Abstract
Extracellular vesicles (EVs) have emerged as important mediators of intercellular communication in cancer, including by conveying tumor-promoting microRNAs between cells, but their regulation is poorly understood. In this study, we report the findings of a comparative microRNA profiling and functional analysis in human glioblastoma (GBM) that identifies miR-1 as an orchestrator of EV function and GBM growth and invasion. Ectopic expression of miR-1 in GBM cells blocked in vivo growth, neovascularization and invasiveness. These effects were associated with a role for miR-1 in intercellular communication in the microenvironment mediated by EVs released by cancer stem-like GBM cells. An EV-dependent phenotype defined by GBM invasion, neurosphere growth and endothelial tube formation was mitigated by loading miR-1 into GBM-derived EVs. Protein cargo in EVs was characterized to learn how miR-1 directed EV function. The mRNA encoding Annexin A2 (ANXA2), one of the most abundant proteins in GBM-derived EVs, was found to be a direct target of miR-1 control. In addition, EV-derived miR-1 along with other ANXA2 EV networking partners targeted multiple pro-oncogenic signals in cells within the GBM microenvironment. Together, our results showed how EV signalling promotes the malignant character of GBM and how ectopic expression of miR-1 can mitigate this character, with possible implications for how to develop a unique miRNA-based therapy for GBM management.
Keywords: glioblastoma, microRNA, extracellular vesicles, tumor microenvironment, AnnexinA2
INTRODUCTION
Glioblastoma multiforme (GBM), the most common and aggressive primary brain tumor, is diagnosed in approximately 10,000 new cases per year in the USA (1), with a median survival of approximately 14 months (2). GBM is characterized by cellular heterogeneity, rapid proliferation, angiogenesis and extensive invasion (3). Concomitant high proliferation and infiltration constitutes the major challenge for GBM therapy, because even with extensive resection tumor cells are left behind in the brain, leading to recurrence. Angiogenesis constitutes a major advantage to rapidly growing tumors by providing oxygen and nutrients, yet anti-angiogenic treatment may paradoxically increase invasiveness (4). Thus novel therapies aimed at blocking GBM growth, as well as curbing its invasive and angiogenic potential, are needed.
GBM displays a high level of intratumoral cellular heterogeneity, and has been shown to contain subpopulations of tumorigenic cells with stem cell-like properties (5). The tumor vasculature provides a specialized niche for stem-like tumor cells (6). Moreover, these tumors modify normal cells in their environment to promote tumorigenicity in various ways (7) and tumor associated cells such as vascular cells, microglia, peripheral immune cells, and neural precursors, also play important roles in the microenvironment (8). Increasing evidence shows that a crucial mode of cell-cell communication within tumors is the release and uptake of small extracellular vesicles (EVs), that contain a subset of cellular proteins and RNAs (9, 10). EVs are taken up by cells and can change their phenotype by delivery of tumor derived RNAs, including microRNAs (miRs) (11). In recent years, numerous reports suggested an important role for EV communication in GBM, including the transfer of miRs (9, 12, 13).
MiRs are small, non-coding RNAs that act as regulators of gene expression by reducing translation of target mRNAs with partial complementarity in their 3'-untranslated regions (UTRs) (14). MiR deregulation is a general feature of cancer, including GBM, and is associated with both tumor suppressor and oncogenic effects (15, 16).
MiR-1 has been shown to function as a tumor suppressor in several human cancers (17). In this study we report that miR-1 is novel tumor suppressor in GBM. We describe multi-level anti-tumorigenic effects of miR-1 restoration, including signaling by EVs. Multiple oncogenic signaling pathways were inactivated by miR-1, demonstrating the distinctive ability of a single miR to simultaneously inhibit numerous targets, both directly and indirectly. Moreover, we demonstrate that the pro-oncogenic effect of GBM-derived EVs was alleviated by miR-1, which directly targets ANXA2, one of the most abundant EV proteins. ANXA2 is upregulated in various tumor types (18), including GBM (19) where it plays a critical role in driving invasion and progression (20). Our results show that miR-1 delivery reduces GBM tumorigenicity and impairs EV-based microenvironmental communication, suggesting that such communication may be a candidate for therapeutic treatment of GBM.
METHODS
Human Specimens
All tumor samples were obtained as approved by the institutional review board at The Ohio State University (OSU). Surgery was performed by E.A.C or I.N. For each patient, samples of both tumor and adjacent brain devoid of gross tumor were processed for extraction of total RNA (Trizol, Invitrogen) and protein (Cell Lysis Buffer, Cell Signaling) or establishment of patient derived neurospheres.
Cell Culture
U87, U251, U373, Gli36 malignant GBM (American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) with 10% or 2% fetal calf serum (FBS, Sigma). Human brain micro-vessels endothelial cells (HBMVEC) were cultured in endothelial cell basal medium supplemented with 1% endothelial cell growth supplement and 5% FBS. The normal human astrocytes (NHA, Clonetics) were cultured in astrocyte basal medium supplemented with 1% astrocyte growth supplement and 2% FBS (Lonza). Primary human GBM cells X12 were obtained as previously reported (21, 22). For neurosphere culture cells were grown in stem cell medium, consisting of Neurobasal/Glutamax (Invitrogen) supplemented with 1% N2 and 2% B27 (Invitrogen) and 20 ng/mL epidermal growth factor and fibroblast growth factor–2 (PeproTech). Cells were grown as adherent monolayers in poly-l-ornithine and laminin-covered dishes (Invitrogen) and as neurospheres in stem cell medium.
Vector construction and transfection
The lentiviral pCDH-GFP empty vector and miR-1-1 expressing vector was purchased from System Biosciences. The lentiviral constructs along with lentiviral packaging constructs (System Biosciences) were used for establishing U87 and X12 cell lines constitutively overexpressing GFP or GFP/miR-1, according to the manufacturer's instruction. For each construct, three independent clones were created and expression of miR-1 was validated by qPCR. The RFP CD63 lentiviral particles from System Bioscience were used for creating U87 and X12 cell lines overexpressing RFP CD63.
The 3’-UTR encompassing the target sequence for miR-1 of ANXA2 cDNA were cloned into the pMIR-REPORT vector (Ambion). For the mutated construct of the QuickChange Site-Directed Mutagenesis Kit (Stratagene) was used according to manufacturer's protocol to alter the miR-1 seed sequence. Luciferase reporter assays were performed as previously described (23) using luciferase reagent (Promega). EVs loaded with miR (NC or miR-1) were used for luciferase assays at a concentration of500 EVs per cell. Cells were treated with EVs 24h prior to reporter transfection. Transfection (25-75 nmol/L) of negative control (NC) and pre-miR-1 (miR-1) or pre-miR-1 FAM labeled (miR-1 FAM) (Ambion), or pMIR-REPORT was done with Lipofectamine2000 (Invitrogen).
