Abstract
Permethylation is a valuable and widely used tool for the mass spectrometry of carbohydrates, improving sensitivity and fragmentation and increasing the amount of information that can be obtained from tandem mass spectrometric experiments. Permethylation of most glycans is easily performed with sodium hydroxide and iodomethane in dimethyl sulfoxide (DMSO). However, permethylation has not been widely used in the mass spectrometry of glycosaminoglycan (GAG) oligosaccharides, partly because it has required the use of the difficult Hakomori method employing the methylsulfinylmethanide (`dimsyl') base, which has to be made in a tedious process. Additionally, the Hakomori method is not as effective as the sodium hydroxide method in making fully methylated derivatives. A further problem in the permethylation of highly sulfated oligosaccharides is their limited solubility in DMSO. This paper describes the use of the triethylammonium counterion to overcome this problem, as well as the application of the sodium hydroxide method to make permethylated heparin disaccharides and their workup to yield fully methylated disaccharides for electrospray ionization mass spectrometry. The ease, speed, and effectiveness of the described methodology should open up permethylation of GAG oligosaccharides to a wider circle of mass spectrometrists and enable them to develop further derivatization schemes in the effort to rapidly elucidate the structure of these important molecules. Permethylation may also provide new ways of separating GAG oligosaccharides in LC/MS, their increased hydrophobicity making them amenable for reversed-phase chromatography without the need for ion pairing reagents.
Permethylation is widely used in carbohydrate analysis (i) as the first step in glycosyl linkage analysis[1] and (ii) to improve sensitivity and fragmentation in mass spectrometry of oligosaccharides.[2] On the other hand, permethylation has not found widespread use in the glycosaminoglycan field, although good results have been obtained with the Hakomori method.[3,4] This method employs the potassium methylsulfinyl base, which is neither commercially available nor easy to make, and this may be partially responsible for the relative unpopularity of glycosaminoglycan (GAG) oligosaccharide permethylation. Another reason may be the immense progress that has been made in the mass spectrometry of underivatized GAG oligosaccharides.[2,5–7] Nevertheless, permethylation offers significant benefits to the mass spectrometry of sulfated carbohydrates. It leads to improved sensitivity,[8] provides information on the position of the sulfate on the sugar chain,[3,9] as well as on linkage positions. Furthermore, it has been demonstrated that permethylated oligosaccharides are more susceptible to cross-ring cleavage than native oligosaccharides.[10] This feature may provide information useful to determine the sulfate positions on individual monosaccharide residues. Additionally, in case of loss of sulfate, with a permethylated sample it is apparent whether the loss occurred during derivatization or during ionization. Because of their increased hydrophobicity permethylated GAG oligosaccharides may also provide improved separation efficiency in liquid chromatography/mass spectrometry (LC/MS) by making them amenable to reversed-phase chromatography.
The sodium hydroxide method of Ciucanu and Kerek[11] and its modification by Anumula and Taylor[12] have rendered permethylation a simple, rapid, and effective tool so that it is now the method of choice in the mass spectrometry of N- and O-linked glycans. However, the Ciukanu/Kerek method has been reported to be unsuitable for the production of permethylated GAG oligosaccharides because even after purification they contained too much salt.[3] Apparently, the problem in that study lay in the isolation of the permethylated glycans, rather than in the effectiveness of the derivatization.
We have now found that the Anumula modification[12] of the sodium hydroxide method yields permethylated GAG oligosaccharides in sufficient purity to give good mass spectra with electrospray ionization (ESI) and nanospray ionization without any detectable loss of sulfate during the derivatization. It is not clear why the salt was not sufficiently removed in the study by Dell et al.,[3] as the authors used a similar cleanup procedure to the one described here. It is possible that the fast atom bombardment mass spectrometric (FAB-MS) ionization they used was more sensitive to salt contamination than the ESI-MS used here.
In our studies of the permethylation of sulfated oligosaccharides we have observed that due to their ionic character they tend to be only sparingly soluble in dimethyl sulfoxide (DMSO). We have found that the conversion of the disaccharide sodium salts into their triethylammonium salts provides a quick and straightforward way to overcome this problem.
EXPERIMENTAL
Materials
All experiments were carried out with Δ4,5-unsaturated heparin disaccharide standards purchased from Dextra Laboratories (UK). All other reagents were from Sigma-Aldrich (Milwaukee, WI, USA). C18 Sep-Paks were from Waters (Franklin, MA, USA).
