Abstract
Titania (TiO2)-based nanocomposites subjected to light excitation are remarkably effective in eliciting microbial death. However, the mechanism by which these materials induce microbial death and the effects that they have on microbes are poorly understood. Here, we assess the low dose radical-mediated TiO2 photocatalytic action of such nanocomposites and evaluate the genome/proteome-wide expression profiles of Pseudomonas aeruginosa PAO1 cells after two minutes of intervention. The results indicate that the impact on the gene-wide flux distribution and metabolism is moderate in the analysed time span. Rather, the photocatalytic action triggers the decreased expression of a large array of genes/proteins specific for regulatory, signalling and growth functions in parallel with subsequent selective effects on ion homeostasis, coenzyme-independent respiration and cell wall structure. The present work provides the first solid foundation for the biocidal action of titania and may have an impact on the design of highly active photobiocidal nanomaterials.
The incorporation of biocidal agents into engineered polymer-based nanocomposites has led to the development of versatile antimicrobial materials that are useful for a wide variety of packaging, biomedical and general use applications1,2,3,4,5. The development of such materials is difficult because thermodynamic and kinetic barriers inhibit the dispersal of inorganic, often hydrophilic, nanoparticles in hydrophobic polymer matrixes. Antimicrobial nanocomposites based on titania (TiO2) have been actively investigated in recent years. Upon photoactivation of the oxide component, the biocidal action is a result of the modulation of charge (electron-hole) carriers at the interface of the external surface of the material, yielding potent and long-lasting capabilities when the dispersion of the inorganic phase and organic-inorganic interfacial contact are optimally achieved6,7,8,9,10,11,12. Titania has substantial advantages over both chemical (NO, H2O2, small organic molecules) and metal (typically Ag)-based systems13,14. First, titania nanoparticles have a broad spectrum of activity against microorganisms, including Gram-negative and positive-bacteria and fungi, which is of particular importance for multiple drug resistant strains13,14. Second, and more importantly, titania-polymer nanocomposites are intrinsically environmentally friendly and exert a non-contact biocidal action. Therefore, no release of potentially toxic nanoparticles (with unpredictable effects on human health) to the media is required to achieve disinfection capabilities6,7,10,11,15,16.
A critical point for addressing the biocidal capabilities of titania-based nanomaterials concerns the mechanism by which cell viability is lost and cell death occurs. While this has been addressed to some degree with Ag-based biocidal materials15,16,17,18, reports on titania-based biocides are scarce19,20. Little is known about the mechanisms of titania-induced biocidal activity beyond the relatively well-studied initial oxidative attack on the outer/inner cell membrane of the microorganism19,20 as well as alterations of Coenzyme A-dependent enzyme activities21 and damage to DNA via hydroxyl radicals22. Therefore, there is a need for basic knowledge linking the action of photocatalytic-induced biocidal entities with their biological effects; this knowledge would allow us to achieve the full disinfection power of TiO2-based systems. Such studies would define the nature (random vs. non-random) of the photocatalytic reaction, the cell systems involved in the response and the degree of reversibility of the action of TiO2.
In this paper we aimed to i) analyse and quantify the radical species released from the biocidal films and ii) assess their role in cell damage and death. Using a joint chemical and biological approach, we attempt to address the current limitations in exploring the link between the photobiocidal action of titania and the metabolic and transport properties of the microorganism. The investigation was carried out using a clinically relevant bacterium, Pseudomonas aeruginosa strain PAO16. We found that the cells attempted to counteract the TiO2 (or other photoactive nanomaterials)-mediated action by triggering a broad array of regulatory responses without substantial loss of metabolic function within the tested time scale. Moreover, the study provides clear indications about the selective effect of TiO2-photo catalysis in specific, cell-critical systems.
Results
Nanocomposites: charged carriers with biocidal action
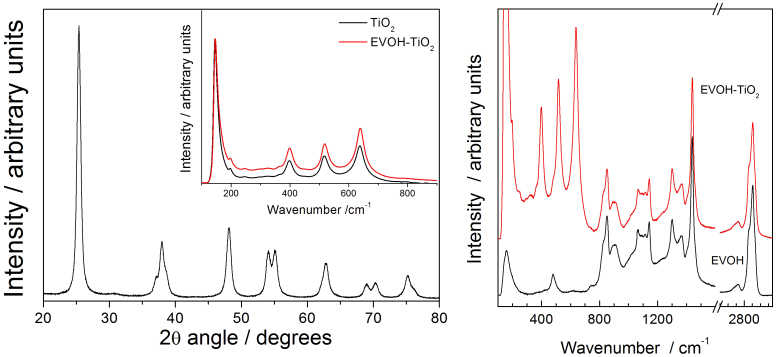
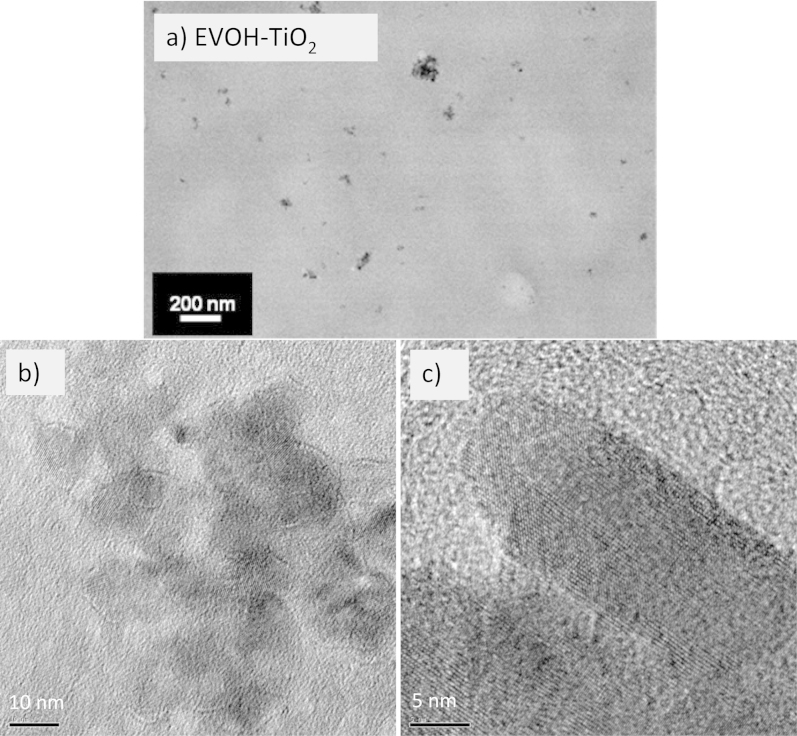
Fig. 1 displays the X-ray diffraction (XRD) and Raman spectra of the bare TiO2 component, which showed an anatase structure with a primary particle size of ca. 9 nm and a Brunauer Emmett Teller (BET) area of ca. 104 m g−1. The dispersion into the ethylene-vinyl alcohol (EVOH) copolymer matrix to a concentration of 2 wt.% (previously found to provide optimum killing rates)6 was carried out by a simple but effective melting compounding method. This process generated aggregates with an average diameter of ca. 90 nm, as evidenced by the transmission electron microscopy (TEM) image included in Fig. 2. Micro-Raman studies (also presented in Fig. 1) showed that both the oxide (inset at left) and the polymer (main Raman plot at right) materials were essentially unaltered after the process of incorporation of titania into the polymer matrix. The physicochemical properties of these materials were therefore preserved in the nanocomposite material. The high resolution TEM (HR-TEM) study presented in Fig. 2 provides evidence that anatase aggregates are composed of oxide nanoparticles interwoven with the organic component at a nanometre scale. A detailed view of the inorganic-organic interface is also presented in Fig. 2. The crystal planes of the inorganic nanoparticles are observed together with the more disordered (essentially amorphous, according to the electron diffraction patterns) polymer matrix surrounding the oxide nanoparticles. The oriented, parallel preferential stacking of both components at the interface layers is direct evidence of the perfect matching between the components in our nanocomposite film.
