Abstract
Exome sequencing was performed in three index cases with bone marrow failure and neurological dysfunction and whose parents are first-degree cousins. Homozygous truncating mutations were identified in ERCC6L2 in two of the individuals. Both of these mutations affect the subcellular localization and stability of ERCC6L2. We show here that knockdown of ERCC6L2 in human A549 cells significantly reduced their viability upon exposure to the DNA-damaging agents mitomycin C and Irofulven, but not etoposide and camptothecin, suggesting a role in nucleotide excision repair. ERCC6L2-knockdown cells also displayed H2AX phosphorylation, which significantly increased upon genotoxic stress, suggesting an early DNA-damage response. Intriguingly, ERCC6L2 was seen to translocate to the mitochondria and the nucleus in response to DNA damage, and ERCC6L2 knockdown induced intracellular reactive oxygen species (ROS). Treatment with the ROS scavenger N-acetyl cysteine attenuated the Irofulven-induced cytotoxicity in ERCC6L2-knockdown cells and abolished ERCCGL2 traffic to the mitochondria and nucleus in response to this DNA-damaging agent. Collectively, these observations identify a distinct bone-marrow-failure syndrome due to mutations in ERCC6L2, a gene implicated in DNA repair and mitochondrial function.
Introduction
Bone-marrow-failure syndromes are a heterogeneous group of life-threatening disorders characterized by the inability of the bone marrow to make an adequate number of mature blood cells.1,2 They can be variable in their severity and can affect either one or all of the hematopoietic lineages. In some cases, individuals can be classified into recognized syndromes, such as Fanconi anemia (FA [MIM 227650])3 and dyskeratosis congenita (DC [MIM 305000]).4
In these diseases, the categorization of affected individuals has been facilitated by the recognition of characteristic features, such as the diagnostic mucocutaneous symptoms of DC (abnormal skin pigmentation, nail dystrophy, and leukoplakia). Alternatively, specific genetic and/or functional defects can be used in diagnosis, exemplified by the characteristically increased chromosomal breakage observed in FA individuals. However, there are also cases where the bone marrow failure is associated with one or more extrahematopoietic abnormalities but does not fit into a recognized syndrome and the underlying genetic and functional basis is thus unknown.
The recent availability of next-generation sequencing technology is making it possible to elucidate the genetic basis and pathophysiology of uncharacterized human diseases. From our repository of bone-marrow-failure cases, we chose three genetically uncharacterized index cases in whom to perform exome sequencing with the aim of identifying variants in a common gene. They had trilineage (erythroid, myeloid, and megakaryocytic) bone marrow failure and came from consanguineous families; all three also had developmental delay characterized by learning disability, and two out of the three cases also had microcephaly (Table 1). We hypothesized that this approach would enrich for homozygous disease-causing mutations.
Table 1.
Features of Index Cases Who Underwent Exome Sequencing in Comparison to Those of FA, CS, and DC
| Features |
Index Cases in This Study |
|||||
|---|---|---|---|---|---|---|
| Case 1 (Family 1) | Case 2 (Family 2) | Case 3 | FA | CS | DC | |
| Gender | male | female | female | male and female | male and female | male and female |
| Age at presentation (years) | 12 | 19 | 9 | - | - | - |
| Ethnic origin | French | Pakistani | Pakistani | varied | varied | varied |
| First-cousin parents | yes | yes | yes | some | some | some |
| Trilineage bone marrow failure | yesa | yesb | yesc | yes | no | yes |
| Learning difficulties and/or developmental delay | yes | yes | yes | yes | yes | yes |
| Microcephaly | yes | yes | no | yes | yes | yes |
| Cutaneous photosensitivity | no | no | no | no | some | some |
| Cancer | no | no | no | yes | no | yes |
| Mucocutaneous features | no | no | no | some | some | yes |
| Other clinical features | yesd | yese | yesf | yesg | yesh | yesi |
| Chromosomal breakage in PB lymphocytes after treatment with DEB or MMC | normal | normal | normal | increased | normal | normal |
| Telomere length | normal | short | short | short | ? | very short |
Abbreviations are as follows: CS, Cockayne syndrome; DC, dyskeratosis congenita; DEB, diepoxybutane; FA, Fanconi anemia; MMC, mitomycin C; PB, peripheral blood; and ?, unknown.
Peripheral-blood analysis at presentation showed hemoglobin at 99 g/l, white cell count at 2.4 × 109/l, platelets at 9 × 109/l, and very hypocellular bone marrow (Figure 1E).
Peripheral-blood analysis at presentation showed hemoglobin at 93 g/l, white cell count at 5.3 × 109/l, platelets at 33 × 109/l, and hypocellular bone marrow with features of dysplasia.
Peripheral-blood analysis at presentation showed hemoglobin at 102 g/l, white cell count at 2.3 × 109/l, platelets at 128 × 109/l, and hypocellular bone marrow.
“Abnormal facies” and ear abnormalities.
Floppy in infancy.
Cleft palate and craniosynostosis.
Variable other features, including skin, gastrointestinal, and renal abnormalities.
Variable other features, including skin photosensitivity.
Variable other features, including immune deficiency and cerebellar hypoplasia.
Material and Methods
Exome Sequencing
Peripheral-blood samples were obtained with written consent under the approval of our local research ethics committee (London – City and East). DNA extracted from these samples was submitted to the Beijing Genomics Institute for exome sequencing. Ten micrograms of genomic DNA was supplied and, after passing quality control, was subjected to Agilent SureSelect library preparation and exome enrichment before being sequenced on the Illumina GAII system. Sequencing data were processed through the Illumina pipeline. Variants were called with the Genome Analysis Toolkit (GATK) v.2.7.4. All samples were called jointly with an additional data set of 1,005 locally sequenced exomes with unrelated conditions (University College London Exomes [UCL-ex] Consortium) after BAM file reduction as implemented by GATK using default options. We used the Illumina TruSeq target region (± 200 bp on each side) for variant calling. We followed the GATK best practices and implemented variant recalibration with separate models for SNPs and indels. We excluded read depth from our recalibration model owing to the large read-depth variability generated by the heterogeneous capture kits used in the multiple studies that form UCL-ex. Variants with PASS filter and the top-level recalibration tranche (VQSRTrancheSNP99.00to99.90) were retained. We used a variant-quality threshold of 30 and a genotype (i.e., sample-based) Phred-scaled quality threshold of 20, with the exception of heterozygous calls, for which we found the error model overly permissive and for which we used a more stringent genotype Phred quality threshold of 40. After variant calling, we used ANNOVAR to perform annotation, which included allele frequencies obtained from the NHLBI Exome Sequencing Project. We used the Ensembl genes and transcripts to annotate the functional effect of exonic variants. Splice variants were flagged within 8 bp of the exon-intron junction. All variants identified were validated by Sanger sequencing on a 3130xl Genetic Analyzer with a BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems).