Conversion into triethylammonium salts
The heparin disaccharides (50 mg each) were dissolved in water (50 μL per disaccharide), loaded onto a Dowex 50W×8 column in the triethylammonium form (~0.7 mL in a Pasteur pipette), and eluted with water. The collected sample was freeze-dried.
Permethylation
The sample was completely dissolved in 300 μL of dry DMSO by vortexing briefly, then 360 μL of sodium hydroxide suspension in DMSO[12] was added, followed by 100 μL of iodomethane. The mixture was then vortexed for 5 min and sonicated for 10 min at room temperature. The reaction was quenched by adding 2 mL of water, and the excess iodomethane was removed by sparging with nitrogen. The product mixture was loaded onto a C18 Sep-Pak cartridge, the cartridge was washed with 3 mL of water, and the permethylated disaccharides were eluted with 2 mL of 50% MeOH.
Permethylation followed by saponification
The permethylation was carried out exactly as described above, but after removal of iodomethane one drop of 1 M NaOH was added (final pH 12.5). After 1 h at room temperature, the mixture was passed through a Dowex 50W×8 column (triethylammonium form) and then loaded onto a C18 Sep-Pak, previously equilibrated with a buffer of 10 mM NEt3 and 12 mM AcOH. The column was washed with 2 mL of the same buffer and 3 mL of water, followed by elution with 3 mL of 50% MeOH.
Reduction, followed by permethylation and saponification
The mixture of disaccharides (67 μg D2S6 and 40 μg D0A0 in 100 μL H2O) was treated with 14 μL of conc. NH4OH and 1 mg of NaBH4 for 18 h at room temperature. The mixture was neutralized with 3 drops of glacial AcOH, 200 μL MeOH/AcOH (9:1) was added, and the mixture was dried down under a stream of nitrogen. After repeating the addition and evaporation of MeOH/AcOH two more times, the mixture was evaporated three times with MeOH and dissolved in 1 mL of H2O. The mixture was converted into triethylammonium salts, permethylated, and saponified as described above.
Mass spectrometry
Direct infusion ESI-MS was carried out on a Thermo LCQ ion trap instrument using a 1:1 mixture of water/MeOH as spray solvent. Samples were introduced by direct infusion at 10 μL/min using a Harvard syringe pump. Scans were averaged over several minutes. The volume infused during the data collection for the individual experiments is specified in the figure legends. The capillary temperature was 200°C, and the absolute value of the spray voltage was 3.5 kV in both positive and negative ion mode.
After completion of the main part of this work we repeated some of the experiments on a recently acquired LTQ-Orbitrap instrument and obtained equivalent results.
RESULTS
Conversion into triethylammonium salts
Due to their anionic character, sulfated oligosaccharides in general do not readily dissolve in DMSO, the solvent that has been shown to give the best results in permethylation reactions. This problem has previously been observed in the methylation of sulfated fucans, where it has been solved by converting the sodium salt into the triethylammonium salt, which is less polar.[13] Similarly, we have found that the solubility of GAG oligosaccharides is improved dramatically if the sodium counterions are first exchanged with triethylammonium by passage through the triethylammonium form of a strong cation-exchange resin.
For the next step, the triethylammonium salts are dissolved in DMSO. It is important that they are dissolved completely before the sodium hydroxide base is added because the latter will immediately revert the GAGs back to the sodium salts, which would not go into solution. However, if the triethylammonium salts are already completely dissolved before addition of the base, a clear solution is maintained.
Permethylation
Permethylation is carried out by first adding an anhydrous suspension of NaOH in DMSO, followed by iodomethane. After completion of the permethylation, which is accelerated by vigorous mixing (vortexing and sonication), the reaction is quenched by addition of water. We found that the amounts of reagents and the reaction time critically influence the outcome of the reaction through the pH in the resulting aqueous solution. With the conditions given in the Experimental section, the pH of the solution is between 3 and 4, which is counterintuitive since the methylation reaction is conducted under basic conditions. The change in proton concentrations is due to reaction of NaOH with excess iodomethane, producing methanol, and likely by reaction of iodomethane with DMSO, producing dimethyl sulfide and hydroiodic acid.[14] The slightly acidic pH of the aqueous solution prevents saponification of the methyl ester formed by methylation of the uronic acids in the GAG oligosaccharides. In some cases it may be advantageous to deliberately carry out the saponification, for example if it is desired to preserve a negative charge on oligosaccharides that do not carry any sulfates in a mixture with those that are sulfated. However, it is preferable to start this process with the acidic solution and add a carefully controlled amount of base, rather than adding more base before the permethylation, in which case it would be difficult to avoid degradation of oligosaccharide due to the peeling reaction.
