Abstract
BACKGROUND
Several genome-wide association studies (GWAS) of prostate cancer (PCa) have identified many single nucleotide polymorphisms (SNPs) that are significantly associated with PCa risk in various racial groups. The objective of this study is to evaluate which of these SNPs are associated with PCa risk in Chinese men and estimate their strength of association.
METHODS
All SNPs that were reported to be associated with PCa risk in GWAS from populations of European, African American, Japanese, and Chinese descent were evaluated in 1,922 PCa cases and 2,175 controls selected from the Chinese Consortium for Prostate Cancer Genetics (ChinaPCa). A logistic regression analysis was used to estimate allelic odds ratios (ORs) of these SNPs for PCa.
RESULTS
Among the 53 SNPs, 50 were polymorphic in the Chinese population. Of which, 10 and 24 SNPs were significantly associated with PCa risk in Chinese men at P < 0.001 and <0.05, respectively. These 24 significant SNPs included 17, 5, and 2 SNPs that were originally discovered in European, Japanese, and Chinese descent, respectively. The estimated ORs ranged from 1.10 to 1.49 and the direction of association was consistent with previous studies. When ORs were estimated separately for PCa with Gleason score ≤7 and ≥8, a marginally significant difference in ORs was found only for two of the 24 SNPs (P = 0.02 and 0.04).
CONCLUSION
About half of PCa risk-associated SNPs identified in GWAS of various populations are associated with PCa risk in Chinese men. Information on PCa risk-associated SNPs and their ORs may facilitate risk assessment of PCa risk in Chinese men.
Keywords: prostate cancer, SNPs, genome-wide association, Chinese
INTRODUCTION
With an estimated 914,000 new cases and 258,000 deaths every year globally [1], prostate cancer (PCa) is a major public health concern worldwide. The highest incidence and mortality rates are found in the Western developed countries, whereas the lowest incident and mortality rates are reported in Asians. In China, while the incidence is relatively low, it is increasing rapidly, especially in developed metropolitan areas [2]. For instance, in Shanghai, the incidence of PCa rose from 1.6/100,000 person-years in 1972 to 7.7/100,000 person-years in 2000 [3].
The etiology of PCa is largely unknown and is likely to be multifactorial. Genetic susceptibility is a major risk factor for PCa, estimated to account for 42% variation of the disease [4]. Since 2006, more than 50 genomic regions that harbor inherited variants associated with PCa risk have been identified from multiple genome-wide association studies (GWAS) in populations of European, African American, Japanese, and Chinese descent [5–26]. These PCa risk-associated variants are common in respective populations and typically confer modest to moderate risk, with estimated odd ratios (ORs) ranging from 1.04 to 1.82. However, they have a stronger cumulative effect on PCa risk [27–30]. Risk assessment using combinations of these genetic variants are able to significantly discriminate individual’s risk to PCa, independent of family history and levels of prostate specific antigen (PSA). Several studies have demonstrated the clinical utility of these genetic variants in predicting outcomes of initial and repeat prostate biopsies in populations of European descent [31–32].
The clinical utility of genetic variants in assessing genetic risk to PCa relies strongly on the validity of PCa risk associated single nucleotide polymorphisms (SNPs) and their magnitude of risk to PCa. While PCa risk-associated SNPs and their ORs are well established in populations of European descent, there is limited information regarding these two critical components in the Chinese population. The objective of this study is to evaluate which of the ~50 known PCa risk-associated SNPs identified from various populations are associated with PCa risk in Chinese men and to estimate their strength of association.
METHODS
Study Subjects
The subjects included in this study are part of a Chinese Consortium for Prostate Cancer Genetics (ChinaPCa), which was initiated in 2010 and was designed to understand genetic determinants of PCa risk in Chinese men. Due to relatively low incidence of PCa in China and the lack of a national cancer registry, a consortium effort to recruit PCa patients was necessary to achieve a sufficiently large sample size. Any hospital in China that was interested in joining ChinaPCa and was approved by the Institutional Ethic Review Board was welcomed to the consortium. Inclusion criteria were pathology confirmation of PCa (prevalent or incident cases), availability of blood samples, and Han Chinese ethnicity. Because PSA screening for PCa is uncommon in China, most of the PCa patients were diagnosed due to PCa related symptoms. The ChinaPCa also included control subjects from two community-based studies, including male subjects from Taizhou, Jiangsu Province and the Pudong District in Shanghai. Both populations are located in the South-East of China. Control subjects were defined as men without a diagnosis of PCa. Age and other factors were not matched for PCa patients. Serum levels of prostate-specific antigen (PSA) are available for all control subjects.
This study was approved by the Institutional Review Board at Fudan University and each participating hospital.
SNP Selection and Genotyping
All established PCa risk-associated SNPs from published GWAS of PCa in populations of European, African American, Japanese, and Chinese descent were selected for study. They include 42, 1, 8, and 2 PCa risk-associated SNPs in populations of European [5–22], African [23], Japanese descent [24–25], and Chinese descent [26], respectively. The information of these SNPs is presented in Supplementary Table I.
Two subsets of cases and controls from ChinaPCa were evaluated for these SNPs. The first subset included 1,417 cases and 1,008 controls from Pudong District, Shanghai, as part of a GWAS using Illumina Human OmniExpress Bead Chips [26]. For SNPs that were not included in the GWAS chip, they were imputed based on haplotype data from the 1000 Genomes Project CHB + JPT subjects (Phase I integrated data version 3, released Mar 2012) using the IMPUTE2.2.2 computer program. A posterior probability of >0.90 was applied to call imputed genotypes. Genotype data were not available for three of the targeted SNPs (rs16902094, rs620861, and rs5945619) in this subset of subjects because they were not included in the GWAS chip and were not successfully imputed.
The second subset included 505 PCa cases and 1,167 controls from Taizhou, Jiangsu Province. SNP genotyping in this subset was performed using MassARRAY iPLEX (Sequenom, Inc., San Diego, CA) at the Fudan-VARI Center for Genetic Epidemiology at Fudan University. Duplicates from two subjects and two water samples (negative controls) were included in each 96-well plate for genotyping quality control. Four SNPs (rs10993994, rs7127900, rs130067, and rs10936632) were not evaluated in this subset of subjects because they failed quality control (missing data >5% or Hardy–Weinberg equilibrium P-value <0.001).
Statistical Methods
The allelic ORs and 95% confidence intervals (CI) for each SNP were estimated first in each of the two subsets of cases and controls using a logistic regression analysis assuming an additive model. A meta-analysis was then performed to obtain the pooled estimate of OR and its CI. Heterogeneity of OR between the two subsets was tested using the Q-statistic for heterogeneity and the I2 statistic, which measures the proportion of total variance in estimated ORs due to heterogeneity. The I2 statistic provides the degree of heterogeneity while the Q-statistic only provides the presence or absence of heterogeneity. A value of the I2 statistic >50% indicates a high degree of heterogeneity in estimated ORs between study populations. If there was evidence for heterogeneity in OR estimates (P < 0.05 for Q-statistic or I2 statistic >50%), a random effect was used for meta-analysis to calculate the pooled OR and 95% CI; otherwise, a fixed effect was used. Forest plots are provided to visually present the OR and 95% CI for each SNP. Two criteria were used to determine the significance of SNPs in this study. The first is a P of 0.001 to ensure a Type I error of 5% in the study when taking 50 independent tests into consideration. The second is a liberal criterion, with P of 0.05.
OR and 95% CI were also estimated separately for cases with Gleason score ≤7 and ≥8 by comparing to the control subjects.
All analyses and forest plots in this study were performed using R software.
RESULTS
The characteristics of study subjects in the two subsets are presented in Table I. Most of the PCa patients (79.8%) in this study had clinically significant disease, defined as serum PSA levels ≥20 ng/ml, T3 or higher, N+, M+, or Gleason score ≥8. In control subjects, 49 (2%) had serum PSA ≥4 ng/ml; they were not included in the association test.
TABLE I.