H&E Staining and Immunohistochemistry
All sections were stained with hematoxylin and eosin (H&E) and immunohistochemistry. We incubated glycol-methacrylate-embedded sections in Tris-buffered saline (TBS) for 1 hr and further subjected them to epitope unmasking by using a heat-induced epitope-retrieval system with citrate buffer (Vector labs) to avoid crosslinking of the proteins formed by formaldehyde fixation without decalicification. We blocked sections with 5% goat serum in PBS to completely avoid the background staining. Primary-antibody incubations diluted in blocking solution (1:500) were carried out overnight at 4°C, subsequently washed, and then incubated with secondary antibody Alexa Fluor 488 (Invitrogen) for 1 hr at room temperature. Slices were washed in TBS containing 0.1% Tween 20, mounted with vectashield mounting reagent (Vector Labs) on a cover glass, and subjected to fluorescence imaging.
Cell Culture and Plasmid Transfection
Human A549, human embryonic kidney 293, HeLa, and 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (v/v) fetal bovine serum (FBS; HyClone), 100 IU/ml penicillin, and 100 mg/ml streptomycin (Invitrogen). All cells were maintained at 37°C in a humidified incubator with 5% CO2. For ectopic expression studies, both 293T and A549 cells were transfected with the appropriate plasmids by electroporation with a Genepulser MX (Biorad) at 400V, 125 μF, and ∞ ohms in optiMEM (Invitrogen).
Immunocytochemistry, Imaging, and Colocalization Studies
In brief, after appropriate treatment, cells grown on coverslips were washed in warm PBS and fixed in 4% paraformaldehyde. After fixation, cells were permeabilized with 0.1% Triton X-100 (TX100) in PBS, quenched in 50 mM NH4Cl, and blocked in 10% goat serum and 1% BSA in PBS containing 0.05% TX100 for 1 hr. Cells were incubated in the primary ERCC6L2 antibody (Abcam) and the corresponding goat anti-rabbit secondary antibody conjugated to Alexa Fluor 488/568 (Invitrogen) in blocking solution for 1 hr separately. Cells were washed three times in PBS containing 0.05% TX100 between primary and secondary antibody incubations and mounted with vectashield containing DAPI (Vector Labs). In cells expressing GFP-tagged wild-type (WT) and variant forms of ERCC6L2, endoplasmic reticulum (ER) staining was performed with mouse BiP antibody (Calbiochem). Autophagy vacuoles were stained with LC3β antibody (Santa Cruz). For lysosomes, A549 cells were stained with LysoTracker Red DND-99 (Invitrogen) at a 100 nM/ml concentration; mitochondria were labeled with MitoTracker Orange CMTM Ros (Invitrogen) at 37°C before fixation. Images were collected with an LSM710 laser scanning confocal microscope (Olympus) under relevant laser excitation, and the emitted signals were visualized with ZEN software (Zeiss). For colocalization analysis, the background over noncellular regions was subtracted prior to determination of Pearson’s correlation coefficient (r2) in a minimum of 12 cells in each of three independent experiments. Image processing was limited to contrast enhancement.
siRNA Transfection, Real-Time PCR, and Immunoblotting Densitometry
For small interfering RNA (siRNA) studies, A549 cells were seeded out in 6-well plates at 3.0 × 105 cells per well in antibiotic-free DMEM containing 10% fetal calf serum (Invitrogen) and were transfected the following day with Lipofectamine RNAiMAX (Invitrogen). A pool of two siRNAs (SASI_Hs02_00305873 and SASI_Hs01_00164316, Sigma-Aldrich) was used at a 30 nM final concentration for ERCC6L2 knockdown, and a nontarget siRNA (AllStars Negative Control siRNA, QIAGEN) was used as a negative control. Mock-transfected cells were treated identically but without any siRNA. Knockdown of ERCC6L2 expression was confirmed by quantitative real-time PCR. RNA was extracted with the RNeasy Mini Kit (QIAGEN), and cDNA was synthesized with SuperScript II Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed with TaqMan probes Hs02758991_g1 for ERCC6L2 and Hs00418541_m1 for GAPDH, and reactions were run on an ABI 7500 thermal cycler (Applied Biosystems). For immunoblotting, 30 μg of BSA-quantified protein was loaded, transferred, and analyzed with a WesternBreeze Chromogenic Kit (Invitrogen) and antibodies against ERCC6L2 (Abcam) and β-actin (Abcam), the latter of which was used as a loading control. We performed semiquantitative analysis of raw immunoblots by scanning the colorimetric blots at 600 dpi resolution to TIFF format files, which we subjected to densitometry analysis software (GelPro) to identify quantitative changes in protein levels.
Cell Viability and ROS Evaluation
After 48 hr exposure to ERCC6L2-siRNA-lipofection complexes (SASI_Hs02_00305873 and SASI_Hs01_00164316, Sigma-Aldrich) and nontarget siRNA (1027280, QIAGEN), medium was replaced with DMEM containing 10% FBS supplemented with mitomycin C (MMC), camptothecin (CPT), etoposide (ETP), and Irofulven over a specified dose range in the presence or absence of N-acetyl cysteine (NAC) (all purchased from Sigma-Aldrich). After further 48 hr incubation at 37°C, cells were washed in PBS, propidium iodide was added to the cell suspension to a final concentration of 5 μg/ml, and the cells were analyzed for staining on an LSRII Flow Cytometer (Becton Dickinson). For the evaluation of basal reactive oxygen species (ROS), cells were labeled with ROS marker dihydroethidium (DHE, Invitrogen) at a 5 μM concentration, and the fluorescence was measured by flow cytometry. Real-time changes in ROS fluorescence upon Irofulven treatment were measured at 37°C with standard 520 nm excitation and 610 nm emission at 2 min intervals for a minimum of 60 min with the use of a FLUROstar Optima plate reader (BMG Labtech). All readings were normalized to the basal level (time = 0). We calculated statistical significance by comparing the linear regression of the two curves with GraphPad Prism 5 software (GraphPad).