One may also carry out the reduction of the oligosaccharide mixture to be analyzed prior to permethylation. This can be advantageous in applications where the presence of α- and β-reducing ends interferes with the analysis, such as in high-performance liquid chromatography (HPLC), where anomeric species lead to peak broadening or splitting.
Although we did not determine the absolute efficiency of the permethylation, a significant difference in permethylation efficiency between the various disaccharides would have manifested itself in the presence of undermethylated species. Since no such species were detected by MS (see below), we conclude that the efficiency of the permethylation is nearly 100% for all disaccharides.
Sample cleanup for ESI-MS
Unlike in the case of neutral carbohydrates, sulfated sugars cannot be solvent-extracted after permethylation because the sulfates will make them water-soluble. As described previously,[3] permethylated esters can conveniently be isolated by passage through a C18 Sep-Pak cartridge, from which they elute with 50% methanol. Initially, we desalted the product mixture before Sep-Pak purification by passage through a cation-exchange resin, but found this to be unnecessary later on.
If the permethylated oligosaccharides were saponified, only the less sulfated were retained on the C18 cartridge, while the more highly sulfated ones eluted in the water wash. Nevertheless, retention could be accomplished by running the Sep-Pak in ion-pairing mode. For this, we first passed the mixture through a cation-exchange column in triethylammonium form before loading onto the Sep-Pak, which had previously been equilibrated with triethylammonium acetate. In this way all the permethylated oligosaccharides eluted in the 50% methanol fraction.
Mass spectrometry
In the following description, we use structural abbreviations for the permethylated disaccharides based on a recently introduced structure code for GAG oligosaccharides.[15] This code uses a four-character alphanumeric combination for each disaccharide subunit. Letters designate the isomeric characteristics of each monosaccharide, and numbers refer to their sulfation patterns. Table 1 lists the definitions of each letter/number of the monosaccharide at the non-reducing end of the disaccharide, and Table 2 lists the corresponding definitions for the hexosamine residue. It should be noted that the upper and lower cases of the letters designating the aminosugars are used to identify glucosamine and galactosamine, respectively. Each permethylated disaccharide is designated by the code for the underivatized disaccharide, followed by the number of methyl groups in parentheses. In those cases where one less than the highest possible number of methyls is given it is implied that it is the carboxylic acid group that is not methylated (i.e. the permethylated and saponified disaccharides).
Table 1.
Disaccharide structure code letters and numbers for the non-reducing end residue
| Descriptor | Sulfation | ||
|---|---|---|---|
| U | undesignated uronic acid | 0 | no sulfates |
| D | Δ4,5-unsaturated uronic acid | 2 | 2-O-sulfation |
| G | glucuronic acid | 3 | 3-O-sulfation |
| I | iduronic acid | 6 | 6-O-sulfation |
| g | galactose |
Table 2.
Disaccharide structure code letters and numbers for the hexosamine residue
| Descriptor | Sulfation | ||
|---|---|---|---|
| A | N-acetylglucosamine | 0 | no sulfates |
| a | N-acetylgalactosamine | 3 | 3-O-sulfation |
| S | N-sulfoglucosamine | 4 | 4-O-sulfation |
| 6 | 6-O-sulfation | ||
| 9 | 3,6-O-disulfation | ||
| 10 | 4,6-O-disulfation |
Mass spectra of the permethylated disaccharides could be obtained in both negative and positive ion mode. The spectra were more complex in the negative ion mode with the appearance of various ionization and sodiation states. In the spectrum of permethylated D2S6 (D2S6(5)) (Fig. 1) we observed singly, doubly, and triply charged ions with zero, one, or two sodium ions. We also detected loss of MeOH (M/2–16) and loss of SO3 (M/2–40). This sulfate loss must have occurred during ionization and not during permethylation, otherwise there would be an additional methyl group. The spectrum of the mixture containing D2A6(5), D0A0(7), D2S0(6), and D0S0(7) (Fig. 2) showed singly and doubly charged ions for D2A6(5), a singly charged ion for D0S0(7), and a doubly charged ion for D2S0(6), but D0A0(7) was not detected, because it does not have any ionizable groups. The saponified form of the latter (D0A0(6)) could be detected, however, in the spectrum of the mixture of permethylated and saponified disaccharides (Fig. 3), as it now had the free carboxylate group. In fact, ions from all permethylated disaccharides were observed in this spectrum. Minor ions resulting from in-source loss of SO3 were also detected.