In vivo studies
Female immunodeficient mice were purchased from Taconic. For all studies the mice were housed in animal facility at the OSU in accordance with all NIH regulations. All protocols were approved by the OSU Institutional Animal Care and Use Committee. In vivo studies were performed as previously described (24) (see Supplemental Experimental Procedures). Tumors from flank and brains were placed in 4% paraformaldehyde for 24 h, then in 30% sucrose for 48 h. Sections of 20 μm were evaluated for Ki67 (Abcam), cleaved caspase-3 (Cell Signaling), CD31 (BD Pharminogen) and Lectin (Invitrogen) immunostaining or green/red fluorescence. For quantification of staining/fluorescence three sections per tumor were analyzed.
In vitro 2D and 3D assays
3D spheroid migration assay in collagen matrix and its quantification were performed as previously described (23). The vessel-forming ability of HBMVEC was characterized in vitro using a Matrigel assay (25) (see Supplemental Experimental Procedures). Propidium iodide exclusion and flow cytometry-based cell-cycle analysis was carried out using the Becton Dickinson FACSCalibur system.
Purification of EVs
To isolate EVs, U87 and X12 cells were cultured for 2 days in EV free medium without antibiotics. The conditioned media were collected and EVs were isolated by differential centrifugation as previously described (26) (see Supplemental Experimental Procedures).
Proteomic analysis
All mass spectra were acquired at the Bioproximity LLC. Proteins were prepared for digestion using the filter-assisted sample preparation (FASP) method (27) (see Supplemental Experimental Procedures).
Protein extraction and Western blot analysis was done as described previously (28). Representative images from two or three independent experiments are shown. Antibodies used were as follows: ANXA2 (1:1000, Santa Cruz), CD133 (1:1000, Amersham), BMI1 and GFAP (1:1000, Millipore) Akt and pAKT Ser473, ERK and pERK Thr202/Tyr204Y, JNK and pJNK Thr183/Tyr185, MET and p-MET Tyr1234/1235, EGFR, PDGFRA, SUZ12, FASN (1:2000, Cell Signaling), YWHAZ and CD63 (1:1000, Santa Cruz), CD9 (1:500, Novus) and a Tubulin (1:10,000, Sigma Aldrich).
Microscopy
All fluorescent and light microscopy based assays were monitored using a Zeiss LSM510 confocal microscope system (Carl Zeiss Inc.).Ultrathin frozen sections and immunogold labeled CD63 antibody were prepare in Cellular Neuroscience Core Laboratory. The Transmission Electron microscopy Tecnai G2 Spirit BioTWIN or with AMT 2k CCD camera was used to analyze EVs stained with immunogold labeled anti CD63 antibody in Electron Microscopy Facility at Harvard Medical School.
Quantitative PCR
Total RNA was extracted using Trizol (Invitrogen) and treated with RNase-free DNase (Qiagen). Mature and pri-miR expression analysis by qPCR was carried out using the miR real-time PCR detection kit (Applied Biosystems) as described (23). Messenger RNA expression analysis was carried out using Power SYBR Green (Applied Biosystems). RNA concentration was quantified using Nanodrop RNA 6000 nano-assays and analyzed using Agilent 2100 Bioanalyzer total and Pico RNA platform.
Data and Statistical Analysis
All microscopy-based assays were edited/quantified using ImageJ (http://rsb.info.nih.gov.ezpprod1.hul.harvard.edu/ij/), including the Analyze Skeleton plugin and Analyze Particles function of binary images with automatic threshold. Pathway analysis was performed using Ingenuity Pathways Analysis (IPA, http://www.ingenuity.com/) and David Functional Annotation tools (http://david.abcc.ncifcrf.gov/). Results are expressed as mean ± SD, and differences were compared using the 2-tailed Student's t-test. Statistical analyses were performed using Microsoft Office Excel 2010 or Graph Pad Prism 6 software. P < 0.05 was considered statistically significant (single asterisks in the Figures), and P < 0.01 was strongly significant (double asterisks).
RESULTS
MiR-1 is downregulated in human GBMs and in GBM cells during migration in vitro
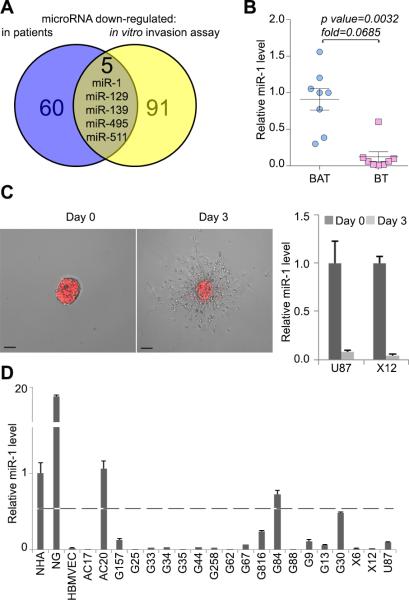
Our previously published microarray study indicated that miR-1 levels were significantly reduced when compared to adjacent brain tissue from the same individual (29). More recently, our group also demonstrated, that the expression of miR-1 was shut off over a three day in vitro spheroid migration assays (23). In fact, miR-1 was one of five miRs downregulated in both arrays (Fig.1A). To validate the microarray data from the first study (29), we tested miR-1 expression in a panel of eight patient GBM samples with matched adjacent brain by quantitative real time (qPCR) (Fig.1B). This confirmed that there was significant downregulation of miR-1 in GBM compared to surrounding brain. The microarray data from the second study (23) was also validated by showing that miR-1 was downregulated in U87 and X12 GBM cells during a three days migration assay (Fig.1C). Finally, we assessed miR-1 expression in non-malignant astrocytes, neuro-glia, and endothelial brain microvasculature, as well as in a broad panel of GBM and brain tumor-derived cell lines (primary GBM stem-like cells from grade III and IV tumors, and established GBM cell lines). Expression was moderate in astrocytes, high in neuro-glia and very low in endothelial cells. In 17 out of 20 malignant cell lines, miR-1 expression was low or undetectable (Fig.1D). The identification of low levels of miR-1 in tumor tissue and tumor cells, as well as a reduction in migrating cells strongly suggests that miR-1 may play a tumor suppressive role in GBM.
FIGURE 1. MiR-1 is downregulated in patient GBM specimens and in GBM cells during migration in vitro.
A. Venn diagram depicting the overlap between miRs identified as downregulated in GBM specimens compared to matching (i.e. from the same individual) adjacent brain (blue) and miRs identified as downregulated in GBM cells in time course of a migration assay (yellow).
B. Relative expression of miR-1 was validated by qPCR in GBM brain tumor (BT) specimens versus matching brain adjacent to tumor (BAT) (n = 8). Values are expressed as mean relative miR-1 expression level ± SD.
C. Representative images of RFP-X12 GBM cells in the migration assay at day 0 and day 3 are shown. RNA was extracted from multiple spheroids for subsequent analysis. Scale bars: 200μm. Relative expression of miR-1 was validated by qPCR in U87 and X12 GBM cell lines at day 0 and day 3 of migration in the spheroid assay. Values are expressed as mean relative miR-1 expression level ± SD.