Subcellular Fractionation
Subcellular fractionation was performed according to a previously described protocol5 with a slight modification. In brief, 1 × 106 DMSO-treated A549 cells and cells treated for 3 hr with MMC (33 nM) or Irofulven (100 nM) were lysed in ice-cold HEPES containing 5 μg/ml digitonin (Sigma Aldrich) and centrifuged at 3,000 rpm for extraction of cytoplasmic protein. Cell pellets were resuspended in 1% Triton-X-114 in ice-cold PBS containing a cocktail of protease and phosphatase inhibitors and centrifuged at 14,000 rpm at 4°C for pelleting nuclei; the resulting supernatants consisting of membrane proteins were mixed in sample buffer. The pellet-containing nuclei were briefly washed three times in ice-cold radioimmunoprecipitation assay buffer containing a cocktail of protease and phosphatase inhibitors (Roche); sonication followed twice for 10 s at 50% pulse. The final mixture was shaken gently on ice for 15 min, and the protein supernatant was obtained by centrifugation of lysates at 14,000 × g for 15 min. Fractionated lysates were verified with antibodies against cytoplasmic GAPDH (Abcam), nuclear TATA binding protein (Abcam), and the ER membrane protein BiP (Calbiochem) by immunoblotting as described above.
Isolation of Mitochondrial Enriched Lysates
Extracts enriched for mitochondrial proteins were obtained according to a previously described protocol.6 In brief, after treatment with MMC or Irofulven, cell pellets were resuspended in ten packed cell volumes of buffer A (1 mM TrisHCl, pH 7.4, 0.13 M NaCl, 5 mM KCl, and 7.5 mM MgCl2), pelleted, and resuspended again in six packed cell volumes of homogenization buffer B (10 mM Tris-HCl, pH 6.7, 10 mM KCl, 0.15 mM MgCl2, 1 mM PMSF, and 1 mM DTT). Cells were homogenized on ice, and the homogenate was transferred into a tube containing one packed cell volume of 2 M sucrose solution and mixed gently. The cell mixture was centrifuged at 1,200 × g for 5 min for pelleting unbroken cells, nuclei, and large debris. The supernatant was transferred to another tube and centrifuged at 7,000 × g for 10 min for pelleting mitochondria. The resulting supernatant was saved as soluble cytoplasmic lysate 1. The mitochondrial pellet was resuspended in three packed cell volumes of mitochondrial suspension buffer C (10 mM TrisHCl, pH 6.7, 0.15 mM MgCl2, 0.25 M sucrose, 1 mM PMSF, and 1 mM DTT) and centrifuged again at 9,500 × g for 5 min for repelleting the mitochondria. The resulting supernatant was mixed with cytoplasmic lysate 1, obtained above. The pelleted mitochondria were mixed with 1× sample buffer boiled at 95°C for 5 min and subjected to immunoblotting analysis. Mitochondrial fractionated lysates were verified with mitochondrial COXIV (Abcam) by immunoblotting as described above.
γH2AX Analysis
For evaluating the DNA-damage response, cells were processed for immunocytochemistry as described above with an antibody specific to γH2AX, and the corresponding goat anti-rabbit secondary antibody was conjugated to Alexa Fluor 488 (Invitrogen) in blocking solution for 1 hr separately. Images were acquired with a Leica Epi fluorescence microscope with MetaMorph (Molecular Devices) image capture software under relevant filters. Captured images were processed for Image J analysis for acquiring γH2AX quantification, and the quantified florescence value obtained from each field of view was normalized to the number of cells stained by DAPI.
Statistics
In bar graphs, a Student’s t test was used for determining significant differences between control and experimental groups. The significant change in ROS (Figure 5A; n = 3) and data are plotted ± SEM. In line graphs, for each experimental data set, linear regression was conducted for determining the best-fit line describing the data from each independent experiment. The overall significance of cytotoxicity (e.g., Figure 3A and B) or in ROS accumulation rate (e.g., Figure 5B) was then determined with a Student two-tailed t test of slopes of the regression lines from each data set (n = 3 independent experiments performed in octuplicate). Where indicated, statistical analysis was performed with GraphPad Prism software, and a p value < 0.05 was considered statistically significant.
Figure 5.
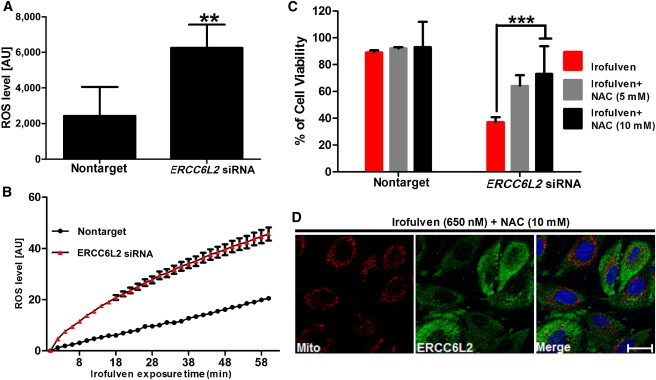
The Relationship among ERCC6L2, ROS, and Genotoxic Stress
(A) Compared to cells transfected with nontarget siRNA, A549 cells transfected with ERCC6L2 siRNA showed an increase in intracellular ROS levels. Error bars represent the SEM of three independent experiments performed in triplicate (∗∗p < 0.01).
(B) Changes in ROS levels upon Irofulven stimulation were monitored over time. Error bars represent the SEM of eight readings in three independent experiments.
(C) Analysis with fluorescence-activated cell sorting revealed reduced cytotoxicity of Irofulven in ERCC6L2-knockdown cells in the presence of NAC in a concentration-dependent manner. Error bars represent the SEM from three independent experiments.
