Abstract
The TRIM family of genes is largely studied because of their roles in development, differentiation and host cell anti-viral defenses; however, roles in cancer biology are emerging. Loss of heterozygosity of the TRIM3 locus in approximately 20% of human glioblastomas raised the possibility that this NHL containing member of the TRIM gene family might be a mammalian tumor suppressor. Consistent with this, reducing TRIM3 expression increased the incidence of and accelerated the development of PDGF-induced glioma in mice. Furthermore, TRIM3 can bind to the cdk inhibitor p21WAF1/CIP1. Thus, we conclude that TRIM3 is a tumor suppressor mapping to chromosome 11p15.5 and that it can block tumor growth by sequestering p21 and preventing it from facilitating the accumulation of cyclin D1-cdk4.
Keywords: glioma, p21, PDGF, stem/progenitor, TRIM3
Introduction
Glioblastomas (GBM) are amongst the most common central nervous system tumors, comprising approximately 25% of adult brain cancers [1]. Despite advances in diagnosis and therapy, the prognosis for patients with malignant glioma remains poor. While a number of studies have stratified gliomas into distinct groups based on molecular and biochemical profiles [2-5], the significance of many of these differences to understanding or treating the disease has yet to be established.
One third of GBMs are associated with chronic PDGF signaling and have a pro-neural signature. These tumors are characterized by dysregulation of the Rb signaling pathway, which contributes to excessive cell proliferation, a hallmark of this subtype of glioma. Nevertheless, the genetic heterogeneity within this group suggests that additional changes contribute to the pathogenesis of these tumors and there are likely multiple subtypes, perhaps reflecting different cells of origin [6, 7]
Loss of heterozygosity in chromosomal region 11p15.5 in approximately 20% of human GBM raised the possibility that TRIM3 could be a potential tumor suppressor [8]. The TRIM family members contain a structurally conserved amino-terminal RING motif followed by one or two B-boxes and a coiled coil domain [9]. Distinct subclasses of this family are defined by the presence of other domains following this conserved unit. A few of the members containing SPRY domains, PHD/BROMO domains, or TM domains have been implicated in human cancer [10]. TRIM3 is in the NHL sub-group, which contains three other mammalian paralogs (TRIM2, TRIM32 and TRIM71) and orthologs in both D. melanogaster (Brat, Mei-P26, Abba, and Wech), and C. elegans (Ncl-1, and Nhl-1, Nhl-2, Nhl-3, Lin41) (reviewed in [11]). Of these, only the fly ortholog brat has previously been implicated as a growth suppressor with a role preventing tumor-like proliferation of secondary neuroblasts [12].
Expression of brat is largely restricted to the embryonic peripheral nervous system, ventral nerve cord and developing brain of the fly. Loss of function alleles, including those that eliminate the NHL domain are associated with increased numbers of secondary neuroblasts. Brat promotes asymmetric division that produces ganglion mother cells which ultimately divide one more time to produce neurons [13-16]. How the brat mutation leads to increased numbers of secondary neuroblasts is not particularly well understood [17]. Like other TRIM-NHL proteins brat can interact with Ago1, an miRISC effector protein (reviewed in [11]). Brat, TRIM32 and mei-P26 also regulate the post-transcriptional accumulation of myc [14, 18-21], and Brat can interact with numb and prospero to control mRNA localization as well [15, 16, 18]. The contribution of each mechanism to tumor suppression however is not clear.
The RCAS-PDGF/nestin-TvA mouse model recapitulates the morphologic hallmarks, the dependence on tumor microenvironment, and the stem and progenitor cell behaviors of human proneural PDGF-associated GBM [22-27]. Additionally, it models the molecular pathways that govern proliferation, differentiation, cell migration, DNA repair and tumor cell-microglia interaction in the microenvironment [28-32]. Here we show that TRIM3 has a tumor suppressive role in this model, and it acts in part by regulating the availability of the cdk inhibitor p21, which promotes cyclin D1-cdk4 nuclear accumulation.
Results
TRIM3, a potential tumor suppressor in human GBM
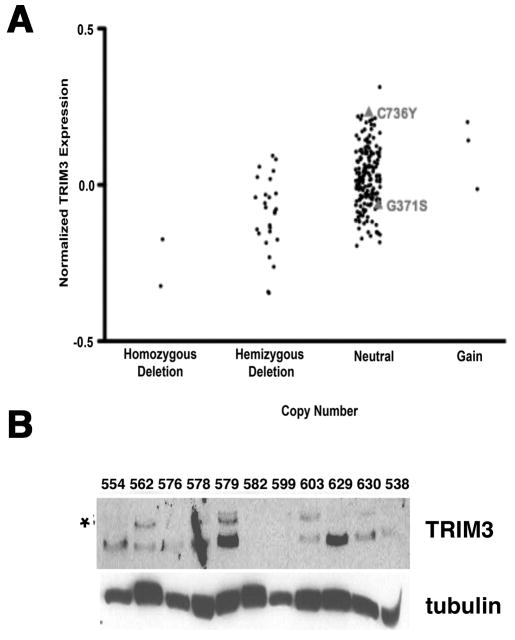
TRIM3 is encoded in the chromosomal locus 11p15.5. Loss of heterozygosity encompassing this region is observed in approximately 20% of human GBM, suggesting that TRIM3 might be a tumor suppressor gene [8]. Supporting this hypothesis of TRIM3 function, the loss of a Drosophilia ortholog of the TRIM-NHL family, brat, results in an increase in the number of secondary neuroblasts at the expense of ganglion mother cells. As comparable progenitors are found in the developing brain of mammals (reviewed in [12]) we re-analyzed 203 tumors in the TCGA study [2, 33] for TRIM3 expression, copy number, and mutation. Twenty-six of these tumors had loss of heterozygosity, two of these tumors had homozygous deletion, and three had low-level amplification of this locus. mRNA expression was correlated to gene expression (Fig. 1A). Additionally, in ninety-one tumors in which the locus was completely sequenced we identified two non-recurrent missense mutations (Fig. 1A). One of these, C736Y, is in the NHL domain and the other, G371S, is in the ABP/filamin domain.
