Abstract
Metastatic solid tumors are aggressive and mostly drug resistant leading to few treatment options and poor prognosis as seen with clear cell renal cell carcinoma (ccRCC) and triple negative breast cancer (TNBC). Therefore the identification of new therapeutic regimes for the treatment of metastatic disease is desirable. ccRCC and TNBC cell lines were treated with the HDAC inhibitor romidepsin and the methyltransferase inhibitor decitabine, two epigenetic modifying drugs approved by the FDA for the treatment of various hematologic malignancies. Cell proliferation analysis, flow cytometry, quantitative PCR and immuno-blotting techniques were utilized to evaluate the antitumor synergy of this drug combination and identify the re-expression of epigenetically silenced tumor suppressor genes. Combinatorial treatment of metastatic TNBC and stage 4 ccRCC cell lines with romidepsin/decitabine leads to synergistic inhibition of cell growth and induction of apoptosis above levels of individual drug treatments alone. Synergistic re-expression of the tumor suppressor gene secreted frizzled-related protein one (sFRP1) was observed in combinatorial drug treated groups. Silencing sFRP1 (shRNA) prior to combinatorial drug treatment demonstrated that sFRP1 mediates the growth inhibitory and apoptotic activity of combined romidepsin/decitabine. Furthermore, addition of recombinant sFRP1 to ccRCC or TNBC cells inhibits cell growth in a dose-dependent manner through the induction of apoptosis identifying that epigenetic silencing of sFRP1 contributes to renal and breast cancer cell survival. Combinatorial treatment with romidepsin and decitabine in drug resistant tumors is a promising treatment strategy. Moreover, recombinant sFRP1 may be a novel therapeutic strategy for cancers with suppressed sFRP1 expression.
Keywords: Triple negative breast cancer, clear cell renal cell carcinoma, methyltransferase inhibitor, histone deacetylase inhibitor, secreted frizzled-related protein 1
Introduction
Cancer is a multistep process facilitated by the accumulation of genetic abnormalities resulting in genomic instability and the mutation of tumor suppressor and oncogenic genes. Furthermore, epigenetic changes in cancer lead to modulations of gene expression through mechanisms of DNA methylation and histone deacetylation. The hypermethylation of cytosines in areas of rich CpG islands, and deacetylation of histones which facilitate a tighter formation of chromatin, contribute to the inappropriate silencing of gene expression. Histone deactylase (HDAC) inhibitors such as trichostatin A (TSA) and romidepsin (FK228), and methyltransferase inhibitors such as decitabine (DAC; 5-aza-2′-deoxycytodine) are capable of reversing these epigenetic events and suppressing the cancer phenotype. The HDAC inhibitor romidepsin exhibits antitumor properties in human cell lines both in vitro and in vivo (1). Studies have identified that romidepsin treatment of tumor cells leads to inhibition of angiogenesis and cell growth, while inducing apoptosis, cell death and cell differentiation (2-6). Romidepsin was approved by the FDA for the treatment of cutaneous T-cell lymphoma in 2009, and for peripheral T-cell lymphoma (PTCL) in 2011. It continues to be actively investigated as an anti-cancer therapeutic for both hematological and solid malignancies.
Methyltransferase inhibitors are analogues of cytosine that incorporate into the DNA during replication before covalently linking with DNA methyltransferases (DNMTs) leading to global loss of gene methylation (7). Treatment of cancer cell models with the methyltransferase inhibitor decitabine leads to suppression of growth and apoptosis through re-expression of silenced genes and the activation of p53 and p21Waf1/Cip1 (8-10). Studies have identified that decitabine causes G2 arrest, reduces clonogenic survival, and inhibits growth while causing DNA fragmentation and activating the ATM and ATR DNA repair pathways (11). In 2006 decitabine was FDA approved for the treatment of myelodysplastic syndromes.
Constitutive activation of the Wnt signaling pathway as a mechanism for cancer development was first identified in colon cancer (12). The binding of secreted Wnt family members to Frizzled receptor complexes on the cell surface leads to activation of downstream gene targets through either the canonical/β-catenin pathway or one of the non-canonical/β-catenin independent pathways (13). Composition of the Wnt/Frizzled complex governs which of these pathways are activated. Canonical Wnt signaling influences genes associated with cell proliferation, survival and invasion (14), whilst non-canonical pathways regulate those involved in cell adhesion, migration and cytoskeletal reorganization (15). sFRP1, secreted frizzled-related protein 1, functions as a negative regulator of Wnt signaling by sequestering Wnt proteins and heterodimerizing with Frizzled to form non-functional receptor complexes. However in colorectal, ovarian, lung, hepatocellular, kidney and breast cancer, hypermethylation of the sFRP1 promoter and subsequent loss of expression has been identified allowing aberrant Wnt signaling (14, 16-20).
Renal cell carcinoma (RCC) is the third most prevalent urological cancer, and is the 10th most common cause of cancer death in men and 9th in women (21). Clear cell renal cell carcinoma (ccRCC) is the largest subtype of RCC and accounts for approximately ∼80% of renal cancers. Breast cancer is the most common cancer in women with triple negative breast cancer (TNBC) accounting for approximately 15% of newly diagnosed cases. TNBCs are associated with poor prognosis, a higher mitotic index and younger age (22). In ccRCC and breast cancer, early diagnosis and treatment dramatically increase median survival rates as when metastatic, these cancers are mostly aggressive and drug resistant. Development of metastatic disease in ccRCC patients reduces the 5 year survival rate to less than 10% (23) and in TNBC reduces survival to around 18 months (24). Therefore there is a dire need for new chemotherapeutic drug therapies in these drug resistant cancers.
Strategies to re-express epigenetically silenced genes are attractive therapeutic options in drug resistant ccRCC and TNBC. To this end we treated primary site, metastatic ccRCC and TNBC cell lines with mono- and synergistic combinatorial treatments of romidepsin and decitabine inhibiting proliferation and inducing cell death via apoptosis. We identify that the tumor suppressor gene sFRP1 is re-expressed with combinatorial treatment and that silencing of sFRP1 plays a prominent role in survival of ccRCC and TNBC cells. Together these data suggest that romidepsin and decitabine in combination may be a promising therapeutic drug regimen for the treatment of ccRCC and TNBC.