Figure 1.
Right: X-ray Diffraction (main view) and Raman spectra (inset) of TiO2 alone or at the nanocomposite (e.g., polymer contribution subtracted). Left: Raman spectra of the TiO2-EVOH nanocomposite material and the EVOH polymer reference.
Figure 2. Slide views of the TiO2-EVOH nanocomposite film.
(a) TEM general view (darker zones, TiO2; lighter zones, EVOH); (b) HR-TEM detail showing a TiO2 aggregate; and (c) HR-TEM view of the organic-inorganic interface (interference fringes are evident in the TiO2 component while an amorphous state of the EVOH polymer is observed at the interface).
The effectiveness of this system in eliminating a series of microorganisms, including two Gram-negative (Escherichia coli and P. aeruginosa) and two Gram-positive (Staphylococcus aureus and Enterococcus faecalis) bacterial species and a yeast species (Pichia jadinii), was previously investigated and is summarised in Supplementary Table S16. The system is particularly effective with P. aeruginosa strain PAO1, but there is no obvious correlation between the cell wall characteristics of the bacteria and the killing capacity as high and low death rates are observed for specific microorganisms of Gram-positive or Gram-negative bacteria. Concerning P. aeruginosa, we can highlight the exceptional biocidal action of the nanocomposite, with the rates and time response competing quite favourably with those of chemicals or Ag-based disinfection technologies1,2,3,4,5,6,13,14,15,16.
To ascertain the mechanism of action of the composite upon photocatalysis, we first analyzed the specific charge carrier species involved in the system using electron paramagnetic resonance (EPR). Using 5,5-dimethyl-1-pyrroline N-oxide (DMPO) as a trapping molecule in water and ethanol, we tracked the formation of reactive species after light excitation4,6,12,19,20. Representative spectra are presented in Supplementary Fig. S1, which shows the exclusive formation of OH· radicals in water and the absence of other signals related to oxygen species, even after 20 min of prolonged light treatment. This result clearly demonstrates that only reactive species reached the surface of the polymer-TiO2 film and subsequently interacted with the neighbouring media and microorganisms. Moreover, nanocomposite films also ensure the absence of cell damage/death induced by phagocytosis, a situation typically observed for powder TiO2 samples19,20.
From detailed measurements (see the Methods section), we estimated an initial rate of OH· radical formation of 1.0 × 10−8 mol-radical s−1 mg−1. In our experimental assay we used 20 ng of nanomaterial per 109 cells, which means that an initial rate of 2 × 10−3 nmol OH· radical s−1 for 109 cells and a total amount of ~0.24 nmol OH· radicals were produced in our 2-min assays. This amount is comparable to that found in vivo when cells are exposed to stress conditions favouring oxidative damage23,24; however, we should note that the type of cells and experimental conditions just mentioned are not comparable to those described in the present study.
Genome- and proteome-wide responses of P. aeruginosa PAO1 to TiO2-photocatalysis
No statistically significant differences (p ≤ 0.001) in transcription patterns were discernible in samples of PAO1 cell suspensions irradiated with UV light (UV-alone) in the presence or absence of EVOH (UV + EVOH) reference films (not shown), which is in agreement with the fact that in both control conditions (e.g., UV-alone and UV + EVOH), no differences in the biocidal activity were observed after two minutes of intervention6. Accordingly, the expression values in the presence of UV + EVOH were considered hereafter for comparative purposes. In contrast, the levels of 165 transcripts (approximately 3.0% of the total transcripts) increased from 5.5 to 224-fold and those of 151 transcripts (approximately 2.7% of the total transcripts) decreased from 5.4 to 117-fold in samples of PAO1 cell suspensions containing TiO2-coated EVOH particles (Supplementary Table S2).
Integration of the expression data (Supplementary Table S2) can be performed to further constrain the available PAO1 genome-scale constraint-based model and to develop context-specific models25,26. This integration organises the data in terms of the underlying knowledge embedded in the model structure and might help to better understand the on-going biological process. However, post-transcriptional and post-translational regulation and modifications, as well as allosteric effects, prevent the direct use of transcript levels as a proxy for reaction fluxes in the available model. Here we used the integration algorithm iMAT to find a flux distribution resembling, as close as possible, the measured gene expression levels, while allowing for biomass production27. In this case, the predicted biomass production compatible with the expression data indicates that the impact on the gene-wide flux distribution and the metabolism itself is moderate (approximately below 17%) in this time span.
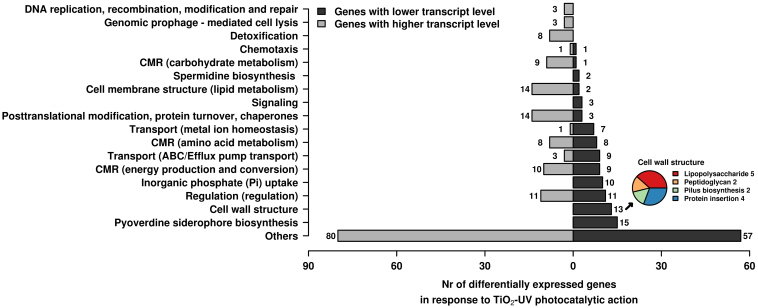
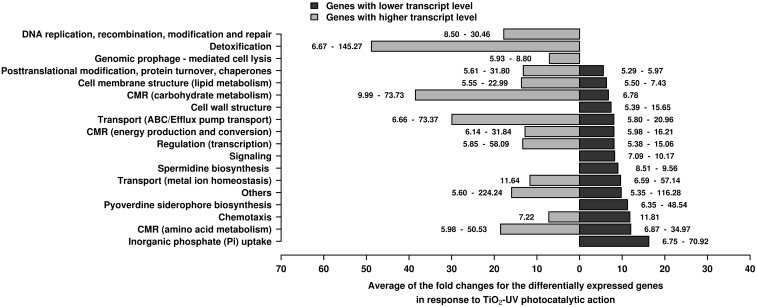
The limited reduction in the maximum growth rate points to the fact that the main factors leading to a decreased survival rate of P. aeruginosa (when confronted to TiO2-based biocidal materials)6 are not directly caused by the loss of metabolic robustness. The differentially expressed genes identified on the arrays (Supplementary Table S2) were classified according to biological categories for interpreting the data. As shown in Fig. 3, fourteen general biological categories were defined, with the number of differentially expressed genes assigned to 11 out of 14 categories being extensively altered as a consequence of TiO2-UV treatment. Fig. 4 further summarises the average expression levels of the genes assigned to each of the corresponding biological categories.
Figure 3. Distribution by function of genes expressed at higher (light grey colour) and lower (dark grey colour) transcript level in response to low doses of TiO2-UV treatment compared to the control (UV + EVOH) condition.
The number of genes assigned to each category was calculated on the basis of the gene expression levels specifically shown in Supplementary Table S2. Abbreviation as follows: CMR, central metabolic reactions. For the cell wall biogenesis pathway, the number of under-expressed genes encoding proteins involved in lipopolysaccharide and peptidoglycan metabolisms, pilus biosynthesis and protein insertion are specifically shown (inset at the right down corner).