Figure 1. TRIM3 expression is reduced in some human GBM.
(A) Expression of TRIM3 mRNA (y-axis) relative to the DNA copy number (x-axis) in 203 GBM samples. Data were downloaded from MSKCC's Cancer Genomics Data Portal (http://cbio.mskcc.org/cancergenomics-dataportal) and samples were separated by the DNA copy number status of 11p15.5 (the TRIM3 containing locus). Deletion and gain calls were determined by the RAE algorithm [55]. Of the 91 samples that were subjected to DNA sequencing, two that exhibited a neutral DNA copy number calls of the 11p15.5 locus contained missense mutations in TRIM3. These mutations, G317S and C736Y and their mRNA expression are indicated in the chart. (B) Immunoblot. The expression of TRIM3 was examined in extracts prepared from 11 individual tumors (indicated above each lane). Tubulin is a loading control. The asterik marks the TRIM3 band.
To determine if the expression of TRIM3 was correlated to the presence of the gene, we looked for TRIM3 protein expression by immunoblotting eleven protein extracts prepared from fresh surgical resections in which the 11p15.5 locus was intact. Strong expression of TRIM3 was seen in only four (Fig. 1B). We identified TRIM3 by comparing its migration with a paired sample of extracts prepared from T98G cells, a human PDGF-transformed glioma cell line, in which TRIM3 was knocked down by a lentiviral shRNA. The nature of the faster migrating anti-TRIM3 reactive band is not clear. This suggests that TRIM3 expression might be reduced more frequently in human GBM than suggested by the genomic data.
TRIM3 is a tumor suppressor in mice
The RCAS-PDGF-HA/nestin-TvA model recapitulates a subtype of human proneural GBMs driven by PDGF signaling. To determine if TRIM3 was a tumor suppressor we wanted to reduce its expression and ask if that would lead to increased tumor development or accelerate tumor progression in response to PDGF. We first identified RCAS vectors encoding distinct shRNAs that reduced the expression of TRIM3 in YH/J12 cells (Fig. 2A). YH/J12 are spontaneously immortalized cells derived by infecting primary brain cultures obtained from nestin-TvA mice with RCAS viral vectors expressing PDGF-HA. These cells are a good ex vivo surrogate of tumor behavior as they recapitulate the cellular and molecular events during differentiation [25] and proliferation [28, 30] associated with this disease. Notably, reducing TRIM3 correlated with increased proliferation (Fig. 2B).
Figure 2. Reducing TRIM3 increases cell proliferation and p21 accumulation.

(A) Immunoblot. YH/J12 cells were infected with a lentiviral vector expressing either a scramble shRNA (scram) or TRIM3 shRNAs (E6, E7; Openbiosystems). 48 hours after infection, cells were selected in puromycin for three days, harvested, and extracts prepared. The amount of TRIM3 and p21 was measured by immunoblot as shown in left panel. Tubulin serves as a loading control. (B) Flow cytometry. The DNA profile in TRIM3 shRNA transfected cells is shown in the right panel. The percentage of S+G2+M phase cells is shown. This experiment was repeated at least twice with similar results.
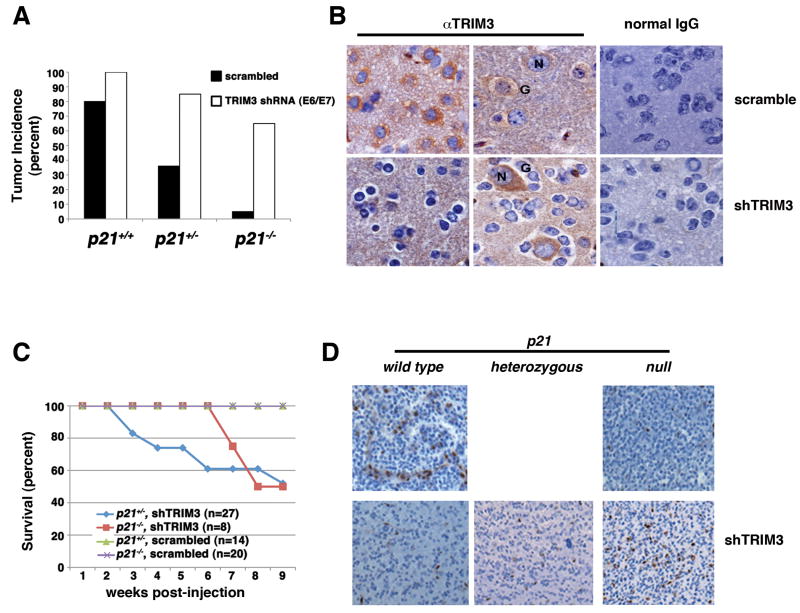
We subsequently introduced these vectors into nestin-TvA mice with and without RCAS-PDGF-HA. Consistent with the fact that PDGF is the driving oncogene in this model, disease never developed in mice in the absence of RCAS-PDGF-HA. On the other hand, 80% of the mice that were infected with RCAS-PDGF-HA and a scrambled shRNA vector developed tumors, and 100% developed tumors when challenged with RCAS-PDGF-HA and the TRIM3 shRNA vectors (Fig. 3A). TRIM3 protein expression was clearly reduced in the glial cells of the tumor, but not in the trapped neurons (Fig. 3B). This confirmed the delivery of the shRNA and the cell type specificity of the RCAS system, as well as the specificity of the antibodies to detect TRIM3 by immunohistochemistry. However, although these differences were significant (p<0.05), due to the strong tumor promoting ability of the RCAS-PDGF-HA virus alone it was difficult to unequivocally conclude that reducing TRIM3 enhanced tumorigenicity.
Figure 3. TRIM3 is a tumor suppressor that antagonizes PDGF-induced gliomagenesis in the mouse brain.