Materials and Methods
Reagents
Romidepsin was generously provided by Celgene Corporation (Summit, NJ) and the National Cancer Institute (NCI). Decitabine (5-aza-2′-deoxycytidine) was purchased from Sigma-Aldrich (St. Louis, MO) and recombinant human sFRP1 from R&D Systems (Minneapolis, MN).
Cell line verification
Genomic DNA was used for short tandem repeat (STR) analysis of all cell lines at John Hopkins University Fragment Analysis Facility using the StemElite service in July 2010. Furthermore, KIJ265T cells were verified by AmpF/STR Identifiler analysis (Applied Biosystems, Foster City, CA). The renal origin of KIJ265T cells was validated by Immunohistochemistry (Supplemental Figure 1). Cells were fixed using 2% paraformaldehyde (Sigma), permeabilized using 1% Triton X-100 (Sigma), and blocked with diluent containing background reducing components (Dakocytomation, Denmark) prior to staining with primary antibody. Control slides were prepared by excluding primary antibody during staining. Primary antibodies included RCC-Ma (Cell Marque Corporation, Rocklin, CA), podocin (ABCAM, Cambridge, MA), gamma glutamyl transpeptidase (Lifespan Biosciences, Seattle, WA), PAX2 (Lifespan), and aquaporin2 (Santa Cruz, Santa Cruz, CA). The KIJ265T cells was identified to be VHL mutant (Exon 2 c.407T>C; protein modification of p.F136S) by DNA sequencing.
Cell Culture
The A498, MDA-231 and BT20 cell lines (ATCC, Manassas, VA) and KIJ265T cells (derived from primary tumor site stage 4 human ccRCC patient tissue) were maintained in DMEM medium (Cellgro, Herndon, VA) supplemented with 10% FBS (Hyclone, Logan, UT) and penicillin-streptomycin-amphotericin B (PSA) (Mediatech, Winchester, VA) at 37°C with 5% CO2.
Drug Treatments and proliferation Assays
Cells were plated (1×105 per well) in 12-well plates (Midwest Scientific, St. Louis, MO) and treatments carried out in triplicate. For mono-therapeutic treatments, cells were treated for 72hrs with a dose range of 0.01μM to 10μM decitabine or 0.01nM to 100nM romidepsin. DMSO was used for vehicle control. Cells were trypsinized and counted using a Coulter Particle Counter (Beckman, Brea, CA). For combination dose-outs, cells were treated with a dose range of 0.1, 1, or 10μM decitabine. After 48hrs, romidepsin was added in combination to the decitabine at a dose range of 0.5nM to 7.5nM for a further 24hrs. Appropriate mono-therapeutic and DMSO treatments were included and administered in accordance with combinatorial time-points. An optimal combinatorial dose of 1μM decitabine for 72hrs with the addition of 5nM romidepsin for the last 24hrs was utilized in further treatments.
For treatment of cells with recombinant human sFRP1, MDA-231 and KIJ265T cells were seeded in 96-well culture plates at 5000 cells/well in 100μl supplemented DMEM overnight. Cells were washed and 100μl of serum free DMEM containing the final concentration of sFRP1 added for 6hrs. Serum was re-introduced to these wells to a final concentration of 2% FBS and incubated for 72hrs.
RNA Isolation, RT-PCR, and Quantitative PCR
RNA was isolated using the RNAqueous Midi Kit (Ambion, Austin, TX), tested for O.D. 260/280 mRNA ratios between 1.8 and 2.0 and agarose gel verified for purity. RNA was reverse transcribed using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems). For subsequent real-time PCR quantification Applied Biosystems' assay-on-demand 20× primers and TaqMan® FAM™ dye-labeled probes for sFRP1 (Hs00610060_m1), RhoB (Hs00269660_s1), p21 (Hs00355782_m1), TβRIII (Hs00234257_m1), GATA3 (Hs00231122_m1), and GAPDH (Hs99999905_m1) were used. Samples were normalized to GAPDH and ΔΔCT methods used to calculate fold expression changes (25).
Protein Expression Analysis
Cells were seeded (1×107) on 100mm plates (Midwest Scientific) and treated with the optimal combinatorial dose. Cells were lysed using M-PER reagent containing phosphatase inhibitors (Pierce, Rockford, Il) and protease inhibitor cocktail (Roche, Mannheim, Germany). Quantification and transfer to 0.2μM Immobilon Psq membranes were carried out as previously described (26). Membranes were incubated overnight in primary antibody [β-actin (Sigma-Aldrich), caspase-3, sFRP1 or PARP (Cell Signaling, Boston, MA)] before treatment with secondary species-specific horseradish peroxidase-labeled antibodies (Jackson Immunoresearch, West Grove, PA) for 45 minutes at room temperature. Supersignal chemiluminescent kit (Pierce) was used for detection.
Cell Death Analysis via Flow Cytometry
Cells were treated with the optimal combinatorial therapy with appropriate control groups. After 72 hour treatment, adhered and floating cells were collected and re-suspended in 1× cold binding buffer (BD Pharmingen, San Jose, CA) at 1×106 cells per ml. Cells were propidium iodide (BD Pharmingen) stained for 10 minutes and FACS analysis performed using Accuri C6 flow cytometer (Accuri, Ann Arbor, MI). Unstained cells were used as controls for setting the cell population parameters.
Methylation-Specific PCR
Genomic DNA was isolated from the four cell lines and treated with sodium bisulfite using the EZ DNA Methylation-Startup kit (Zymo Research, Irvine, CA) according to manufacturer's instructions. Purified bisulfite-treated DNA was amplified with primers specific for methylated or unmethylated sFRP1 at an annealing temperature of 58°C as previously described (14, 27). Positive control templates were bisulfite-treated Universal Methylated Human DNA. Sequence analysis of the methylation patterns of ten clones for each cell line, treated with DMSO or combinatorial drug treatments, using previously published primers (14) were performed at ATCG, Inc. (Wheeling, IL).