(D) Immunocytochemistry on A549 cells treated with Irofulven (650 nM) in the presence of NAC (10 mM). Images display MitoTracker (red), ERCC6L2 (green), and DAPI (blue). Panels are representative of images taken from different fields of view in three separate experiments. The scale bar represents 30 μm.
Figure 3.
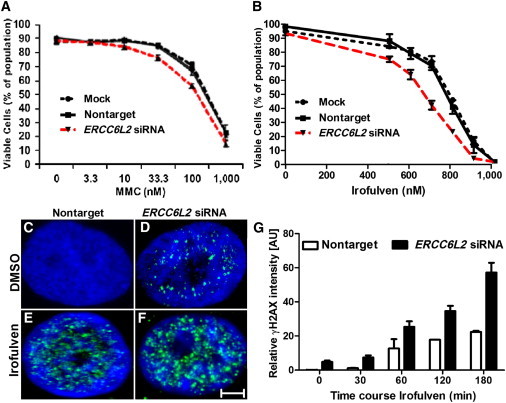
ERCC6L2 Plays a Role in the DNA-Damage-Response Pathway
(A and B) Compared to nontarget-siRNA-transfected cells, ERCC6L2-knockdown cells showed reduced survival after 48 hr treatment with MMC and Irofulven in a dose-dependent manner. Error bars represent the SEM obtained from three independent experiments (one-way ANOVA with Tukey’s test).
(C and D) Compared to nontarget-siRNA-transfected cells, ERCC6L2-knockdown cells revealed γH2AX foci at basal level.
(E and F) γH2AX foci were higher in number in ERCC6L2-knockdown cells than in nontarget-siRNA-transfected cells after treatment with Irofulven. Panels are representative of images taken from different fields of view displaying γH2AX (green) and DAPI (blue). The scale bar represents 5 μm.
(G) The bar graph represents the quantified γH2AX fluorescence value obtained from each individual field of view and normalized to the number of cells stained by DAPI (n > 50) at each individual time point. Error bars represent the SEM (Mann-Whitney test), derived from data obtained from five fields of view at each individual time point from two independent experiments.
Results
Exome Sequencing Identifies Truncating Mutations in ERCC6L2
Exome sequencing revealed that two (cases 1 and 2, Table 1) of the three cases we studied with bone marrow failure and developmental delay had homozygous truncating variants in ERCC6L2 (alias RAD26L; RefSeq accession number NM_001010895.2). No other genes with biallelic rare or somewhat rare variants were found to be shared among the three cases. Specifically, the ERCC6L2 variants identified were c.1963C>T (p.Arg655∗) (Figure 1A) and c.1236_1239delAACA (p.Thr413Cysfs∗2) (Figure 1B). To our knowledge, the c.1236_1239del variant has not been reported in any publicly available database, but c.1963C>T is present in the NHLBI Exome Variant Server at a frequency of 6 in 12,998 alleles, although not in the homozygous state. No loss-of-function (LOF) variant in ERCC6L2 was detected in an additional data set of 1,005 locally sequenced exomes (UCL-ex Consortium). Owing to this rarity of LOF variants (<1 in 2,000 alleles), the presence of biallelic LOF variants in two out of three cases is extremely unlikely (chi-square, p < 10−10). On that basis, we considered that the ERCC6L2 mutations were the likely cause of disease in these two individuals. No obvious disease-causing variant was identified in the third case.
Figure 1.
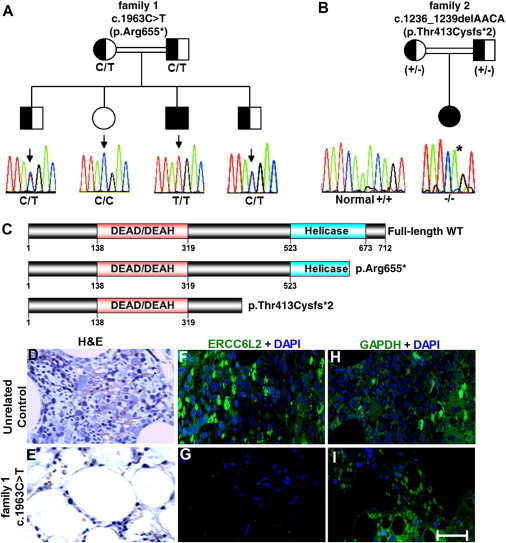
Truncating Mutations in ERCC6L2
(A and B) Shown are two families in which ERCC6L2 mutations segregate as an autosomal-recessive trait. A Sanger sequencing trace and the genotype of each individual are given; inferred genotypes are in parentheses.
(B) For family 2, a plus sign (+) indicates the WT allele and a minus sign (−) indicates the mutant allele. The normal sequencing trace to the left of family 2 comes from an unrelated individual.
(C) The identified ERCC6L2 alterations leading to premature truncation are indicated on a diagram of the protein; functional domains are also annotated.
(D and E) H&E staining of bone marrow trephine biopsies from an unrelated control sample and the index case (case 1) from family 1 reveals the degree of hypoplasia in this individual.
(F and G) Immunostaining on these bone marrow trephine sections revealed the presence of ERCC6L2 in a normal unrelated control, but it was not detected in the affected individual.
(H and I) Positive control staining for GAPDH antigen was observed in both sections. Figures are representative of different images taken from different fields of view. Magnification is 40×. The scale bar represents 50 μm.
Bone Marrow of an Affected Individual Lacks ERCC6L2
ERCC6L2 is located on chromosome 9 (9q 22.32) and spans 14 exons. Expasy prosite analysis of ERCC6L2 revealed an N-terminal DEAH ATP-helicase domain and a catalytic helicase C-terminal domain (Figure 1C). Pfam analysis indicated that it belongs to the Snf2 family of helicase-like proteins, which are involved in transcription regulation, DNA repair, DNA recombination, DNA translocation, and chromatin unwinding.7 ERCC6L2 is 712 amino acids in length and has a predicted molecular weight of 81 kDa. Immunoblotting on human cell lines revealed a 72 kDa band (Figure S1A, available online), and the specificity of the antibody was verified by two siRNAs targeting the ERCC6L2 sequence at two different regions. Both siRNAs decreased the expression of mRNA, as analyzed 48 hr posttransfection by real-time PCR (Figure S1B), and reduced protein levels, as detected by immunoblotting, confirming the efficiency of this antibody in detecting ERCC6L2 (Figure S1C). Immunostudies on A549 cells revealed the presence of ERCC6L2 in both cytoplasmic and nuclear compartments (Figures S1D–S1H).