(A) Tumor incidence. Wild type, p21 heterozygous and p21 knockout animals were infected as indicated and tumor incidence scored over 12 weeks. Animals infected with RCAS-TRIM3 shRNA E6 and RCAS-TRIM3 shRNA E7 were combined. (B) TRIM3 immunohiostochemistry in mouse tumors. Endogenous TRIM3 was detected with a monoclonal antibody from Becton-Dickinson followed by DAB staining and hematoxylin counter-staining (brown deposits). Normal IgG was used as control. Representative neurons (N) and oligodendroglioma cells (G) are indicated. This was carried out on at least five different mice. Representative images at 1000X magnification are shown. (C) Kaplan-Meier survival curve. Mice were infected as described in panel B and sacrificed when morbid. (D) Representative images of Id1 immunohistochemistry in tumors that arose in mice. Magnification, 400X. The p21 status is indicated above each panels and TRIM3 manipulation to the right of the panels.
We reasoned that we might be better able to unmask a tumor suppressive activity of TRIM3 by using a sensitized background. The tumor promoting activity of PDGF is directly proportional to p21 gene dosage [30]. Thus, we repeated this analysis using p21 heterozygous mice and p21 null mice. Whereas 33% of the p21 heterozygous mice co-infected with RCAS-PDGF-HA and a non-specific shRNA vector developed tumors, 80% of the nestin-TvA;p21+/- mice co-infected with RCAS-TRIM3 shRNA and RCAS-PDGF-HA developed tumors (p<0.004) (Fig. 3A). Furthermore, there was no morbidity in p21 heterozygous mice expressing PDGF-HA and the scrambled shRNA; however, a significant amount of morbidity was observed in p21 heterozygous animals expressing PDGF-HA and the shRNA against TRIM3 (Fig. 3C). We also found that tumor incidence increased in p21 deficient mice expressing PDGF when TRIM3 was reduced with the shRNA vectors (Fig. 3A). Nevertheless, the incidence of tumors was significantly lower than that observed in the wild type or heterozygous animals (p<0.01), and the morbidity associated with the tumors was delayed (Fig. 3C). Thus, TRIM3 is a bona fide tumor suppressor and its loss could co-operate with expression of PDGF to increase the frequency of tumor development and accelerate the morbidity of afflicted animals.
The status of p21 affects the nature of the tumors arising in TRIM3 deficient mice challenged with PDGF
The difference in the rate of morbidity associated with the development of proneural GBMs when TRIM3 was reduced in the p21 heterozygous and p21 deficient animals was puzzling. Recently, Barrett and colleagues [6] showed that tumor cells derived from the RCAS-PDGF-HA/nestin-TvA model could be sorted into two groups based on their Id1 staining. Those that highly expressed Id1 had high self-renewal capacity and generated slow growing tumors when engrafted into hosts relative to the cells with low Id1 expression, which had limited self-renewal capacity. Thus given the differences in the onset of morbidity in p21 heterozygous and p21 null mice in which TRIM3 was reduced we looked to see if the Id1 staining of the tumors was different. We found strong Id1 staining in 3/3 tumors arising in p21 null mice whereas only scattered staining was observed in 6/6 p21 heterozygous and wild type mice. A representative image from each cohort is shown in Figure 3D. Thus, the nature of the tumors that arise in the animals that have p21 is distinct from that in animals that lacked p21, albeit all were proneural GBMs based on morphological and immunohistochemical criteria as defined by Ciznadijia and colleagues [28] (data not shown).
Reducing the expression of TRIM3 increases accumulation of p21
It then occurred to us that the proneural GBMs in this model were exquisitely dose dependent with respect to p21 gene dosage [30]; thus, tumor promoting activity in the p21 heterozygous mice associated with the loss of TRIM3 might be due to increased accumulation of p21. Consistent with this, p21 was barely detectable in extracts from nestin-TvA;p21+/- mice infected with RCAS-PDGF-HA alone, but was easily detected in extracts derived from tumor bearing nestin-TvA;p21+/- mice co-infected with the RCAS-PDGF-HA and RCAS-TRIM3 shRNA vectors, even after accounting for oligodendrocyte contribution by normalizing levels to olig2 expression (Fig. 4A). Additionally, p21 protein accumulated in YH/J12 cells in which TRIM3 protein was reduced following transduction of these shRNA vectors (Fig. 2A), or following transfection with two different siRNAs (Fig. 4B). Thus, reducing TRIM3 increases the amount of p21.
Figure 4. Reducing TRIM3 increases the accumulation of p21.
(A) Immunoblot from mouse tumors. Whole brain extract was prepared and immunoblotted for olig2 to normalize oligodendrocyte contribution and p21. N: Normal brain; P: Brain from a p21 heterozygous mouse infected with RCAS-PDGF and RCAS-scrambled shRNA; 1,2,3: three random p21 heterozygous mouse brains infected with RCAS-PDGF and RCAS-TRIM3 shRNA. (B) Immunoblot. YH/J12 cells were transfected with either a control siRNA (scr) or a TRIM3 siRNA Smartpool (Dharmacon), and extracts were prepared 24 and 48 hours later as indicated above each lane. The amount of TRIM3 and p21 were determined by immunoblot. Tubulin serves as a loading control.
TRIM3 binds to p21
We next wanted to know if TRIM3 could interact with p21. We could coimmunoprecipitate TRIM3 and p21 at endogenous levels from densly grown cultures of YH/J12 cells (Fig. 5A). We could also coprecipitate the proteins when full length TRIM3 was over-expressed in YH/J12 or T98G cells, but not when mutants of TRIM3 lacking either the NHL domain or the RBCC domain were expressed to an even higher level (Fig. 5B). Additionally, we could reconstitute the interaction of TRIM3 with p21 using recombinant proteins produced in E. coli (Fig. 5C). Thus, TRIM3 interacts with p21, and stable interaction depends on multiple domains in both the conserved RBCC motif and in the ABP/filamin/NHL domains.