Characteristics of Study Subjects
| Subset 1 (GWAS) | Subset 2 | |||
|---|---|---|---|---|
| Variables | Cases (N = 1,417) | Controls (N = 1,008) | Cases (N = 505) | Controls (N = 1,167) |
| Age, mean (SD), yeara | 71.3 (8.1) | 62.1 (10.0) | 70.6 (8.3) | 67.0 (6.6) |
| PSA levels, # (%), ng/mlb | ||||
| 0–3.99 | 54 (4.0) | 965 (95.9) | 7 (1.9) | 1,141 (99.3) |
| 4–9.99 | 187 (14.0) | 32 (3.2) | 52 (13.8) | 6 (0.5) |
| 10–19.99 | 305 (22.8) | 6 (0.6) | 74 (19.7) | 2 (0.2) |
| 20–49.99 | 312 (23.3) | 3 (0.3) | 71 (18.9) | 0 |
| 50–99.99 | 187 (14.0) | 0 | 42 (11.2) | 0 |
| ≥100 | 292 (21.9) | 0 | 130 (34.6) | 0 |
| Missing | 80 | 2 | 129 | 18 |
| T-stage, # (%) | ||||
| T1 | 180 (14.7) | N/A | 20 (6.7) | N/A |
| T2 | 547 (44.7) | N/A | 151 (52.2) | N/A |
| T3 | 359 (29.4) | N/A | 85 (29.4) | N/A |
| T4 | 137 (11.2) | N/A | 33 (11.4) | N/A |
| TX | 194 | N/A | 216 | N/A |
| N-stage, # (%) | ||||
| N0 | 786 (68.1) | N/A | 229 (76.3) | N/A |
| N+ | 369 (31.9) | N/A | 71 (23.7) | N/A |
| NX | 262 | N/A | 205 | N/A |
| M-stage, # (%) | ||||
| M0 | 832 (65.4) | N/A | 217 (62.4) | N/A |
| M+ | 440 (34.6) | N/A | 131 (37.6) | N/A |
| MX | 145 | N/A | 157 | N/A |
| Gleason score, # (%) | ||||
| ≤7 | 809 (60.1) | N/A | 184 (55.4) | N/A |
| ≥8 | 537 (39.9) | N/A | 148 (44.6) | N/A |
| Missing | 71 | N/A | 173 | N/A |
Age at diagnosis for cases or at recruitment for controls.
Serum PSA levels were measured at diagnosis for cases orobtained at recruitment for controls.
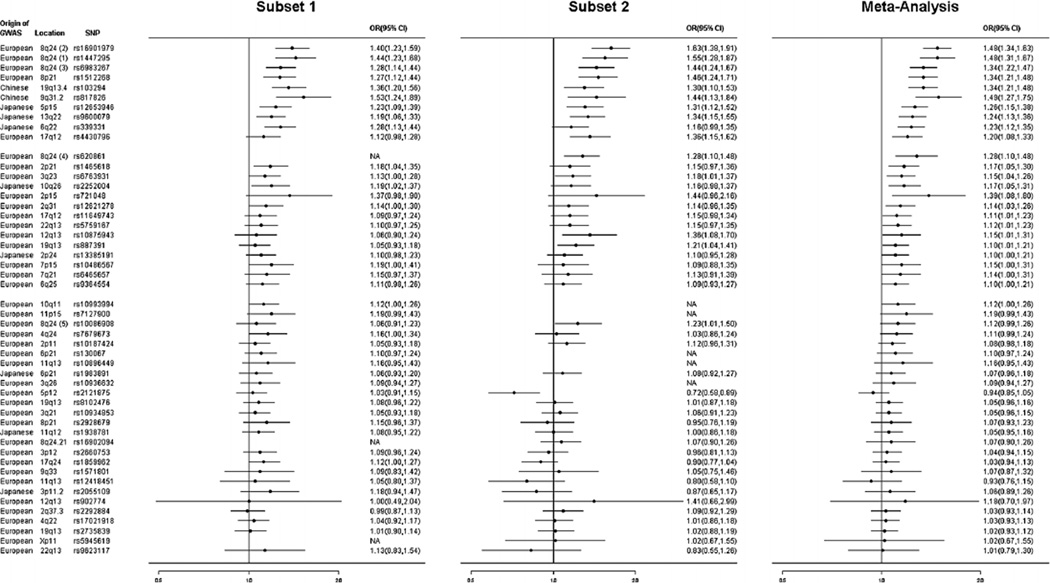
Among the 53 SNPs, three SNPs were not polymorphic (rs7210100 reported from African descent, rs4962416 and rs5919432 reported from the European descent) and therefore were not tested for their association with PCa risk. For the 50 polymorphic SNPs, 10 were significantly associated with PCa risk in Chinese men at P < 0.001 (Table II); they included five SNPs originally reported in subjects of European descent, three SNPs originally reported in subjects of Japanese descent, and two SNPs originally reported in subjects of Chinese descent. In addition, 14 additional SNPs were significantly associated with PCa risk in Chinese men at P < 0.05 (Table II); they included 12 SNPs originally reported in subjects of European descent and two SNPs originally reported in subjects of Japanese descent.
TABLE II.
Results of Association Test in Chinese Men for Reported PCa Risk-Associated SNPs From Genome-Wide Association Studies of Populations in European, African, Japanese, and Chinese Descent
| Subset1 | Subset 2 | Meta-analysis | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin of GWAS |
Chr | SNPs | BP | Alleles | Risk alleles |
Alele frequency | ORa | P-value | Alele frequency | OR | P-value | P for Q statistic |
I2 | OR | P-value | ||
| Cases | Controls | Cases | Controls | ||||||||||||||
| European | 8q24 (Region 2) | rs16901979 | 128,194,098 | C/A | A | 0.336 | 0.265 | 1.4 (1.23–1.59) | 2.74E−07 | 0.359 | 0.256 | 1.63 (1.38–1.91) | 4.84E−09 | 0.1506 | 51.6 | 1.48 (1.34–1.63) | 2.33E−14 |
| European | 8q24 (Region 1) | rs1447295 | 128,554,220 | C/A | A | 0.196 | 0.147 | 1.44 (1.23–1.68) | 5.45E−06 | 0.218 | 0.153 | 1.55 (1.28–1.87) | 4.60E−06 | 0.5543 | 0 | 1.48 (1.31–1.67) | 1.54E−10 |
| European | 8q24 (Region 3) | rs6983267 | 128,482,487 | T/G | G | 0.467 | 0.411 | 1.28 (1.14–1.44) | 3.71E−05 | 0.519 | 0.428 | 1.44 (1.24–1.67) | 1.25E−06 | 0.211 | 36.08 | 1.34 (1.22–1.47) | 4.55E−10 |
| European | 8p21 | rs1512268 | 23,582,408 | G/A | T | 0.324 | 0.277 | 1.27 (1.12–1.44) | 2.42E−04 | 0.351 | 0.271 | 1.46 (1.24–1.71) | 3.42E−06 | 0.1873 | 42.48 | 1.34 (1.21–1.48) | 8.26E−09 |
| Chinese | 19q13.4 | rs103294 | 59,489,660 | T/C | C | 0.301 | 0.242 | 1.36 (1.2–1.56) | 3.64E−06 | 0.292 | 0.241 | 1.3 (1.1–1.53) | 2.03E−03 | 0.6465 | 0 | 1.34 (1.21–1.48) | 3.15E−08 |
| Chinese | 9q31.2 | rs817826 | 109,196,121 | T/C | C | 0.107 | 0.075 | 1.53 (1.24–1.89) | 5.52E−05 | 0.112 | 0.081 | 1.44 (1.13–1.84) | 3.64E−03 | 0.6982 | 0 | 1.49 (1.27–1.75) | 8.26E−07 |
| Japanese | 5p15 | rs12653946 | 1,948,829 | C/T | T | 0.410 | 0.362 | 1.23 (1.09–1.39) | 6.70E−04 | 0.421 | 0.358 | 1.31 (1.12–1.52) | 5.63E−04 | 0.5472 | 0 | 1.26 (1.15–1.38) | 1.54E−06 |
| Japanese | 13q22 | rs9600079 | 72,626,140 | G/T | T | 0.487 | 0.444 | 1.19 (1.06–1.33) | 3.96E−03 | 0.510 | 0.437 | 1.34 (1.15–1.55) | 1.17E−04 | 0.2073 | 37.1 | 1.24 (1.13–1.36) | 3.5E−06 |
| Japanese | 6q22 | rs339331 | 117,316,745 | T/C | T | 0.687 | 0.635 | 1.28 (1.13–1.44) | 9.20E−05 | 0.685 | 0.653 | 1.16 (0.99–1.35) | 7.42E−02 | 0.332 | 0 | 1.23 (1.12–1.35) | 2.91E−05 |
| European | 17q12 | rs4430796 | 33,172,153 | T/C | A | 0.744 | 0.725 | 1.12 (0.98–1.28) | 9.29E−02 | 0.766 | 0.706 | 1.36 (1.15–1.62) | 4.08E−04 | 0.0733 | 68.81 | 1.2 (1.08–1.33) | 5.15E−04 |