Figure 4. Differential expression of genes at higher (light grey colour) and lower (dark grey colour) transcript levels, distributed by function, in response to low doses of TiO2-UV treatment compared to the control (UV + EVOH) condition.
Expression levels for genes belonging to each category, under TiO2-coated EVOH particles with UV activation, were summed, and the average values were calculated and compared to the control condition; the expression range (maximum and minimum) is specifically indicated in the bars. Detailed transcript expression levels per gene are provided in Supplementary Table S2. Abbreviations as in Fig. 3, with light and dark grey colours indicating the expression levels of genes expressed at higher and lower levels, respectively.
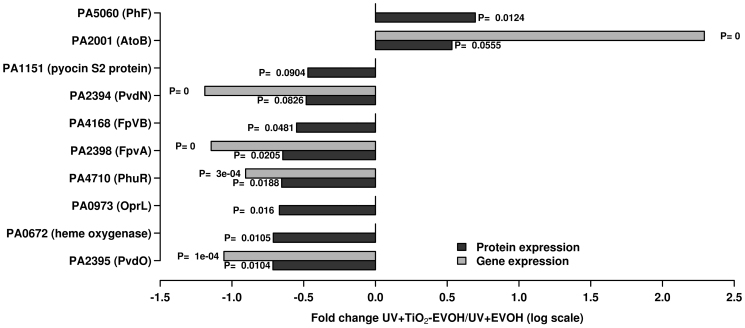
To confirm the expression levels and the roles of key relevant genes as indicated by the array analysis (see details below), the protein expression profiles of bacterial cells on control (UV + EVOH) and TiO2-coated EVOH particles with UV activation were evaluated. Statistical comparison was performed between triplicates. Across all experiments, a total of 1137 proteins were identified, and among them 722 proteins were quantified with at least two peptides, representing 13.4% of the theoretical PAO1 proteome. Compared to the control (UV + EVOH) condition, only a minor number of proteins were differentially detected at a statistically significant level (p ≤ 0.1) in cells treated with TiO2-coated EVOH particles (Fig. 5 and Supplementary Table S3). First, it should noted that there is still a need to improve the proteomic approaches applied to studies on the biocidal effects of titania for assessing statistical significance, as titania exhibited a profound biocidal action under the conditions of our short-term assay (2-min intervention). In spite of this, the examination of the expression levels of those proteins confirms many of the effects caused by the titania at the gene level. We describe each of these effects below.
Figure 5. Expression level (as fold changes) observed for proteins as quantified by shotgun proteomics as a consequence of the TiO2 photocatalytic action, compared to control samples (UV + EVOH).
The corresponding p-values (for triplicates) are shown. For comparison, the expression levels (with p-values) of genes encoding such proteins are also indicated (for details see Supplementary Table S2).
TiO2-UV treatment selectively affects cell structure components
Altered expression of a number of genes encoding enzymes involved in the cell wall and cell membrane composition and integrity was found (Figs. 3 and 4). In total, 14 genes encoding enzymes for the metabolism of lipids essential for cell membrane structure were found to be over-expressed (from 5.6 to 23.0-fold), whereas only two were expressed at a lower level (from 5.5 to 7.4-fold). The over-expressed genes include those encoding an acetyl-CoA synthetase, two CoA transferases, five (hydroxyl) acyl-CoA dehydrogenases, two (keto) acyl-CoA thiolases, a 2,4-dienoyl-CoA reductase, an acetyl-CoA acetyl transferase, a short-chain dehydrogenase and a fatty acid oxidation complex subunit alpha (Supplementary Table S2). In contrast to the above observations, the genes essential for cell wall structure, including those encoding proteins involved in lipopolysaccharide and peptidoglycan metabolism, pilus biosynthesis and protein insertion (13 genes in total), were expressed at significantly lower levels (from 5.4 to 15.1-fold) (Figs. 3 and 4). These genes include i) a 4-amino-4-deoxy-L-arabinose transferase, a glycosyl transferase family 1 protein, a nucleotide sugar dehydrogenase, an ADP-L-glycero-D-manno-heptose-6-epimerase and a membrane protein, possibly involved in lipopolysaccharide metabolism; ii) a cell shape-determining protein and a membrane-bound lytic murein transglycosylase, possibly involved in peptidoglycan metabolism; iii) two pilus assembly proteins, involved in pilus biosynthesis; and iv) two outer membrane proteins, a membrane translocase and a cell wall peptidase, possibly involved in cell wall integrity (Supplementary Table S2). Lower transcription levels of the pmrAB regulatory genes (7.5- and 6.7-fold, respectively), which are known to be involved in regulating proteins involved in cell wall composition and resistance28, provided further strong evidence for possible deficiencies at the level of the cell wall.
The expression of certain key genes was also confirmed by shotgun proteomics. Acetyl-CoA acetyltransferase was significantly over-expressed (PA2001, approximately 3.4-fold) whereas the peptidoglycan-associated lipoprotein (OprL; PA0973) was found to be expressed at a significantly lower level (approximately 4.7-fold). Taken together, the results unambiguously demonstrate that after the initiation of the intervention, cells may have significant deficiencies in the cell wall components, including lipopolysaccharides, peptidoglycan, pili and proteins. To compensate for these deficiencies, cells seem to react by activating the set of genes and proteins that produce lipids for the cell membrane found immediately below the cell wall. Clearly, radicals disrupt the first cell defence barrier (the cell wall), which provides strength and rigidity, while the cells react by activating a second defence barrier (the cell membrane), which provides support for the cytoskeleton.
Cells react to TiO2-UV treatment by activating detoxification and repair mechanisms
Detoxification (from 6.7- to 145.3-fold) and stress-related genes, such as those assigned to posttranslational modification, protein turnover, chaperones (from 5.6- to 31.8-fold) and DNA replication, recombination, modification and repair (from 8.5- to 30.1-fold), were expressed at significantly higher levels in TiO2-UV treated cells than in control cells (Figs. 3 and 4). They include genes encoding: i) three alkyl hydroperoxide reductases, two catalases, a periplasmic ankyrin-like protein required for optimal catalase activity and resistance to H2O2, a glutathione peroxidase, a xenobiotic reductase, a thioredoxin reductase, a ferredoxin, a cyanide insensitive terminal oxidase and a cytochrome c550, which is likely involved in electron transport and protection from superoxide radicals, H2O2 and alkyl hydroperoxides; ii) two DNA repair proteins and two regulators of DNA damage repair systems, which contribute to the biological cycle of DNA damage and repair; and iii) a Zn-dependent protease, three ATP-dependent proteases, six chaperone proteins, and a co-chaperone involved in Fe-S biogenesis, which is most likely involved in protein folding and related activities (Supplementary Table S2). Only a peptidyl-prolyl cis-trans isomerase, a peptidase M4 family protein and a ClpA/B-type protease, which may be involved in posttranslational modification and protein turnover, and chaperones were found among those genes that were expressed at a lower level. This result confirms that the small amount of radical species produced in the 2-min time span provokes an intensive stress response in cells, which react by activating the set of proteins that are directly involved in protein folding, DNA repair and detoxification.