Luciferase Reporter Assay
KIJ265T and MDA231 cells were transfected with 0.5ug/well TOPflash luciferase reporter construct (kind gift from Dr. Bert Vogelstein, Johns Hopkins University), 0.5ug/well sFRP1 expression vector (kind gift from Dr. Jeffrey Rubin, NIH/NCI) or pcDNA3.1 empty vector control (Invitrogen, Carlsbad, CA) and 10ng/well control Renilla luciferase reporter using Fugene 6 transfection reagent (Roche). Cells were collected 48hrs post transfection and reporter activity measured using the Dual Luciferase Kit (Promega, Madison, WI). Data is normalized to renilla luciferase and reported as mean ± S.D. of replicates of three.
Lentivirus and Infections
MISSION shRNA pLKO.1 constructs (Sigma-Aldrich) were used to make self-inactivating shRNA lentiviruses for sFRP1 [target sequence 5′- CGAGATGCTTAAGTGTGACAA-3′ (clone NM_003012.3-758s1c1)], and a non-target random scrambled sequence control (SHC002). For virus transduction, 2×107 MDA-231 or KIJ265T cells were incubated with lentivirus plus 5μg/ml polybrene (American Bioanalytical, Natick, MA) for 24hrs. Clones were identified by puromycin (Fisher Scientific) selection.
Statistical Analysis
Data are presented as the mean ± SD and comparisons of treatment groups were analyzed by 2–tailed paired Student's t test. Data for comparison of multiple groups are presented as mean ± SD and were analyzed by ANOVA. p<0.05 was considered statistically significant.
Results
Single drug therapy
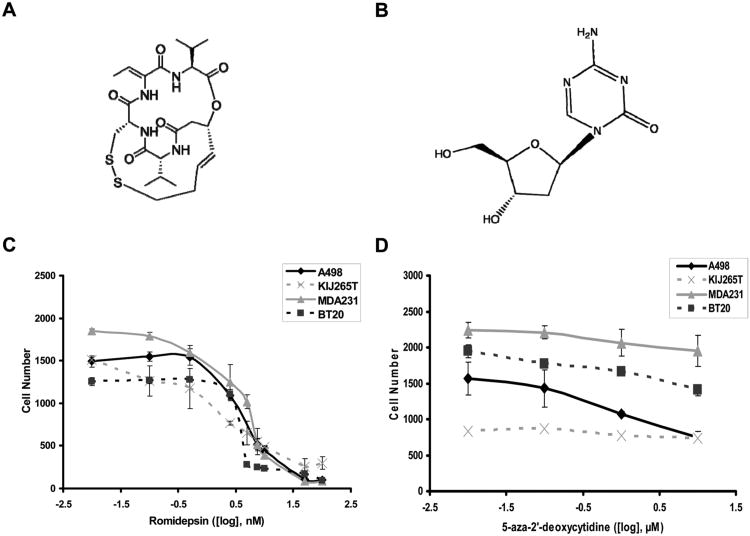
Individual 72 hour drug treatments of romidepsin (chemical structure Figure 1A) or decitabine (chemical structure Figure 1B) were evaluated in two ccRCC stage 4 cell lines (A498 and KIJ265T) and two TNBC cell lines (MDA231 and BT20) for their ability to inhibit cell proliferation. Romidepsin was identified as a potent inhibitor of cell proliferation in all cell lines producing significant inhibition in the 2.5-100nM range (Figure 1C). Treatment with decitabine had minimal effect on cell proliferation and was unable to provide an IC50 in the 0.01-10μM drug range (Figure 1D). These data identify romidepsin as a potent inhibitor of cell proliferation in ccRCC and TNBC cell lines.
Figure 1.
Dose response of ccRCC and TNBC cell lines to a 72 hour exposure of romidepsin or decitabine. A498, KIJ265T, MDA-231, and BT-20 cells received a treatment range of (C) 0.01nM to 100nM romidepsin (chemical structure A) and (D) 0.01μM to 10μM decitabine (5-aza-2′deoxycytidine) (chemical structure B). Treatments were applied to cells for 72hrs prior to collection.
Combinational therapy induces synergistic responses in ccRCC and TNBC cell lines
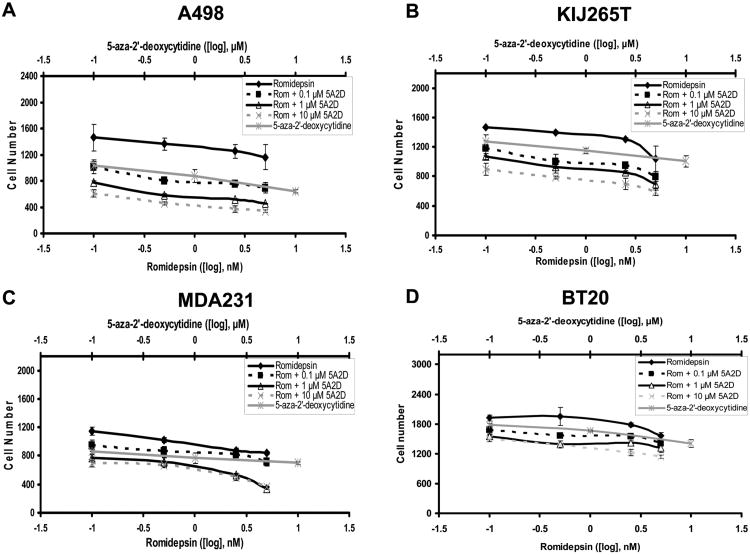
Combinatorial treatments of romidepsin and decitabine were evaluated for their ability to inhibit cell proliferation (Figure 2). Drug doses identified in the single drug exposure experiments were evaluated in these investigations. Romidepsin doses were standardized to 0.1, 0.5, 2.5 and 5nM, with 5nM being a dose that induced ∼50% cell death 72hrs post treatment. Since decitabine had little effect upon cell proliferation, doses of 0.1, 1 and 10μM were chosen for combinatorial treatments. Treatment protocols for these studies evaluated the response of the cells to either a 24 hour treatment of romidepsin alone, a 72 hour treatment of decitabine alone or treatment for 72hrs of decitabine with the final 24hrs in combination with romidepsin. Please note, in these combinatorial therapy treatments the cells were exposed to romidepsin for 24hrs and not 72hrs as in the single drug exposures (Figure 1). In all cell lines combinational drug treatment induced greater inhibition of cell proliferation than single drug treatments alone.