| European | 8q24 (Region 4) | rs620861 | 128,335,673 | G/A | G | 0.610 | 0.551 | 1.28 (1.1–1.48) | 1.63E−03 | 1 | 0 | 1.28 (1.1–1.48) | 1.63E−03 | ||||
| European | 2p21 | rs1465618 | 43,407,453 | A/G | T | 0.744 | 0.714 | 1.18 (1.04–1.35) | 1.35E−02 | 0.757 | 0.730 | 1.15 (0.97–1.36) | 1.17E−01 | 0.7909 | 0 | 1.17 (1.05–1.3) | 3.54E−03 |
| European | 3q23 | rs6763931 | 142,585,523 | C/T | A | 0.367 | 0.336 | 1.13 (1–1.28) | 4.64E−02 | 0.371 | 0.334 | 1.18 (1.01–1.37) | 3.80E−02 | 0.6844 | 0 | 1.15 (1.04–1.26) | 4.38E−03 |
| Japanese | 10q26 | rs2252004 | 122,834,699 | G/T | C | 0.768 | 0.739 | 1.19 (1.02–1.37) | 2.23E−02 | 0.752 | 0.723 | 1.16 (0.98–1.37) | 8.71E−02 | 0.8402 | 0 | 1.17 (1.05–1.31) | 4.42E−03 |
| European | 2p15 | rs721048 | 62,985,235 | G/A | A | 0.038 | 0.028 | 1.37 (0.98–1.9) | 6.58E−02 | 0.039 | 0.027 | 1.44 (0.96–2.16) | 7.85E−02 | 0.8458 | 0 | 1.39 (1.08–1.8) | 1.14E−02 |
| European | 2q31 | rs12621278 | 173,019,799 | A/G | A | 0.749 | 0.724 | 1.14 (1–1.3) | 5.60E−02 | 0.743 | 0.717 | 1.14 (0.96–1.35) | 1.29E−01 | 0.9806 | 0 | 1.14 (1.03–1.26) | 1.47E−02 |
| European | 17q12 | rs11649743 | 33,149,092 | C/T | G | 0.687 | 0.671 | 1.09 (0.97–1.24) | 1.60E−01 | 0.683 | 0.652 | 1.15 (0.98–1.34) | 9.14E−02 | 0.6438 | 0 | 1.11 (1.01–1.23) | 3.15E−02 |
| European | 22q13 | rs5759167 | 41,830,156 | G/T | G | 0.730 | 0.710 | 1.1 (0.97–1.25) | 1.54E−01 | 0.717 | 0.688 | 1.15 (0.97–1.35) | 1.01E−01 | 0.6862 | 0 | 1.12 (1.01–1.23) | 3.29E−02 |
| European | 12q13 | rs10875943 | 47,962,277 | T/C | C | 0.853 | 0.850 | 1.06 (0.9–1.24) | 4.95E−01 | 0.887 | 0.852 | 1.36 (1.08–1.7) | 7.69E−03 | 0.0774 | 67.93 | 1.15 (1.01–1.31) | 3.56E−02 |
| European | 19q13 | rs887391 | 46,677,464 | T/C | T | 0.605 | 0.595 | 1.05 (0.93–1.18) | 4.60E−01 | 0.627 | 0.581 | 1.21 (1.04–1.41) | 1.42E−02 | 0.1408 | 53.9 | 1.1 (1.01–1.21) | 3.66E−02 |
| Japanese | 2p24 | rs13385191 | 20,751,746 | G/A | G | 0.461 | 0.438 | 1.1 (0.98–1.23) | 1.14E−01 | 0.471 | 0.447 | 1.1 (0.95–1.28) | 2.04E−01 | 0.9775 | 0 | 1.1 (1–1.21) | 4.17E−02 |
| European | 7p15 | rs10486567 | 27,943,088 | T/C | G | 0.148 | 0.127 | 1.19 (1–1.41) | 4.89E−02 | 0.144 | 0.134 | 1.09 (0.88–1.35) | 4.39E−01 | 0.5313 | 0 | 1.15 (1–1.31) | 4.29E−02 |
| European | 7q21 | rs6465657 | 97,654,263 | C/T | C | 0.875 | 0.859 | 1.15 (0.97–1.37) | 1.00E−01 | 0.863 | 0.849 | 1.13 (0.91–1.39) | 2.68E−01 | 0.8744 | 0 | 1.14 (1–1.31) | 4.77E−02 |
| European | 6q25 | rs9364554 | 160,753,654 | C/T | C | 0.678 | 0.657 | 1.11 (0.98–1.26) | 8.86E−02 | 0.685 | 0.667 | 1.09 (0.93–1.27) | 3.04E−01 | 0.824 | 0 | 1.1 (1–1.21) | 4.83E−02 |
| European | 10q11 | rs10993994 | 51,219,502 | T/C | T | 0.511 | 0.484 | 1.12 (1–1.26) | 5.89E−02 | 1 | 0 | 1.12 (1–1.26) | 5.89E−02 | ||||
| European | 11p15 | rs7127900 | 2,190,150 | G/A | G | 0.895 | 0.878 | 1.19 (0.99–1.43) | 6.17E−02 | 1 | 0 | 1.19 (0.99–1.43) | 6.17E−02 | ||||
| European | 8q24 (Region 5) | rs10086908 | 128,081,119 | T/C | T | 0.830 | 0.820 | 1.06 (0.91–1.23) | 4.85E−01 | 0.847 | 0.818 | 1.23 (1.01–1.5) | 4.40E−02 | 0.2365 | 28.63 | 1.12 (0.99–1.26) | 7.61E−02 |
| European | 4q24 | rs7679673 | 106,280,983 | A/C | C | 0.211 | 0.186 | 1.16 (1–1.34) | 5.02E−02 | 0.203 | 0.198 | 1.03 (0.86–1.24) | 7.63E−01 | 0.3293 | 0 | 1.11 (0.99–1.24) | 8.39E−02 |
| European | 2p11 | rs10187424 | 85,647,808 | A/G | T | 0.626 | 0.612 | 1.05 (0.93–1.18) | 4.28E−01 | 0.654 | 0.627 | 1.12 (0.96–1.31) | 1.41E−01 | 0.4944 | 0 | 1.08 (0.98–1.18) | 1.29E−01 |
| European | 6p21 | rs130067 | 31,226,490 | T/G | G | 0.328 | 0.307 | 1.1 (0.97–1.24) | 1.40E−01 | 1 | 0 | 1.1 (0.97–1.24) | 1.40E−01 | ||||
| European | 11q13 | rs10896449 | 68,751,243 | A/G | G | 0.097 | 0.084 | 1.16 (0.95–1.43) | 1.43E−01 | 0.000 | 0.038 | NA | 4.32E−04 | 1 | 0 | 1.16 (0.95–1.43) | 1.43E–01 |
| Japanese | 6p21 | rs1983891 | 41,644,405 | C/T | T | 0.345 | 0.332 | 1.06 (0.93–1.2) | 3.89E–01 | 0.330 | 0.313 | 1.08 (0.92–1.27) | 3.55E–01 | 0.8522 | 0 | 1.07 (0.96–1.18) | 2.13E–01 |
| European | 3q26 | rs10936632 | 171,612,796 | A/C | A | 0.269 | 0.253 | 1.09 (0.94–1.27) | 2.59E–01 | 1 | 0 | 1.09 (0.94–1.27) | 2.59E–01 | ||||
| European | 5p12 | rs2121875 | 44,401,302 | T/G | A | 0.483 | 0.473 | 1.03 (0.91–1.15) | 6.50E–01 | 0.413 | 0.495 | 0.72 (0.58–0.89) | 1.93E–03 | 0.0034 | 88.33 | 0.94 (0.85–1.05) | 2.72E–01 |
| European | 19q13 | rs8102476 | 43,427,453 | A/G | C | 0.374 | 0.360 | 1.08 (0.96–1.22) | 2.08E–01 | 0.382 | 0.380 | 1.01 (0.87–1.18) | 8.85E–01 | 0.5046 | 0 | 1.05 (0.96–1.16) | 2.84E–01 |