TiO2-UV treatment selectively affects a restricted set of respiratory components
The eight genes found to be over-expressed (from 6.1 to 31.8-fold; Supplementary Table S2) within the energy production and conversion category include those encoding four (ubi)quinol dependent oxygen uptake cytochrome c terminal oxidase components (exaB, 31.8-fold; cioA, 16.0; cioB, 7.5-fold; PA1551, 6.1-fold), five NAD(P)+/NADH oxidoreductases (xenB, 24.0-fold; exaC, 11.9-fold; pauC, 7.9-fold; bauC, 6.3-fold; fadH1, 6.2-fold) and an ethanol oxidation quinoprotein (exaA, 73.7-fold)29. In contrast, the genes expressed at lower levels (from 6.0 to 16.1-fold; Supplementary Table S2) include those encoding six cbb3-type cytochrome C oxidase subunits (coax, 6.0-fold; ccoN2, 16.2-fold; coxB, 8.1-fold; coIII, 6.1-fold; PA0107, 6.7-fold; PA0111, 8.3-fold), an electron transfer flavoprotein (PA5400, 6.5-fold) and two oxidoreductases (adhA, 14.3-fold; dgcA, 9.4-fold).
These results provide evidence of the selective effect of TiO2 photocatalysis on distinct components of the respiratory chain and the electron transfer chain. Whereas cytochrome oxidases requiring reduced and electron rich ubiquinol coenzymes for respiration are expressed at a higher level, those supporting coenzyme-independent oxygen uptake30 (cbb3-like) were expressed at a lower level. It is plausible that the antioxidant action of (ubi)quinol and its higher capacity to exchange more electrons in the respiration redox cycle might correspond with the activation of proteins using those molecules compared to the cbb3-type of cytochromes. In addition, the fact that the dnr gene regulator was also found to be under-expressed (14-8-fold) suggests that the dissimilatory nitrate respiration31 is also negatively affected after the initiation of TiO2-UV treatment.
Cells react to TiO2-UV treatment by increasing the one-carbon pool through non-oxidative pathways
Within the amino acid metabolism pathway, genes encoding proteins for glycine production from serine (glyA2, 50.5-fold) and its utilisation via the glycine cleavage system H protein (gcvH2, 10.0-fold), glycine dehydrogenase (gcvP2, 39.1-fold) and glycine cleavage system aminomethyltransferase (gcvT2, 7.7-fold) were significantly over-expressed, whereas those of the glycine betaine to sarcosine pathway (gbcA, 19.5-fold; gbcB, 17-fold; dgcB, 7.8-fold) and the sarcosine to glycine pathway (soxABD, 13.2-, 11.0- and 11.7-fold, respectively) were expressed at significantly lower levels than the control. This is of particular significance because reactions supported by glyA2, gcvT2 and gcvH2 are known to provide the largest part of the one-carbon units to the cells through tetrahydrofolate and 5,10-methylenetetrahydrofolate32.
Accordingly, it is plausible that when confronted with rather short treatment times and modest radical levels, cells react by activating the catabolic fate of glycine to the one-carbon pool for synthetic purposes (Supplementary Fig. S2). Alternatively, the fact that 5,10-methenyltetrahydrofolate acts as a chromophore in enzymes that repair UV-B-induced DNA lesions in a light-dependent manner33 might also explain the activation of reactions producing such molecules. The attenuation of the glycine betaine to glycine pathway may be explained by the fact that SoxABD sarcosine oxidases produce H2O2 as a subproduct during the oxidative demethylation of sarcosine34, which could potentially be harmful to cells under the strongly oxidative stress conditions induced by TiO2 photocatalysis. To investigate this possibility, we monitored the relative concentrations of some of the most representative metabolites involved in this pathway (Supplementary Fig. S2). As shown in Table 1, we found that dimethylglycine (5.3-fold) and sarcosine (4.1-fold), from the pathway that converts glycine betaine to glycine, were less abundant in cells treated with TiO2-coated EVOH particles with UV activation compared to control cells (UV + EVOH), in agreement with the lower expression levels of the gbcAB, dgcB and soxABC genes (see above). In contrast, serine (5.2-fold) and tetrahydrofolate (15.5-fold), from the pathway that converts glycine to serine, were significantly more abundant in the cells treated with TiO2-coated EVOH particles with UV activation, which also agrees with the higher expression level of the glyA2 gene (see above).
Table 1. Metabolomic target analysis of key chemical species participating in the one-carbon pool pathway. The separation and quantification was performed as described in the Methods section. The area of the peak (calculated on the basis of appropriate standards) as determined in triplicate assays (with standard deviation) by CE-TOF-MS is given.
| Peak area (arbitrary units) | ||
|---|---|---|
| Metabolite | UV + TiO2-coated EVOH | UV + EVOH |
| L-Serine | 91348931 ± 3081018 | 17539253 ± 446069 |
| Sarcosine | 20606119 ± 275529 | 83962799 ± 4251813 |
| Dimethyl glycine | 28050873 ± 901772 | 1.48e8 ± 7147982 |
| Tetrahydrofolate | 19764 ± 1072 | 1272 ± 159 |
TiO2-UV treatment decreases the ability of cells to assimilate and transport iron and inorganic phosphate (Pi)
With regards to the functions related to ion biogenesis (including proteins with roles in transport, binding and catalysis), major differences were found in the expression of genes related to iron homeostasis (7 genes) and Pi uptake (10 genes), which were expressed at significantly lower levels compared to the control (Supplementary Table S2 and Figs. 3 and 4). These include the complete set of genes constituting the Pho regulon: three regulatory proteins (phoBRU; from 24.6 to 15.6-fold), four transporters and porins (pstASCB, from 71.0 to 5.8-fold; oprO, 9.2-fold) and an alkaline phosphatase (phoA, 6.7-fold). These data suggest that P. aeruginosa cells will be highly deficient in phosphorus uptake and metabolism. It should also be noted that the Pho regulon has been reported to influence biofilm formation capacity and pathogenicity35.
The lower ability of cells to assimilate and transport iron was further evidenced by the decreased expression of twelve siderophore pyoverdine synthesis genes (pvdAEFHGNQO, opmQ, PA2411, PA2393, from 48.5 to 6.4-fold), three pyoverdine/transferrin/lactoferrin receptors (phuR, 8.0-fold; vreA, 8.3-fold; PA4675, 6.6-fold), three iron transport proteins (icmP, PA1673, PA4358; from 57.1 to 8.0-fold) and a haem degradation protein (PA4709, 6.9-fold), which catalyses the final step in heme degradation in vitro36. Additionally, an oxygen-independent coproporphyrinogen III oxidase (hemN), also involved in the biosynthesis of heme groups37, was expressed at significantly lower levels (34.6-fold). This suggests that TiO2 photocatalysis most likely decreases the capacity to produce siderophores and thus decreases the ability to assimilate and transport iron, an essential ion for cell growth and survival. It should be recalled that pyoverdine has also been demonstrated to be important for bacterial virulence and biofilm development38,39. In addition, it is plausible that TiO2 photocatalysis lowers the cell capacity to synthesise and degrade heme components required for the correct activity of a number of heme-proteins and to acquire iron ions. This possibility was further demonstrated by examining the shifts in the protein expression patterns in cells treated with TiO2-coated EVOH particles with UV activation compared to control cells (UV + EVOH). Most notably, there was as significant reduction in the expression of pyoverdine biosynthesis-related proteins including PvdO (PA2395; approximately 5.2-fold) and PvdN (PA2394; ~3.0-fold), the ferripyoverdine receptors FpvA (PA2398; approximately 4.4-fold) and FpvB (PA4168; approximately 3.5-fold), a TonB-dependent haemoglobin/transferrin/lactoferrin receptor family protein PhuR (PA4710; approximately 4.5-fold) and a heme oxygenase (PA0672; approximately 5.2-fold).