Figure 2.
Combinatorial dose response to decitabine (5A2D) and romidepsin (Rom) in ccRCC and TNBC cell lines. (A) A498, (B) KIJ265T, (C) MDA-231, and (D) BT-20 cells were incubated with a dose of 0.1, 1, or 10μM decitabine for 48hrs. Romidepsin (0.5 to 7.5nM) was added in combination to the decitabine for a further 24hrs. Data is presented as proliferation curves with monotherapeutic controls included.
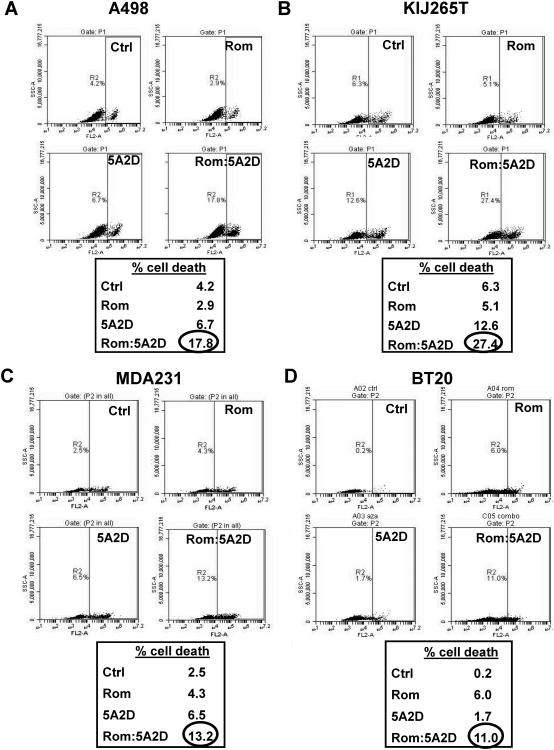
Experimental cell lines were further evaluated for drug induced cell death. Propidium iodide staining of cell lines treated with 5nM romidepsin or 1μM decitabine alone or in combination identified synergistic induction of cell death in the combination drug therapy group (Figures 3). In ccRCC KIJ265T cells, combinatorial treatment induced cell death 21.1% above DMSO controls (Figure 3B). For the other cell lines, A498, MDA231 and BT20, cell death with combination treatment was induced 13.6%, 10.7% and 10.8% respectively compared to DMSO controls. Single drug exposures did not consistently induce cell death in the tested cell lines.
Figure 3.
Synergistic induction of cell death in decitabine (5A2D) and romidepsin (Rom) combinatorial treated ccRCC and TNBC cell lines. (A) A498, (B) KIJ265T, (C) MDA231 and (D) BT20 cell lines treated with monotherapeutic doses of decitabine or romidepsin were analyzed versus combination treatment for drug effects leading to cell death, analyzed via flow cytometry. Only combinatorial treatment was sufficient to induce cell death.
Combinational treatment therapy induces apoptosis
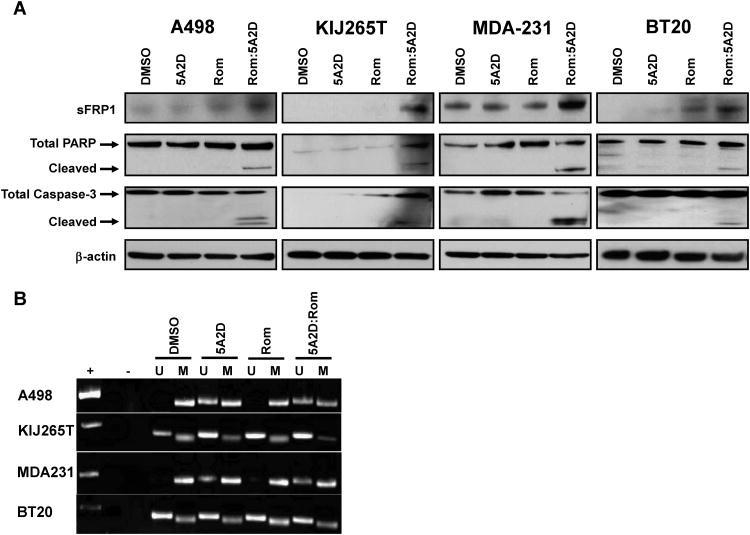
Drug treated cells were analyzed for markers of apoptosis to elucidate mechanisms of cell death. Total proteins from cells treated under the optimal dosing regime, with single or combinatorial therapy demonstrated cleavage of caspase-3 and PARP only in the combinatorial drug treated groups (Figure 4A). These events were absent in single drug exposure and control groups indicating that in combination these drugs are potent inducers of apoptosis.
Figure 4.
Analysis of sFRP1 expression and markers of apoptosis in ccRCC and TNBC cell lines treated with 1μM decitabine (5A2D) and 5nM romidepsin (Rom). (A) Protein lysates from treated A498, KIJ265T, MDA-231, and BT-20 cells were probed for PARP and caspase-3 cleavage identifying synergistic apoptotic escalation with combinatorial treatment. Accumulation of sFRP1 expression was observed with combination treatment. (B) Cell lines were examined for methylation status of the sFRP1 promoter and the ability of single and combinatorial drug treatments to modulate these methylation events via methylation specific PCR utilizing defined primers for the amplification of methylated or unmethylated sequences of sFRP1 (M=methylated, U= unmethylated).
sFRP1 expression is induced by combinational therapy
To elucidate the molecular events taking place in cells treated in combination with romidepsin and decitabine, we analyzed molecular targets we have previously identified to be directly or indirectly affected by epigenetic silencing in cancer. Expression levels of protein and RNA from the cell lines were examined for RhoB (28, 29), p21Waf1/Cip1 (29), the type III transforming growth factor-β receptor (TβRIII) (30), GATA3 (30) and sFRP1 (14) after single or combinational treatments (data not shown). Of these molecular targets, sFRP1 showed consistent synergistic up-regulation of RNA and protein expression across all cell lines treated with combinatorial therapy (Figure 4A and Table 1). Methylation-specific PCR established that the sFRP1 promoter was hypermethylated in all experimental cell lines. Treatment with decitabine alone or in combination with romidepsin led to demethylation of the sFRP1 promoter region (Figure 4B) verifying that re-expression of sFRP1 at the RNA and protein level is due to the reversal of epigenetic silencing. Bisulfite DNA sequencing confirmed a global decrease of sFRP1 promoter methylation in all cell lines after combinatorial treatment compared to DMSO treated controls (Supplemental Figure 2).