D. Relative expression of miR-1 was validated by qPCR in: normal human astrocytes (NHA), neuroglia (NG), human brain microvasculature endothelial cells (HBMVEC) and collection of GBM primary stem cells and established cell line U87. Values are expressed as mean relative miR-1 expression level ± SD. Dashed line shows 50% of expression measured in astrocytes.
Overexpression of miR-1 in GBM cells reduces tumorigenicity, angiogenesis and invasion in vivo
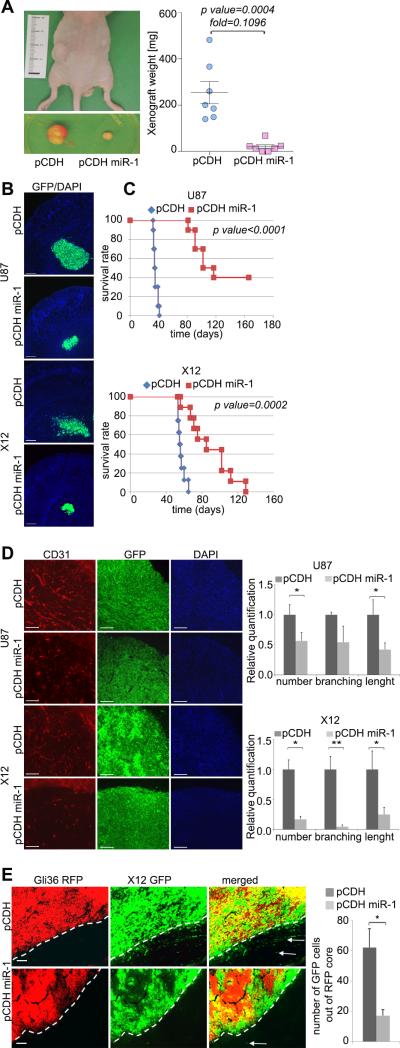
Based on the above findings, we hypothesized that miR-1 may act as a tumor suppressor. To investigate the function of miR-1 in glioblastoma we stably expressed miR-1 using a lentiviral expression vector. U87 flank xenografts expressing miR-1 were significantly smaller than controls (Fig.2A) and there was significant reduction in the proliferation index (Supplementary Fig.S1A). The decreased growth was not due to apoptosis because cleaved Caspase-3 was not significantly elevated in cells expressing miR-1 (Supplementary Fig.S1B). Interestingly, the recruitment of endothelial cells measured by CD31 staining was significantly decreased, suggesting that impaired angiogenesis is at least partially responsible for the observed growth inhibition mediated by miR-1 in vivo (Supplementary Fig.S1C). Next, we evaluated GBM cell tumorigenicity by intracranial implantation of U87 and X12 cells stably expressing miR-1. These tumors were significantly smaller than controls (Fig.2B). Survival analysis demonstrated a significantly better outcome of animals injected with miR-1 expressing cells. In U87 cells we observed a substantial subgroup of long-term survivors (Fig.2C). Analysis of neovascularization was performed on symptomatic animals at 5 weeks (U87) or 7 weeks (X12) after tumor implantation. MiR-1 expressing tumors displayed reduced CD-31 staining, with shorter average branch length and fewer prominent branches (Fig.2D). These results were confirmed by lectin staining (Supplementary Fig.S1D). Finally, tumors formed by invasive X12 cells were smaller and also less diffusely infiltrative when miR-1 was expressed (Fig.2B). To quantify invasiveness in vivo, non-invasive RFP-Gli36 cells were co-injected with invasive X12 cells stably expressing GFP alone or GFP/miR-1. Control X12 cells readily migrated out of the RFP positive tumor core, while those expressing miR-1 were predominantly localized intratumorally (Fig.2E). These results thus link miR-1 to a tumor suppressor role by inhibition of growth, invasiveness and angiogenesis of GBM.
FIGURE 2. Overexpression of miR-1 in GBM cells mitigates tumorigenicity, reduces invasiveness and angiogenesis in vivo.
A. Representative images of tumor xenografts before and after excision from mice injected with U87 cells stably expressing pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 vector (pCDH miR-1). Tumor mass was quantified (n = 7), and the data are expressed as mean ± SD.
B. Representative images of intracranial tumors formed by U87 (2 weeks after injection) and X12 (3 weeks after injection) GBM cells stably expressing pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 vector (pCDH miR-1). Scale bars: 250μm.
C. Kaplan–Meier survival curve of animals injected with GBM tumor cells U87 (upper) and X12 (lower). Cells were stably expressing pCDH-GFP control vector (pCDH) (U87: n = 10; X12: n= 8) or pCDH-GFP miR-1 vector (pCDH miR-1) (U87: n = 10; X12: n= 9). Log-rank test was used to compare the survival probabilities between the two groups.
D. Angiogenesis was determined by CD31 (red) and DAPI (blue) staining of intracranial tumors formed by U87 (5 weeks after injection) and X12 (7 weeks after injection) GBM cells stably expressing pCDH GFP control vector (pCDH) or pCDH-GFP miR-1 vector (pCDH miR-1). Scale bars: 150μm. Data are expressed as mean ±SD, *P < 0.05, **P < 0.01.
E. Invasiveness in vivo was determined by co-injection of non-invasive RFP-Gli36 cells with invasive GFP-X12 cells stably expressing either pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 vector (pCDH miR-1). Scale bars: 100μm. Data are expressed as mean ±SD, *P < 0.05.
Overexpression of miR-1 in GBM cells impairs neurosphere formation and migration through multiple effectors
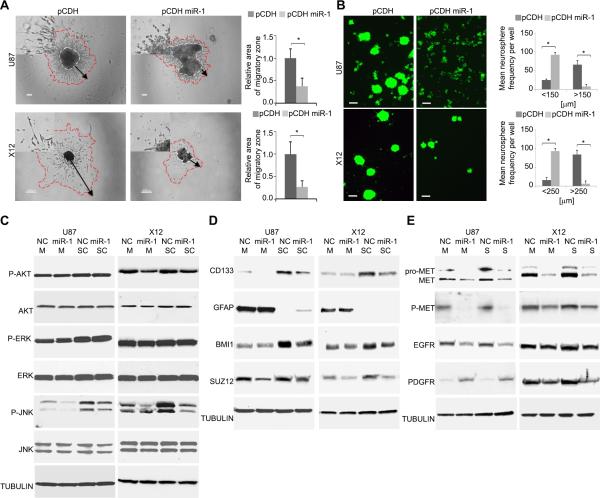
We became interested in exploring the effect of mR-1 on GBM cell invasion. We first employed in vitro adhesion assays and observed that miR-1 expressing cells were poorly adhesive, (Supplementary Fig.S2A) and cell-cell adhesiveness assays quantified the significance of this phenotype (Supplementary Fig.S2B). Indeed, miR-1 expressing cells displayed impaired cell-cell attachment in 2D (Supplementary Fig.S2C) and 3D assays (Supplementary Fig.S2D). Moreover, miR-1 expression reduced infiltration in a 3D collagen matrix with cells displaying a cuboidal, non-invasive morphology (Fig.3A with insets). Reduced migration was also confirmed by a wound healing assay (Supplementary Fig.S2E). In contrast to the effects on migration, there was no effect of miR-1 on the cell cycle (with the exception of a small but non-significant increase in the apoptotic fraction in U87 cells (Supplementary Fig.S2F). Interestingly, GBM cells cultured in stem-like conditions had significantly impaired neurosphere formation upon miR-1 expression, both in stably infected (Fig.3B) and transiently transfected cells (Supplementary Fig.S2G). Therefore, miR-1 expression reduced GBM cell to cell and cell to matrix adhesion leading to reduced neurosphere sizes and inhibited in vitro invasion.