Figure 1.
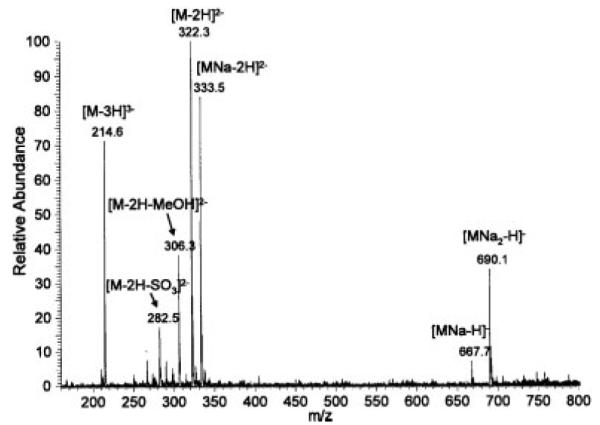
Negative ion mode ESI mass spectrum of permethylated heparin disaccharide D2S6. A 40-μL aliquot of the Sep-Pak eluate was used for direct infusion and the signal was averaged over the time of the infusion.
Figure 2.
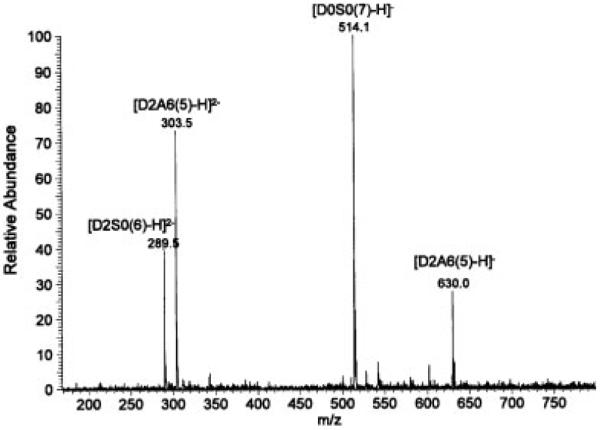
Negative ion mode ESI mass spectrum of permethylated heparin disaccharides D2A6, D0S0, D2S0, and D0A0. Infusion volume: 30 μL.
Figure 3.
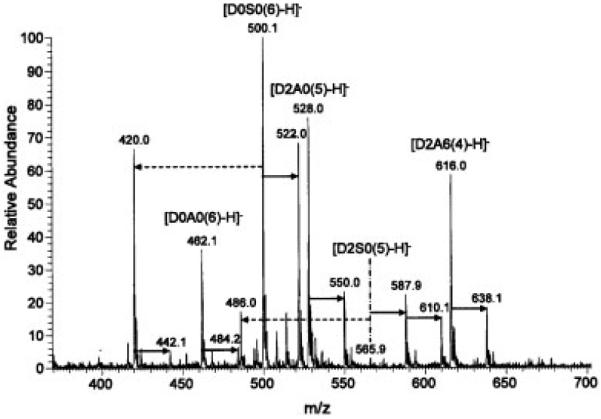
Negative ion mode ESI mass spectrum of permethylated and saponified heparin disaccharides D2A6, D0S0, D2A0, D2S0, and D0A0. Solid arrows indicate difference of 22 (Na salt); dashed arrows indicate loss of SO3. Infusion volume: 50 μL.
The spectrum of reduced, permethylated, and saponified heparin disaccharides D2S6red(7) and D0A0red(9) (Fig. 4) showed the expected ions, in addition to some residual ester due to incomplete saponification. The degree of in-source desulfation appeared to be higher than in the spectra of the non-reduced compounds.
Figure 4.