TiO2-UV treatment decreases cell-to-cell communication
The spermidine biosynthesis pathway was significantly attenuated in the 2-min time span due to the decreased expression of an S-adenosylmethionine decarboxylase (speD, 9.6-fold) and a spermidine synthase (speE, 8.5-fold) (Supplementary Table S2 and Figs. 3 and 4). Recently, new functions for spermidine on the cell surface have been proposed. Namely, spermidine produces polyamines as an organic polycation to bind lipopolysaccharide (LPS) and to stabilise and protect the cell wall against oxidative damage40. This suggests that TiO2 photocatalysis lowers the production capacity not only of cell wall components (see above) but also of molecules involved in their binding. Recent results also strongly suggest an important role for spermidine as a signalling regulator, leading us to suggest that TiO2 photocatalysis affects cell signalling41,42. This hypothesis was supported by the significantly decreased expression of genes involved in the synthesis of quorum-sensing signal molecules (Supplementary Table S2 and Figs. 3 and 4), such as homoserine lactone and pyocyanin, which have been shown to play a role in diverse functions such as pathogenesis, biofilm development, stress resistance and cell survival43,44,45. These included genes encoding an N-acyl-L-homoserine lactone synthetase (rhlI, 10.2-fold), a lactonising lipase (lipA, 8.0-fold lower) and a pyocyanin synthesis protein (ambB, 7.1-fold).
The fact that there was significant reduction of the expression of a pyocianin superfamily protein (PA1151; approximately 3.0-fold) and a pyocin S2 protein (PA1151; approximately 3.0-fold) in cells treated with TiO2-coated EVOH particles with UV activation compared to control cells (UV + EVOH) supports the above mentioned hypothesis: that TiO2 photocatalysis strongly decreases the ability to produce signalling molecules. This could impact the survival of P. aeruginosa through decreasing cell-to-cell communication as well as the activation of a wide range of target genes involved in virulence determinants whose expression and/or regulation depends on those molecules43,44,45.
TiO2-UV treatment affects growth-essential genes and operon-linked regulatory genes
Within the carbohydrate metabolism pathway, a set of nine genes was over-expressed, whereas only one displayed decreased expression (Supplementary Table S2 and Figs. 3 and 4). In contrast, a gene encoding a poly(3-hydroxyalkanoic acid) synthase (phaC2) was expressed at lower level (6.8-fold); this enzyme is involved in the production of polyhydroxyalkanoates and is produced by Pseudomonas species to store carbon and energy sources46. This might suggest that cells tend to react to TiO2-UV treatment by decreasing their capacity to store carbon sources as cells demand the mobilisation of carbon sources rather than carbon storage under stress conditions. This hypothesis was further supported by proteomic data demonstrating that a polyhydroxyalkanoate synthesis protein PhaF (PA5060) was more highly expressed (~5.0-fold) in cells treated with TiO2-coated EVOH particles with UV activation than control cells (UV + EVOH). This protein has been shown to behave as a negative regulator of the phaC gene (i.e., the expression of phaF gene decreases the expression rate of the phaC)47. Accordingly, the higher expression level of the PhaF protein agrees with the significantly lower expression (6.8-fold) of phaC2 as evidenced by transcriptome data.
Notably, the expression of a pyruvate dehydrogenase (PA3416) was also found to be significantly higher (10.1-fold) in TiO2-UV treated samples, which suggests that PAO1 cells also have a higher capacity to transform pyruvate into acetyl-CoA, thus linking the glycolysis pathway to the citric acid cycle and releasing energy via NADH. Furthermore, in silico analyses, as well as genome-scale transposon mutant studies, have shown that the acetyl-CoA carboxylase subunit accD is essential for growth48,49. Therefore, lower expression (5.5-fold) of acetyl-CoA under TiO2 photocatalysis might also be linked to increased cell death. Moreover, the expression of a number of regulatory genes potentially involved in morphogenesis and virulence was significantly decreased including hpaR (8.0-fold) and gntR (11.2-fold)50. Finally, a set of three genes (PA0622, 5.9-fold; PA0623, 6.5-fold; PA0627, 8.8-fold) encoding proteins presumptively involved in genomic prophage-mediated cell death and lysis, as well as a prtN gene (7.8-fold) that encodes a related transcriptional activator51, were expressed at significantly higher levels as a consequence of TiO2 photocatalysis. Our interpretation of this result is that prophage-mediated cell lysis is rapidly induced after the initiation of TiO2-UV treatment, as previously reported during H2O2-induced oxidative stress51.
Finally, the fact that 72 over- and 54 under-expressed genes encoded uncharacterised proteins suggests that these proteins may play yet unknown roles in radial defence and metabolic alterations. Interestingly, 48 highly expressed genes were components of 14 operons (containing at least 3 genes each) whose expression, according to ProdoNet (http://www.prodonet.tu-bs.de/query.php), is controlled by the transcriptional regulatory genes anr, PA4493 and PA5344, none of which were found to be differentially expressed in our experimental conditions. In contrast, genes whose expression levels were lowered by the TiO2 photocatalysis were mostly under the control of 25 different genes encoding transcriptional regulators. Notably, of these 25 genes, the haem biosynthesis and denitrification enzyme regulator Dnr, the ferric uptake regulator PvdS and the quorum-sensing regulator RhlR showed significant decrease in their transcript levels (14.8-, 48.5- and 6.7-fold, respectively). This finding is of special significance because these genes regulate operons consisting of 20, 41 and 16 genes (http://www.prodonet.tu-bs.de/query.php), thus indicating a rapid and concerted regulatory deactivation as a direct consequence of TiO2 photocatalysis.
Discussion
Recent studies using silver- or zinc-containing nanomaterials have recently suggested that the toxicity of these materials partially originates from membrane damage and disruption of ion homeostasis52,53. Here, by integrating transcription profiling with genome-scale constraint-based modelling, we found that the impact of radical-mediated TiO2 photocatalysis on genome-wide flux distribution and the metabolism itself is limited and thus does not directly affect PAO1 cell activity in the time frame measured. Rather, gene chip experiments and shotgun proteomics in combination with chemical species identification and quantification support the notion that PAO1 cells respond by attempting to increase their capacity to protect and repair DNA and proteins in addition to promoting the coenzyme-dependent respiratory chain, mobilisation of carbon resources and one-carbon pools, as well as the cycle of fatty acid β-oxidation (the latter being essential for maintenance of membrane integrity). Furthermore, cells react to TiO2 photocatalysis by improving membrane integrity to compensate for the strong alterations caused in the cell wall that covers the cell membrane. Additionally, we observed the activation of metabolic pathways involving production and utilisation of antioxidant coenzymes, such as (ubi)quinol (used by cytochrome oxidases) and 5,10-methenyltetrahydrofolate (produced by GlyA2 and GcvT2 proteins), with the corresponding attenuation of those producing oxidative sub-products, such as H2O2 (produced by Sox enzymes), most likely to compensate for the strong radical stress pressure inside the cell. Supplementary Fig. S3 summarises the major effects in P. aeruginosa cells exposed to TiO2 photocatalysis.