Figure 5. TRIM3 interacts with p21.
(A) Endogenous TRIM3 and p21 interact in cells. Cell lysates were processed for reciprocal immunoprecipitation using Santa Cruz antibody p21M19 and a Genway antibody to TRIM3. Immunoprecipated protein was blotted with the antibodies indicated on the left of each panel. Normal mouse IgG (RαM) was used as negative control for the immunoprecipitation. This experiment was repeated at least three times. (B) Immunoblot. myc-tagged TRIM3, a myc-tagged NHL only domain, or a myc-tagged TRIM3 mutant in which the NHL domain was removed were expressed in YH/J12 cells. The level of expression of these constructs was determined by immunoblot (labeled input: myc). p21 was immunoprecipitated from these extracts and the associated myc-TRIM3 was detected by immunoblot (labeled myc and p21). This experiment was repeated at least three times. (C) Recombinant purified GST-tagged TRIM3 or GST (indicated above the panel) was incubated with 6XHis tagged p21 (His-p21) and re-isolated on glutathione sepharose. Coomassie staining was used to detect the amount of precipitated GST and GST fusion protein. The amount of p21 (in micromol) added is indicated at the bottom of each lane, and p21 was detected by immunoblot. This experiment was repeated at least three times with independent preparations of the recombinant proteins with similar results.
Over-expression of TRIM3 induces growth arrest
Other NHL containing TRIMs inhibit cell proliferation when over-expressed in various cell types, both those in which they are endogenously expressed and in heterologous cell types (reviewed in [11]). Thus, we looked at the effect of manipulating TRIM3 expression in the PDGF-transformed glial tumor cells. When we reduced the level of TRIM3 in YH/J12 cells the fraction of cycling cells increased and the amount of p21 increased (Fig. 2). Conversely BrdU incorporation was reduced significantly when TRIM3 was over-expressed (Fig. 6A). When TRIM3 was over-expressed to similar levels in p21-deficient YH/J12 cells, it did not reduce proliferation; however, proliferation in these cells is so poor it is difficult to reach a conclusion about the significance of this (data not shown). Cells in which TRIM3 was over-expressed exited the cell cycle and accumulated in G0/G1 phase (Fig. 6B). However, we could not determine if p21 levels were changed because only a fraction of the cells express TRIM3 and we could not identify any reliable immunohistochemical reagents specific for mouse p21. Thus, there is a reciprocal relationship between the expression of TRIM3 and the fraction of cells in S phase.
Figure 6. TRIM3 induces growth arrest.
(A) Immunofluorescence. YH/J12 cells were transfected with either a mock vector or a myc-TRIM3 expression plasmid and cells were labeled with 20μM BrdU for two hours, forty-eight hours after transfection. A representative image of the cells is shown in the top panel (green, αmyc, TRIM3; red, αBrdU, S-phase cell; blue, DAPI, DNA). BrdU incorporation is quantitated at the bottom of the panel and was determined from at least three independent experiments (mean ± standard deviation), for each of which 100-200 cells were counted. (B) TRIM3 induces G0/G1 arrest in human T98G glioma cells. T98G cells were transfected with myc-TRIM3 and 48 hours later were fixed and stained with propidium iodide (DNA content) and anti-myc. In this representative transfection, 14.7% of the cells were transfected with the myc-TRIM3 expression plasmid and these were uniformly arrested at G0/G1. The remaining cells were still distributed throughout the cell cycle. This experiment was repeated twice with similar results.
The ability of TRIM3 to bind to p21 and to reduce cell proliferation are inseparable
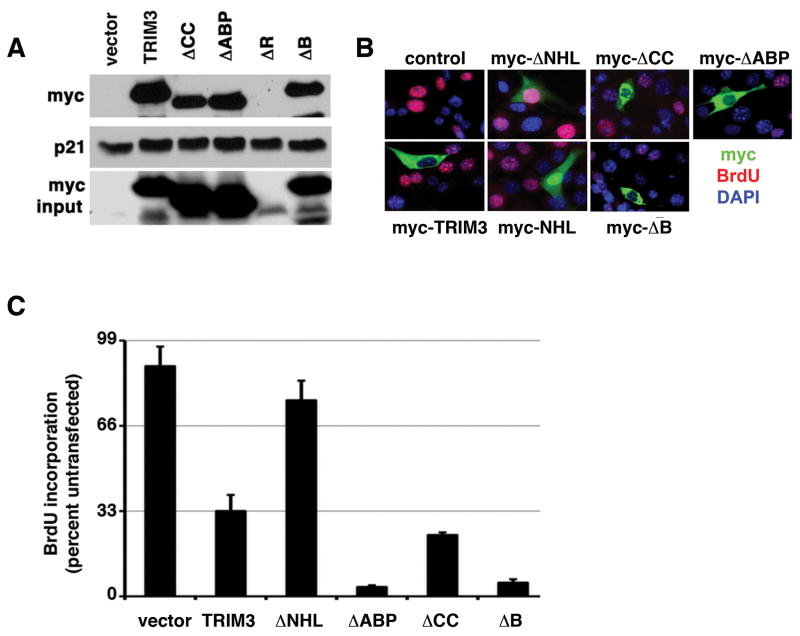
To determine if the growth suppressive activity of TRIM3 correlated with its ability to bind p21 we tried to generate a mutant that would separate these activities. To accomplish this, we deleted each conserved domain and asked if these mutants could bind p21 when expressed in YH/J12 cells. All of the mutants with the exception of RING mutants expressed well (Fig. 7A). A mutant lacking the NHL domain was unable to bind to p21 (Fig. 5B), but the wild type protein and the mutants individually lacking the B-box, coiled-coil or ABP domain could bind p21 (Fig. 7A). All these proteins accumulated in the cytosol (Fig. 7B). We could also detect binding of the RING mutant to p21 by using recombinant protein in vitro (data not shown). Thus, the NHL domain of TRIM3 was necessary but not sufficient for p21 binding (Fig. 5B).