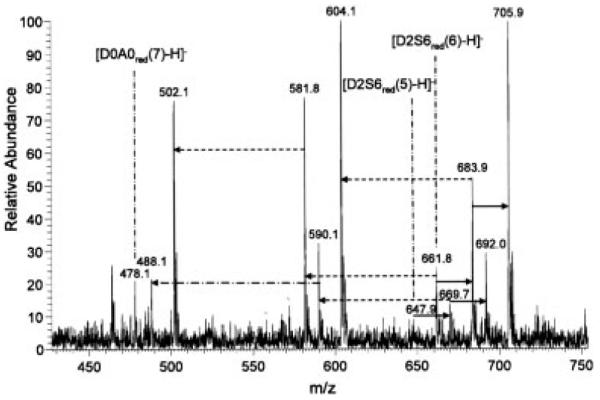
Negative ion mode ESI mass spectrum of reduced, permethylated, and saponified heparin disaccharides D2S6 and D0A0. Solid arrows indicate difference of 22 (Na salt); dashed arrows indicate loss of SO3, dash-dot arrows indicate loss of SO3Na. Infusion volume: 50 μL.
The spectra of permethylated heparin disaccharides in positive ion mode were less complex than those in negative ion mode. Although all disaccharides could be detected, it was apparent that the more highly sulfated disaccharides were harder to ionize and gave significantly less intense ions than those with less sulfates. This is clearly seen in Fig. 5, which shows the positive ion mode mass spectrum of an equimolar mixture of four permethylated heparin disaccharides. Only singly charged, monosodiated species were seen, and their peak intensity varied widely according to the number of sulfates. In-source loss of sulfate was also more pronounced in the highly sulfated disaccharides, likely because of the better ionizability of the resulting products (Fig. 6). In both positive and negative ion mode, we observed in-source sulfate loss only from N-sulfated disaccharides.
Figure 5.
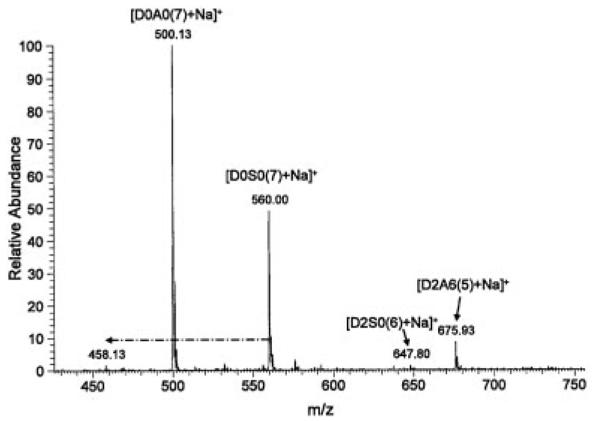
Positive ion mode ESI mass spectrum of permethylated heparin disaccharides D2A6, D0S0, D2S0, and D0A0. Dash-dot arrows indicate loss of SO3Na. Infusion volume: 10 μL.
Figure 6.
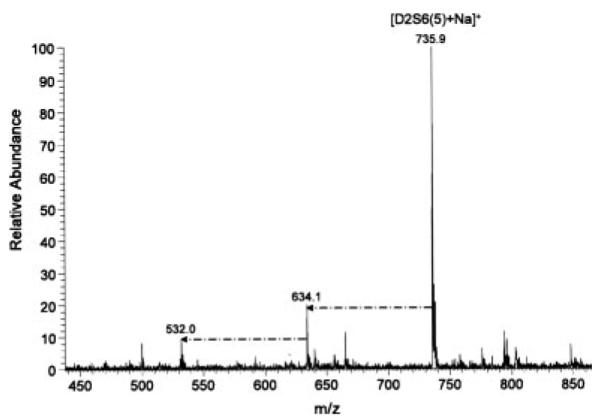
Positive ion mode ESI mass spectrum of permethylated heparin disaccharide D2S6. Dash-dot arrows indicate loss of SO3Na. Infusion volume: 10 μL.
The spectra shown were acquired using about 1 μg of infused material, although we were able to obtain usable spectra with amounts in the range of 1–10 ng (data not shown) using the LCQ instrument. With more advanced instruments, such as the LTQ, even lower amounts should be detectable.