This study provides evidents that cells exposed to TiO2 photocatalysis enxibit i) rapid cell inactivation at the regulatory and signalling levels, ii) a strong decrease of the coenzyme-independent respiratory chains, iii) a lower ability to assimilate and transport iron and phosphorous and iv) a lower capacity for the biosynthesis and degradation of heme (Fe-S cluster) groups. These activities, together with the extensive cell wall modifications, are the main factors that explain the high biocidal performance of TiO2-based nanomaterials. However, it is not known which of these responses precipitate subsequent events. Future dynamic profiling assays are needed to further study these responses. Nonetheless, the fact that titania confers profound biocidal action, even at extremely low doses, under our temporally short assay conditions (2-min intervention), causes dynamic profiling at shorter and longer assay times to be technically challenge due to technical difficulties and extensive cell death and damage, respectively. It should noted that TiO2-based materials possess a high affinity for phospho-proteins and phospho-peptides54,55, and that phosphorylation of proteins plays a role in the regulation of signalling pathways and the control of enzymatic activities by an “on/off” switch mechanism56. Therefore, the fact that after the initiation of TiO2-UV treatment cell lysis is rapidly induced, suggests that this material could be used in situ to bind and enrich phosphoproteins to further explore the level of post-transcriptional protein modification at given times, as a molecular measure of altered protein activities. Whatever the case, the broad-spectrum action at the genetic level illustrated herein allows an investigation of the most significant properties of the photocatalytic disinfection process: universality (i.e., the absence of weakness against any type of microorganisms), and irreversibility (i.e., avoiding cell reparation opportunities and slowing down regulatory/signalling networks). In addition, the high efficiency demonstrated against a clinically relevant pathogen indicates the adequacy of TiO2-based polymer films for a series of technological applications, such as hospital tools and furniture, but also food preservation or wastewater treatment.
Methods
Synthesis of nanocomposite films and basic characterization
The TiO2 component was prepared using a microemulsion synthetic route and calcined at 723 K for 2 h6. A commercially available EVOH (Solvay), containing a nominal 71 mol% vinyl alcohol content, was used as polymeric matrix in the preparation of EVOH-TiO2 nanocomposite films with an inorganic content of 2 wt.%. These novel materials were prepared by melt blending in a Haake Minilab twin-screw extruder. An optimal processing window was selected to optimise the final dispersion of nanoparticles and to avoid EVOH degradation (temperature at 195°C, a rotor speed of 100 rpm and a mixing time of 15 min). Prior to this stage, nanoparticles and the EVOH copolymer were sonicated (Sonics VC505) and stirred to homogenise the batch. After this processing, the specimens were compression-moulded in a Collin press between hot plates at 210°C and at a pressure of 1.5 MPa for 5 min to obtain films of approximately 100 microns. A quench was applied to the different films from the melt to room temperature.
X-ray diffraction (XRD) patterns were recorded with a Bruker D8 Advance apparatus using Cu Kα radiation and a 20-step program of ca. 0.02 degrees per second. Micro-Raman spectra were acquired with an inVia Raman microscope using a near-IR diode laser (785 nm) with a nominal 2-nm resolution. Transmission electron microscopy experiments were also carried out at room temperature with a 200 Kv JEM-2000 FX JEOL microscope. The samples were embedded in Spurr resine (low viscosity epoxy, cured at 60°C for 48 h) to obtain parallel cuts of the film surface in thin (80 μm) sections by ultramicrotomy (Reichert-Jung Ultracut E), which were picked up on copper grids and coated with a thin layer of carbon graphite (MED 010 Balzers evaporator) to improve the heat capacity.
Electron paramagnetic resonance experiments
The EPR measurements were performed with a Bruker ER200D spectrometer operating in the X-band and calibrated with a 2,2-diphenyl-1-picrylhydrazyl (DPPH) standard. For the DMPO spin trapping EPR experiments, the nanocomposite samples were immersed in a solvent. An aqueous or methanolic solution (0.01 M) of DMPO spin trap (supplied by Fluka-Aldrich-Sigma Chemical Co.; St Louis, MO, USA) was prepared and kept on ice during the whole set of experiments. Bidistilled water (Elix-10) or methanol (HPLC grade; Fluka-Aldrich-Sigma Chemical Co.) was used for the preparations. Equal volumes (100 μl each) of the DMPO solution and water or methanol were mixed in an EPR flat quartz cell under atmospheric air and irradiated at different times through a filter with a cut-off at ca. 220 nm with a UV-B (280 nm) light and were then immediately transferred to the spectrometer cavity for EPR analysis. A small decay in radical concentration (of ca. 5% on average) was observed in the dark during the course of spectrum recording. The spectra were obtained at 20°C at ca. 9.75 GHz microwave frequency, 19.5 mW microwave power, 100 kHz modulation frequency, 2 G modulation amplitude and 2–4 × 105 spectrometer gain; no significant signal saturation was observed under these conditions. Blank experiments were also performed over mixtures of 100 μl of the DMPO solution and 100 μl of water or ethanol to confirm the absence of radical formation in the absence of the nanocomposite film under the employed conditions.
Bacterial strains, media and chemicals
TiO2-EVOH composites and the P. aeruginosa PAO1 strain were used in the present study6. The PAO1 strain was maintained at 37°C on Luria Bertani (LB) agar plates. Overnight cultures were grown with agitation in 250-ml Erlenmeyer flasks at 37°C in LB and used to inoculate 10-ml portions of pre-warmed LB (10 times diluted) in 250-ml Erlenmeyer flasks to obtain an initial optical density at 600 nm (OD600) of 0.05. The resulting cultures were grown at 37°C with agitation until the exponential phase (OD600 0.3) and subsequently used for the photochemical cell assays. An appropriate amount of this culture was diluted in pre-warmed LB (10 times diluted) to achieve a final cell concentration of 109 CFU ml−1. The oxide-cell slurry (3 ml final volume) was placed in the UV-B spectrometer chamber (UVIKON 930) and irradiated with a UV-B light of 280 nm. Identical cultures treated with either non-composite EVOH lacking TiO2 or UV-B alone served as the controls. After 2 min of irradiation, three aliquots of 1000 μl were immediately collected and subjected to RNA, protein and metabolite isolation as described below. For each experiment, the culturing of bacteria, exposure to the TiO2-nanocomposite and subsequent RNA, protein and metabolite isolation was performed in triplicate on the same day.
Gene chip experiments
All samples for the RNA measurements were stabilised with RNAprotect bacteria reagent (Qiagen GmbH, Hilden, Germany). The samples were mixed with this reagent, incubated for 5–15 min at room temperature and centrifuged for 15 min at 4000 × g and 4°C. The supernatant was discarded, and the pellet frozen in −70°C. In a deep freezer, the RNA was expected to remain intact for up to one month. Prior to RNA extraction, the samples were thawed on ice. RNA isolation was performed using the RNAeasy Mini Kit (Qiagen GmbH) according to manufacturer's instructions. The concentration of the RNA was assessed using a Nanodrop system (NanoDrop, Wilmington, DE), and the integrity was estimated by the 28S/18S ratio using an Agilent 2100 bioanalyser (Agilent, Palo Alto, CA). The microarray analysis was performed at the Microarray facility of the Helmholtz Centre for Infection Research (Branschweig, Germany). Then, 1 μg of RNA was amplified using a MessageAmp II-Bacteria kit (Ambion Inc., Austin, TX) according to the manufacturer's instructions and used in the microarray analysis. The microarray analysis was performed with the Affymetrix P. aeruginosa GeneChip as described in the Supplementary Methods section.