FIGURE 3. Overexpression of miR-1 in GBM cells impairs cell-cell adhesion, spheroid migration and neurosphere formation by affecting multiple effectors.
A. Migration of GBM cells was measured by a spheroid dispersal assay. Representative images of spheroid migration of U87 (upper panels) and X12 (lower panels) GBM cells transiently transfected with either negative control miR (NC) or miR-1. Scale bars: 100μm (upper panels) and 200μm (lower panels). The insets in all panels are magnified × 2.5. Migratory zones were quantified after indicated time, expressed as mean ± SD. *P < 0.05.
B. Neurosphere formation capacity was determined by a self-renewal assay. Representative images of U87 and X12 cells stably expressing pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 vector (pCDH miR-1). Scale bars: 200μm. Neurosphere frequency were quantified after 72h, expressed as mean ± SD. *P < 0.05.
C. Cellular signaling was monitored by Western blot analysis of U87 and X12 cell lines cultured as monolayer (M) or stem cell-like neurospheres (SC). Cells were transiently transfected with either negative control miR (NC) or miR-1. Cell lysates were blotted with anti-phospho-specific antibodies, and compared with total kinase antibodies. Tubulin was used as a loading control.
D. Stemness was monitored by Western blot analysis of U87 and X12 cell lines cultured as monolayer (M) or stem cell-like neurospheres (SC). Cells were transiently transfected with either negative control miR (NC) or miR-1. Cell lysates were blotted with indicated antibodies. Tubulin was used as a loading control.
E. Expression of cellular receptors was monitored by Western blot analysis of U87 and X12 cell lines cultured as monolayer (M) or stem cell-like neurospheres (SC). Cells were transiently transfected with either negative control miR (NC) or miR-1. Cell lysates were blotted with anti-phospho-specific antibodies and with total protein antibodies. Tubulin was used as a loading control.
The above results showed that miR-1 expression led to a variety of phenotypic changes. We thus hypothesized that miR-1 could be de-regulating multiple signaling pathways linked to GBM progression. We thus investigated the status of such oncogenic signaling in GBM cells transfected with miR-1 and cultured either as monolayers or in “stem-like” neurosphere conditions. We observed reduced level of phospho AKT (X12 only) but no changes in phospho-ERK levels (Fig.3C). However phospho-JNK levels were consistently reduced, especially in stem-like conditions (where basal phospho-JNK levels were elevated). Stem-like culture conditions caused a significant increase in levels of the putative glioma stem cell marker CD133, as well as an increase in stem cell self-renewal factorsBMI1 and SUZ12. Levels of the astrocyte lineage marker GFAP were significantly reduced under these neurosphere culture conditions. MiR-1 transfection abrogated these effects of neurosphere culture, and reduced levels of CD133, BMI1 and SUZ12, while increasing levels of differentiation marker GFAP (Fig.3D). Interestingly, cells transferred from monolayer culture to stem-like conditions had significantly reduced endogenous expression of miR-1 (Supplementary Fig.S2H). Finally, we analyzed the effect of miR-1 expression on cellular receptors known to play a crucial role in GBM cell biology. We observed elevated levels of and the known miR-1 target MET (30, 31) under neurosphere conditions. As expected, pro-MET, MET and phospho-MET levels were significantly reduced by miR-1 expression. Also EGFR level was diminished in miR-1 expressing cells, while PDGFR level was increased in U87 and reduced in X12 cells, suggesting cell type specific effects (Fig.3E). These results thus indicated that miR-1 inhibited multiple signaling pathways, associated with “stemness” of GBM stem-like cells.
MiR-1 expression blocks EV stimulation of GBM invasion and growth
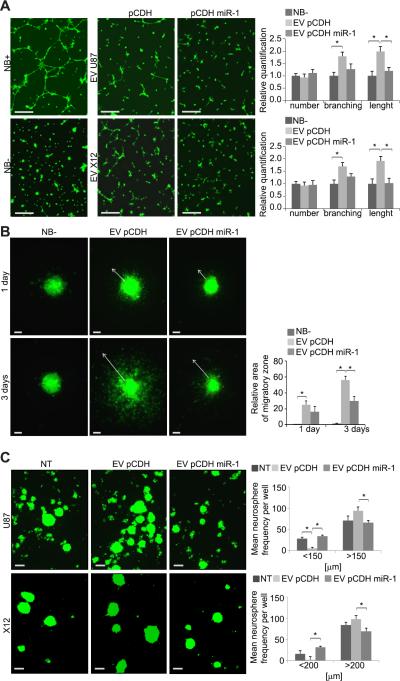
The findings showing reduced invasion and angiogenesis in miR-1 expressing cells suggested that it influences the tumor microenvironment. Thus we hypothesized that miR-1 could be acting on intercellular communication via EVs which have been shown to be important mode of release biomolecules by GBM cells (13). To investigate the role of paracrine communication via EVs in neovascularization, we used a tube formation assay using recipient brain microvascular endothelial cells (HBMVEC) (32). These cells form tube-like structures in growth factors supplemented medium (Fig.4A, left panels). HBMVEC treated with GBM neurospheres-derived EVs (donor cells) formed significantly longer and more branched tube-like structures than controls. This effect was significantly reduced in the presence of EVs collected from miR-1 expressing neurospheres compared with controls (Fig.4A, right panels).
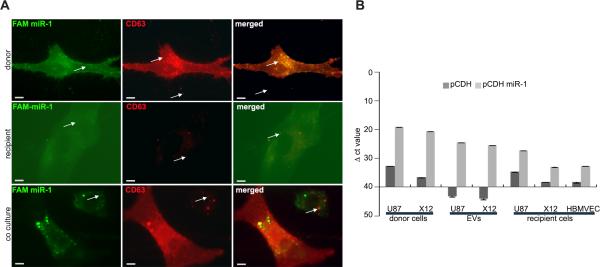
FIGURE 4. The EV-dependent phenotype is mitigated by miR-1.
A. Tube-like formation of HBMVEC in unsupplemented (NB-) and supplemented (NB+) Neurobasal medium and upon the presence of EVs was monitored by 3-D Matrigel assay. EVs were collected from U87 (EV U87) and X12 (EV X12) cells stably infected with either pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 expressing vector (pCDH miR-1). Representative images are shown. Scale bars: 250μm. Data are expressed as mean ±SD, *P < 0.05.