Table 1.
Induction of sFRP1 RNA expression observed with combinatorial treatment. Statistically significant increases in RNA expression (*P<0.05 Student's t-test) are bold-faced, with the highest re-expression values observed in combinatorial treatment groups designated with an asterisk.
| sFRP1 | Ctrl CT value |
Ctrl Fold Change |
5A2D Fold Change |
Rom Fold Change |
Rom:5A2D Fold Change |
|---|---|---|---|---|---|
| A498 | 38.5 | 1.0 ± 0.1 | 11.1 ± 2.9 | 1.4 ± 0.9 | 35.3 ± 2.5* |
| KIJ265T | 34.3 | 1.1 ± 0.5 | 2.5 ± 0.7 | 20.6 ± 1.1 | 62.4 ± 6.6* |
| MDA231 | 39.2 | 1.0 ± 0.2 | 95.4 ± 15.9 | 4.8 ± 3.4 | 1302.6 ± 49.5* |
| BT20 | 27.7 | 1.0 ± 0.1 | 2.6 ± 0.1 | 1.4 ± 0.1 | 3.6 ± 0.4* |
Silencing of dual treatment induced sFRP1 leads to gain of cell survival
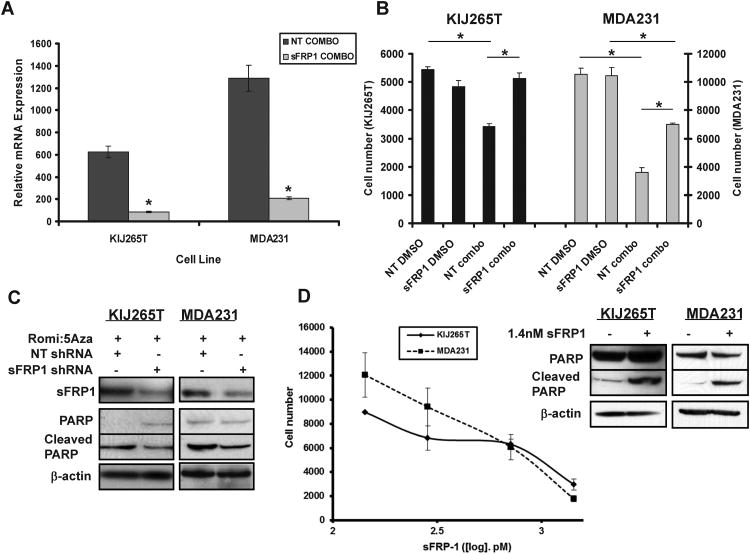
Since sFRP1 is a tumor suppressor gene we hypothesize that re-expression of sFRP1 in dual therapy treated cancer cells is sufficient to modulate cell survival. Therefore, endogenous levels of sFRP1 were shRNA silenced in MDA231 and KIJ265T cell lines. Cells were treated with 5nM romidepsin and 1μM decitabine in combination and the expression of sFRP1 RNA message evaluated (Figure 5A). Combinatorial treatment induced expression of sFRP1 in the KIJ265T and MDA231 non-target cell lines ∼600 and ∼1300 fold respectively, when compared to non-treated, non-target controls. shRNA silencing of sFRP1 before combinatorial treatment reduced this expression by 6 fold or greater in KIJ265T and MDA231 cells. Loss of inducible sFRP1 was observed to reduce the effects of combinatorial treatment on KIJ265T and MDA231 cells. Following combinatorial treatment, the growth of KIJ265T sFRP1 shRNA silenced cells was unchanged compared to shRNA non-target DMSO controls (Figure 5B). Thus, the loss of inducible sFRP1 completely abolished the growth inhibitory effects of combinatorial treatment in KIJ265T cells. Combinatorial drug responses (romidepsin 5nM, decitabine 1μM) in sFRP1 shRNA silenced cells were 35% and 50% less responsive in KIJ265T and MDA231 cells than shRNA non-target treated controls (Figure 5B). Total cell protein analysis showed that sFRP1 shRNA silenced cells had reduced levels of PARP cleavage identifying that cell survival after combinatorial treatment was due to an inhibited apoptotic response (Figure 5C). Therefore, sFRP1 is a target of romidepsin/decitabine treatment and its re-expression plays a significant and critical role in the inhibition of cell growth/induction of cell death when these drugs are administered in combination.
Figure 5.
Loss of sFRP1 facilitates cell survival in ccRCC and TNBC cell lines treated in combination with decitabine (5A2D) and romidepsin (Rom). (A) Real-time PCR of KIJ265T and MDA-231 cells infected with sFRP1 shRNA demonstrate a significant decrease in sFRP1 re-expression with combinatorial drug treatment vs. control (*P<0.05 Student's t-test). (B) Resulting loss of sFRP1 re-expression yielded an increase in cell survival of KIJ265T and MDA-231 cells when treated with combinatorial dose of 1μM 5A2D and 5nM Rom. (C) Attenuation of apoptosis in KIJ265T and MDA-231 sFRP1 knockdown cells was demonstrated via reduction in PARP cleavage versus non-target controls with combination treatment. (D) Decreased proliferation of KIJ265T and MDA-231 parent cell lines was observed in a dose-dependent manner when treated with recombinant human sFRP1 and verified to be through the induction of apoptosis as seen by PARP cleavage, after a single dose of 1.4nM sFRP1.