Genome/transcriptome-scale metabolic reconstruction
Genome sequences contain comprehensive information about the genetic elements present in an organism. Recently, Oberhardt et al.25, constructed a genome-scale constraint-based (CB) model for the metabolic processes of P. aeruginosa by integrating genome information with the knowledge stored in reaction databases, strain-specific knowledge and manual curation. A comparative analysis with the closely related, albeit non-pathogenic, organism P. putida led to the development of a more accurate model26, which contains information on 1,086 genes (approximately 20% of the genome) and 1,136 reactions. To integrate the expression values with the CB model, we used the iMAT algorithm, which uses discrete levels for gene expression values: low, medium and high37. High gene expression levels are associated with high fluxes through the corresponding reactions, whereas low levels are associated with no flux. We used the fold change in gene expression in the presence or absence of the TiO2-EVOH composite film to perform this classification with a threshold of 2.0 for high and 0.5 for low. The iMAT algorithm iteratively searches for a new set of predictions of metabolic flux through the enzymes that fall, whenever possible, within the range of flux values linked to the levels of the associated genes but that simultaneously are able to lead to biomass production (with the composition described by Oberhardt et al.25, on LB medium). Simulations were performed on a PC using the Cobra Toolbox57 and the iMAT implementation tool from the Tiger Toolbox58 with Matlab (MathworksInc., Natick, MA, USA). Linear optimisations were performed utilising the CPLEX (IBM, Armonk, NY, USA) and Gurobi (http://www.gurobi.com/) linear programming solvers.
Protein mass spectrometric analysis
The samples were centrifuged for 1 min at 16,000 × g and 4°C. The supernatant was discarded, and the bacterial cell pellet was lysed in 1.2 ml of BugBuster® Protein Extraction Reagent (Novagen, Darmstadt, Germany) for 30 min at 4°C with further disruption by sonication (using a pin Sonicator® 3000; Misonix) for 2.5 min (10 watts) on ice (5 cycles × 0.5 min). The extracts were then centrifuged for 5 min at 12,000 × g to separate the cell debris and intact cells. Finally, the supernatants were carefully aspirated (to avoid disturbing the pellet) and transferred to a new tube. The full description of the methods used for protein digestion and tagging with iTRAQ-4-plex® reagent, liquid chromatography and mass spectrometer analysis and data analysis is available in the Supplementary Methods section. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository59 with the dataset identifier PXD000732.
Metabolomic analysis by CE-TOF-MS
The samples were centrifuged for 1 min at 16,000 × g and 4°C, and the cell pellets immediately mixed with 1.2 ml of cold (−80°C) HPLC-grade methanol (Sigma Chemical Co.; St. Louis, MO, USA). The samples were then stored at −80°C for 60 min. Then, the samples were vortex-mixed (for 10 seg) and sonicated for 30 s (10 watts) on ice (5 cycles × 0.5 min). After sonication, the pellet was removed by centrifugation at 16,000 × g for 20 min at 4°C and the methanol solution was stored at −80°C until use. Immediately, 1.2 ml milli-Q H2O was added to the cell pellet, and, after re-suspension, the samples were sonicated for 30 s (10 watts) on ice (5 cycles × 0.5 min). After sonication, the pellet was removed by centrifugation at 16,000 × g for 20 min at 4°C and the water solution was separated. Finally, equal volumes (1 ml) of each of the methanol and H2O solutions were mixed and stored at −80°C until analysis. The full description of the methods used for CE-TOF-MS analysis is available in the Supplementary Methods section.
Author Contributions
M.F., M.F.-G. and A.K. designed the project. M.F. and M.F.-G. wrote the main manuscript text, and M.S.D. and R.B. prepared the figures. M.S.D. and V.A.P.M.S. analysed and wrote the manuscript part regarding the transcription profiles combined with a genome-scale, constraint-based metabolic model. R.B. and M.F. analysed the gene and protein expression data. A.K. prepared and performed the physicochemical analysis of the TiO2 nanocomposites. A.K. and M.F. carried out photo-killing test and prepare samples serving as raw materials for omic tests. S.C., I.Z. and J.P.A. performed and interpreted the proteomic data. D.R. and C.B. performed and interpreted the chemical analysis. All authors participated in the discussion and commented on the paper.
Supplementary Material
Supplementary Information
Acknowledgments
The authors gratefully acknowledge the financial support provided by grants CTQ2010-14872/BQU, PRI-PIBJP-2011-0914 and BIO2011-25012. A.K. thanks the “Ministerio de Economia y Competitividad” MINECO (Spain) for a Ramon y Cajal Fellowship. D.R. received a fellowship from the Spanish Ministry of Economy and Competitiveness (formerly MICINN).
References
- Appendini P. & Hotchkiss J. H. Review of antimicrobial food packaging. Innov. Food Sci. Emerg. 3, 113–126 (2002). [Google Scholar]
- Noimark S., Dunnill C. W. & Wilson I. P. The role of surfaces in catheter-associated infections. Chem. Soc. Rev. 38, 3435–3448 (2009). [DOI] [PubMed] [Google Scholar]
- Rai M., Yada A. & Gade V. A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 27, 76–83 (2009). [DOI] [PubMed] [Google Scholar]
- Dunlop C. P. et al. Inactivation of clinically relevant pathogens by photocatalytic coatings. J. Photochem. Photobiol. A 216, 303 (2010). [Google Scholar]
- Duncan T. V. Applications of nanotechnology in food packaging and food safety: barrier materials, antimicrobials and sensors. J. Colloid Interface Sci. 363, 1–24 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubacka A. et al. High-performance dual-action polymer-TiO2 nanocomposite films via melting processing. Nano Letters 7, 2529–2534 (2007). [DOI] [PubMed] [Google Scholar]
- Cerrada M. L. et al. Self-sterilized EVOH-TiO2 nanocomposites: interface effects on biocidal properties. Adv. Funct. Mater. 18, 1949–1960 (2008). [Google Scholar]
- Hamming L. M., Qiao R. & Messersmith P. B. Effects of dispersion and interfacial modification on the macroscale properties of TiO(2) polymer matrix nanocomposites. Compos. Sci. Technol. 69, 1880–1886 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan J., Wang S., Hu Z. & Zhang L. Synthesis techniques, properties and applications of polymer nanocomposites. Curr. Org. Synth. 9, 114–136 (2012). [Google Scholar]
- Kubacka A. et al. Surface-enhanced Raman scattering from uniform gold and silver nanoparticle-coated substrates. J. Phys. Chem. C 113, 9182–9190 (2009). [Google Scholar]
- Kubacka A. et al. Boosting TiO2-anatase antimicrobial activity: Polymer-oxide thin films. Appl. Catal. B 89, 441–447 (2009). [Google Scholar]
- Kubacka A., Ferrer M. & Fernández-García M. Kinetics of photocatalytic disinfection in TiO2-containing polymer thin films: UV and visible light performance. Appl. Catal. B 121–122, 230–248 (2012). [Google Scholar]
- Wiener J. et al. Multiple antibiotic–resistant Klebsiella and Escherichia coli in nursing homes. J. Am. Med. Assoc. 281, 517–523 (1999). [DOI] [PubMed] [Google Scholar]
- Josset S., Keller N., Lett M. C., Ledoux M. J. & Keller V. Numeration methods for targeting photoactive materials in the UV-A photocatalytic removal of microorganisms. Chem. Soc. Rev. 37, 744–755 (2008). [DOI] [PubMed] [Google Scholar]
- Asahani P. V., Mun G. L. K., Handi M. P. & Valiyaveetil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano 3, 279–290 (2009). [DOI] [PubMed] [Google Scholar]
- Llorens A., Lloret E., Picouet P. A., Trbjevich R. & Fernández A. Metallic-based micro and nanocomposites in food contact materials and active food packaging. Trends Food Sci. Technol. 24, 19–29 (2012). [Google Scholar]
- Elechiguerra J. L. et al. Interaction of silver nanoparticles with HIV-1. J. Nanobiotechnol. 3, 6–15 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok C.-N. et al. Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J. Proteome Res. 5, 916–924 (2006). [DOI] [PubMed] [Google Scholar]