Figure 7. Domain deletion mutants cannot separate the TRIM3-growth suppressive activity from its ability to bind to p21.
(A) p21 binding. This is arranged as described in the legend to figure 5B. ΔABP, a mutant lacking the ABP domain; ΔCC, a mutant lacking the coiled coil domain; ΔB, a mutant lacking the B-box domain, and ΔR is a mutant lacking the RING domain. (B) Immunofluorescence. As decribed in the legend to figure 6A, YH/J12 cells were transfected with the indicated vectors (C) BrdU quantification. The percentage of BrdU incorporating YH/J12 cells expressing the indicated alleles was determined from at least three independent experiments (mean ± standard deviation), for each of which 100-200 cells were counted.
We next looked at the ability of these mutants to induce growth arrest. Whereas the NHL deficient mutant was unable to induce growth arrest, the B-box deficient, coiled-coil deficient, and the ABP-deficient mutants could (Fig. 7C). The G371S and C736Y missense mutants that we detected in the genomic sequencing of the locus were also expressed well in cells, could bind p21, and could induce growth arrest (data not shown). Because the NHL-alone mutant accumulated in the nucleus, and the RING mutant was poorly expressed we did not examine their affect on cell proliferation.
Cyclin-cdk binding sequesters p21 from TRIM3
Given that p21 is a cyclin D-cdk4 assembly factor in the RCAS-PDGF-HA/nestin-TvA model [30], and TRIM3 can bind to p21 and suppress cell proliferation, we wanted to know if cyclin-cdks affected the interaction of TRIM3 and p21. We found that a mutant of p21 deficient in binding cyclin-cdk complexes bound better to TRIM3 than wild type p21 in extracts containing cyclin-cdk complexes (Fig. 8A). However the wild type p21 and the non-cyclin-cdk binding mutant bound equivalently if cyclin-cdk complexes were depleted from the extract (Fig. 8B). Thus, TRIM3 binds better to p21 that was not physically associated with cyclin-cdk complexes, and TRIM3 and cyclin-cdk complexes might compete with each other for p21.
Figure 8. The association of TRIM3 with p21 is reduced in the presence of cyclin-cdk complexes binding to p21.
(A) Immunoblot. GST-p21 and GST-p21cy(-) were produced and purified from E. coli and the amount indicated above each lane (ng) was incubated with 0.5mg total cell extract obtained from myc-TRIM3 transfected 293T cells. myc-TRIM3 was subsequently immunoprecipitated and the amount of TRIM3 and p21 determined by immunoblotting as indicated to the right of each panel. (B) Extracts from myc-TRIM3 expressing 293T cells were passed through either p13-sepharose to deplete cyclin-cdk comlexes or sepharose CL-4B (mock). On the left, the amount of cyclin A, cyclin D1, cdk2 and cdk4 was measured by immunoblot to assess whether the depletion was successful. On the right, 12.5ng of recombinant substrate indicated above each lane was incubated with 0.25mg of the depleted extracts and recovered as described.
Discussion
The evidence presented here strongly supports the interpretation that TRIM3 is a mammalian tumor suppressor whose loss can increase the incidence and promote the development of GBM. This is the first time that a member of the NHL-domain containing TRIM family has been demonstrated to play a role in tumorigenesis in mammals.
Taxonomic classification of GBM is an active area and the amplification or loss of PDGFR, EGFR, CDKN2C, PTEN, MET, CDKN2A, CDK6, CDK4, MDM2, MDM4 and PTPRD are frequently associated with different subclasses of glioma. Additionally, tumors can be subdivided on the expression of a mesenchymal or proneural signature, or on specific chromosomal abnormalities such as 1p and 19q deletions. These classifications might relate to malignancy or therapeutic possibilities (reviewed in [2, 7]). However, we were unable to detect any relationship between established copy number alterations, the presence of a proneural or mesenchymal signature, or 1p/19q deletions when we attempted to correlate TRIM3 protein expression to these parameters in a small set of 40 tumors (unpublished data). Thus, expression of TRIM3 may classify GBM in a unique way and may provide insight into prognostic and therapeutic subgroups of human proneural GBM.
Loss of the TRIM-NHL proteins brat and mei-P26 in flies leads to excessive stem cell proliferation and tumor-like accumulation of neuroblasts and ovarian stem cells, respectively [10, 11]. However, there were no spontaneous tumor phenotypes reported in mammalian TRIM-NHL deficient mice, including TRIM3 [34]. Nevertheless, as we have shown, mammalian TRIM3 can suppress tumor development induced by overexpression of PDGF in nestin-positive cells in the brain of mice.
Interestingly, the nature of the tumor induced by reducing TRIM3 was dependent on the status of p21. Reducing TRIM3 in p21 deficient mice gives rise to tumors that retain “stem-like” expression of Id1, but reducing TRIM3 in p21 proficient mice gives rise to tumors that retain a “progenitor-like” expression of Id1. Although the precise relationship and mechanisms of action of TRIM3 in mammalian glial stem and progenitor cells needs to be defined, there may be therapeutic value to determining the expression of these proteins in human tumors. GBM arising from Id1high expressing “stem-like” tumor cells may be less sensitive to irradiation than GBM arising from Id1low “progenitor-like” cells. Thus, patients who are low for TRIM3 and high for p21 might have better outcome when using modalities such as ionizing radiation.
How does TRIM3 suppress tumor growth? Other members of the TRIM-NHL family have been reported to have roles in miRNA processing and post-transcriptional regulation of myc [11, 14, 20, 21, 35]. These TRIM-NHL proteins interact with Ago1, but we have not been able to detect an interaction of TRIM3 with Ago1, at least in the p21 positive tumor cells. However, in these cells we could confirm the interaction of TRIM3 with actinin-4 and myosin V [36], and we could demonstrate that TRIM3 can interact with p21 in an NHL-dependent manner. Thus, we propose that in these cells TRIM3 sequesters p21 away from cyclin D1-cdk4 reducing proliferation. Given the similar role of p21 in cytokine induced proliferation of bone marrow progenitors (reviewed in [37, 38]) and glial progenitors [30] it seems likely that TRIM3 may play a similar tumor suppressive role in some types of leukemia as well.