Human recombinant sFRP1 inhibits cell proliferation
To verify that epigenetic silencing of sFRP1 is vital for TNBC and ccRCC cell survival, we re-introduced recombinant human sFRP1 to the cells and examined effects on cell proliferation and apoptotic cell death. Escalating doses of recombinant sFRP1 (142.8pM–1.4nM) led to a dose-dependent decrease in KIJ265T and MDA231 cell proliferation (Figure 5D) identifying that low nM doses of sFRP1 are capable of inhibiting cancer cell growth. This inhibition was verified to be through induction of apoptosis 28 hours after a single dose of 1.4nM recombinant sFRP1 (Figure 5D). Re-expression of sFRP1 by transient transfection in KIJ265T and MDA231 cells inhibited Wnt activated β-catenin/TCF mediated transcription as measured by the TOPflash reporter assay (Supplemental Figure 3A) indicative that sFRP1 attenuates oncogenic Wnt signaling in ccRCC and TNBC cells. These data demonstrate that romidepsin and decitabine in combination are a potential drug therapy option for the treatment of ccRCC and TNBC, and that this mechanism of activity is predominantly through synergistic re-expression of sFRP1.
Discussion
Despite recent progress made in therapeutics for ccRCC and TNBC, there is an urgent need for more effective therapies (31, 32). Our preclinical studies identify that combinatorial treatment of ccRCC and TNBC cell lines with decitabine and romidepsin leads to inhibition of cell proliferation and the induction of apoptosis as measured by caspase-3 and PARP cleavage. Combinatorial epigenetic treatment induces the re-expression of the Wnt signaling pathway antagonist sFRP1, an event that is not seen with single drug treatments alone, identifying that the epigenetic silencing of sFRP1 plays a prominent role in the onset and progression of clear cell renal cell carcinoma and triple negative breast cancer. Furthermore, treatment of ccRCC and TNBC cells with low doses of exogenous sFRP1 produced dose-dependent inhibition of cancer cell growth (142.8pM–1.4nM) through induction of apoptosis. It is our belief that these findings reveal potential treatment options for those patients diagnosed with advanced ccRCC and TNBC. Since sFRP1 is a secreted protein, these data also suggest that sFRP1 re-expression could be used as a molecular biomarker of response to this combinatorial therapeutic regimen.
We and others have published that the promoter of sFRP1 is epigenetically silenced in all stages of ccRCC suggesting that silencing of sFRP1 is an early event in ccRCC carcinogenesis (14, 33, 34). In our earlier publication, re-expression of sFRP1 within a ccRCC cell line with no endogenous sFRP1 expression led to reduced tumor growth in vitro and in vivo. Recently this was confirmed by others who also identified that loss of sFRP1 in RCC cells increases their tumorigenic potential and that methylation of sFRP1 in RCC was associated with reduced patient survival although this was not independent of tumor stage, size and grade (35). In breast cancer, methylation silencing of sFRP1 is also associated with poor patient survival (27) while studies in breast cancer cells examining the role of sFRP1 on carcinogenesis have identified similar findings to those observed in ccRCC. Expression of sFRP1 in many breast cancer cell lines leads to decreased proliferation potential, decreased ERK1/2 activity, induced apoptosis and suppressed colony formation (36, 37), while loss of sFRP1 enhances breast cancer growth via increases in cell viability (37). More importantly xenograft models using the breast cancer cell line MDA-MB-231 identified that re-expression of sFRP1 led to inhibited xenograft outgrowth and lung metastasis in vivo (38). Taken together, these findings identify sFRP1's critical role as a tumor suppressor in breast and renal cancer.
Furthermore, we have previously identified that a number of downstream targets of the Wnt signaling pathway are over-expressed in patient ccRCC tissue samples and that re-expression of sFRP1 in ccRCC cell lines inhibits their expression (14). There is also evidence that in both RCC and breast cancer epigenetic silencing of other Wnt antagonists occurs, implicating a role for aberrant Wnt signaling in tumorigenesis. In primary breast tumors methylation silencing of sFRP1, 2, 5 and DKK1 was identified to be present in 40%, 77%, 71% and 17% of all tissues respectively (37). Loss of sFRP5 in breast cancer is associated with reduced overall survival (39) although loss of sFRP2 showed no correlation with patient outcome (40). In ccRCC, sFRP1, sFRP2, sFRP4, sFRP5 and DKK3 are frequently silenced in tumor tissues (14, 41-43). Re-expression of sFRP5 in RCC cells inhibits anchorage independent colony formation, cell invasion and induces apoptosis suggesting that like sFRP1, sFRP5 is a tumor suppressor gene (41). Further analysis is required so that we can evaluate the role of these antagonists of Wnt signaling within our experimental renal and breast cancer cell lines. More importantly it needs to be ascertained whether combinational treatment with romidepsin and decitabine leads to the re-expression of these epigenetically silenced Wnt antagonists and the roles that they play on cell proliferation and apoptosis. Care must also be taken to identify that re-expression of these soluble Wnt antagonists does not lead to the activation of Wnt signaling as previously observed (44, 45). Clearly, multiple soluble Wnt antagonists are silenced in ccRCC and TNBC delineating the importance of Wnt up-regulation in tumor promotion. Thus, it makes sense that re-expression of sFRP1 has a profound effect upon cell proliferation and cell survival since sFRP1 is known to bind and antagonize multiple Wnts as well as the Wnt membrane binding partners, Frizzleds leading to Wnt/β-catenin signaling repression (18).
Decitabine and romidepsin have been approved by the FDA for treatment of myelodysplastic syndrome (MDS) and cutaneous T-cell and peripheral T-cell lymphoma respectively. Very few clinical trials exist that examine the combinatorial effects of these two agents on human cancers. One clinical trial (NCT00037817) used a 72 hour IV infusion of decitabine followed by a 4 hour IV infusion of romidepsin for the treatment of patients with pulmonary and pleural malignancies while a recently completed phase II clinical trial assessed decitabine/romidepsin combinatorial treatments in patients with relapsed or refractory leukemia, myelodysplastic syndromes, or myeloproliferative disorders (NCT00114257). Findings from both of these studies have yet to be published. Phase 1 clinical trials examining individual treatment of romidepsin in RCC patients at a dose of 9.1mg/m2 led to a partial response in one patient after two 21-day cycles of treatment and was maintained for a further 6 cycles (4). Treatment with decitabine at a dose of 75mg/m2 resulted in tumor stabilization in one RCC patient (46) and at dose levels of 0.15 and 0.25mg/kg stable disease was observed in three patients (47). Although no clinical trials have been published with these individual agents in breast cancer, recent research identifies that poor prognosis breast cancers express high levels of deoxycytidine kinase (DCK) whose phosphorylation is needed for the pharmacological activity of decitabine. Screening for expression of DCK would allow for the selection of breast cancer patients that are more likely to respond to nucleoside analog treatments (48). From our findings, we identify that combinatorial treatment with decitabine and romidepsin is effective at inducing cell death in ccRCC and TNBC cell lines and therefore, clinical trials of this combination in patients with these solid tumors would be of interest.