- Kiwi J. & Nadtochenko V. Evidence for the mechanism of photocatalytic degradation of the bacterial wall membrane at the TiO2 interface by ATR-FTIR and laser kinetic spectroscopy. Langmuir 21, 4631–4641 (2005). [DOI] [PubMed] [Google Scholar]
- Foster H. A., Ditta I. B., Varghese S. & Steele A. Photocatalytic disinfection using titanium dioxide: spectrum and mechanism of antimicrobial activity. Appl. Microbiol. Biotechnol. 90, 1847–1868 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumaga T., Tomada R., Nakajima T. & Wake H. FEBS Microbiol. Lett. 29, 211–214 (1985). [Google Scholar]
- Gogniat G., & Dukan S. TiO2 photocatalysis causes DNA damage via fenton reaction-generated hydroxyl radicals during the recovery period. Appl. Environ. Microbiol. 73, 7740–7743 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. W., Spiegelhalder B. & Bartsch H. Generation of reactive oxygen species by the faecal matrix. Gut 46, 225–232 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielefeldt-Ohmann H. & Babiuk L. A. In vitro generation of hydrogen peroxide and of superoxide anion by bovine polymorphonuclear neutrophilic granulocytes, blood monocytes, and alveolar macrophages. Inflammation 8, 251–275 (1984). [DOI] [PubMed] [Google Scholar]
- Oberhardt M. A., Puchałka J., Fryer K. E., Martins dos Santos V. A. & Papin J. A. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J. Bacteriol. 190, 2790–2803 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt M. A., Puchałka J., Fryer K. E., Martins dos Santos V. A. & Papin J. A. Reconciliation of genome-scale metabolic reconstructions for comparative systems analysis. PLoS Comput Biol. 7, e1001116 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili T. M. N., Herrgård M. J. & Palsson B. Ø. Network-based prediction of human tissue-specific metabolism. Nat. Biotechnol. 26, 1003–1010 (2008). [DOI] [PubMed] [Google Scholar]
- Schurek K. N. et al. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 4345–4351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert M. & Görisch H. Cytochrome c550 is an essential component of the quinoprotein ethanol oxidation system in Pseudomonas aeruginosa: cloning and sequencing of the genes encoding cytochrome c550 and an adjacent acetaldehyde dehydrogenase. Microbiology 145, 471–481 (1999). [DOI] [PubMed] [Google Scholar]
- Kulajta C., Thumfart J. O., Haid S. & Daldal F. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. J. Mol. Biol. 355, 989–1004 (2006). [DOI] [PubMed] [Google Scholar]
- Castiglione N., Rinaldo R., Giardina G. & Cutruzzolà F. The transcription factor DNR from Pseudomonas aeruginosa specifically requires nitric oxide and haem for the activation of a target promoter in Escherichia coli. Microbiology 155, 2838–2844 (2009). [DOI] [PubMed] [Google Scholar]
- Piper M. D., Hong S. P., Ball G. E. & Dawes I. W. Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 275, 30987–30995 (2000). [DOI] [PubMed] [Google Scholar]
- Oberpichler I. et al. A photolyase-like protein from Agrobacterium tumefaciens with an iron-sulfur cluster. PLoS One 6, e26775 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya Y., Zuihara S. & Imanaka T. Active site analysis and stabilization of sarcosine oxidase by the substitution of cysteine residues. Appl. Environ. Microbiol. 61, 367–370 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad A., Jensen V., Becker T. & Häussler S. The Pho regulon influences biofilm formation and type three secretion in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 1, 488–494 (2009). [DOI] [PubMed] [Google Scholar]
- Ratliff M., Zhu W., Deshmukh R., Wilks A. & Stojiljkovic I. Homologues of neisserial heme oxygenase in gram-negative bacteria: degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 183, 6394–6403 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer G., Reichelt J., Jahn D. & Heinz D. W. Structure and function of enzymes in heme biosynthesis. Protein Sci. 19, 1137–1161 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont I. L. & Martin L. W. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149, 833–842 (2003). [DOI] [PubMed] [Google Scholar]
- Imperi F., Tiburzi F., Fimia G. M. & Visca P. Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 12, 1630–1642 (2010). [DOI] [PubMed] [Google Scholar]
- Johnson L., Heidi M., Uliana K., Yan S. & Shawn L. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J. Bacteriol. 194, 813–826 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukabe Y. et al. Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol. 45, 712–722 (2004). [DOI] [PubMed] [Google Scholar]
- Zhou L., Wang J. & Zhang L. H. Modulation of bacterial Type III secretion system by a spermidine transporter dependent signaling pathway. PLoS One 2, e1291 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson M. K. et al. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92, 9427–9431 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel K. et al. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147, 3249–3262 (2001). [DOI] [PubMed] [Google Scholar]
- Kawaguchi T., Chen Y. P., Norman R. S. & Decho A. W. Rapid screening of quorum-sensing signal N-acyl homoserine lactones by an in vitro cell-free assay. Appl. Environ. Microbiol. 74, 3667–3671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisano A., Overhage J. & Rehm B. H. The polyhydroxyalkanoate biosynthesis genes are differentially regulated in planktonic- and biofilm-grown Pseudomonas aeruginosa. J. Biotechnol. 133, 442–452 (2008). [DOI] [PubMed] [Google Scholar]
- Prieto M. A., Bühler B., Jung K., Witholt B. & Kessler B. PhaF, a polyhydroxyalkanoate-granule-associated protein of Pseudomonas oleovorans GPo1 involved in the regulatory expression system for pha genes. J. Bacteriol. 181, 858–868 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M. A. et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100, 14339–14344 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewenza S. et al. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 15, 583–589 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K. et al. hpaR, a putative marR family transcriptional regulator, is positively controlled by HrpG and HrpX and involved in the pathogenesis, hypersensitive response, and extracellular protease production of Xanthomonas campestris pathovar campestris. J. Bacteriol. 189, 2055–2062 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb S. J. et al. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185, 4585–4592 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernousova S. & Epple M. Silver as antibacterial agent: ion, nanoparticle, and metal. Angew. Chem. Int. Ed. 52, 1636–1653 (2013). [DOI] [PubMed] [Google Scholar]
- Reyes V. C., Li M., Hoek E. M., Mahendra S. & Damoiseaux R. Genome-wide assessment in Escherichia coli reveals time-dependent nanotoxicity paradigms. ACS Nano 6, 9402–9415 (2012). [DOI] [PubMed] [Google Scholar]
- Thingholm T. E., Jørgensen T. J., Jensen O. N. & Larsen M. R. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 1, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- Richardson B. M., Soderblom E. J., Thompson J. W. & Moseley M. A. Automated, reproducible, titania-based phosphopeptide enrichment strategy for label-free quantitative phosphoproteomics. J. Biomol. Tech. 24, 8–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers R., Kraiczek K., de Jong A. P., Mohammed S. & Heck A. J. Exploring the human leukocyte phosphoproteome using a microfluidic reversed-phase-TiO2-reversed-phase high-performance liquid chromatography phosphochip coupled to a quadrupole time-of-flight mass spectrometer. Anal. Chem. 82, 824–832 (2010). [DOI] [PubMed] [Google Scholar]
- Schellenberger J. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P. A., Kyla A. L. & Jason A. P. TIGER: Toolbox for integrating genome-scale metabolic models, expression data, and transcriptional regulatory networks. BMC Syst. Biol. 5, 147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino J. A. et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41(D1), D1063–1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information