Whilst the importance of growth promoting activities of Kip-family cdk inhibitors has met with skepticism, four lines of evidence suggest that these proteins play a significant growth-promoting role in some subtypes of cancer. First, mouse studies document the causal importance of cdk inhibitors for tumor progression [39-42]. Second, p21 is required for cytokine induced proliferation in bone marrow derived progenitor cells and glial progenitor cells [30, 37, 38]. Third, correlative studies in human tissue samples suggest that the presence of cdk inhibitors can be associated with more aggressive disease [43-50]. Fourth, our understanding of how tyrosine phosphorylation disrupts the inhibitory activity but preserves cyclin-cdk binding activity provides a molecular understanding of this switch in behavior [51]. Ultimately, the RCAS-PDGF-HA/nestin-TvA model allowed us to assess the crucial contribution that p21, the tyrosine phosphorylation of p21, and cyclin D1 and cdk4 make to tumor progression [28-30].
However, the ability of TRIM3 to interact with p21 only accounts for part of its growth suppressive activity, because reducing TRIM3 could promote tumor development in p21 deficient mice as well. Although the tumors that arise in p21 deficient mice are morphologically similar to those that arose in p21 proficient mice, they were clearly distinct with regards to their Id1 expression status. Given that Id1 is a proxy for the stem and progenitor nature of the cell, and p21 activities change from growth suppressor to growth promoter as cells undergo this transition, then understanding how TRIM3 works in these cells may allow us to understand how cell cycle regulators integrate with the machinery of asymmetric divisions and drive cancer.
Materials and Methods
Cell culture and vector production
The PDGF-transformed primary glial cell line YH/J12 was described previously [30]. T98G and Chicken DF-1 cell lines were purchased from ATCC and cultured according to their directions.
The RCAS-PDGF-HA expression plasmids were described previously [30]. Mouse TRIM3 cDNA was kindly provided by Dr. Andrea Ballabio [52]. myc-TRIM3 expression vector was constructed by cloning the mouse TRIM3 cDNA into pCMV-Myc vector (Clontech). myc-TRIM3 deletion constructs were generated by PCR-based DNA mutagenesis or in vitro site-directed mutagenesis as described previously [30]. The deletions encompassed the following amino acid sequences (ΔNHL, 421-734; NHL, 1-420; ΔABP, 290-420; ΔCC, 150-280; ΔB, 115-146, and ΔR, 22-62) Lentiviral TRIM3 shRNA were purchased from OpenBiosystems. RCAS-TRIM3 shRNA vectors were created using the Gateway in vitro recombination system as described previously [30]. Lentiviral vectors expressing these TRIM3 shRNA were prepared as described previously [53]. RCAS vectors were prepared in DF1 cells as described previously [30].
Flow Cytometry and BrdU incorporation
Flow cytometry analysis and BrdU incorporation assay were performed as described previously [30, 54].
Protein extracts, immunoblot and immunoprecipitation
Protein extracts from cells were prepared as we previously described [30]. Brains were removed from euthanized mice, snap frozen in liquid nitrogen, ground to a fine powder and extracts prepared as described [28].
Immuoblot and immunoprecipitation assays were carried out as we previously described [30]. The following antibodies were used: p21 (M19 and C19, Santa Cruz), Flag (M2, Sigma), c-myc (9E10, Santa Cruz), HA (Y11, Santa Cruz), tubulin (Santa Cruz), TRIM3 (BD and Genway) and olig2 (Millipore).
Preparation of E. coli produced recombinant protein
GST or His-tagged recombinant proteins were produced in E coli using GST Gene Fusion System (Amersham pharmacia biotech) and Probind™ Purification System (Invitrogen) respectively, according to manufactures instructions.
In vitro p21-TRIM3 binding assays
0.78μM of recombinant GST or GST-TRIM3 were combined with increasing amounts of His-p21 in a 60μl reaction containing 50mM Tris-Cl pH 7.5, 5mM MgCl2, 1mM DTT, 2mM PMSF, 10μg/ml leupeptin, 10μg/ml aprotinin, 10μg/ml soy bean trypsin inhibitor. Following a 30 minute incubation at 37°C, GST or GST-TRIM3 was captured on a 30μl bed volume of Glutathione-agarose beads and washed three times with the reaction buffer prior to adding sample buffer. Glutathione bound proteins were resolved be SDS-PAGE and GST and GST-TRIM3 detected by Coomassie blue staining, and the coprecipitating p21 was detected by immunoblot using p21 antibody (F-5 mouse monoclonal, Santa Cruz) at a 1:1000 dilution.
Histology and immunohistochemistry in mouse tumor tissue
Mouse brain sections were prepared and processed as described previously [30]. The Trim3 Antibody (BD) was used as recommended by the provider. The anti-mouse Id1 antibody was obtained from Biocheck, Inc. (BCH-1/37-2) and used as described by Barrett and colleagues.
Acknowledgments
We thank Marta Kovatcheva and other members of the Koff lab, Pengbo Zhou (Cornell University Medical School), John Petrini (MSKCC) and (MSKCC) for comments on this manuscript, and Max Chan Liu (The Browning School) for his assistance with Id1 staining and image acquisition. This work was supported by the Memorial Sloan-Kettering Cancer Center Core Grant (P30CA08748) and grants to Andrew Koff (CA89563). Funding was also provided by the Brain Tumor Center (YL, DC) and the Golfers Against Cancer Foundation (AK).
Footnotes
AUTHOR CONTRIBUTIONS: YL carried out the experiments identifying TRIM3 and measuring the effect of manipulating TRIM3 on p21 in cells. AMP, TO, NPG, CB, and ECH analyzed TRIM3 expression in human tumors and human tumor extract. RR, NY, DC, EH, HEB and PT provided experimental assistance or reagents throughout the course of this work. AK directed the research.
