 1993;13:285–299. doi: 10.1523/JNEUROSCI.13-01-00285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg JF. Chicago: The University of Chicago; 1981. The mammalian radiations: an analysis of trends in evolution, adaptation and behavior. [Google Scholar]
- Finlay BL, Clancy B. Chronology of the development of the mouse visual system. In: Chalupa L, Williams RW, editors. Eye, retina and visual system of the mouse. Cambridge, MA: MIT; 2008. pp. 257–265. [Google Scholar]
- Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Hersman MN, Darlington RB. Patterns of vertebrate neurogenesis and the paths of vertebrate evolution. Brain Behav Evol. 1998;52:232–242. doi: 10.1159/000006566. [DOI] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB, Nicastro N. Developmental structure of brain evolution. Behav Brain Sci. 2001;24:263–308. doi: 10.1017/S0140525X01003958. [DOI] [PubMed] [Google Scholar]
- Flatt T, Heyland A. Mechanisms of life history evolution. Oxford, UK: Oxford UP; 2011. [Google Scholar]
- Garcia-Lopez R, Pombero A, Martinez S. Fate map of the chick embryo neural tube. Dev Growth Differ. 2009;51:145–165. doi: 10.1111/j.1440-169X.2009.01096.x. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Ontogeny and phylogeny. Cambridge, Massachusetts: Harvard UP; 1977. [Google Scholar]
- Harris HA. The foetal growth of the sheep. J Anat. 1937;71:516–527. [PMC free article] [PubMed] [Google Scholar]
- Hawkes K. Slow life histories and human evolution. In: Hawkes K, Paine RR, editors. The evolution of human life history. Santa Fe, NM: School of American Research; 2006. pp. 95–126. [Google Scholar]
- Herculano-Houzel S. 2010 Coordinated scaling or cortical and cerebellar numbers of neurons. Front Neuroanat. 2010;4:12. doi: 10.3389/fnana.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaniuk AN, Dean KM, Nelson JE. A mosaic pattern characterizes the evolution of the avian brain. Proc Biol Sci. 2004;271(Suppl4):S148–151. doi: 10.1098/rsbl.2003.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerison H. Evolution of the brain and intelligence. New York: Academic; 1973. [Google Scholar]
- Kesslak JP, Nagata SF, Lott I, Nalcioglu O. Magnetic resonance imaging analysis of age-related changes in the brains of individuals with Down's syndrome. Neurology. 1994;44:1039–1045. doi: 10.1212/wnl.44.6.1039. [DOI] [PubMed] [Google Scholar]
- Kingsbury MA, Gleason ED, Ophir AG, Phelps SM, Young LJ, Marler CA. Monogamous and promiscuous rodent species exhibit discrete variation in the size of the medial prefrontal cortex. Brain Behav Evol. 2012;80:4–14. doi: 10.1159/000339247. [DOI] [PubMed] [Google Scholar]
- Kostovic I, Rakic P. Cytology and time of origin of interstitial neurons in the white matter in infant and adult human and monkey telencephalon. J Neurocytol. 1980;9:219–242. doi: 10.1007/BF01205159. [DOI] [PubMed] [Google Scholar]
- Le Magueresse C, Monyer H. GABAergic interneurons shape the functional maturation of the cortex. Neuron. 2013;77:388–405. doi: 10.1016/j.neuron.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Locke JL, Bogin B. Language and life history: a new perspective on the development and evolution of human language. Behav Brain Sci. 2006;29:259–325. doi: 10.1017/s0140525x0600906x. [DOI] [PubMed] [Google Scholar]
- Lumsden A. The cellular basis of segmentation in the developing hindbrain. Trends Neurosci. 1990;13:329–335. doi: 10.1016/0166-2236(90)90144-Y. [DOI] [PubMed] [Google Scholar]
- Lumsden A. Segmentation and compartition in the early avian hindbrain. Mech Dev. 2004;121:1081–1088. doi: 10.1016/j.mod.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Passingham RE. Rates of brain development in mammals including man. Brain Behav Evol. 1985;26:167–175. doi: 10.1159/000118773. [DOI] [PubMed] [Google Scholar]
- Puelles L. A segmental morphological paradigm for understanding vertebrate forebrains. Brain Behav Evol. 1995;46:319–337. doi: 10.1159/000113282. [DOI] [PubMed] [Google Scholar]
- Puelles L, Rubenstein JL. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
- Raznahan A, Greenstein D, Lee NR, Clasen LS, Giedd JN. Prenatal growth in humans and postnatal brain maturation into late adolescence. Proc Natl Acad Sci U S A. 2012;109:11366–11371. doi: 10.1073/pnas.1203350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reep RL, Darlington RB, Finlay BL. The limbic system in mammalian brain evolution. Brain Behav Evol. 2007;70:57–70. doi: 10.1159/000101491. [DOI] [PubMed] [Google Scholar]
- Robson SL, van Schaik CP, Hawkes K. The derived features of human life history. In: Hawkes K, Paine RR, editors. The evolution of human life history. Santa Fe, NM: School of American Research; 2006. pp. 17–44. [Google Scholar]
- Ross CF, Kirk EC. Evolution of eye size and shape in primates. J Hum Evol. 2007;52:294–313. doi: 10.1016/j.jhevol.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Schuppli C, Isler K, van Schaik CP. How to explain the unusually late age at skill competence among humans. J Hum Evol. 2012;63:843–850. doi: 10.1016/j.jhevol.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, Evans A, Rapoport J, Giedd J. Intellectual ability and cortical development in children and adolescents. Nature. 2006;440:676–679. doi: 10.1038/nature04513. [DOI] [PubMed] [Google Scholar]
- Smaers JB, Dechmann DK, Goswami A, Soligo C, Safi K. Comparative analyses of evolutionary rates reveal different pathways to encephalization in bats, carnivorans, and primates. Proc Natl Acad Sci U S A. 2012;109:18006–18011. doi: 10.1073/pnas.1212181109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Liu L, Edwards SV, Wu S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci U S A. 2012;109:14942–14947. doi: 10.1073/pnas.1211733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CF. An evolutionary scaling law for the primate visual system and its basis in cortical function. Nature. 2001;411:193–195. doi: 10.1038/35075572. [DOI] [PubMed] [Google Scholar]
- Workman AD, Gaffney CD, Choi BA, Charvet CJ, Clancy B, Darlington RB, Finlay BL. Translating time: extending a model of the timing of early neurodevelopmental events in mammals to cross-vertebrate structural, physiological and behavioral postnatal maturation. Soc Neurosci Abstr. 2011;37:638.05. [Google Scholar]
- Workman AD, Charvet CJ, Gaffney CD, Clancy B, Darlington RB, Finlay BL. Translating time: unexpected developmental characteristics of standard mammalian models. Soc Neurosci Abstr. 2012;38 639:05. [Google Scholar]
- Yopak KE, Lisney TJ, Darlington RB, Collin SP, Montgomery JC, Finlay BL. A conserved pattern of brain scaling from sharks to primates. Proc Natl Acad Sci U S A. 2010;107:12946–12951. doi: 10.1073/pnas.1002195107. [DOI] [PMC free article] [PubMed] [Google Scholar]