H&E staining revealed that compared to those of an unrelated control, bone marrow trephine sections obtained from the individual homozygous for the c.1963C>T mutation were hypocellular (Figures 1D and 1E). Immunohistochemistry on these sections showed no detectable staining for ERCC6L2 in the sample from the affected individual, but it did show clear positive cells in the unrelated control (Figures 1F and 1G); both sections stained positive for GAPDH (Figures 1H and 1I). These results indicate that truncating mutations might have an impact on ERCC6L2 stability.
Mislocalization and Degradation of ERCC6L2 Variants
To investigate the impact of truncating mutations on ERCC6L2 stability, we expressed both N-terminal GFP-fused WT and mutant plasmid cDNA constructs in human cell lines. In human A549 cells, WT GFP-ERCC6L2 revealed a continuous diffuse pattern throughout the cytoplasm and nucleus (Figure 2A). In contrast, both truncated forms of GFP-ERCC6L2 showed marked aggregate-like structures (Figures 2B and 2C). The distinct aggregate pattern of the ERCC6L2 variants was not cell-type specific given that it was also seen in 293T cells (Figure S2).
Figure 2.
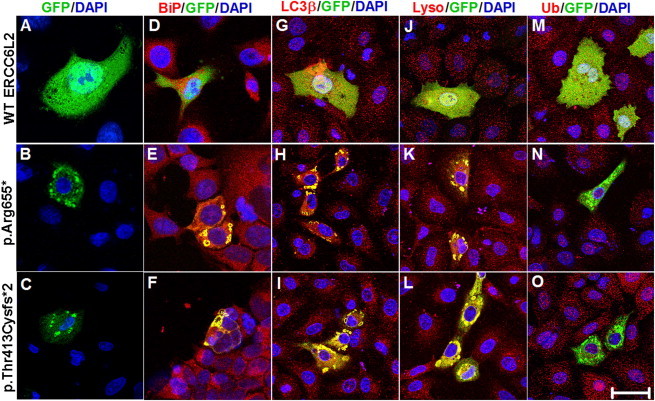
Truncating Mutations Affect the Localization and Degradation of ERCC6L2
(A) Confocal-microscopy images demonstrate predominant cytoplasmic and nuclear localization of WT GFP-ERCC6L2 in A549 cells.
(B and C) Both truncating variants of ERCC6L2 formed aggregates.
(E–L) Colocalization in yellow was observed for both of the altered ERCC6L2 proteins with BiP antibody, which binds to the ER protein BiP (E and F), LC3β antibody, which localizes to autophagic vacuoles (H and I), and LysoTracker, which stains lysosomes (K and L). The colocalization observed was not an artifact of cross-channel noise or bleed from a compliment channel. The Pearson correlation coefficients for colocalization revealed r2 > 0.5 when measured for the entire cell with ZEN software (Zeiss).
(D, G, and J) No colocalization was observed between WT ERCC6L2 and any of these organelle markers (Pearson correlation coefficients r2 < 0.08).
(M–O) Neither the WT nor altered forms of ERCC6L2 showed any colocalization with ubiquitin, as evidenced by the green staining pattern (Pearson correlation coefficient r2 < 0.03). All panels are representative of images taken from different fields of view in three separate experiments. Images display DAPI (blue), GFP-tagged ERCC6L2 (green), and organelle-labeling markers (red). The scale bar represents 30 μm.
Further characterization of these variant ERCC6L2 aggregates in A549 cells revealed that they were localized to the ER (Figures 2D–2F), autophagic vacuoles (Figures 2G–2I), and lysosomes (Figures 2J–2L), as observed by costaining with specific cell organelle markers. However, no colocalization was observed with ubiquitin (Figures 2M–2O). Consistent with the lack of ERCC6L2 staining in the affected individual’s bone marrow, these studies demonstrate that both truncating alterations in ERCC6L2 impair its normal localization and thus result in aggregation in the ER and likely degradation through ER-associated autophagy.8,9
ERCCL2 Knockdown Reduces Cell Viability after Genotoxic Stress
Until now, the function of ERCC6L2 has not been defined experimentally, but it has been shown to have a ubiquitous expression profile (Figure S3). Among the Snf2 family of proteins, it shares the greatest peptide sequence homology with the chromatin-remodelling and DNA-repair proteins ALC1, CHD1, CHD4, RAD54, ATRX, and ERCC6 (Figures S4A and S4B). Mutations in ATRX (MIM 300032) have been linked to X-linked mental retardation most often accompanied by alpha-thalassemia (ATRX syndrome [MIM 301040]),10 and mutations in ERCC6 (MIM 609413) have been shown to cause Cockayne syndrome (CS [MIM 133540]), characterized clinically by dwarfism, microcephaly, cachexia, and neurodegeneration.11–13 Functional studies on RAD54, ALC1, CHD1, and CHD4 in eukaryotic cells revealed their role in facilitating DNA repair through chromatin remodelling.14 Loss of ATRX has been shown to cause genomic instability and altered DNA-damage response.15 ERCC6 has also been shown to play an important role in a subpathway of nucleotide excision repair (NER), known as transcription-coupled NER (TC-NER), and also in mitochondrial function.16–21 Considering the homology of these genes and the overlap of the clinical features seen in our individuals (specifically learning difficulties and microcephaly), we speculate that ERCC6L2 might also play a role in the DNA-damage response.
