Abstract
Summary: Experimental MSn mass spectral libraries currently do not adequately cover chemical space. This limits the robust annotation of metabolites in metabolomics studies of complex biological samples. In silico fragmentation libraries would improve the identification of compounds from experimental multistage fragmentation data when experimental reference data are unavailable. Here, we present a freely available software package to automatically control Mass Frontier software to construct in silico mass spectral libraries and to perform spectral matching. Based on two case studies, we have demonstrated that high-throughput automation of Mass Frontier allows researchers to generate in silico mass spectral libraries in an automated and high-throughput fashion with little or no human intervention required.
Availability and implementation: Documentation, examples, results and source code are available at http://www.biosciences-labs.bham.ac.uk/viant/hammer/.
Contact: m.viant@bham.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Mass spectrometry (MS)-based metabolomics is a rapidly developing field that aims to detect and measure a variety of small biological molecules (metabolites) over a wide dynamic range (Dettmer et al., 2007). Although thousands of metabolites are typically detected in an untargeted metabolomics study of a biological sample, their subsequent identification represents the most significant bottleneck in the discovery of new biochemical knowledge (Dunn et al., 2013; Kind and Fiehn, 2010; Wishart, 2011). In many cases, multiple empirical formulae and/or putative chemical structures are reported for each observed mass feature (or more strictly mass-to-charge ratio, m/z). Multistage (MSn) mass spectrometry, which is an experimental technique to collect in-depth fragmentation data related to the chemical structure of metabolites, is often applied to increase accuracy and specificity in metabolite annotation and identification. However, experimental MSn mass spectral libraries currently do not adequately cover the search space for all metabolites present in complex biological samples, as authentic chemical standards are not available for all metabolites. The generation of in silico MSn libraries is anticipated to greatly improve the success rate of annotation of metabolites detected in metabolomics studies, when experimentally acquired MSn data of authentic chemical standards are unavailable. Commercial software packages such as Mass Frontier and ACD/MS Fragmenter predict in silico fragmentation patterns to assist the interpretation of experimental MSn data (Krauss et al., 2010; Oppermann et al., 2012). Additionally, a freely available tool named MetFrag has been developed previously (Wolf et al., 2010). This tool selects candidate structures from a compound library based on the molecular ion m/z and generates in silico substructures to subsequently annotate fragment peaks. MetFrag does not provide the ability to create in silico patterns or libraries independent of the use of experimental data; however, it has been proven to be an efficient approach to annotate fragmentation mass spectra derived from MS experiments.
Mass Frontier is a well-known software package that has been used by more than a thousand academic institutions and companies throughout the world for the management, evaluation and interpretation of mass spectra. It includes three resources that assist the user to robustly predict in silico fragmentation patterns: a set of general fragmentation rules, a fragmentation library comprising ∼150 000 fragmentation mechanisms collected from the scientific literature and user-specified mechanisms. However, Mass Frontier cannot readily perform batch processing on the scale required to generate comprehensive in silico libraries for thousands of metabolites.
Here, we present a freely available software package, named HAMMER (High-throughput AutoMation of Mass frontiER), to automatically control Mass Frontier software to perform in silico fragmentation in an automated and high-throughput matter. This package allows the user to readily generate large-scale in silico fragmentation libraries and perform mass spectral matching to mass spectrometry-derived data.
2 METHODS AND IMPLEMENTATION
HAMMER is developed under the Python language and supports Windows 7 and XP (both 32 bit and 64 bit). Its software requirements include Java 6/7, Open Babel (O'Boyle et al., 2011), Sikuli (Yeh et al., 2009) and Mass Frontier 7. A desktop PC with 2.66 GHz CPU and 4 GB memory was used as the operating platform.