| European | 3q21 | rs10934853 | 129,521,063 | C/A | A | 0.441 | 0.428 | 1.05 (0.93–1.18) | 4.54E–01 | 0.434 | 0.421 | 1.06 (0.91–1.23) | 4.79E–01 | 0.9295 | 0 | 1.05 (0.96–1.15) | 3.04E–01 |
| European | 8p21 | rs2928679 | 23,494,920 | G/A | A | 0.134 | 0.120 | 1.15 (0.96–1.37) | 1.24E–01 | 0.124 | 0.129 | 0.95 (0.76–1.19) | 6.65E−01 | 0.1982 | 39.61 | 1.07 (0.93–1.23) | 3.45E−01 |
| Japanese | 11q12 | rs1938781 | 58,671,686 | T/C | G | 0.328 | 0.309 | 1.08 (0.95–1.22) | 2.50E−01 | 0.322 | 0.322 | 1 (0.86–1.18) | 9.72E−01 | 0.4936 | 0 | 1.05 (0.95–1.16) | 3.51E−01 |
| European | 8q24.21 | rs16902094 | 128,320,346 | A/G | A | 0.726 | 0.713 | 1.07 (0.9–1.26) | 4.55E−01 | 1 | 0 | 1.07 (0.9–1.26) | 4.55E−01 | ||||
| European | 3p12 | rs2660753 | 87,193,364 | C/T | T | 0.310 | 0.287 | 1.09 (0.96–1.24) | 1.88E−01 | 0.290 | 0.299 | 0.96 (0.81–1.13) | 6.07E−01 | 0.2229 | 32.7 | 1.04 (0.94–1.15) | 4.67E−01 |
| European | 17q24 | rs1859962 | 66,620,348 | T/G | G | 0.426 | 0.399 | 1.12 (1–1.27) | 5.20E−02 | 0.394 | 0.420 | 0.9 (0.77–1.04) | 1.57E−01 | 0.0209 | 81.27 | 1.03 (0.94–1.13) | 5.07E−01 |
| European | 9q33 | rs1571801 | 123,467,194 | G/T | T | 0.053 | 0.050 | 1.09 (0.83–1.42) | 5.40E−01 | 0.054 | 0.051 | 1.05 (0.75–1.46) | 7.93E−01 | 0.8587 | 0 | 1.07 (0.87–1.32) | 5.23E−01 |
| European | 11q13 | rs12418451 | 68,691,995 | G/A | A | 0.052 | 0.049 | 1.05 (0.8–1.37) | 7.43E−01 | 0.053 | 0.065 | 0.8 (0.58–1.1) | 1.67E−01 | 0.2061 | 37.45 | 0.93 (0.76–1.15) | 5.24E−01 |
| Japanese | 3p11.2 | rs2055109 | 87,550,022 | C/T | C | 0.080 | 0.069 | 1.18 (0.94–1.47) | 1.44E−01 | 0.064 | 0.073 | 0.87 (0.65–1.17) | 3.60E−01 | 0.1083 | 61.22 | 1.06 (0.89–1.26) | 5.33E−01 |
| European | 12q13 | rs902774 | 51,560,171 | G/A | G | 0.993 | 0.993 | 1 (0.49–2.04) | 9.94E−01 | 0.991 | 0.988 | 1.41 (0.66–2.99) | 3.69E−01 | 0.5188 | 0 | 1.18 (0.7–1.97) | 5.36E−01 |
| European | 2q37.3 | rs2292884 | 238,107,965 | A/G | A | 0.717 | 0.714 | 0.99 (0.87–1.13) | 9.25E−01 | 0.735 | 0.718 | 1.09 (0.92–1.29) | 3.19E−01 | 0.3988 | 0 | 1.03 (0.93–1.14) | 5.96E−01 |
| European | 4q22 | rs17021918 | 95,781,900 | C/T | C | 0.662 | 0.653 | 1.04 (0.92–1.17) | 5.60E−01 | 0.663 | 0.661 | 1.01 (0.86–1.18) | 9.10E−01 | 0.787 | 0 | 1.03 (0.93–1.13) | 5.96E−01 |
| European | 19q13 | rs2735839 | 56,056,435 | G/A | A | 0.391 | 0.384 | 1.01 (0.9–1.14) | 8.22E−01 | 0.382 | 0.377 | 1.02 (0.88–1.19) | 7.70E−01 | 0.9285 | 0 | 1.02 (0.93–1.12) | 7.18E−01 |
| European | Xp11 | rs5945619 | 51,241,672 | A/G | T | 0.932 | 0.931 | 1.02 (0.67–1.55) | 9.17E−01 | 1 | 0 | 1.02 (0.67–1.55) | 9.17E−01 | ||||
| European | 22q13 | rs9623117 | 38,782,065 | T/C | C | 0.040 | 0.035 | 1.13 (0.83–1.54) | 4.38E−01 | 0.032 | 0.038 | 0.83 (0.55–1.26) | 3.86E−01 | 0.2471 | 25.36 | 1.01 (0.79–1.3) | 9.18E−01 |
| European | 10q26 | rs4962416 | 126,686,862 | A/G | G | Not polymorphic | |||||||||||
| African | 17q21.32 | rs7210100 | 44,791,748 | G/A | A | Not polymorphic | |||||||||||
| European | Xq12 | rs5919432 | 66,938,275 | A/G | A | Not polymorphic | |||||||||||
Allelic odds ratio (OR) and 95% confidence interval (95% CI).
The OR of each SNP for PCa was estimated for the risk alleles identified in previous studies. For the 24 significant SNPs, the estimated ORs in the Chinese population were all >1.0 (i.e., consistent with previous studies) and ranged from 1.10 to 1.49 (Table II and Fig. 1). The top three SNPs that confer the strongest risk for PCa in the Chinese population were rs16901979 at 8q24 (Region 2), OR = 1.48, 95% CI: 1.34–1.63, P-value = 2.33 × 10−14, rs1447295 at 8q24 (Region 1), OR = 1.48, 95% CI: 1.31–1.67, P-value = 1.54 × 10−10, and rs817826 at 9q31, OR = 1.49, 95% CI: 1.27–1.75, P-value = 8.26 × 10−7. For the remaining 26 SNPs that were not significantly associated with PCa risk in the Chinese population, all but two SNPs had OR estimates >1.0.
Fig. 1.
Forest plots of PCa risk-associated SNPs identified from GWAS of various populations with PCa risk in Chinese men.
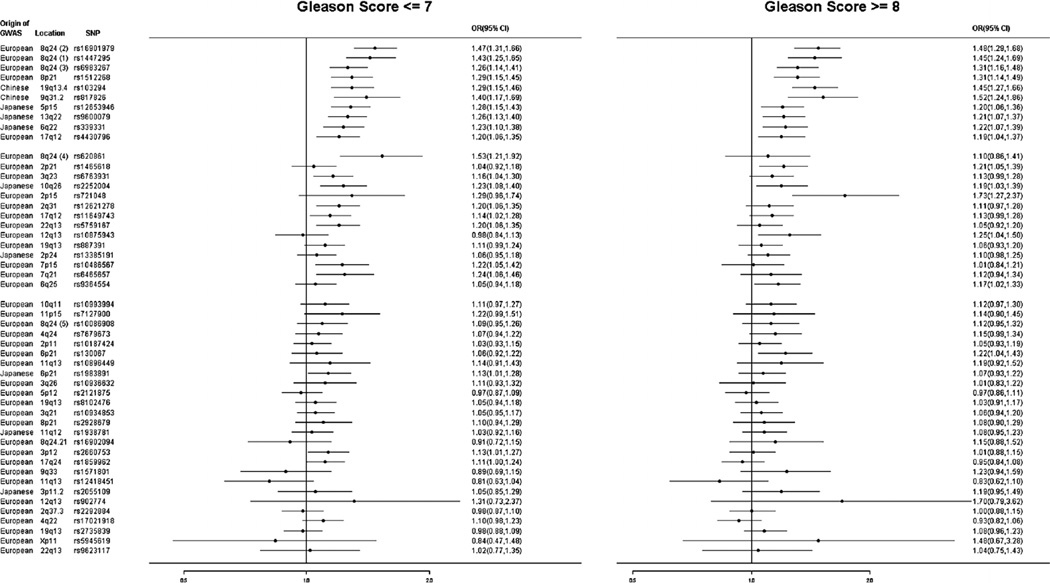
We also estimated ORs for PCa separately for high-grade PCa (Gleason score ≥8) and for low-grade PCa (Gleason score ≤7; Table III and Fig. 2). For the 24 SNPs that were significantly associated with PCa risk in the meta-analysis, significantly different ORs between the two types of PCa were found only for two SNPs (P < 0.05). One of these two SNPs was rs620861 at 8q24 (Region 4), in which the association was stronger for PCa of Gleason Score ≤7 (OR = 1.53, 95% CI: 1.21–1.92) than that of Gleason score ≥8 (OR = 1.10, 95% CI: 0.86–1.41), P = 0.04. The other SNP was rs10875943 at 12q13, in which the association was stronger for PCa of Gleason Score ≥8 (OR = 1.25, 95% CI: 1.04–1.50) than that of Gleason score ≤7 (OR = 0.98, 95% CI: 0.84–1.13, P = 0.02.