B. Migration of GBM stem-like neurospheres upon the presence of NB- medium neurospheres upon the presence of EVs was measured by spheroid dispersal assay.
Representative images of non-transfected X12 spheroid migration after indicated time are shown. EVs were collected from X12 cells stably expressing pCDH-GFP control vector (EV pCDH) or pCDH-GFP miR-1 vector (EV pCDH miR-1). Scale bars: 100μm. Migratory zones were quantified after indicated time, expressed as mean ± SD. *P < 0.05.
C. Neurosphere formation capacity in the presence of EVs was determined by self-renewal assay. Representative images of GFP-labeled U87 and X12 cells either non-treated (NT) or cultured in the presence of EVs are shown. EVs were collected from corresponding cell lines stably expressing pCDH-GFP control vector (EV pCDH) or pCDH-GFP miR-1 vector (EV pCDH miR-1). Scale bars: 200μm. Neurosphere frequency were quantified after 72h, expressed as mean ± SD. *P < 0.05.
Next, we tested the effect of EVs on migration of stem-like neurospheres. We found that the presence of EVs (collected from donor neurospheres) strongly promoted recipient cell migration in spheroid assays (Fig.4B). This effect was significantly diminished using EVs collected from miR-1 expressing donor cells, both in terms of distance travelled by the cells and number of cells leaving the spheroid core (Fig.4B, Supplementary Fig. S3A). We also found that EVs derived from control neurosphere donor cells promoted neurosphere formation of recipient cells; while EVs collected from miR-1 expressing donor cells had no significant effect (Fig.4C). Expression of miR-1 in recipient cells also prevented EV promotion of neurosphere formation (Supplementary Fig. S3B). Thus expression of miR-1 prevents EV stimulation of angiogenesis, invasion and neurosphere formation in recipient cells.
As a control, we confirmed that miR-1 expression did not alter expression of the EV marker CD63 (Supplementary Fig. S3C). Interestingly, miR-1 expression did lead to a small but significant decrease in EV size (Supplementary Figure S3D), but miR-1 expression did not alter the number of EVs secreted by GBM cells (Supplementary Figure S3E). Additional characterization of EVs released by GBM stem-like cells showed that EV fractions lacked larger RNAs compared to total cellular RNA (Supplementary Fig. S3F); and EVs collected from miR-1 expressing cells showed increased levels of RNAs smaller than 40nt (Supplementary Fig. S3G). Therefore, we conclude that observed effects of EVs derived from miR-1 expressing donor cells on recipient cells were more likely due to such EVs cargo and not to a difference in EV quantity or release from donor cells.
MiR-1 is transferred between GBM cells by EV transport
We next asked whether miR-1 itself could be transferred by GBM-derived EVs. To visualize EV transfer we created cell lines expressing RFP-CD63 and used them as donor cells. After treatment with EVs derived from RFP-CD63 expressing cells we detected RFP inside GFP-labeled recipient cells (Supplementary Fig. S4A). To visualize miR-1 transfer, we used U87 cells co-transfected with FAM-labeled miR-1 and RFP-labeled CD63 as EV donor cells. We observed that a substantial fraction of FAM-miR-1 co-localized with RFP-CD63, but also that a fraction of FAM-miR-1 remained in the donor cells cytoplasm (Fig.5A, upper panels). EVs released by FAM-miR-1/RFP-CD63 transfected cells were then added to recipient, non-fluorescent U87 cells. We observed partial co-localization of FAM-miR-1 with RFP-CD63 in the cytoplasm of recipient cells (Fig.5A, middle panels; Supplementary Fig.S4B). A similar result was observed in co-culture experiments (Fig.5A, lower panels) strongly suggesting that miR-1 can be transferred between cells through EVs. To survey the transfer of miR-1 from donor to recipient cells, we performed qPCR analysis on RNA extracted from donor cells (U87, X12), as well as their corresponding EVs and recipient cells. We observed a significant increase of mature miR-1 levels in all tested samples (Fig.5B), but the primary transcript was not detected in recipient cells (Supplementary Fig. S4C). This suggests that the observed increase of mature miR-1 levels in recipient cells is due to EV transfer, not to increased endogenous miR-1 expression. These observations support the notion that the miR-1 dependent paracrine effect is at least partially mediated via EV transfer.
FIGURE 5. MiR-1 is transferred between GBM cells by EV transport.
A. Uptake of GBM-derived EVs was monitored using co-labeled EVs and miR. Donor U87 cells were stably infected with lentiviral particles of RFP-CD63 and transiently transfected with FAM labeled miR-1. Representative images show partial co-localization of miR-1 (green) and EVs (red) in donor cells (upper panels) in recipient U87 cells treated with EVs from donor cells (middle panels) and in donor and recipient cells in co-culture (lower panels). Scale bars, 50 μm. Arrows indicate FAM signal alone (left panels), RFP signal alone (middle panels) and both signals co-localized (right panels).
B. Expression of miR-1 in donor cells, EVs and recipient cells was validated by qPCR. RNA was isolated from: donor cells (U87, X12) stably infected with either pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 expressing vector (pCDH miR-1), EVs collected from donor cells, and recipient U87, X12 and HBMVEC cells upon the treatment with EVs. Data is shown as the mean raw ΔCt value ± SD.
MiR-1 overexpression affects the EV protein cargo by direct and indirect targeting
The EV-mediated phenotype observed in recipient cells suggested an active role of the EV molecular cargo. Since microRNAs modulate the cellular proteome, we characterized the EV protein cargo in miR-1 expressing cells by global mass-spectrometry analysis of EVs. We identified 1038 proteins of which 462 were significantly downregulated and 11 upregulated in miR-1 expressing U87 cells compared with controls (Fig.6A, Table S1). “RNA binding” and “Vesicles” were among the most abundant functional annotations among these proteins and enzymes and transporters were among the most prominent types of proteins found in GBM EVs (Supplementary Fig. S5A, B). The most abundant networks identified were cancer-related and cell survival was among the most significant molecular process connected to proteins found in GBM EVs (Table S2). Among identified proteins, 205 were putative targets of miR-1 and among these 84 were downregulated in EVs collected from miR-1 expressing cells (Supplementary Fig. S5C, D). This was confirmed for three putative targets by Western analysis, which showed that ANXA2, FASN and 14-3-3ζ were significantly reduced in EVs collected from miR-1 expressing cells (Fig.6B). These results thus showed that miR-1 expression led to changes in the EV proteome, linked to cancer-related signaling networks.