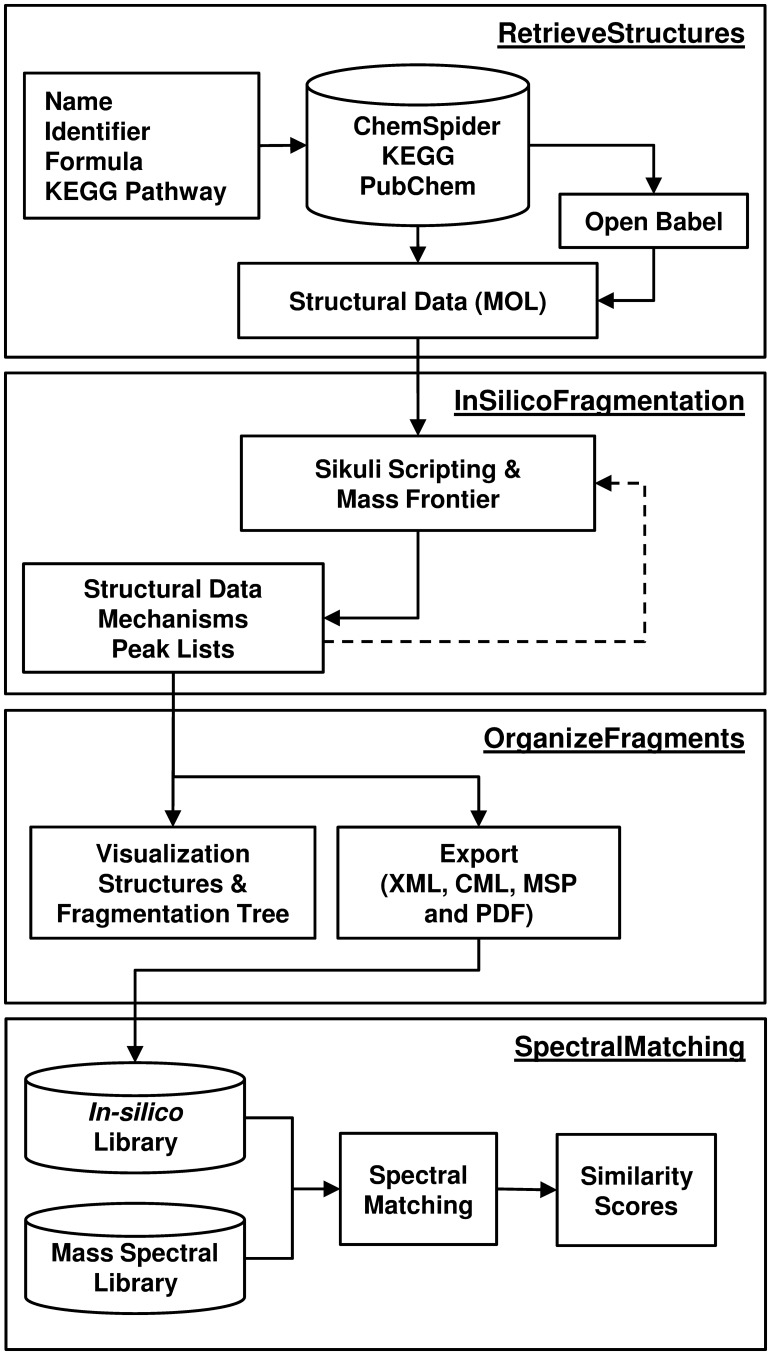
As shown in Figure 1, HAMMER consists of four modules:
RetrieveStructures retrieves chemical structures from a compound database of interest such as ChemSpider, Kyoto Encyclopedia of Genes and Genomes (KEGG) and PubChem. The retrieval is based on a submitted plain text file that includes a list of compounds or chemicals (with specific names, database identifier or chemical formula). Additionally, metabolites within specific KEGG pathways can be downloaded based on a KEGG pathway identifier. Candidate entries found in the databases of interest are automatically downloaded, verified and converted into .mol format (required by Mass Frontier) or other formats such as SMILES, InChI or empirical formula. Open Babel is used to perform structure verification and conversion. A report is generated to summarize the information retrieved for each compound (e.g. number of candidates, database IDs and URL to the specific database entry).
InSilicoFragmentation automatically controls the operation of Mass Frontier by using the open-source visual scripting software Sikuli. Sikuli allows control of software when no application programming interface is available, which is the case for many commercial software packages. It readily uses image recognition of graphical user interface (GUI) elements to operate software. HAMMER contains a Sikuli standalone runtime. As a result, the InSilicoFragmentation module works without any setup, otherwise simple configuration is required. Two versions of InSilicoFragmentation are provided: Windows 7 and Windows XP. We have separated the GUI images (or patterns) and search regions from the source code itself. This allows the user to modify the script more easily if required (e.g. applying a different version of Mass Frontier, see online manual for details). The structure file (.mol) of each compound is separately imported into Mass Frontier, and in silico fragmentation is performed applying user-defined fragmentation settings (see online manual for details). Structural and m/z information for each fragment is collected for each in silico reaction and if required (i.e. for MSn) is used for the next fragmentation step. This use allows closed-loop operation of Mass Frontier to perform ‘multistage’ in silico fragmentation. In silico fragments, their corresponding unique m/z values, chemical structures (in each stage) and fragmentation mechanisms are exported for further analysis and visualization.
OrganiseFragments uses the in silico fragmentation results as its input. The fragments are organized systematically in separate folders according to the compound of interest, and a 2D chemical structure image file in.png format is generated for each fragment. The in silico fragmentation results are exportable in several formats, such as in extensible mark-up language and plain text formats, as well as chemical mark-up language and National Institute of Standards and Technology spectra library files (NIST-MSP) for further analysis. These formats are compatible with common mass spectrometry databases and software packages. The hierarchical relationships of the fragments are parsed to an extensible mark-up language file that can easily be accessed using scripting languages. Additionally, a hierarchical tree visualization (as a .pdf) is generated to allow visual comparison or interpretation.
SpectralMatching computes a score, using the pMatch algorithm, to evaluate the probability of a match between an experimental fragmentation spectrum and an in silico fragmentation pattern in a library (Ye et al., 2010). The pMatch algorithm is a spectral matching algorithm originally developed for mass spectrometry-based protein identification (see Supplementary Information for details of the algorithm). The SpectralMatching module uses MSP files (see previous module), experimental and in silico, as its input. For each experimental fragmentation spectrum, a report is generated, which includes detailed information on the matching and annotation.