TABLE III.
Estimated OR for PCa With Gleason Score ≤7 or ≥8 in Chinese Men
| Allele frequency | Risk for PCa of Gleason score ≤7 |
Risk for PCa of Gleason score ≥8 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origin of GWAS |
Chr | SNP | BP | Risk allele |
Controls (N = 2,126) |
Gleason score ≤7 (N = 993) |
Gleason score ≥8 (N = 684) |
ORa | P-value | OR | P-value |
P for different ORs |
| European | 8q24 (Region 2) | rs16901979 | 128,194,098 | A | 0.26 | 0.34 | 0.34 | 1.47 (1.31–1.66) | 5.67E−11 | 1.48 (1.29–1.68) | 7.03E−09 | 9.86E−01 |
| European | 8q24 (Region 1) | rs1447295 | 128,554,220 | A | 0.15 | 0.20 | 0.20 | 1.43 (1.25–1.65) | 3.20E−07 | 1.45 (1.24–1.69) | 3.39E−06 | 9.11E−01 |
| European | 8q24 (Region 3) | rs6983267 | 128,482,487 | G | 0.42 | 0.48 | 0.48 | 1.26 (1.14–1.41) | 1.80E−05 | 1.31 (1.16–1.48) | 0.00001526 | 6.15E−01 |
| European | 8p21 | rs1512268 | 23,582,408 | T | 0.27 | 0.33 | 0.33 | 1.29 (1.15–1.45) | 1.37E−05 | 1.31 (1.14–1.49) | 0.00007238 | 8.98E−01 |
| Chinese | 19q13.4 | rs103294 | 59,489,660 | C | 0.24 | 0.29 | 0.31 | 1.29 (1.15–1.46) | 2.68E−05 | 1.45 (1.27–1.66) | 4.83E−08 | 1.29E−01 |
| Chinese | 9q31.2 | rs817826 | 109,196,121 | C | 0.08 | 0.10 | 0.11 | 1.4 (1.17–1.69) | 2.56E−04 | 1.52 (1.24–1.86) | 4.10E−05 | 4.72E−01 |
| Japanese | 5p15 | rs12653946 | 1,948,829 | T | 0.36 | 0.42 | 0.40 | 1.28 (1.15–1.43) | 9.79E−06 | 1.2 (1.06–1.36) | 4.05E−03 | 3.81E−01 |
| Japanese | 13q22 | rs9600079 | 72,626,140 | T | 0.44 | 0.50 | 0.49 | 1.26 (1.13–1.4) | 2.33E−05 | 1.21 (1.07–1.37) | 2.11E−03 | 5.81E−01 |
| Japanese | 6q22 | rs339331 | 117,316,745 | T | 0.64 | 0.69 | 0.69 | 1.23 (1.1–1.38) | 3.91E−04 | 1.22 (1.07–1.39) | 2.76E−03 | 9.25E−01 |
| European | 17q12 | rs4430796 | 33,172,153 | A | 0.71 | 0.75 | 0.75 | 1.2 (1.06–1.35) | 4.04E−03 | 1.19 (1.04–1.37) | 1.35E−02 | 9.77E−01 |
| European | 8q24 (Region 4) | rs620861 | 128,335,673 | G | 0.55 | 0.65 | 0.58 | 1.53 (1.21–1.92) | 2.90E−04 | 1.1 (0.86–1.41) | 4.34E−01 | 4.36E−02 |
| European | 2p21 | rs1465618 | 43,407,453 | T | 0.72 | 0.73 | 0.76 | 1.04 (0.92–1.18) | 4.87E−01 | 1.21 (1.05–1.39) | 9.94E−03 | 7.94E−02 |
| European | 3q23 | rs6763931 | 142,585,523 | A | 0.34 | 0.37 | 0.36 | 1.16 (1.04–1.3) | 7.99E−03 | 1.13 (0.99–1.28) | 5.98E−02 | 6.99E−01 |
| Japanese | 10q26 | rs2252004 | 122,834,699 | C | 0.73 | 0.77 | 0.76 | 1.23 (1.08–1.4) | 2.18E−03 | 1.19 (1.03–1.39) | 2.23E−02 | 7.37E−01 |
| European | 2p15 | rs721048 | 62,985,235 | A | 0.03 | 0.04 | 0.05 | 1.29 (0.96–1.74) | 9.60E−02 | 1.73 (1.27–2.37) | 4.52E−04 | 9.36E−02 |
| European | 2q31 | rs12621278 | 173,019,799 | A | 0.72 | 0.76 | 0.74 | 1.2 (1.06–1.35) | 3.72E−03 | 1.11 (0.97–1.28) | 1.32E−01 | 3.56E−01 |
| European | 17q12 | rs11649743 | 33,149,092 | G | 0.66 | 0.69 | 0.69 | 1.14 (1.02–1.28) | 2.23E−02 | 1.13 (0.99–1.28) | 7.51E−02 | 8.47E−01 |
| European | 22q13 | rs5759167 | 41,830,156 | G | 0.70 | 0.74 | 0.71 | 1.2 (1.06–1.35) | 3.08E−03 | 1.05 (0.92–1.2) | 4.74E−01 | 9.36E−02 |
| European | 12q13 | rs10875943 | 47,962,277 | C | 0.85 | 0.85 | 0.88 | 0.98 (0.84–1.13) | 7.60E−01 | 1.25 (1.04–1.5) | 1.53E−02 | 1.62E−02 |
| European | 19q13 | rs887391 | 46,677,464 | T | 0.59 | 0.61 | 0.60 | 1.11 (0.99–1.24) | 6.87E−02 | 1.06 (0.93–1.2) | 3.84E−01 | 5.21E−01 |
| Japanese | 2p24 | rs13385191 | 20,751,746 | G | 0.44 | 0.46 | 0.47 | 1.06 (0.95–1.18) | 2.69E−01 | 1.1 (0.98–1.25) | 1.18E−01 | 5.97E−01 |
| European | 7p15 | rs10486567 | 27,943,088 | G | 0.13 | 0.16 | 0.13 | 1.22 (1.05–1.42) | 1.03E−02 | 1.01 (0.84–1.21) | 8.99E−01 | 6.63E−02 |
| European | 7q21 | rs6465657 | 97,654,263 | C | 0.85 | 0.88 | 0.87 | 1.24 (1.06–1.46) | 7.51E−03 | 1.12 (0.94–1.34) | 2.01E−01 | 3.38E−01 |
| European | 6q25 | rs9364554 | 160,753,654 | C | 0.66 | 0.67 | 0.69 | 1.05 (0.94–1.18) | 4.09E−01 | 1.17 (1.02–1.33) | 2.10E−02 | 1.57E−01 |
| European | 10q11 | rs10993994 | 51,219,502 | T | 0.48 | 0.51 | 0.51 | 1.11 (0.97–1.27) | 1.26E−01 | 1.12 (0.97–1.3) | 1.28E−01 | 8.73E−01 |
| European | 11p15 | rs7127900 | 2,190,150 | G | 0.88 | 0.90 | 0.89 | 1.22 (0.99–1.51) | 6.19E−02 | 1.14 (0.9–1.45) | 2.67E−01 | 5.99E−01 |
| European | 8q24 (Region 5) | rs10086908 | 128,081,119 | T | 0.82 | 0.83 | 0.84 | 1.09 (0.95–1.26) | 2.25E−01 | 1.12 (0.95–1.32) | 1.64E−01 | 7.64E−01 |
| European | 4q24 | rs7679673 | 106,280,983 | C | 0.19 | 0.20 | 0.22 | 1.07 (0.94–1.22) | 3.29E−01 | 1.15 (0.99–1.34) | 6.91E−02 | 4.01E−01 |
| European | 2p11 | rs10187424 | 85,647,808 | T | 0.62 | 0.63 | 0.63 | 1.03 (0.93–1.15) | 5.56E−01 | 1.05 (0.93–1.19) | 4.30E−01 | 8.07E−01 |
| European | 6p21 | rs130067 | 31,226,490 | G | 0.31 | 0.32 | 0.35 | 1.06 (0.92–1.22) | 4.46E−01 | 1.22 (1.04–1.43) | 1.42E−02 | 8.71E−02 |
| European | 11q13 | rs10896449 | 68,751,243 | G | 0.08 | 0.09 | 0.09 | 1.14 (0.91–1.43) | 2.66E−01 | 1.19 (0.92–1.52) | 1.79E−01 | 7.44E−01 |
| Japanese | 6p21 | rs1983891 | 41,644,405 | T | 0.32 | 0.35 | 0.34 | 1.13 (1.01–1.28) | 3.53E−02 | 1.07 (0.93–1.22) | 3.27E−01 | 4.47E−01 |
| European | 3q26 | rs10936632 | 171,612,796 | A | 0.25 | 0.27 | 0.25 | 1.11 (0.93–1.32) | 2.57E−01 | 1.01 (0.83–1.22) | 9.50E−01 | 3.52E−01 |
| European | 5p12 | rs2121875 | 44,401,302 | A | 0.49 | 0.48 | 0.48 | 0.97 (0.87–1.09) | 6.32E−01 | 0.97 (0.86–1.11) | 6.81E−01 | 9.99E−01 |
| European | 19q13 | rs8102476 | 43,427,453 | C | 0.37 | 0.38 | 0.38 | 1.05 (0.94–1.18) | 3.50E−01 | 1.03 (0.91–1.17) | 6.14E−01 | 7.83E−01 |
| European | 3q21 | rs10934853 | 129,521,063 | A | 0.42 | 0.44 | 0.44 | 1.05 (0.95–1.17) | 3.46E−01 | 1.06 (0.94–1.2) | 3.48E−01 | 9.19E−01 |
| European | 8p21 | rs2928679 | 23,494,920 | A | 0.12 | 0.14 | 0.13 | 1.1 (0.94–1.29) | 2.29E−01 | 1.08 (0.9–1.29) | 4.10E−01 | 8.41E−01 |
