Abstract
Summary: We report on the development of the high-throughput screening (HTS) Navigator software to analyze and visualize the results of HTS of chemical libraries. The HTS Navigator processes output files from different plate readers' formats, computes the overall HTS matrix, automatically detects hits and has different types of baseline navigation and correction features. The software incorporates advanced cheminformatics capabilities such as chemical structure storage and visualization, fast similarity search and chemical neighborhood analysis for retrieved hits. The software is freely available for academic laboratories.
Availability and implementation: http://fourches.web.unc.edu/
Contact: fourches@email.unc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
With the growing number of academic centers (such as UCLA’s Molecular Screening Shared Resources—MSSR, NIMH’s Psychoactive Drug Screening Program—PDSP or Molecular Libraries Screening Center Network—MLSCN) conducting high-throughput screening (HTS) comes the need for processing HTS results efficiently. Some of these centers have the potential to screen up to 200 000 compounds against several panels of targets (e.g. GPCRs, kinases). As a result, significant amounts of screening data are generated, which require extensive management and analysis. Several commercial software packages (e.g. Spotfire by TIBCO, Vortex by DOTMATICS) are available to conduct these data analysis and mining tasks, but these are relatively expensive, have fixed sets of features and are not necessarily suited for academic structures and small biotech companies. As a consequence, there is a need for freely available and easily customizable software capable of visualizing and analyzing HTS results with an eye toward more extensive and chemical analysis using advances in cheminformatics for HTS data analysis (Kümmel and Parker, 2011).
2 FEATURES IMPLEMENTED IN NAVIGATOR
Different streams of information are involved and produced in HTS platforms. The current version of HTS Navigator offers several important capabilities for data storage, analysis and visualization.
2.1 Loading of multiple output files from different plate readers
The Plate Loader module processes and organizes collections of output files generated by multiple plate readers with different file format. Information encoded in the files such as the multiple series of plates with the measured responses, assay and target information, replicate plates (or runs) and the associated compounds are automatically extracted. Users can select different types of configurations (e.g. one target/multiple compounds per plate, one compound/multiple targets per plate or multiple compounds/multiple targets per plate). Chemical information (chemical IDs, names and structures—MOL and SDF files) and additional external data such as target full names, assay information or users’ comments can also be uploaded. In addition, the Plate Designer module gives users the opportunity to create their own plate setup (e.g. four specific wells on each plate with reference compounds or a given row or column for target baselines only).
2.2 Create, append and update HTS data matrix
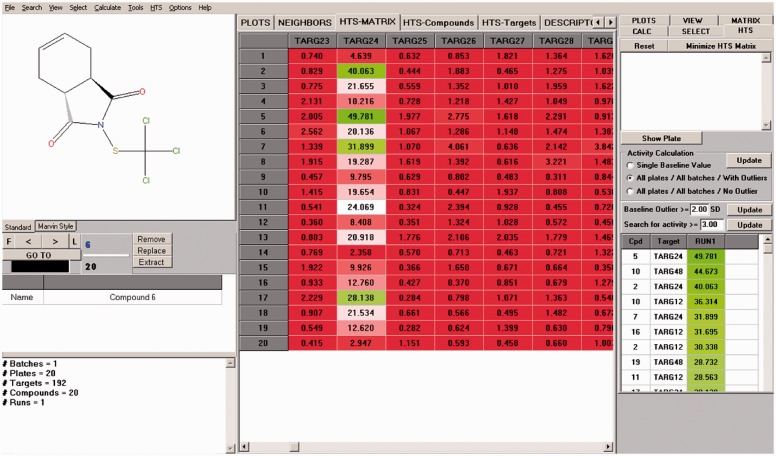
Navigator allows users to upload a large number of plates within the same project, which drastically facilitates raw data processing. As shown in Figure 1, Navigator automatically computes the HTS data matrix (i.e. tested chemicals versus targets) with different normalization and curation options. Each time the user uploads new plates, the HTS data matrix is updated and the graphic user interface is dynamically refreshed.
Fig. 1.
Main window of the HTS Navigator program divided into three parts: (left) chemical information and properties of the current HTS project, (central) HTS data matrix (row = compound, column = target) colored by compounds potencies and (right) baseline controls as well as the HTS hit picker
2.3 Navigation and identification of screening hits
Browsing the whole set of tested compounds is facilitated with the navigation panel (Fig. 1, left side) and a direct access to the distribution of responses across all targets (Supplementary Fig. S1A). Compounds with high experimental responses are easily detected. At any time, users have direct access to all information related to the selected case (target, compound, run, plate, potency with different options for baselines). Moreover, the HTS Hit Picker located on the right panel of Navigator (Fig. 1) automatically identifies all compounds with an experimental response higher or equal to a user-defined threshold (e.g. 10-fold activity increase, IC50 < 100 nM) so that users can rapidly analyze the most active compounds without spending a lot of time browsing the HTS matrix manually. Dual thresholds can also be activated for filtering out compounds with dual-selective polypharmacology.