CONFLICT OF INTEREST: The authors have no conflict of interest.
References
- 1.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nature reviews Cancer. 2010 May;10(5):319–31. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 2.Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, et al. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PloS one. 2009;4(11):e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008 Sep 26;321(5897):1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer cell. 2010 Jan 19;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitucci M, Hayes DN, Miller CR. Gene expression profiling of gliomas: merging genomic and histopathological classification for personalised therapy. British journal of cancer. 2011 Feb 15;104(4):545–53. doi: 10.1038/sj.bjc.6606031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett LE, Granot Z, Coker C, Iavarone A, Hambardzumyan D, Holland EC, et al. Self-renewal does not predict tumor growth potential in mouse models of high-grade glioma. Cancer cell. 2012 Jan 17;21(1):11–24. doi: 10.1016/j.ccr.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012 Mar 30;149(1):36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulay JL, Stiefel U, Taylor E, Dolder B, Merlo A, Hirth F. Loss of heterozygosity of TRIM3 in malignant gliomas. BMC cancer. 2009;9:71. doi: 10.1186/1471-2407-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nature reviews Microbiology. 2005 Oct;3(10):799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 10.Hatakeyama S. TRIM proteins and cancer. Nature reviews Cancer. 2011 Nov;11(11):792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 11.Wulczyn FG, Cuevas E, Franzoni E, Rybak A. MiRNA need a TRIM regulation of miRNA activity by Trim-NHL proteins. Advances in experimental medicine and biology. 2010;700:85–105. [PubMed] [Google Scholar]
- 12.Reichert H. Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development. Results and problems in cell differentiation. 2011;53:529–46. doi: 10.1007/978-3-642-19065-0_21. [DOI] [PubMed] [Google Scholar]
- 13.Arama E, Dickman D, Kimchie Z, Shearn A, Lev Z. Mutations in the beta-propeller domain of the Drosophila brain tumor (brat) protein induce neoplasm in the larval brain. Oncogene. 2000 Aug 3;19(33):3706–16. doi: 10.1038/sj.onc.1203706. [DOI] [PubMed] [Google Scholar]
- 14.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006 Mar 24;124(6):1241–53. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 15.Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Developmental cell. 2008 Apr;14(4):535–46. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Developmental cell. 2006 Apr;10(4):441–9. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 17.Januschke J, Gonzalez C. Drosophila asymmetric division, polarity and cancer. Oncogene. 2008 Nov 24;27(55):6994–7002. doi: 10.1038/onc.2008.349. [DOI] [PubMed] [Google Scholar]
- 18.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006 Jul;133(14):2639–48. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 19.Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, et al. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Developmental cell. 2006 Dec;11(6):775–89. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Herranz H, Hong X, Perez L, Ferreira A, Olivieri D, Cohen SM, et al. The miRNA machinery targets Mei-P26 and regulates Myc protein levels in the Drosophila wing. The EMBO journal. 2010 May 19;29(10):1688–98. doi: 10.1038/emboj.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009 Mar 6;136(5):913–25. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bleau AM, Hambardzumyan D, Ozawa T, Fomchenko EI, Huse JT, Brennan CW, et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell stem cell. 2009 Mar 6;4(3):226–35. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charles N, Ozawa T, Squatrito M, Bleau AM, Brennan CW, Hambardzumyan D, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell stem cell. 2010 Feb 5;6(2):141–52. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charles NA, Holland EC. TRRAP and the maintenance of stemness in gliomas. Cell stem cell. 2010 Jan 8;6(1):6–7. doi: 10.1016/j.stem.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes & development. 2001 Aug 1;15(15):1913–25. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz AM, Amankulor NM, Pitter K, Helmy K, Squatrito M, Holland EC. Astrocyte-specific expression patterns associated with the PDGF-induced glioma microenvironment. PloS one. 2012;7(2):e32453. doi: 10.1371/journal.pone.0032453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer research. 2004 Jul 15;64(14):4783–9. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- 28.Ciznadija D, Liu Y, Pyonteck SM, Holland EC, Koff A. Cyclin D1 and cdk4 mediate development of neurologically destructive oligodendroglioma. Cancer research. 2011 Oct 1;71(19):6174–83. doi: 10.1158/0008-5472.CAN-11-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hukkelhoven E, Liu Y, Yeh N, Ciznadija D, Blain SW, Koff A. Tyrosine phosphorylation of p21 facilitates the development of proneural glioma. The Journal of biological chemistry. 2012 Sep 24; doi: 10.1074/jbc.M112.366542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Yeh N, Zhu XH, Leversha M, Cordon-Cardo C, Ghossein R, et al. Somatic cell type specific gene transfer reveals a tumor-promoting function for p21(Waf1/Cip1) The EMBO journal. 2007 Nov 14;26(22):4683–93. doi: 10.1038/sj.emboj.7601886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.See WL, Heinberg AR, Holland EC, Resh MD. p27 deficiency is associated with migration defects in PDGF-expressing gliomas in vivo. Cell Cycle. 2010 Apr 15;9(8):1562–7. doi: 10.4161/cc.9.8.11259. [DOI] [PubMed] [Google Scholar]