| Japanese | 11q12 | rs1938781 | 58,671,686 | G | 0.32 | 0.32 | 0.33 | 1.03 (0.92–1.16) | 5.75E−01 | 1.08 (0.95–1.23) | 2.33E−01 | 5.37E−01 |
| European | 8q24.21 | rs16902094 | 128,320,346 | A | 0.71 | 0.69 | 0.74 | 0.91 (0.72–1.15) | 4.29E−01 | 1.15 (0.88–1.52) | 3.08E−01 | 1.69E−01 |
| European | 3p12 | rs2660753 | 87,193,364 | T | 0.30 | 0.32 | 0.30 | 1.13 (1.01–1.27) | 3.32E−02 | 1.01 (0.88–1.15) | 9.35E−01 | 1.18E−01 |
| European | 17q24 | rs1859962 | 66,620,348 | G | 0.41 | 0.44 | 0.40 | 1.11 (1–1.24) | 5.18E−02 | 0.95 (0.84–1.08) | 4.41E−01 | 2.91E−02 |
| European | 9q33 | rs1571801 | 123,467,194 | T | 0.05 | 0.04 | 0.06 | 0.89 (0.69–1.15) | 3.64E−01 | 1.23 (0.94–1.59) | 1.28E−01 | 4.04E−02 |
| European | 11q13 | rs12418451 | 68,691,995 | A | 0.06 | 0.05 | 0.05 | 0.81 (0.63–1.04) | 1.00E−01 | 0.83 (0.62–1.1) | 1.88E−01 | 9.24E−01 |
| Japanese | 3p11.2 | rs2055109 | 87,550,022 | C | 0.07 | 0.07 | 0.08 | 1.05 (0.85–1.29) | 6.52E−01 | 1.19 (0.95–1.49) | 1.23E−01 | 3.19E−01 |
| European | 12q13 | rs902774 | 51,560,171 | G | 0.99 | 0.99 | 0.99 | 1.31 (0.73–2.37) | 3.69E−01 | 1.7 (0.79–3.62) | 1.67E−01 | 5.57E−01 |
| European | 2q37.3 | rs2292884 | 238,107,965 | A | 0.72 | 0.71 | 0.72 | 0.98 (0.87–1.1) | 7.53E−01 | 1 (0.88–1.15) | 9.60E−01 | 7.73E−01 |
| European | 4q22 | rs17021918 | 95,781,900 | C | 0.66 | 0.68 | 0.64 | 1.1 (0.98–1.23) | 1.11E−01 | 0.93 (0.82–1.06) | 2.80E−01 | 2.82E−02 |
| European | 19q13 | rs2735839 | 56,056,435 | A | 0.38 | 0.38 | 0.40 | 0.98 (0.88–1.09) | 6.82E−01 | 1.08 (0.96–1.23) | 2.16E−01 | 1.58E−01 |
| European | Xp11 | rs5945619 | 51,241,672 | T | 0.93 | 0.92 | 0.95 | 0.84 (0.47–1.48) | 5.38E−01 | 1.48 (0.67–3.28) | 3.28E−01 | 2.19E−01 |
| European | 22q13 | rs9623117 | 38,782,065 | C | 0.04 | 0.04 | 0.04 | 1.02 (0.77–1.35) | 9.19E−01 | 1.04 (0.75–1.43) | 8.21E−01 | 9.04E−01 |
| European | 10q26 | rs4962416 | 126,686,862 | Not polymorphic | ||||||||
| African | 17q21.32 | rs7210100 | 44,791,748 | Not polymorphic | ||||||||
| European | Xq12 | rs5919432 | 66,938,275 | Not polymorphic | ||||||||
Allelic odds ratio (OR) and 95% confidence interval (95% CI).
Fig. 2.
Forest plots of PCa risk-associated SNPs identified from GWAS of various populations with risk to PCa of Gleason score ≤7 and ≥8 in Chinese men.
DISCUSSION
Genetic susceptibility to PCa is well established [4]. GWAS of PCa in the past 5 years have identified more than 50 regions in the genome that were associated with PCa risk in various racial populations [6–26]. The potential clinical utility of these PCa risk-associated SNPs in predicting outcomes of initial and repeat prostate biopsy have been reported in populations of European descent [31–32]. However, such utility in the Chinese population has not been established, largely due to a lack of information on PCa risk-associated SNPs in Chinese and the degree of risk they confer. This study attempted to fill the gap by cataloging SNPs that are associated with PCa risk and the OR of these SNPs in Chinese. With the information of the 24 PCa risk-associated SNPs and their ORs, it is now possible to calculate a genetic score based on these SNPs for Chinese men and to assess their genetic risk for PCa.
Risk assessment of PCa using inherited genetic markers may be particularly important in Chinese men because family history of PCa, a commonly used marker for genetic risk to PCa in western countries, is essentially uninformative in China. The percentage of men with family history of PCa is extremely low in China because the disease was rarely detected in this country in past decades, likely due to low adoption of PSA screening and low life expectancy. Other issues such as small family size due to the family planning policies in China may further limit the value of family history in the future. Genetic score derived from PCa risk-associated SNPs, on the other hand, does not rely on information from relatives and therefore may overcome these limitations.
Several papers in the last 2 years reported PCa risk-associated SNPs in Chinese men. In a case–control study of Chinese men (1,108 cases and 1,525 controls) selected from the ChinaPCa, Liu et al. [33] studied the first 33 PCa risk-associated SNPs initially discovered from GWAS of European descent and found 11 of them were associated with PCa risk at P < 0.05. Eight of these 11 were also implicated in the current study. The statistical evidence for association of the other three SNPs was weaker in our current study of larger sample size. In another case–control study of Chinese men (1,524 cases and 2,169 controls) also selected from the ChinaPCa, Wang and colleagues assessed association of the first five PCa risk-associated SNPs identified from GWAS of a Japanese population and found three of which were associated with PCa risk at P < 0.05 [34]. These three SNPs were also implicated in this current study. Finally, in a multi-stage GWAS of PCa in Chinese men, Xu et al. [26] identified two SNPs that were significantly associated with PCa risk at a genome-wide significance level. This Chinese GWAS also reported significant associations of 12 SNPs initially reported from GWAS of European descent (N = 8) and Japanese descent (N = 4) in subjects of the first-stage GWAS (1,417 cases and 1,008 controls). All 12 of these SNPs were implicated in this current study. It is noted that the study subjects between the current study and the previous three studies overlapped; therefore, the findings are not completely independent and the similarities are not surprising. The current study, however, represents the most comprehensive analysis for all established PCa risk-associated SNPs to date from various racial populations in a larger number of Chinese men. As a result, it provided evidence for the largest number of PCa risk-associated SNPs in Chinese.