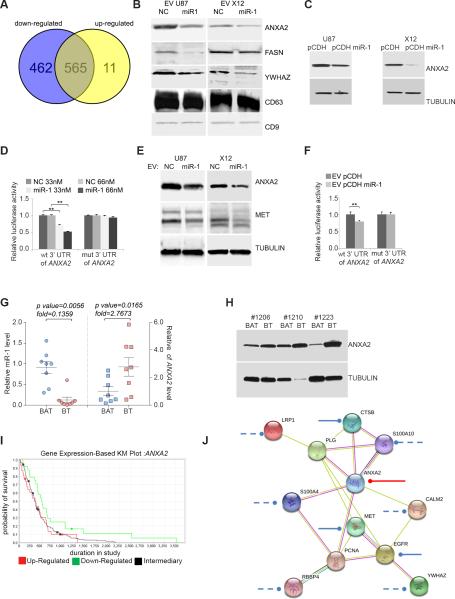
FIGURE 6. MiR-1 directly targets EV ANXA2 overexpressed in human GBM.
A. Venn diagram depicting differential protein composition of EV's derived from U87 cells stably infected with pCDH-GFP miR-1 expressing vector (downregulated: blue, upregulated: yellow) compared to EVs derived from U87 cells stably infected with pCDH-GFP control vector.
B. EV proteins were validated by Western blotting analysis. EVs were derived from U87 and X12 cells lines stably infected with either pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 expressing vector (pCDH miR-1). EV lysates were blotted with specific antibodies against indicated proteins. CD9 and CD63 were used as a loading control.
C. Effects of miR-1 on the expression of ANXA2 were validated by Western blotting analysis. U87 and X12 GBM cell lines were stably expressing pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 expressing vector (pCDH miR-1). Cell lysates were blotted with antibodies against ANXA2. Tubulin was used as a loading control.
D. Direct targeting of ANXA2 3′UTR by miR-1 was validated using a luciferase/3’UTR reporter assay. COS7 cells were co-transfected with luciferase/ANXA2 wild type 3′UTR reporter vector (wt) and33 nM or 66 nM negative control miR (NC) or miR-1. A reporter vector with a mutated miR-1 binding site in the ANXA2 3′UTR (mut) was used as a control. Luciferase levels are expressed as mean relative to controls ± SD; **P < 0.01.
E. Effects of EV-carried miR-1 on the expression of ANXA2 and MET were validated by Western blotting analysis. U87 and X12 cells were exposed to the presence of EVs derived from corresponding cells transiently transfected with either negative control miR (NC) or miR-1 oligonucleotides. Cell lysates were blotted with antibodies against ANXA2, MET and tubulin as a loading control.
F. Direct targeting of ANXA2 3′UTR by EV-carried miR-1 was validated using luciferase/3’UTR reporter assay. U87 and X12 cells were exposed to the presence of EVs derived from corresponding cells transiently transfected with either negative control miR (NC) or miR-1 and after 24h transfected with luciferase/ANXA2 3′UTR wild type (wt) and mutant (mut) reporter vector. Luciferase levels are expressed as mean relative to controls ± SD; **P < 0.01.
G. Relative expression of miR-1 (left) and ANXA2 mRNA (right) levels were validated by qPCR in GBM brain tumor (BT) specimens vs. matching brain adjacent to tumor (BAT) (n = 8). Values are expressed as mean relative miR-1 expression level ± SD.
H. ANXA2 protein level was validated in GBM by Western blotting analysis. Cell lysates from GBM brain tumor (BT) specimens vs. matching brain adjacent to tumor (BAT) (n = 3) were blotted with antibodies against ANXA2. Tubulin was used as a loading control.
I. Association of ANXA2 expression with patient survival (Kaplan–Meier [KM] plot) in GBM. Data was obtained from The Cancer Genome Atlas. Upregulated (n = 31), downregulated (n = 24), and intermediary samples (n = 156) were analyzed. Up- vs. downregulated P = 0.05; up-regulated vs. intermediary P = 0.7134; down-regulated vs. intermediary P = 0.0147.
J. MiR-1 dependent targeting of EV ANXA2 network. ANXA2 partners were selected from EV proteins carried differentially in miR-1-dependent manner. Experimentally validated miR-1 targeting of ANXA2 is shown as a red line; targeting based on published data are shown as a solid blue lines, putative targeting based on target prediction software only are shown as a dashed lines. Networking was analyzed with STRING software.
Interestingly, ANXA2 was identified as the most abundant EV protein (Table 1) and its mRNA 3’UTR contains a conserved miR-1 target site (Supplementary Fig. S5E). ANXA2 was previously implicated as an important pro-oncogenic factor in GBM, promoting proliferation, invasion and angiogenesis (20). The observed miR-1 dependent phenotype was thus consistent with ANXA2 downregulation, providing a biologic rationale for exploring a link between miR-1 and ANXA2. We thus proceed to verify the link between miR-1 and ANXA2. The level of ANXA2 in two cell lines stably expressing miR-1 was significantly reduced compared with that of controls (Fig.6C). Similarly downregulation of miR-1 was observed in a panel of five cell lines transfected with miR-1 (Supplementary Fig. S5F). Direct targeting of ANXA2 by miR-1 was demonstrated using a luciferase reporter assay with the 3’UTR of ANXA2 and, in addition, mutagenesis of the predicted miR-1 binding site abolished the suppression of luciferase by miR-1(Fig.6D, Supplementary Fig. S5E). ANXA2 in conditioned medium was detected almost exclusively in the EV compartment (Supplementary Fig. S5G) and stem-like culture conditions caused a significant increase in ANXA2 levels, which was reflected in its EV content. This was strongly reduced by miR-1 expression (Supplementary Fig. S5H). This data thus confirmed that ANXA2 was a direct mRNA target of miR-1 and that miR-1 expression resulted in decreased ANXA2 levels in EVs.
TABLE 1.
ANXA2 is the most abundant protein in GBM-derived EVs.
| Top deregulated molecules indentified in EVs | ||
|---|---|---|
| Name | Hits NC/miR-1 | base 2 log of ratio |
| ANXA2 | 1679/232 | 2.9 |
| MYH9 | 709/107 | 2.7 |
| FASN | 99/1 | 5.6 |
| UBA1 | 59/0 | 5.9 |
| PRSS3 | 1/22 | −3.5 |
| IGHG4 | 0/11 | −3.6 |
| COL1A2 | 0/8 | −3.2 |
| LGALS7/LGALS7B | 0/6 | −2.8 |
To further demonstrate the functional consequences of EV-mediated miR-1 transfer, we analyzed its effect on ANXA2 in recipient cells. When purified EVs collected from miR-1 expressing cells were added to recipient cells, there was a significant reduction of ANXA2 and MET proteins, when compared to treatment with control EVs (Fig.6E). Since the endogenous level of mRNAs for these genes did not significantly change (data not shown), the observed downregulation was most likely due to expected microRNA-mediated effects during translation. The functionality of miR-1 in recipient cells was demonstrated by a luciferase assay with the ANXA2 3’UTR reporter (Fig.6F), showing a significant decrease in luciferase activity in cells treated with EVs from miR-1 expressing cells. The effect was abolished by mutations in the miR-1 site of the ANXA2 3’UTR. These results strongly suggested that miR-1 delivered by EVs is functional and can reduce ANXA2 levels in recipient cells by direct targeting of ANXA2 3’UTR.
