Abstract
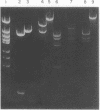
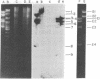
Genes encoding thymidylate synthetase from Bacillus subtilis bacteriophages were cloned in Escherichia coli. Chimeric plasmids pCD1 and pCD3 were constructed from site-specific endonuclease digests of bacteriophage phi3T DNA cloned in pMB9 in E. coli. Similar cloning techniques with bacteriophage beta22 DNA yielded chimeric plasmids pCD4, pCD5, and pCD6. Endonuclease digests of DNA from pCD1 and pCD3 propagated in E. coli or from DNA isolated from bacteriophage phi3T propagated in B. subtilis transformed B. subtilis from Thy- to Thy+. Intact DNA from bacteriophage beta22, endonuclease digests of beta22 DNA, and a chimeric plasmid (pCD5) composed only of the thybeta22 gene and pMB9 did not transform B. subtilis from Thy- to Thy+ even though pCD5 could transform Thy- E. coli to Thy+. However, if the thybeta22 fragment from pCD5 was introduced into another chimeric plasmid, pCD2, that contains a region of homology to the chromosome of B. subtilis in addition to pMB9, transformation of Thy- clones of B. subtilis was possible. Furthermore, Southern hybridization analyses of the digests of chromosomal DNA from the Thy+ transformants established that the entire chimeric plasmid was incorporated into the chromosome of B. subtilis. Treatment of these plasmids with site-specific endonucleases abolished transformation. These results indicated that the entire chimeric plasmid can be incorporated into the chromosome of B. subtilis by a Campbell-like model. Therefore, an additional mechanism for transformation exists whereby plasmids can be integrated if sufficient chromosomal homology is maintained.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Boylan R. J., Mendelson N. H., Brooks D., Young F. E. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J Bacteriol. 1972 Apr;110(1):281–290. doi: 10.1128/jb.110.1.281-290.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidoff-Abelson R., Dubnau D. Kinetic analysis of the products of donor deoxyribonucleate in transformed cells of Bacillus subtilis. J Bacteriol. 1973 Oct;116(1):154–162. doi: 10.1128/jb.116.1.154-162.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C. H., Wilson G. A., Young F. E. Transformation of Bacillus subtilis and Escherichia coli by a hybrid plasmid pCD1. Gene. 1977 Mar;1(2):153–167. doi: 10.1016/0378-1119(77)90026-9. [DOI] [PubMed] [Google Scholar]
- Ehrlich S. D., Bursztyn-Pettegrew H., Stroynowski I., Lederberg J. Expression of the thymidylate synthetase gene of the Bacillus subtilis bacteriophage Phi-3-T in Escherichia coli. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4145–4149. doi: 10.1073/pnas.73.11.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R. S., Young F. E., Wilson G. A. Effect of site-specific endonuclease digestion on the thyP3 gene of bacteriophage phi 3T and the thyP11 gene of bacteriophage rho11. Gene. 1977 Mar;1(2):169–180. doi: 10.1016/0378-1119(77)90027-0. [DOI] [PubMed] [Google Scholar]
- Lovett P. S. Plasmid in Bacillus pumilus and the enhanced sporulation of plasmid-negative variants. J Bacteriol. 1973 Jul;115(1):291–298. doi: 10.1128/jb.115.1.291-298.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler I., Halvorson H. O. Transformation of Escherichia coli and Bacillus subtilis with a hybrid plasmid molecule. J Bacteriol. 1977 Jul;131(1):374–377. doi: 10.1128/jb.131.1.374-377.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Wilson G. A., Young F. E. Recognition sequence of specific endonuclease BamH.I from Bacillus amyloliquefaciens H. Nature. 1977 Jan 6;265(5589):82–84. doi: 10.1038/265082a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tucker R. G. Acquisition of thymidylate synthetase activity by a thymine-requiring mutant of Bacillus subtilis following infection by the temperate phage phi 3. J Gen Virol. 1969 Jun;4(4):489–504. doi: 10.1099/0022-1317-4-4-489. [DOI] [PubMed] [Google Scholar]
- Williams M. T., Young F. E. Temperate Bacillus subtilis bacteriophage phi 3T: chromosomal attachment site and comparison with temperate bacteriophages phi 105 and SPO2. J Virol. 1977 Feb;21(2):522–529. doi: 10.1128/jvi.21.2.522-529.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. A., Williams M. T., Baney H. W., Young F. E. Characterization of temperate bacteriophages of Bacillus subtilis by the restriction endonuclease EcoRI: evidence for three different temperate bacteriophages. J Virol. 1974 Oct;14(4):1013–1016. doi: 10.1128/jvi.14.4.1013-1016.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. A., Young F. E. Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. J Mol Biol. 1975 Sep 5;97(1):123–125. doi: 10.1016/s0022-2836(75)80028-3. [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Wilson G. A., Young F. E. Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. J Bacteriol. 1975 Jan;121(1):296–304. doi: 10.1128/jb.121.1.296-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehle C. O., Doi R. H. Differential expression of bacteriophage genomes in vegetative and sporulating cells of Bacillus subtilis. J Virol. 1967 Oct;1(5):935–947. doi: 10.1128/jvi.1.5.935-947.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]