To test this, we attempted to mimic the truncating mutations by using siRNA to knock down ERCC6L2 expression in human A549 cells. After successful knockdown (Figure S1B), we exposed the cells to the following clastogens: (1) MMC, a compound that causes interstrand crosslinks that can be repaired by the FA pathway or NER,22 (2) Irofulven, a compound that specifically induces DNA adducts recognized by TC-NER, but not global genome NER,23 and (3) CPT and (4) ETP, which induce double-stranded DNA breaks via inhibition of topoisomerase I and II, respectively. After 48 hr exposure, cell viability was assessed by flow cytometry. Compared to mock-transfected cells or cells transfected with nontarget siRNA, cells transfected with ERCC6L2 siRNA showed significantly reduced viability after exposure to MMC (unpaired Student’s two-tailed t tests, p < 0.01, Figure 3A) and Irofulven (unpaired Student’s two-tailed t tests, p < 0.001, Figure 3B) in a dose-dependent manner. With CPT or ETP, no difference was observed in cell survival between cells transfected with ERCC6L2 siRNA and cells transfected with nontarget siRNA (Figure S5). The consistent increase in MMC and Irofulven sensitivity suggests a role for ERCC6L2 in the repair of interstrand crosslinks and specifically in TC-NER of DNA damage. We note that there is evidence suggesting a role for TC-NER in the repair of MMC-induced interstrand crosslinks.24 Despite the fact that ERCC6L2-knockdown cells also showed increased susceptibility to ETP and, to a lesser extent, CPT, a similar pattern was also observed in cells treated with nontarget siRNA. These results prevent us from concluding that ERCC6L2 also plays a role in DNA-repair pathways activated upon topoisomerase inhibition.
Knockdown of ERCC6L2 Induces a DNA-Damage Response
Snf2 protein complexes have been shown to be involved in the recruitment of γH2AX, a phosphorylated form of histone 2A, to sites of DNA damage.25 Given the increased sensitivity of ERCC6L2-knockdown cells to MMC and Irofulven, we wanted to investigate the role of ERCC6L2 with respect to this marker of DNA damage. Immunostaining with a γH2AX-specific antibody revealed the presence of discrete foci in ERCC6L2-knockdown cells in the absence of any genotoxic stress (Figures 3C and 3D). Compared to nontarget-siRNA-transfected cells, cells treated with Irofulven for 3 hr at a low concentration (100 nM) showed a further significant increase in the level of γH2AX (Figures 3E and 3F). The kinetics of the increase in γH2AX was more pronounced in ERCC6L2-knockdown cells than in nontarget-siRNA-transfected cells over time, suggesting an early DNA-damage response (Figure 3G). Collectively, these results indicate that ERCC6L2 plays a role in the DNA-damage-response pathway and that its depletion sensitizes cells to genotoxic agents.
ERCC6L2 Traffics to the Mitochondria and Nucleus after Genotoxic Stress
To further characterize the response of ERCC6L2 to DNA-damaging agents, we performed immunocytochemistry on A549 cells. Compared to the normal diffuse cytosolic and nuclear staining pattern seen in control DMSO-treated cells (Figure S6A), a distinct perinuclear accumulation of ERCC6L2 was seen in the cells treated with both MMC and Irofulven (Figures S6B and S6C). Immunoblotting on subcellular fractionated protein lysates obtained from these cells revealed a dramatic translocation of ERCC6L2 from cytosolic to membraneous compartments and, to a lesser extent, toward the nucleus after treatment with MMC or Irofulven for 3 hr (Figures 4A and 4B).
Figure 4.
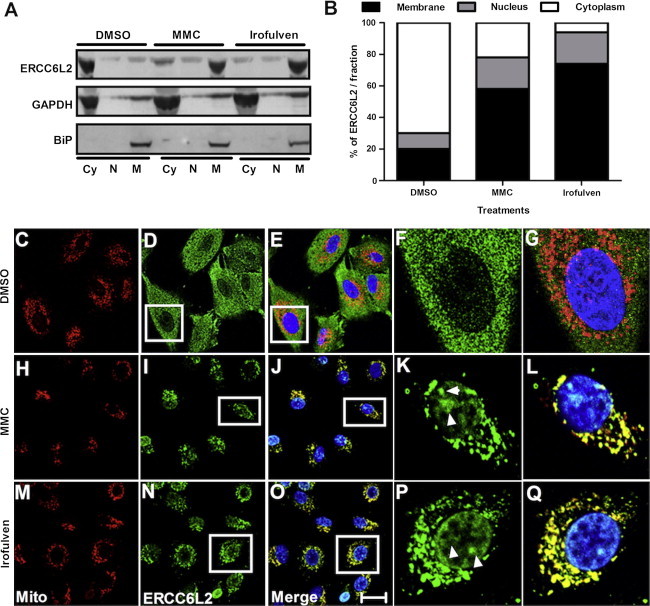
ERCC6L2 Traffics to the Mitochondria and Nucleus after Genotoxic Stress
(A) Immunoblotting was performed on subcellular fractionated protein lysates after MMC and Irofulven treatment with ERCC6L2 antibody. Abbreviations are as follows: Cy, cytosolic; N, nuclear; and M, membraneous. Subcellular fractionation was verified by GAPDH as a cytoplasmic marker and BiP as a membrane marker.
(B) The increased membraneous and nuclear localization of ERCC6L2 is represented graphically by extrapolation of the values acquired from densitometry analysis.
(C–Q) Confocal images show ERCC6L2 localization to the mitochondria and nucleus after genotoxic stress. Nuclear localization of ERCC6L2 (white arrows) showed a correlation with DAPI staining after MMC (I and K) and Irofulven (N and P) treatment but to a lesser extent with DMSO (D and F). Colocalization of ERCC6L2 to the mitochondria is represented in yellow for MMC (J and L) and Irofulven (O and Q) treatment, but not DMSO (E and G) treatment. The colocalization observed was not an artifact of cross-channel noise or bleed from a compliment channel. For colocalization analysis, the Pearson correlation coefficients (r2) of the relative distribution of the two channels in mitochondrial regions of the cells revealed r2 > 0.482 after MMC treatment, r2 > 0.739 after Irofulven treatment, and r2 < 0.03 after DMSO treatment. Images display MitoTracker (red), ERCC6L2 (green), and DAPI (blue). Panels are representative of images taken from different fields of view in three separate experiments. The scale bar represents 30 μm.