To demonstrate the clinical relevance of miR-1/ANXA2 targeting in GBM, we established that ANXA2 mRNA levels were elevated in patient GBM samples and were also significantly inversely correlated with miR-1 (Fig.6G). Moreover, Western analysis showed a significant upregulation of ANXA2 protein in brain tumor samples (Fig.6H). Also, according to Oncomine and TCGA databases, GBMs are characterized by high expression of ANXA2 mRNA (Supplementary Fig. S5I) and its low expression is significantly associated with better outcome (Fig.6I). Thus, these data strongly implicate ANXA2 as a significant player in GBM biology and identify miR-1 as an important direct regulator of ANXA2, both in tumor cells and in the tumor microenvironment.
DISCUSSION
Despite considerable progress in recent years, the regulation of signaling in the GBM microenvironment is not well understood (7). EV signaling via intercellular transport of proteins and RNAs including miRs is a recently discovered mechanism by which tumor cells, including GBM, interact with their surroundings (12, 33). .MicroRNAs have been implicated in GBM pathophysiology and therapy (34-36) but there are a limited number of studies showing the functional consequences of microRNA reintroduction to targeted cells in the context of the tumor microenvironment (37). As EVs are capable of transporting nucleic acids (13, 38) and proteins as a part of physiological intercellular communication, the approach of microRNA overexpression in donor cell may be utilized for their delivery. Here we provide novel evidence that reintroduction of miR-1, important in anti-proliferative, anti-angiogenic and anti-invasive action for GBM, also contributes to microenvironmental remodeling by direct targeting of the GBM cells’ EV cargo. We also demonstrate for the first time that this remodeling is likely mediated by miR-1 targeting of ANXA2. These studies thus characterize GBM's EV-mediated paracrine signaling network and identify some of the factors, such as miR1/ANXA2, that are relevant for known observed GBM phenotypes.
Tumor suppressive functions of miR-1 have been demonstrated in numerous human malignancies (17, 39-41). Its action was associated with direct targeting of several oncogenes, including MET (30) – one of the crucial oncogenic drivers in GBM. However the role of miR-1 in GBM has never been explored. Here we cross-referenced the results of two microRNA microarray analyses and found that the expression of miR-1 is significantly decreased in human GBM patient samples compared with normal brain and is also downregulated in migrating cells, suggesting a role in GBM growth and invasiveness. Upregulation of miR-1 expression in GBM has far reaching consequences for tumor phenotype, including diminished neurosphere formation capacity in vitro tumor growth, angiogenesis and invasiveness in vivo. Unexpectedly, we observed significantly reduced endothelial cell recruitment and neovascularization in vivo suggesting microenvironmental alterations. Such a complex phenotype is likely to be mediated by multiple effectors. Restoration of miR-1 expression in GBM cells is biologically consistent with our understanding of cancer as a disease of multiple aberrant signaling pathways that would thus require intervention at multiple levels (42). The fact that a single miR simultaneously downregulates a broad set of mRNA targets with potential pro-oncogenic properties provides a broad modulatory mechanism that may operate in other cancers as well as GBM. Among a panel of oncogenic signaling kinases, the reduction of JNK activity, shown to play a crucial role in maintenance of self-renewal and tumorigenicity of GBM stem cells (43, 44), was apparent. Also, stem cell factors known to play a vital role in GBM stem cell maintenance such as CD133 or members of Polycomb Repressor Complexes, (36, 45), were significantly reduced upon miR-1 expression. Downregulation of MET and EGFR upon upregulation of miR-1, underlines its strong anti-tumorigenic effects. Here, for the first time we describe targeting of MET by miR-1 in GBM. In silico analysis suggested that the observed changes in the molecular milieu of miR-1 expressing GBM cells are likely consequences of both direct and indirect effects.
The dramatic effect of miR-1 expression on blood vessel formation in vivo suggested complex tumor microenvironment rearrangements. This most likely was due to the altered secretion of pro-angiogenic factors and/or by the active release of overexpressed miR-1 into the microenvironment. Intercellular communication by EVs was recently shown to contribute to horizontal cellular transformation, phenotypic reprograming, and functional re-education of recipient cells via both local and systemic modification of microenvironment and direct transferring of biomolecules including miRs (46). We characterized EVs released by GBM cells and found that miR-1 overexpressed in donor cells was in fact loaded into EVs and transferred to recipient cells to functionally target its effectors there. Moreover, we demonstrated that miR-1 is capable of microenvironmental modifications by affecting the molecular cargo of EVs. Our results underline the functional role of GBM-derived EVs in tumorigenicity. The global proteomic analysis of EV content revealed the presence of over a thousand proteins, among which ANXA2 was the most abundant. ANXA2 is an important oncogene in numerous cancers (47, 48), including GBM where it high expression promoted growth, angiogenesis and invasiveness, and correlated with poorer patient outcomes (20, 49). Interestingly, ANXA2 was among the most downregulated proteins in EVs collected from miR-1 overexpressing cells. Based on the fact that the phenotype observed upon miR-1 expression is consistent with reduced levels of ANXA2, we hypothesized that this protein is not only the direct target of miR-1 but also its major effector in GBM cells. Expression of miR-1 and ANXA2 were inversely correlated in human GBM samples, and miR-1 directly binds to site on the ANXA2 3’UTR leading to repression of ANXA2 protein levels. Furthermore, bioinformatic analysis revealed that miR-1 putatively targets several proteins functionally related to ANXA2 (Fig.6J) in addition to previously published targeting of MET mRNA. MiR-1 deregulated the protein cargo of GBM-derived EVs without significant differences in EV secretion or uptake, suggesting that observed phenotype is in fact mediated by differences in the content of EVs molecular cargo, including ANXA2. These data strongly suggest a mechanistic link between miR-1 and ANXA2 suppression on GBM cells and their microenvironment.
It has been postulated that miR replacement approaches have strong therapeutic potential because of the fact that single miRs can regulate multiple oncogenic pathways that are commonly deregulated in cancer (42). Our report provides novel evidence that miR-1 is inactivated in GBM. Because miR-1 simultaneously targets major components of oncogenic signaling networks (JNK, PRCs, MET, EGFR, ANXA2), miR-1 loss may represent an important step in GBM genesis and/or progression. Moreover, we showed that the pro-oncogenic impact that GBM cells exert on their microenvironment by releasing EVs is alleviated by miR-1. Thus the depletion of cellular and EV ANXA2 points towards miR-1 replacement as an attractive candidate for this therapeutic modality. These findings shed new light on the intricate communication networks in the GBM microenvironment and open new possibilities for therapeutic intervention.
Supplementary Material
Acknowledgments
Financial support: This work was supported by NCI P01 (#108778) (To EAC, FH, RW) and by an institutional sundry fund (to EAC).