We have identified in these studies one epigenetically silenced tumor suppressor gene whose loss of expression plays a significant role in cancer cell survival. We understand that there are many more genes modulated by this drug treatment that we have not investigated. To truly understand the actions of the combinatorial treatment of romidepsin and decitabine in ccRCC and TNBC cell lines, genome wide analysis of gene regulation after treatment must be undertaken (49). We predict that there are a number of drug targetable genes that are re-expressed after combinatorial treatment that possess antitumor activity. We further suggest that the administration of chemotherapeutic or immunotherapeutic agents at the end of epigenetic combinatorial treatment could further increase the inhibition of cell growth or induction of cell death within drug resistant cancer cells. In single drug exposure studies, treatment of the ACHN ccRCC cell line with decitabine reversed resistance to interferon and in combination induced 85% cell apoptosis (50). Thus, the analysis of decitabine/romidepsin modulated genes and the identification of novel signaling pathways could be of clinical relevance for the targeted treatment of ccRCC and TNBC.
Supplementary Material
Acknowledgments
We thank Dr. Derek Radisky for helpful comments related to experimental design and Dr. Marie Becker for careful editing of the manuscript. We also thank ACGT, Inc. (Wheeling, IL) for their assistance with the analysis of methylation patterns.
Grant Support: This work was funded in part from NIH/NCI grants R01CA104505 (J.A.Copland), R01CA104505-05S1 and 2K12CA090628 (M.E.Menefee); a generous gift from the David & Lois Stulberg Endowed Fund for Kidney Cancer Research (J.A.Copland); Kidney Cancer Research at Mayo Clinic in Florida (J.A.Copland); the Breast Cancer Research Foundation (E.A.Perez & J.A.Copland); James C. and Sarah K. Kennedy Mayo Clinic Research Career Development Award for Clinicians (H.W.Tun); The Brenda E. and Roger S. Luca Family Endowment at The Tallahassee Memorial HealthCare Foundation (J.A.Copland, G.Colon-Otero), RITA Foundation (Research Is The Answer; J.A.Copland), Scheidel Foundation (J.A.Copland) and a grant for rare cancers from Dr. Ellis and Dona Brunton (J.A.Copland; G.Colon-Otero).
Abbreviations
- HDAC
histone deactylase
- ccRCC
clear cell renal cell carcinoma
- TNBC
triple negative breast cancer
- sFRP1
secreted frizzled-related protein 1
Footnotes
Conflicts of interest: The authors of this manuscript have no conflicts of interest to disclose.
References
- 1.Ueda H, Nakajima H, Hori Y, Goto T, Okuhara M. Action of FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum no. 968, on Ha-ras transformed NIH3T3 cells. Biosci Biotechnol Biochem. 1994;58:1579–83. doi: 10.1271/bbb.58.1579. [DOI] [PubMed] [Google Scholar]
- 2.Jung M. Inhibitors of histone deacetylase as new anticancer agents. Curr Med Chem. 2001;8:1505–11. doi: 10.2174/0929867013372058. [DOI] [PubMed] [Google Scholar]
- 3.Zhu WG, Lakshmanan RR, Beal MD, Otterson GA. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res. 2001;61:1327–33. [PubMed] [Google Scholar]
- 4.Sandor V, Bakke S, Robey RW, Kang MH, Blagosklonny MV, Bender J, et al. Phase I trial of the histone deacetylase inhibitor, depsipeptide (FR901228, NSC 630176), in patients with refractory neoplasms. Clin Cancer Res. 2002;8:718–28. [PubMed] [Google Scholar]
- 5.Konstantinopoulos PA, Vandoros GP, Papavassiliou AG. FK228 (depsipeptide): a HDAC inhibitor with pleiotropic antitumor activities. Cancer Chemother Pharmacol. 2006;58:711–5. doi: 10.1007/s00280-005-0182-5. [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Cheng H, Kwan W, Lubieniecka JM, Nielsen TO. Histone deacetylase inhibitors induce growth arrest, apoptosis, and differentiation in clear cell sarcoma models. Mol Cancer Ther. 2008;7:1751–61. doi: 10.1158/1535-7163.MCT-07-0560. [DOI] [PubMed] [Google Scholar]
- 7.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–95. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 8.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 9.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 10.Zhu WG, Hileman T, Ke Y, Wang P, Lu S, Duan W, et al. 5-aza-2′-deoxycytidine activates the p53/p21Waf1/Cip1 pathway to inhibit cell proliferation. J Biol Chem. 2004;279:15161–6. doi: 10.1074/jbc.M311703200. [DOI] [PubMed] [Google Scholar]
- 11.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008;28:752–71. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 13.Widelitz R. Wnt signaling through canonical and non-canonical pathways: recent progress. Growth Factors. 2005;23:111–6. doi: 10.1080/08977190500125746. [DOI] [PubMed] [Google Scholar]
- 14.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–9. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 15.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukui T, Kondo M, Ito G, Maeda O, Sato N, Yoshioka H, et al. Transcriptional silencing of secreted frizzled related protein 1 (SFRP 1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005;24:6323–7. doi: 10.1038/sj.onc.1208777. [DOI] [PubMed] [Google Scholar]
- 17.Takada T, Yagi Y, Maekita T, Imura M, Nakagawa S, Tsao SW, et al. Methylation-associated silencing of the Wnt antagonist SFRP1 gene in human ovarian cancers. Cancer Sci. 2004;95:741–4. doi: 10.1111/j.1349-7006.2004.tb03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 19.Shih YL, Hsieh CB, Lai HC, Yan MD, Hsieh TY, Chao YC, et al. SFRP1 suppressed hepatoma cells growth through Wnt canonical signaling pathway. Int J Cancer. 2007;121:1028–35. doi: 10.1002/ijc.22750. [DOI] [PubMed] [Google Scholar]
- 20.Lo PK, Mehrotra J, D'Costa A, Fackler MJ, Garrett-Mayer E, Argani P, et al. Epigenetic suppression of secreted frizzled related protein 1 (SFRP1) expression in human breast cancer. Cancer Biol Ther. 2006;5:281–6. doi: 10.4161/cbt.5.3.2384. [DOI] [PubMed] [Google Scholar]