Although only 10 of the 50 tested SNPs were significantly associated with PCa at P < 0.001 and exceeded study-wise significance after Bonferroni correction, 14 additional SNPs that were associated with PCa risk at P < 0.05 may also be considered as good candidates for risk assessment among Chinese men. These SNPs likely represent true PCa associations because of their statistical evidence and the fact that the direction of association was consistent with previous studies. They may be used for assessment of individual risk for PCa. Although it is possible that some of these SNPs represent false positives, it is known that inclusion of false positive SNPs in risk assessment will not improve or reduce the predictive performance.
Among the 24 SNPs that were significantly associated with PCa risk in Chinese men, 13 were located within known genes, including NXX3.1 (rs1512268 at 8p21), LILRA3 (rs103294 at 19q13), RFX6 (rs339331 at 6q22), HNF1B (rs4430796 and rs11649743 at 17q12), THADA (rs1465618 at 2p21), ZBTB38 (rs6763931 at 3q23), EHBP1 (rs721048 at 2p15), ITGA6 (rs12621278 at 2q31), BIK (rs5759167 at 22q13), JAZF1 (rs10486567 at 7p15), LMTK2 (rs6465657 at 7q21), and SLC22A3 (rs9364554 at 6q25). The exact molecular mechanisms of these genetic variants are unknown. Additional functional studies are warranted.
In addition to common PCa risk associated SNPs discovered from GWAS, a rare but recurrent PCa risk-associated mutation, G84E of HOXB13, was noticed [35]. This gene was also evaluated in 1,518 cases and 1,536 controls selected from ChinaPCa. Although we did not observe the G84E mutation, we found a novel mutation (G125E) in five PCa cases but not in any of the controls [36]. We did not include the mutation in this current study because of its low frequency. However, this high-penetrance mutation should most likely be incorporated into PCa risk assessment for Chinese men.
There are several important limitations in this study. First, the sample size of this study remains limited, which affected statistical power to identify PCa risk-associated SNPs in Chinese men. However, this study nevertheless represents the largest collection of PCa cases in China. Due to the relatively low detection rate of PCa and the lack of a national cancer registry in China, it is a daunting effort to recruit case subjects. We intend to address this limitation by expanding ChinaPCa. Second, due to the open nature of this consortium, the clinical characterization of PCa patients was not consistent across hospitals in this study, which limited our ability to conduct an in-depth analysis based on clinical phenotypes. Third, most of PCa risk-associated SNPs implicated in this study did not distinguish aggressive from indolent disease. This limitation has been commonly encountered in prior genetic studies of PCa as well as other common cancers [37]. More effort should be devoted to the identification of SNPs that are associated with aggressive but not indolent PCa using case–case study designs. Such SNPs would be helpful to address over-diagnosis of indolent PCa.
In conclusion, by systematically evaluating PCa risk-associated SNPs identified from GWAS of various populations in this large Chinese study, we identified 24 PCa risk-associated SNPs in Chinese men and provided estimates of their risk. The information may facilitate risk assessment of PCa in Chinese men for targeted PCa screening, early detection of PCa, and possible chemoprevention.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the subjects included in this study. This work was partially funded by the National Key Basic Research Program Grant 973 (2012CB518300) to Y.S., the National Key Basic Research Program Grant 973 (2012CB518301) to J.X., the Key Project of the National Natural Science Foundation of China (81130047) to J.X., intramural grants from Fudan University “Thousand Talents Program” and Huashan Hospital to J.X., the National Institutes of Health (NCI CA129684) to J.X., National Natural Science Foundation of China (30945204) to Z.M., National Natural Science Foundation of China (30973009) to D.Y.
Footnotes
Additional supporting information may be found in the online version of this article.
REFERENCES
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Hsing AW, Tsao L, Devesa SS. International trends and patterns of prostate cancer incidence and mortality. Int J Cancer. 2000;85(1):60–67. doi: 10.1002/(sici)1097-0215(20000101)85:1<60::aid-ijc11>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 3.Ye DW, Li CL. Epidemiological trends of prostate cancer: Retrospect and prospect. China Oncol. 2007;17:177–180. [Google Scholar]
- 4.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 5.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Bäter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 7.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van-VierssenTrip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 9.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Bälter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Grönberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 10.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 11.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindström S, Adami HO, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF UK Genetic Prostate Cancer Study Collaborators, British Association of Urological Surgeons’ Section of Oncology, UK ProtecT Study Collaborators. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 13.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutchinson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Diver WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Crawford ED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Siddiq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE, Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, Vatten LJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN, Fraumeni JF, Jr, Hunter DJ, Thomas G, Chanock SJ. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J, Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA, Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, Spurdle A, Clements J, Dickinson JL, Maier C, Vogel W, Dörk T, Rebbeck TR, Cooney KA, Cannon-Albright L, Chappuis PO, Hutter P, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, English DR, Leongamornlert DA, Tymrakiewicz M, Morrison J, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Saunders EJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC, Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, Wahlfors T, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM, Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R, Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J, Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F, Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, Stefflova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitkova A, Cao G, Easton DF UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology, UK ProtecT Study Collaborators; PRACTICAL Consortium. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. doi: 10.1038/ng.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P, Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE, Hamdy FC, Donovan JL, Pharoah P, Schumacher F, Henderson BE, Stanford JL, Ostrander EA, Sorensen KD, Dörk T, Andriole G, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, Clements JA, Chambers S, Aitken J, Gardiner RA, Thibodeau SN, Schaid D, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang HW, Lu YJ, Kaneva R, Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tymrakiewicz M, Mahmud N, Guy M, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English DR, Wahlfors T, Tammela TL, Klarskov P, Nordestgaard BG, Røder MA, Tybjærg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wiklund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC, Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitzgerald L, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Meyer A, Serth J, Yeager M, Berndt SI, Marthick JR, Patterson B, Wokolorczyk D, Batra J, Lose F, McDonnell SK, Joshi AD, Shahabi A, Rinckleb AE, Ray A, Sellers TA, Lin HY, Stephenson RA, Farnham J, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, Easton DF, Eeles RA. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Bälter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Grönberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99(24):1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 18.Zheng SL, Stevens VL, Wiklund F, Isaacs SD, Sun J, Smith S, Pruett K, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Johansson JE, Liu W, Kim JW, Chang BL, Duggan D, Carpten J, Rodriguez C, Isaacs W, Grönberg H, Xu J. Two independent prostate cancer risk-associated Loci at 11q13. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1815–1820. doi: 10.1158/1055-9965.EPI-08-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu FC, Sun J, Wiklund F, Isaacs SD, Wiley KE, Purcell LD, Gao Z, Stattin P, Zhu Y, Kim ST, Zhang Z, Liu W, Chang BL, Walsh PC, Duggan D, Carpten JD, Isaacs WB, Grönberg H, Xu J, Zheng SL. A novel prostate cancer susceptibility locus at 19q13. Cancer Res. 2009;69(7):2720–2723. doi: 10.1158/0008-5472.CAN-08-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Zheng SL, Wiklund F, Isaacs SD, Li G, Wiley KE, Kim ST, Zhu Y, Zhang Z, Hsu FC, Turner AR, Stattin P, Liu W, Kim JW, Duggan D, Carpten J, Isaacs W, Grönberg H, Xu J, Chang BL. Sequence variants at 22q13 are associated with prostate cancer risk. Cancer Res. 2009;69(1):10–15. doi: 10.1158/0008-5472.CAN-08-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Zheng SL, Isaacs SD, Wiley KE, Wiklund F, Sun J, Kader AK, Li G, Purcell LD, Kim ST, Hsu FC, Stattin P, Hugosson J, Adolfsson J, Walsh PC, Trent JM, Duggan D, Carpten J, Grönberg H, Isaacs WB. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc Natl Acad Sci USA. 2010;107(5):2136–2140. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amin Al Olama A, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, Giles GG, Severi G, Neal DE, Hamdy FC, Donovan JL, Hunter DJ, Henderson BE, Thun MJ, Gaziano M, Giovannucci EL, Siddiq A, Travis RC, Cox DG, Canzian F, Riboli E, Key TJ, Andriole G, Albanes D, Hayes RB, Schleutker J, Auvinen A, Tammela TL, Weischer M, Stanford JL, Ostrander EA, Cybulski C, Lubinski J, Thibodeau SN, Schaid DJ, Sorensen KD, Batra J, Clements JA, Chambers S, Aitken J, Gardiner RA, Maier C, Vogel W, Dörk T, Brenner H, Habuchi T, Ingles S, John EM, Dickinson JL, Cannon-Albright L, Teixeira MR, Kaneva R, Zhang HW, Lu YJ, Park JY, Cooney KA, Muir KR, Leongamornlert DA, Saunders E, Tymrakiewicz M, Mahmud N, Guy M, Govindasami K, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English D, Virtamo J, Le Marchand L, Campa D, Kaaks R, Lindstrom S, Diver WR, Gapstur S, Yeager M, Cox A, Stern MC, Corral R, Aly M, Isaacs W, Adolfsson J, Xu J, Zheng SL, Wahlfors T, Taari K, Kujala P, Klarskov P, Nordestgaard BG, Røder MA, Frikke-Schmidt R, Bojesen SE, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Rinckleb A, Luedeke M, Herkommer K, Meyer A, Serth J, Marthick JR, Patterson B, Wokolorczyk D, Spurdle A, Lose F, McDonnell SK, Joshi AD, Shahabi A, Pinto P, Santos J, Ray A, Sellers TA, Lin HY, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, Chanock S, Gronberg H, Haiman CA, Kraft P, Easton DF, Eeles RA The UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of OncologyThe UK ProtecT Study Collaborators; The Australian Prostate Cancer Bioresource; The PRACTICAL Consortium. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, Diver WR, Witte JS, Hsing AW, Nemesure B, Rebbeck TR, Cooney KA, Xu J, Kibel AS, Hu JJ, John EM, Gueye SM, Watya S, Signorello LB, Hayes RB, Wang Z, Yeboah E, Tettey Y, Cai Q, Kolb S, Ostrander EA, Zeigler-Johnson C, Yamamura Y, Neslund-Dudas C, Haslag-Minoff J, Wu W, Thomas V, Allen GO, Murphy A, Chang BL, Zheng SL, Leske MC, Wu SY, Ray AM, Hennis AJ, Thun MJ, Carpten J, Casey G, Carter EN, Duarte ER, Xia LY, Sheng X, Wan P, Pooler LC, Cheng I, Monroe KR, Schumacher F, Le Marchand L, Kolonel LN, Chanock SJ, Van Den Berg D, Stram DO, Henderson BE. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, Fujioka T, Nakamura Y, Nakagawa H. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 25.Akamatsu S, Takata R, Haiman CA, Takahashi A, Inoue T, Kubo M, Furihata M, Kamatani N, Inazawa J, Chen GK, Le Marchand L, Kolonel LN, Katoh T, Yamano Y, Yamakado M, Takahashi H, Yamada H, Egawa S, Fujioka T, Henderson BE, Habuchi T, Ogawa O, Nakamura Y, Nakagawa H. Common variants at 11q12, 10q26 and 3p11.2 are associated with prostate cancer susceptibility in Japanese. Nat Genet. 2012;44:426–429. doi: 10.1038/ng.1104. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Mo Z, Ye D, Wang M, Liu F, Jin G, Xu C, Wang X, Shao Q, Chen Z, Tao Z, Qi J, Zhou F, Wang Z, Fu Y, He D, Wei Q, Guo J, Wu D, Gao X, Yuan J, Wang G, Xu Y, Wang G, Yao H, Dong P, Jiao Y, Shen M, Yang J, Ou-Yang J, Jiang H, Zhu Y, Ren S, Zhang Z, Yin C, Gao X, Dai B, Hu Z, Yang Y, Wu Q, Chen H, Peng P, Zheng Y, Zheng X, Xiang Y, Long J, Gong J, Na R, Lin X, Yu H, Wang Z, Tao S, Feng J, Sun J, Liu W, Hsing A, Rao J, Ding Q, Wiklund F, Gronberg H, Shu XO, Zheng W, Shen H, Jin L, Shi R, Lu D, Zhang X, Sun J, Zheng SL, Sun Y. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet. 2012;44:1231–1235. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Bälter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang BL, Isaacs WB, Xu J, Grönberg H. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Sun J, Kader AK, Lindström S,Wiklund F, Hsu FC, Johansson JE, Zheng SL, Thomas G, Hayes RB, Kraft P, Hunter DJ, Chanock SJ, Isaacs WB, Grönberg H. Estimation of absolute risk for prostate cancer using genetic markers and family history. Prostate. 2009;69(14):1565–1572. doi: 10.1002/pros.21002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salinas CA, Koopmeiners JS, Kwon EM, FitzGerald L, Lin DW, Ostrander EA, Feng Z, Stanford JL. Clinical utility of five genetic variants for predicting prostate cancer risk and mortality. Prostate. 2009;69(4):363–372. doi: 10.1002/pros.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akamatsu S, Takahashi A, Takata R, Kubo M, Inoue T, Morizono T, Tsunoda T, Kamatani N, Haiman CA, Wan P, Chen GK, Le Marchand L, Kolonel LN, Henderson BE, Fujioka T, Habuchi T, Nakamura Y, Ogawa O, Nakagawa H. Reproducibility, performance, and clinical utility of a genetic risk prediction model for prostate cancer in Japanese. PLoS ONE. 2012;7(10):e46454. doi: 10.1371/journal.pone.0046454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aly M, Wiklund F, Xu J, Isaacs WB, Eklund M, D’Amato M, Adolfsson J, Gr00F6;nberg H. Polygenic risk score improves prostate cancer risk prediction: Results from the Stockholm-1 cohort study. Eur Urol. 2011;60(1):21–28. doi: 10.1016/j.eururo.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kader AK, Sun J, Reck BH, Newcombe PJ, Kim ST, Hsu FC, D’Agostino RB, Jr, Tao S, Zhang Z, Turner AR, Platek GT, Spraggs CF, Whittaker JC, Lane BR, Isaacs WB, Meyers DA, Bleecker ER, Torti FM, Trent JM, McConnell JD, Zheng SL, Condreay LD, Rittmaster RS, Xu J. Potential impact of adding genetic markers to clinical parameters in predicting prostate biopsy outcomes in men following an initial negative biopsy: Findings from the REDUCE trial. Eur Urol. 2012;62(6):953–961. doi: 10.1016/j.eururo.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F, Hsing AW, Wang X, Shao Q, Qi J, Ye Y, Wang Z, Chen H, Gao X, Wang G, Chu LW, Ding Q, OuYang J, Gao X, Huang Y, Chen Y, Gao YT, Zhang ZF, Rao J, Shi R, Wu Q, Wang M, Zhang Z, Zhang Y, Jiang H, Zheng J, Hu Y, Guo L, Lin X, Tao S, Jin G, Sun J, Lu D, Zheng SL, Sun Y, Mo Z, Xu J. Systematic confirmation study of reported prostate cancer risk-associated single nucleotide polymorphisms in Chinese men. Cancer Sci. 2011;102(10):1916–1920. doi: 10.1111/j.1349-7006.2011.02036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Liu F, Hsing AW, Wang X, Shao Q, Qi J, Ye Y, Wang Z, Chen H, Gao X, Wang G, Chu LW, Ding Q, OuYang J, Gao X, Huang Y, Chen Y, Gao YT, Zhang ZF, Rao J, Shi R, Wu Q, Zhang Y, Jiang H, Zheng J, Hu Y, Guo L, Lin X, Tao S, Jin G, Sun J, Lu D, Zheng SL, Sun Y, Mo Z, Yin C, Zhang Z, Xu J. Replication and cumulative effects of GWAS-identified genetic variations for prostate cancer in Asians: A case–control study in the ChinaPCa consortium. Carcinogenesis. 2012;33(2):356–360. doi: 10.1093/carcin/bgr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.wing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, Gielzak M, Partin AW, Shanmugam V, Izatt T, Sinari S, Craig DW, Zheng SL, Walsh PC, Montie JE, Xu J, Carpten JD, Isaacs WB, Cooney KA. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366(2):141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin X, Qu L, Chen Z, Xu C, Ye D, Shao Q, Wang X, Qi J, Chen Z, Zhou F, Wang M, Wang Z, He D, Wu D, Gao X, Yuan J, Wang G, Xu Y, Wang G, Dong P, Jiao Y, Yang J, Ou-Yang J, Jiang H, Zhu Y, Ren S, Zhang Z, Yin C, Wu Q, Zheng Y, Turner AR, Tao S, Na R, Ding Q, Lu D, Shi R, Sun J, Liu F, Zheng SL, Mo Z, Sun Y, Xu J. A novel germline mutation in HOXB13 is associated with prostate cancer risk in Chinese men. Prostate. 2012 doi: 10.1002/pros.22552. (Online published) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kader AK, Sun J, Isaacs SD, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69(11):1195–1205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.