ERCC6L2 has a distinct 13 amino acid N-terminal sequence that harbors a putative mitochondrial targeting sequence, as predicted by MitoProt software.26 This prompted us to investigate extracts enriched with mitochondria obtained from A549 cells, and indeed these showed an higher level of ERCC6L2 in mitochondria than in the cytoplasm in cells treated with MMC and Irofulven (Figures S6D and S6E). Immunocytochemistry on cells labeled with MitoTracker and DAPI confirmed the predominant localization of ERCC6L2 in the mitochondria (Figures 4C–4Q) and nucleus (Figures 4F, 4K, and 4P) after MMC and Irofulven treatment. These results are particularly intriguing in light of recent data indicating that ERCC6 might act as a sensor of mtDNA damage19–21 and the central role that mitochondrial dysfunction plays in the process of normal human aging.27
ERCC6L2 Knockdown Increases Intracellular ROS
Translocation of ERCC6L2 to the mitochondria in response to DNA-damaging agents led us to investigate its role in mitochondrial function. We used ERCC6L2-knockdown cells and nontarget-siRNA-transfected cells to quantify intracellular ROS by using the ethidium-based probe DHE. After successful knockdown, cells were stained with DHE and subjected to flow cytometry. Compared to cells transfected with nontarget siRNA, ERCC6L2-knockdown cells showed a significant increase in intracellular ROS (Figure 5A). To further investigate the changes in intracellular ROS, we induced genotoxic stress with Irofulven (100 nM) and monitored the change in intracellular ROS over time. After treatment with Irofulven, ERCC6L2-knockdown cells showed a significant increase (p < 0.0001, comparing the linear regression of two curves) in the change of intracellular ROS level over time in comparison to nontarget transfected cells (Figure 5B). These results indicate that depletion of ERCC6L2 results in increased ROS.
The Response of ERCC6L2 to Genotoxic Stress Is ROS Dependent
To investigate a possible role for ROS in sensitizing ERCC6L2-knockdown cells to genotoxic stress, we used NAC, a well-known antioxidant that scavenges ROS. ERCC6L2-knockdown cells and nontarget-siRNA-transfected cells were treated with Irofulven at a cytotoxic concentration (650 nM, refer to Figure 3B) in the presence of NAC at two different concentrations (5 and 10 mM). After 48 hr, flow cytometry analysis revealed that compared to ERCC6L2-knockdown cells treated with Irofulven alone, ERCC6L2-knockdown cells treated with Irofulven and NAC had a significant (p < 0.0001) reduction of cell death in an NAC-dose-dependent manner (Figure 5C). ERCC6L2-knockdown cells also stained negative for γH2AX upon treatment with Irofulven in the presence of NAC (data not shown). In addition, NAC inhibited the Irofulven-induced translocation of ERCC6L2 to mitochondria (Figure 5D) and the nucleus (Figure S7). These results suggest that ERCC6L2 traffic to both mitochondria and nucleus is ROS dependent.
Discussion
In this study, we report on two homozygous truncating ERCC6L2 mutations identified in individuals with neurological dysfunction (developmental delay and microcephaly) and bone marrow failure (cases 1 and 2, Table 1 and Figure 1). These mutations cause impaired subcellular localization and reduced stability of ERCC6L2 (Figure 2). We show that ERCC6L2 acts as a DNA-damage-response protein by exacerbating γH2AX foci formation in response to DNA-damaging agents (Figure 3). Our data also demonstrate that ERCC6L2 traffics to the mitochondria and nucleus in a ROS-dependent fashion in response to DNA damage (Figures 4 and 5).
Given the deficiency of ERCC6L2 in the bone marrow of individuals harboring LOF variants in ERCC6L2, it is reasonable to suggest that their DNA-damage response is impaired. Because a DNA-damage response is required during cell proliferation and tissue maintenance, its persistent impairment would result in a slow accumulation of DNA damage. In addition to showing the dual traffic of ERCC6L2 from the cytosol to the mitochondria and nucleus after genotoxic stress, we have shown a parallel increase in γH2AX foci and intracellular ROS in ERCC6L2-knockdown cells. Increasing ROS levels leading to alteration in cellular homeostasis is considered to be of major pathological significance in neurodegenerative disorders28 and bone marrow failure.29
There is also evidence that the increase in ROS levels and incompetent DNA-damage repair contributes to the pathogenesis of bone-marrow-failure syndromes.30 For example, although FA is primarily characterized by chromosome instability, developmental abnormalities, and cancer susceptibility, it also shows reduced mitochondrial function and increased ROS production.31 CS and the functionally related xeroderma pigmentosum (XP [MIM 278700, 610651, 278720, 278730, 278740, 278760, and 278780]) are also caused by an inability to repair DNA damage, particularly UV-induced DNA crosslinking32 and increased ROS production.32,33 Cells from CS individuals and CS mouse models have also shown increased ROS activity and accumulation of damaged mitochondria as a result of defective mitochondrial clearance by autophagy.19 Recently, studies on DC lymphoblasts have revealed elevated levels of ROS and a pronounced DNA-damage response leading to a proliferative defect and apoptosis.34,35 It is also known that increased ROS levels impair the self-renewal capacity of hematopoietic progenitor cells in bone marrow.31 Taking all this together, we propose that an increase in ROS and the accumulation of DNA damage are the underlying causes of the disease observed in our affected individuals.
Although some features overlap with previously described syndromes, the individuals reported in this study also have significant differences (Table 1). For instance, FA is characterized by abnormal chromosomal breakage, but analysis of peripheral-blood lymphocytes in our affected individuals did not identify such defects. The increased sensitivity to MMC in FA is also much more pronounced than that observed in our ERCC6L2-knockdown assays (Figure 3A) and the primary cells of these individuals (Table 1). Equally, bone marrow failure is not a reported feature of CS, although neurological abnormalities are present in most cases.36 XP, on the other hand, is primarily characterized by hypersensitivity to sunlight and subsequent development of carcinomas, and only some cases present with neurologic disease.36 Interestingly, a recent study reported a variant in ERCC4 (XPF) in a CS individual with overlapping features of XP (severe photosensitivity and abnormal skin pigmentation) and FA (abnormal chromosomal breakage).37 Hoyeraal Hreidarsson syndrome (HHS [MIM 300240]) is characterized by neurological dysfunction, microcephaly, and bone marrow failure and is considered to be a severe form of DC,38 a telomere maintenance disorder,39,40 which itself commonly manifests with mucocutaneous abnormalities and bone marrow failure.4 Our individuals lacked the mucocutaneous features, immunodeficiency, and cerebellar hypoplasia typically seen in DC and HHS and, in comparison to age-matched controls, had variable telomere lengths when measured by multiplex PCR41 (Table 1 and Figure S8). It is also notable that exome sequencing did not identify variations in any of the known genes associated with bone marrow failure in these individuals.