Fig. 1.
Workflow for HAMMER
3 CASE STUDIES
HAMMER was used to generate in silico fragmentation data for two distinct groups of compounds: (i) all 72 metabolites within the ‘phenylalanine metabolism’ KEGG pathway (i.e. map00360) and (ii) the top 200 most prescribed drugs in the USA in 2011 (RxList, 2011). These were selected to demonstrate the applicability and capability of HAMMER across diverse compound sets. Some drugs are composed of multiple compounds, and each compound was separately parsed and imported into Mass Frontier. Additionally, lower mass neutral and charged molecules/atoms (e.g. water and sodium) within these separated drug compounds were excluded from this demonstration dataset (see Supplementary Information for details). This resulted in 151 unique structures for the drugs dataset. See Supplementary Table S1 for specific settings used for both case studies. The run times for the phenylalanine and drug datasets were ∼8.5 and 14.5 h, respectively. On average, ∼3200 fragments were predicted for each compound (see Supplementary Tables S2–S4). Applying the defined Mass Frontier parameter settings, it is expected that MSn data are reported, where n > 2. This relatively high number of fragments illustrates the complexity and diversity of the fragmentation mechanisms. Although, these in silico fragments are predicted based on a set of general fragmentation rules and a fragmentation library comprising ∼150 000 fragmentation mechanisms collected from the scientific literature, the numbers of fragments produced are highly dependent on the complexity of the chemical structures and the parameter settings defined by the user (see Supplementary Information and online manual for details). In silico fragments may be reported, which are false positives; these fragments are not actually created in the MS fragmentation process, or are created but their low stabilities ensure that further and complete fragmentation or decomposition is observed before the ions are detected.
To assess the applicability of each in silico fragmentation library, five experimental fragmentation mass spectra were retrieved from MassBank to perform spectral matching [Supplementary Table S5, Case Study I: acetyl-CoA (KNA00207), capsaicin (WA001605), isobutyryl-CoA (PR100154), N-acetyl-L-phenylalanine (KO002200) and succinic acid (KZ000074). Case study II: amoxicillin (WA001751), digoxin (WA000563), meloxicam (WA002576), naproxen (WA000359) and prednisone (CO000368)] (Horai et al., 2010). All of the 10 real fragmentation spectra were correctly identified based on the data present in these small in silico mass spectral libraries (Supplementary Table S6).
It is important to highlight that the parameter settings used in Mass Frontier to perform in silico fragmentation play an important role in the correct prediction of in silico fragments and mechanisms. Additionally, understanding the experimental parameters used to collect experimental data (e.g. difference in collision energy or type of fragmentation) is also important to maximize metabolite annotation. However, these two challenges are outside the scope of the work presented here.
Both case studies show that HAMMER allows researchers to readily generate in silico MSn libraries and perform batch spectral matching of in silico mass spectral data to MS-derived data in an automated high-throughput fashion with minimal or no human intervention. In addition, we have shown with both case studies the value and applicability of visual scripting in the field of computational biology. We anticipate this software package will be used across a wide range of disciplines including metabolomics, organic synthetic chemistry and pharmaceutical research.
Funding: Royal Society International Exchanges 2011 NSFC funding (61211130120), National Natural Science Foundation of China (NSFC) Joint Fund with Guangdong (U1201256), NSFC funding (61171125 and 61001185), the UK Natural Environment Research Council (NE/I008314/1) and the Systems Science for Health initiative at The University of Birmingham.
Conflict of Interest: R.M. derives income from Mass Frontier sales.
Supplementary Material
REFERENCES
- Dettmer K, et al. Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W, et al. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics. 2013;9:44–66. [Google Scholar]
- Horai H, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- Kind T, Fiehn O. Advances in structure elucidation of small molecules using mass spectrometry. Bioanal. Rev. 2010;2:23–60. doi: 10.1007/s12566-010-0015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss M, et al. LC–high resolution MS in environmental analysis: from target screening to the identification of unknowns. Anal. Bioanal. Chem. 2010;397:943–951. doi: 10.1007/s00216-010-3608-9. [DOI] [PubMed] [Google Scholar]
- O'Boyle N, et al. Open babel: an open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann M, et al. High precision measurement and fragmentation analysis for metabolite identification. Methods Mol. Biol. 2012;860:145–156. doi: 10.1007/978-1-61779-594-7_10. [DOI] [PubMed] [Google Scholar]
- RxList 2011 The Internet Drug Index. Top 200 Drugs U.S. Only. http://www.rxlist.com/script/main/hp.asp (23 December 2013, date last accessed) [Google Scholar]
- Wishart DS. Advances in metabolite identification. Bioanalysis. 2011;3:1769–1782. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- Wolf S, et al. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye D, et al. Open MS/MS spectral library search to identify unanticipated post-translational modifications and increase spectral identification rate. Bioinformatics. 2010;26:i399–i406. doi: 10.1093/bioinformatics/btq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T, et al. Proceedings of the 22nd annual ACM symposium on User interface software and technology. BC, Canada: ACM, Victoria; 2009. Sikuli: using GUI screenshots for search and automation; pp. 183–192. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.