Footnotes
Conflict of interest disclosure: authors declare no conflict of interest
REFERENCES
- 1.Hess KR, Broglio KR, Bondy ML. Adult glioma incidence trends in the United States, 1977-2000. Cancer. 2004;101:2293–9. doi: 10.1002/cncr.20621. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 4.Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23:2411–22. doi: 10.1200/JCO.2005.03.089. [DOI] [PubMed] [Google Scholar]
- 5.Lathia JD, Heddleston JM, Venere M, Rich JN. Deadly teamwork: neural cancer stem cells and the tumor microenvironment. Cell stem cell. 2011;8:482–5. doi: 10.1016/j.stem.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, et al. A perivascular niche for brain tumor stem cells. Cancer cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59:1169–80. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- 8.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer research. 2011;71:4055–60. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Vos KE, Balaj L, Skog J, Breakefield XO. Brain tumor microvesicles: insights into intercellular communication in the nervous system. Cellular and molecular neurobiology. 2011;31:949–59. doi: 10.1007/s10571-011-9697-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature medicine. 2012;18:883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nature communications. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7312–7. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature cell biology. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janga SC, Vallabhaneni S. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv Exp Med Biol. 2011;722:59–74. doi: 10.1007/978-1-4614-0332-6_4. [DOI] [PubMed] [Google Scholar]
- 15.Ferdin J, Kunej T, Calin GA. Non-coding RNAs: identification of cancer-associated microRNAs by gene profiling. Technol Cancer Res Treat. 2010;9:123–38. doi: 10.1177/153303461000900202. [DOI] [PubMed] [Google Scholar]
- 16.Godlewski J, Newton HB, Chiocca EA, Lawler SE. MicroRNAs and glioblastoma; the stem cell connection. Cell Death Differ. 2010;17:221–8. doi: 10.1038/cdd.2009.71. [DOI] [PubMed] [Google Scholar]
- 17.Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lokman NA, Ween MP, Oehler MK, Ricciardelli C. The role of annexin A2 in tumorigenesis and cancer progression. Cancer Microenviron. 2011;4:199–208. doi: 10.1007/s12307-011-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.An JH, Lee SY, Jeon JY, Cho KG, Kim SU, Lee MA. Identification of gliotropic factors that induce human stem cell migration to malignant tumor. Journal of proteome research. 2009;8:2873–81. doi: 10.1021/pr900020q. [DOI] [PubMed] [Google Scholar]
- 20.Zhai H, Acharya S, Gravanis I, Mehmood S, Seidman RJ, Shroyer KR, et al. Annexin A2 promotes glioma cell invasion and tumor progression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:14346–60. doi: 10.1523/JNEUROSCI.3299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer research. 2003;63:5821–8. [PubMed] [Google Scholar]
- 22.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-oncology. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Molecular cell. 2010;37:620–32. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams SP, Nowicki MO, Liu F, Press R, Godlewski J, Abdel-Rasoul M, et al. Indirubins decrease glioma invasion by blocking migratory phenotypes in both the tumor and stromal endothelial cell compartments. Cancer research. 2011;71:5374–80. doi: 10.1158/0008-5472.CAN-10-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothhammer T, Bataille F, Spruss T, Eissner G, Bosserhoff AK. Functional implication of BMP4 expression on angiogenesis in malignant melanoma. Oncogene. 2007;26:4158–70. doi: 10.1038/sj.onc.1210182. [DOI] [PubMed] [Google Scholar]
- 26.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. The Journal of experimental medicine. 1996;183:1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 28.Bronisz A, Sharma SM, Hu R, Godlewski J, Tzivion G, Mansky KC, et al. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Molecular biology of the cell. 2006;17:3897–906. doi: 10.1091/mbc.E06-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer research. 2008;68:9125–30. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 30.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer research. 2008;68:5049–58. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Reid JF, Sokolova V, Zoni E, Lampis A, Pizzamiglio S, Bertan C, et al. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Molecular cancer research : MCR. 2012;10:504–15. doi: 10.1158/1541-7786.MCR-11-0342. [DOI] [PubMed] [Google Scholar]
- 32.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nature medicine. 2012;18:1835–40. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu ZM, Wang YB, Yuan XH. Exosomes from murine-derived GL26 cells promote glioblastoma tumor growth by reducing number and function of CD8+T cells. Asian Pacific journal of cancer prevention : APJCP. 2013;14:309–14. doi: 10.7314/apjcp.2013.14.1.309. [DOI] [PubMed] [Google Scholar]
- 34.Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. The EMBO journal. 2008;27:2616–27. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer research. 2008;68:3566–72. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 36.Peruzzi P, Bronisz A, Nowicki MO, Wang Y, Ogawa D, Price R, et al. MicroRNA-128 coordinately targets Polycomb Repressor Complexes in glioma stem cells. Neuro-oncology. 2013 doi: 10.1093/neuonc/not055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nature cell biology. 2012;14:159–67. doi: 10.1038/ncb2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katakowski M, Buller B, Zheng X, Lu Y, Rogers T, Osobamiro O, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer letters. 2013;335:201–4. doi: 10.1016/j.canlet.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fleming JL, Gable DL, Samadzadeh-Tarighat S, Cheng L, Yu L, Gillespie JL, et al. Differential expression of miR-1, a putative tumor suppressing microRNA, in cancer resistant and cancer susceptible mice. PeerJ. 2013;1:e68. doi: 10.7717/peerj.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic acids research. 2011 doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. The Journal of biological chemistry. 2008;283:33394–405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer research. 2010;70:7027–30. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuda K, Sato A, Okada M, Shibuya K, Seino S, Suzuki K, et al. Targeting JNK for therapeutic depletion of stem-like glioblastoma cells. Scientific reports. 2012;2:516. doi: 10.1038/srep00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon CH, Kim MJ, Kim RK, Lim EJ, Choi KS, An S, et al. c-Jun N-terminal kinase has a pivotal role in the maintenance of self-renewal and tumorigenicity in glioma stem-like cells. Oncogene. 2012;31:4655–66. doi: 10.1038/onc.2011.634. [DOI] [PubMed] [Google Scholar]
- 45.Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:8884–96. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rak J. Extracellular vesicles - biomarkers and effectors of the cellular interactome in cancer. Frontiers in pharmacology. 2013;4:21. doi: 10.3389/fphar.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hedhli N, Falcone DJ, Huang B, Cesarman-Maus G, Kraemer R, Zhai H, et al. The annexin A2/S100A10 system in health and disease: emerging paradigms. Journal of biomedicine & biotechnology. 2012;2012:406273. doi: 10.1155/2012/406273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mussunoor S, Murray GI. The role of annexins in tumour development and progression. The Journal of pathology. 2008;216:131–40. doi: 10.1002/path.2400. [DOI] [PubMed] [Google Scholar]
- 49.Gao H, Yu B, Yan Y, Shen J, Zhao S, Zhu J, et al. Correlation of expression levels of ANXA2, PGAM1, and CALR with glioma grade and prognosis. Journal of neurosurgery. 2013;118:846–53. doi: 10.3171/2012.9.JNS112134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.