- 21.van Spronsen DJ, Mulders PF, De Mulder PH. Novel treatments for metastatic renal cell carcinoma. Crit Rev Oncol Hematol. 2005;55:177–91. doi: 10.1016/j.critrevonc.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Kreike B, van Kouwenhove M, Horlings H, Weigelt B, Peterse H, Bartelink H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantuck AJ, Zisman A, Belldegrun AS. The changing natural history of renal cell carcinoma. J Urol. 2001;166:1611–23. [PubMed] [Google Scholar]
- 24.Berrada N, Delaloge S, Andre F. Treatment of triple-negative metastatic breast cancer: toward individualized targeted treatments or chemosensitization? Ann Oncol. 2010;21(7):vii30–vii5. doi: 10.1093/annonc/mdq279. [DOI] [PubMed] [Google Scholar]
- 25.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 26.Copland JA, Marlow LA, Kurakata S, Fujiwara K, Wong AK, Kreinest PA, et al. Novel high-affinity PPARgamma agonist alone and in combination with paclitaxel inhibits human anaplastic thyroid carcinoma tumor growth via p21WAF1/CIP1. Oncogene. 2006;25:2304–17. doi: 10.1038/sj.onc.1209267. [DOI] [PubMed] [Google Scholar]
- 27.Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, et al. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25:3479–88. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 28.Marlow LA, D'Innocenzi J, Zhang Y, Rohl SD, Cooper SJ, Sebo T, et al. Detailed molecular fingerprinting of four new anaplastic thyroid carcinoma cell lines and their use for verification of RhoB as a molecular therapeutic target. J Clin Endocrinol Metab. 2010;95:5338–47. doi: 10.1210/jc.2010-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marlow LA, Reynolds LA, Cleland AS, Cooper SJ, Gumz ML, Kurakata S, et al. Reactivation of suppressed RhoB is a critical step for the inhibition of anaplastic thyroid cancer growth. Cancer Res. 2009;69:1536–44. doi: 10.1158/0008-5472.CAN-08-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper SJ, Zou H, Legrand SN, Marlow LA, von Roemeling CA, Radisky DC, et al. Loss of type III transforming growth factor-beta receptor expression is due to methylation silencing of the transcription factor GATA3 in renal cell carcinoma. Oncogene. 2010;29:2905–15. doi: 10.1038/onc.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal SK, Childs BH, Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat. 2011;125:627–36. doi: 10.1007/s10549-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banumathy G, Cairns P. Signaling pathways in renal cell carcinoma. Cancer Biol Ther. 2010;10:658–64. doi: 10.4161/cbt.10.7.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, et al. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26:5680–91. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- 34.Awakura Y, Nakamura E, Ito N, Kamoto T, Ogawa O. Methylation-associated silencing of SFRP1 in renal cell carcinoma. Oncol Rep. 2008;20:1257–63. [PubMed] [Google Scholar]
- 35.Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, Brown M, et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29:2104–17. doi: 10.1038/onc.2009.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Autocrine WNT signaling contributes to breast cancer cell proliferation via the canonical WNT pathway and EGFR transactivation. Breast Cancer Res. 2007;9:R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–56. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res. 2009;11:R32. doi: 10.1186/bcr2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, Knuchel R, et al. Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29:991–8. doi: 10.1093/carcin/bgn076. [DOI] [PubMed] [Google Scholar]
- 40.Veeck J, Noetzel E, Bektas N, Jost E, Hartmann A, Knuchel R, et al. Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumor-specific epigenetic marker in human breast cancer. Mol Cancer. 2008;7:83. doi: 10.1186/1476-4598-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawakami K, Yamamura S, Hirata H, Ueno K, Saini S, Majid S, et al. Secreted frizzled-related protein-5 is epigenetically downregulated and functions as a tumor suppressor in kidney cancer. Int J Cancer. 2011;128:541–50. doi: 10.1002/ijc.25357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawamoto K, Hirata H, Kikuno N, Tanaka Y, Nakagawa M, Dahiya R. DNA methylation and histone modifications cause silencing of Wnt antagonist gene in human renal cell carcinoma cell lines. Int J Cancer. 2008;123:535–42. doi: 10.1002/ijc.23514. [DOI] [PubMed] [Google Scholar]
- 43.Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, et al. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–97. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- 44.Wu W, Glinka A, Delius H, Niehrs C. Mutual antagonism between dickkopf1 and dickkopf2 regulates Wnt/beta-catenin signalling. Curr Biol. 2000;10:1611–4. doi: 10.1016/s0960-9822(00)00868-x. [DOI] [PubMed] [Google Scholar]
- 45.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121:737–46. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 46.Abele R, Clavel M, Dodion P, Bruntsch U, Gundersen S, Smyth J, et al. The EORTC Early Clinical Trials Cooperative Group experience with 5-aza-2′-deoxycytidine (NSC 127716) in patients with colo-rectal, head and neck, renal carcinomas and malignant melanomas. Eur J Cancer Clin Oncol. 1987;23:1921–4. doi: 10.1016/0277-5379(87)90060-5. [DOI] [PubMed] [Google Scholar]
- 47.Gollob JA, Sciambi CJ, Peterson BL, Richmond T, Thoreson M, Moran K, et al. Phase I trial of sequential low-dose 5-aza-2′-deoxycytidine plus high-dose intravenous bolus interleukin-2 in patients with melanoma or renal cell carcinoma. Clin Cancer Res. 2006;12:4619–27. doi: 10.1158/1078-0432.CCR-06-0883. [DOI] [PubMed] [Google Scholar]
- 48.Geutjes EJ, Tian S, Roepman P, Bernards R. Deoxycytidine kinase is overexpressed in poor outcome breast cancer and determines responsiveness to nucleoside analogs. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1477-3. [DOI] [PubMed] [Google Scholar]
- 49.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, Beaulieu N, et al. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–9. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.