- 32.See WL, Miller JP, Squatrito M, Holland E, Resh MD, Koff A. Defective DNA double-strand break repair underlies enhanced tumorigenesis and chromosomal instability in p27-deficient mice with growth factor-induced oligodendrogliomas. Oncogene. 2010 Mar 25;29(12):1720–31. doi: 10.1038/onc.2009.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PloS one. 2010;5(2):e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheung CC, Yang C, Berger T, Zaugg K, Reilly P, Elia AJ, et al. Identification of BERP (brain-expressed RING finger protein) as a p53 target gene that modulates seizure susceptibility through interacting with GABA(A) receptors. Proceedings of the National Academy of Sciences of the United States of America. 2010 Jun 29;107(26):11883–8. doi: 10.1073/pnas.1006529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aktas H, Cai H, Cooper GM. Ras links growth factor signaling to the cell cycle machinery via regulation of cyclin D1 and the Cdk inhibitor p27KIP1. Molecular and cellular biology. 1997 Jul;17(7):3850–7. doi: 10.1128/mcb.17.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Q, Sun W, Kujala P, Lotfi Y, Vida TA, Bean AJ. CART: an Hrs/actinin-4/BERP/myosin V protein complex required for efficient receptor recycling. Molecular biology of the cell. 2005 May;16(5):2470–82. doi: 10.1091/mbc.E04-11-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nature reviews Cancer. 2009 Jun;9(6):400–14. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broxmeyer HE. Enhancing engraftment of cord blood cells via insight into the biology of stem/progenitor cell function. Annals of the New York Academy of Sciences. 2012 Aug;1266:151–60. doi: 10.1111/j.1749-6632.2012.06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao H, Ouyang X, Banach-Petrosky W, Borowsky AD, Lin Y, Kim M, et al. A critical role for p27kip1 gene dosage in a mouse model of prostate carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004 Dec 7;101(49):17204–9. doi: 10.1073/pnas.0407693101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J, Kim SS. The function of p27 KIP1 during tumor development. Experimental & molecular medicine. 2009 Nov 30;41(11):765–71. doi: 10.3858/emm.2009.41.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muraoka RS, Lenferink AE, Law B, Hamilton E, Brantley DM, Roebuck LR, et al. ErbB2/Neu-induced, cyclin D1-dependent transformation is accelerated in p27-haploinsufficient mammary epithelial cells but impaired in p27-null cells. Molecular and cellular biology. 2002 Apr;22(7):2204–19. doi: 10.1128/MCB.22.7.2204-2219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viglietto G, Motti ML, Fusco A. Understanding p27(kip1) deregulation in cancer: down-regulation or mislocalization. Cell Cycle. 2002 Nov-Dec;1(6):394–400. doi: 10.4161/cc.1.6.263. [DOI] [PubMed] [Google Scholar]
- 43.Aaltomaa S, Lipponen P, Eskelinen M, Ala-Opas M, Kosma VM. Prognostic value and expression of p21(waf1/cip1) protein in prostate cancer. The Prostate. 1999 Apr 1;39(1):8–15. doi: 10.1002/(sici)1097-0045(19990401)39:1<8::aid-pros2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 44.Bae DS, Cho SB, Kim YJ, Whang JD, Song SY, Park CS, et al. Aberrant expression of cyclin D1 is associated with poor prognosis in early stage cervical cancer of the uterus. Gynecologic oncology. 2001 Jun;81(3):341–7. doi: 10.1006/gyno.2001.6196. [DOI] [PubMed] [Google Scholar]
- 45.Baretton GB, Klenk U, Diebold J, Schmeller N, Lohrs U. Proliferation- and apoptosis-associated factors in advanced prostatic carcinomas before and after androgen deprivation therapy: prognostic significance of p21/WAF1/CIP1 expression. British journal of cancer. 1999 May;80(3-4):546–55. doi: 10.1038/sj.bjc.6690390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ceccarelli C, Santini D, Chieco P, Lanciotti C, Taffurelli M, Paladini G, et al. Quantitative p21(waf-1)/p53 immunohistochemical analysis defines groups of primary invasive breast carcinomas with different prognostic indicators. International journal of cancer Journal international du cancer. 2001 Mar 20;95(2):128–34. doi: 10.1002/1097-0215(20010320)95:2<128::aid-ijc1022>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 47.Cheung TH, Lo KW, Yu MM, Yim SF, Poon CS, Chung TK, et al. Aberrant expression of p21(WAF1/CIP1) and p27(KIP1) in cervical carcinoma. Cancer letters. 2001 Oct 22;172(1):93–8. doi: 10.1016/s0304-3835(01)00624-3. [DOI] [PubMed] [Google Scholar]
- 48.Ferrandina G, Stoler A, Fagotti A, Fanfani F, Sacco R, De Pasqua A, et al. p21WAF1/CIP1 protein expression in primary ovarian cancer. International journal of oncology. 2000 Dec;17(6):1231–5. doi: 10.3892/ijo.17.6.1231. [DOI] [PubMed] [Google Scholar]
- 49.Sarbia M, Gabbert HE. Modern pathology: prognostic parameters in squamous cell carcinoma of the esophagus. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2000;155:15–27. doi: 10.1007/978-3-642-59600-1_2. [DOI] [PubMed] [Google Scholar]
- 50.Winters ZE, Leek RD, Bradburn MJ, Norbury CJ, Harris AL. Cytoplasmic p21WAF1/CIP1 expression is correlated with HER-2/ neu in breast cancer and is an independent predictor of prognosis. Breast cancer research : BCR. 2003;5(6):R242–9. doi: 10.1186/bcr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.James MK, Ray A, Leznova D, Blain SW. Differential modification of p27Kip1 controls its cyclin D-cdk4 inhibitory activity. Molecular and cellular biology. 2008 Jan;28(1):498–510. doi: 10.1128/MCB.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. The EMBO journal. 2001 May 1;20(9):2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009 Jun 12;137(6):1018–31. doi: 10.1016/j.cell.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh N, Miller JP, Gaur T, Capellini TD, Nikolich-Zugich J, de la Hoz C, et al. Cooperation between p27 and p107 during endochondral ossification suggests a genetic pathway controlled by p27 and p130. Molecular and cellular biology. 2007 Jul;27(14):5161–71. doi: 10.1128/MCB.02431-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor BS, Barretina J, Socci ND, Decarolis P, Ladanyi M, Meyerson M, et al. Functional copy-number alterations in cancer. PloS one. 2008;3(9):e3179. doi: 10.1371/journal.pone.0003179. [DOI] [PMC free article] [PubMed] [Google Scholar]