2.4 Baseline visualization and correction factors
As part of the analysis of retrieved HTS hits and more generally the consistency of the different assays used in the HTS platform, Navigator provides users with a complete and easy access to all target baselines obtained within a project. Analyzing baselines is essential to compute accurate responses calculated from the averaged baseline responses by minimizing plate variability and detecting errors. The HTS Baseline Explorer module is illustrated in Supplementary Figure S1B. On the top, there is a mean-centered distribution of target’s baselines across plates, batches and runs. Outlier baselines (user-defined threshold, 2σ by default) are automatically identified and not considered for computing the responses in the HTS matrix. Distribution of compounds’ responses across all plates, batches and runs for the given target is also displayed for a fast identification of the most active compounds with adequate baseline. The HTS baseline explorer dramatically facilitates the identification of incorrect and suspicious baselines and as a consequence false-positive and false-negative hits.
2.5 Navigator is cheminformatics-ready
In addition to processing and analyzing screening data, Navigator incorporates several useful cheminformatics options. A series of filters can be used to identify subsets of compounds with certain properties, e.g. ranges of molecular weight, numbers of H acceptors and donors, octanol-water partition coefficient (logPow) or the intrinsic water solubility (logS) computed by QSAR models previously described (Varnek et al., 2008). Users can also search for a given chemotype via its name (e.g. secondary amine, nitro aromatics, tertiary alcohols) or substructure search. All compounds containing a specific chemical moiety are highlighted and colored in the dataset panel so one can browse HTS results for those compounds only. The sets of compounds can also be visualized using the ADDAGRA dataset graph module (Fourches and Tropsha, 2013). Some key elements of our chemical data curation workflow (Fourches et al., 2010) are directly included in Navigator, especially the neighborhood checker and duplicate finder enabling a fast identification of duplicated structures, isomers and stereoisomers. Chemical similarity is computed based on ISIDA fragments (Varnek et al., 2005) and standard Tanimoto similarity coefficient (Willett, 2010). We recently tested the usefulness of this feature when analyzing a large library of 17 000 compounds bought from different suppliers and tested against the same panel of cytochrome P450 enzymes (Veith et al., 2009). The analysis of some of these duplicates showed serious discrepancies between experimental bioactivity profiles (Fourches et al., 2013). Details concerning this analysis will be published elsewhere.
3 CONCLUSIONS
HTS Navigator has been developed to efficiently process and analyze the diverse data streams produced by HTS platforms. This software was not conceived or designed to compete with commercial software incorporating multiple features demanded by (and adapted for) pharmaceutical companies. Nevertheless, the HTS Navigator includes many important capabilities for managing, processing and analyzing HTS data. Specifically, Navigator is capable of (i) efficiently storing, processing and managing large amounts of chemical and biological data, (ii) automatically identifying hits as well as false-positives and is (iii) cheminformatics-ready with integrated tools for data curation, neighborhood analysis and advanced searches based on chemical similarity and substructural fragments. Importantly, HTS Navigator is freely accessible for academic laboratories and easily customizable for emerging needs.
Funding: NIH (grant GM096967 to A.T.), UNC Faculty Award (to D.F.) and NSF (grant ABI 1147145 to A.T. and D.F.).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Fourches D, Tropsha A. Using graph indices for the analysis and comparison of chemical datasets. Mol. Inform. 2013;32:827–842. doi: 10.1002/minf.201300076. [DOI] [PubMed] [Google Scholar]
- Fourches D, et al. Trust, but verify: on the importance of chemical structure curation in cheminformatics and QSAR modeling research. J. Chem. Inf. Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourches D, et al. The Toxicologist. Reston, VA: Vol. 132, Oxford University Press; 2013. Cheminformatics analysis of compound-Cytochrome P450 interaction profiles; p. 347. [Google Scholar]
- Kümmel A, Parker CN. The interweaving of cheminformatics and HTS. Methods Mol. Biol. 2011;672:435–457. doi: 10.1007/978-1-60761-839-3_17. [DOI] [PubMed] [Google Scholar]
- Varnek A, et al. Substructural fragments: an universal language to encode reactions, molecular and supramolecular structures. J. Comput. Aided Mol. Des. 2005;19:693–703. doi: 10.1007/s10822-005-9008-0. [DOI] [PubMed] [Google Scholar]
- Varnek A, et al. ISIDA-Platform for virtual screening based on fragment and pharmacophoric descriptors. Curr. Comput. Aided Drug Des. 2008;4:191–198. [Google Scholar]
- Veith H, et al. Comprehensive characterization of cytochrome P450 isozyme selectivity across chemical libraries. Nat. Biotechnol. 2009;27:1050–1055. doi: 10.1038/nbt.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett P. Similarity searching using 2D structural fingerprints. Methods Mol. Biol. 2010;672:133–158. doi: 10.1007/978-1-60761-839-3_5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.