CONCLUSIONS
We have shown that the sodium hydroxide permethylation method is suitable for heparin disaccharides. Conversion of the sodium salts into triethylammonium salts aided this process by making the disaccharide readily soluble in DMSO. Fully permethylated disaccharides were easily obtained in sufficient purity for ESI-MS without detectable loss of sulfate during permethylation. All the sulfated permethylated disaccharides could be detected using negative ion mode. Non-sulfated disaccharides were rendered detectable in negative ion mode by saponification of the carboxylate esters. Spectra in positive ion mode detected all permethylated disaccharides applied, but with a strong bias toward less sulfated disaccharides.
Experiments to extend this methodology to larger glycosaminoglycan oligosaccharides are underway in our laboratory. If successful, these may aid in fast structural characterization by combining permethylation with other derivatization schemes, by improving LC/MS selectivity and possibly by providing new ways to effect more extensive cross-ring fragmentations, which are more diagnostic than the more common inter-residue fragmentations.
Acknowledgements
We gratefully acknowledge the National Center for Research Resources (a part of the NIH) for support of the Resource for Integrated Glycotechnology at the University of Georgia (P41RR005351).
REFERENCES
- [1].York WS, Darvill AG, McNeil M, Stevenson TT, Albersheim P, Arthur Weissbach HW. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1986;118:3. [Google Scholar]
- [2].Zaia J. On-line separations combined with MS for analysis of glycosaminoglycans. Mass Spectrom. Rev. 2009;28:254. doi: 10.1002/mas.20200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dell A, Rogers ME, Thomas-Oates JE, Huckerby TN, Sanderson PN, Nieduszynski IA. Fast-atom-bombardment mass-spectrometric strategies for sequencing sulphated oligosaccharides. Carbohydr. Res. 1988;179:7. [Google Scholar]
- [4].Khoo K-H, Morris HR, McDowell RA, Dell A, Maccarana M, Lindahl U. FABMS/derivatisation strategies for the analysis of heparin-derived oligosaccharides. Carbohydr. Res. 1993;244:205. doi: 10.1016/0008-6215(83)85002-2. [DOI] [PubMed] [Google Scholar]
- [5].Schenauer MR, Meissen JK, Seo Y, Ames JB, Leary JA. Heparan sulfate separation, sequencing, and isomeric differentiation: ion mobility spectrometry reveals specific iduronic and glucuronic acid-containing hexasaccharides. Anal. Chem. 2009;81:10179. doi: 10.1021/ac902186h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Saad OM, Leary JA. Heparin sequencing using enzymatic digestion and ESI-MSn with HOST: a heparin/HS oligosaccharide sequencing tool. Anal. Chem. 2005;77:5902. doi: 10.1021/ac050793d. [DOI] [PubMed] [Google Scholar]
- [7].Laremore TN, Leach FE, Iii, Solakyildirim K, Amster IJ, Linhardt RJ, Minoru F. Glycosaminoglycan characterization by electrospray ionization mass spectrometry including Fourier transform mass spectrometry. Methods Enzymol. 2010;478:79. doi: 10.1016/S0076-6879(10)78003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Harvey DJ. Matrix-assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom. Rev. 1999;18:349. doi: 10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- [9].Lei M, Mechref Y, Novotny MV. Structural analysis of sulfated glycans by sequential double-permethylation using methyl iodide and deuteromethyl iodide. J. Am. Soc. Mass Spectrom. 2009;20:1660. doi: 10.1016/j.jasms.2009.05.001. [DOI] [PubMed] [Google Scholar]
- [10].Ashline D, Singh S, Hanneman A, Reinhold V. Congruent strategies for carbohydrate sequencing. 1. Mining structural details by MSn. Anal. Chem. 2005;77:6250. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ciucanu I, Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984;131:209. [Google Scholar]
- [12].Anumula KR, Taylor PB. A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal. Biochem. 1992;203:101. doi: 10.1016/0003-2697(92)90048-c. [DOI] [PubMed] [Google Scholar]
- [13].Stevenson TT, Furneaux RH. Chemical methods for the analysis of sulphated galactans from red algae. Carbohydr. Res. 1991;210:277. doi: 10.1016/0008-6215(91)80129-b. [DOI] [PubMed] [Google Scholar]
- [14].Needs PW, Selvendran RR. Avoiding oxidative degradation during sodium hydroxide/methyl iodide-mediated carbohydrate methylation in dimethyl sulfoxide. Carbohydr. Res. 1993;245:1. [Google Scholar]
- [15].Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat. Methods. 2008;5:291. doi: 10.1038/nmeth0408-291. [DOI] [PubMed] [Google Scholar]