Because there is no precise match between all the features in the cases reported here and other genetically characterized disorders, we propose the presently described disorder as a distinct bone-marrow-failure syndrome. We also believe that our study highlights the combined roles of intracellular ROS and DNA damage in driving bone marrow failure.
Acknowledgments
We would like to thank the families and clinicians who contributed to this research, as well as Martine French for providing bone marrow slides. This work was funded by The Wellcome Trust and Medical Research Council. We would also like to thank Gary Warnes and Ann Wheeler for support and technical assistance in fluorescence-activated cell sorting and imaging.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://www.1000genomes.org
ANNOVAR, www.openbioinformatics.org/annovar
BioGPS, http://www.biogps.org
MitoProt, http://ihg.gsf.de/ihg/mitoprot.html
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
Pfam, http://pfam.sanger.ac.uk
Prosite, http://www.expasy.ch/Prosite
References
- 1.Young N.S., Bacigalupo A., Marsh J.C. Aplastic anemia: pathophysiology and treatment. Biol. Blood Marrow Transplant. 2010;16(1 Suppl):S119–S125. doi: 10.1016/j.bbmt.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dokal I., Vulliamy T. Inherited bone marrow failure syndromes. Haematologica. 2010;95:1236–1240. doi: 10.3324/haematol.2010.025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walne A.J., Dokal I. Advances in the understanding of dyskeratosis congenita. Br. J. Haematol. 2009;145:164–172. doi: 10.1111/j.1365-2141.2009.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holden P., Horton W.A. Crude subcellular fractionation of cultured mammalian cell lines. BMC Res. Notes. 2009;2:243. doi: 10.1186/1756-0500-2-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kluck R.M., Bossy-Wetzel E., Green D.R., Newmeyer D.D. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 7.Flaus A., Martin D.M., Barton G.J., Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yorimitsu T., Nair U., Yang Z., Klionsky D.J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ratnakumar K., Bernstein E. ATRX: the case of a peculiar chromatin remodeler. Epigenetics. 2013;8:3–9. doi: 10.4161/epi.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallery D.L., Tanganelli B., Colella S., Steingrimsdottir H., van Gool A.J., Troelstra C., Stefanini M., Lehmann A.R. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am. J. Hum. Genet. 1998;62:77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laugel V., Dalloz C., Durand M., Sauvanaud F., Kristensen U., Vincent M.C., Pasquier L., Odent S., Cormier-Daire V., Gener B. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum. Mutat. 2010;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 13.Weidenheim K.M., Dickson D.W., Rapin I. Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech. Ageing Dev. 2009;130:619–636. doi: 10.1016/j.mad.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Lans H., Marteijn J.A., Vermeulen W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin. 2012;5:4–18. doi: 10.1186/1756-8935-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovejoy C.A., Li W., Reisenweber S., Thongthip S., Bruno J., de Lange T., De S., Petrini J.H., Sung P.A., Jasin M., ALT Starr Cancer Consortium Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vélez-Cruz R., Egly J.M. Cockayne syndrome group B (CSB) protein: at the crossroads of transcriptional networks. Mech. Ageing Dev. 2013;134:234–242. doi: 10.1016/j.mad.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Licht C.L., Stevnsner T., Bohr V.A. Cockayne syndrome group B cellular and biochemical functions. Am. J. Hum. Genet. 2003;73:1217–1239. doi: 10.1086/380399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aamann M.D., Sorensen M.M., Hvitby C., Berquist B.R., Muftuoglu M., Tian J., de Souza-Pinto N.C., Scheibye-Knudsen M., Wilson D.M., 3rd, Stevnsner T., Bohr V.A. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 2010;24:2334–2346. doi: 10.1096/fj.09-147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheibye-Knudsen M., Ramamoorthy M., Sykora P., Maynard S., Lin P.C., Minor R.K., Wilson D.M., 3rd, Cooper M., Spencer R., de Cabo R. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J. Exp. Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berquist B.R., Canugovi C., Sykora P., Wilson D.M., 3rd, Bohr V.A. Human Cockayne syndrome B protein reciprocally communicates with mitochondrial proteins and promotes transcriptional elongation. Nucleic Acids Res. 2012;40:8392–8405. doi: 10.1093/nar/gks565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamenisch Y., Berneburg M. Mitochondrial CSA and CSB: protein interactions and protection from ageing associated DNA mutations. Mech. Ageing Dev. 2013;134:270–274. doi: 10.1016/j.mad.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Iyer V.N., Szybalski W. A molecular mechanism of mitomycin action: linking of complimentary strands. Proc. Natl. Acad. Sci. USA. 1963;50:355–362. doi: 10.1073/pnas.50.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koeppel F., Poindessous V., Lazar V., Raymond E., Sarasin A., Larsen A.K. Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with XPG expression in solid tumor cells. Clin. Cancer Res. 2004;10:5604–5613. doi: 10.1158/1078-0432.CCR-04-0442. [DOI] [PubMed] [Google Scholar]
- 24.Zheng H., Wang X., Warren A.J., Legerski R.J., Nairn R.S., Hamilton J.W., Li L. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Mol. Cell. Biol. 2003;23:754–761. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan D.P., Owen-Hughes T. Snf2-family proteins: chromatin remodellers for any occasion. Curr. Opin. Chem. Biol. 2011;15:649–656. doi: 10.1016/j.cbpa.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claros M.G., Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 27.Sahin E., DePinho R.A. Axis of ageing: telomeres, p53 and mitochondria. Nat. Rev. Mol. Cell Biol. 2012;13:397–404. doi: 10.1038/nrm3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trushina E., McMurray C.T. Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience. 2007;145:1233–1248. doi: 10.1016/j.neuroscience.2006.10.056. [DOI] [PubMed] [Google Scholar]
- 29.Rossi D.J., Jamieson C.H., Weissman I.L. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 30.Pang Q. HSCs: stressing out over ROS. Blood. 2011;118:2932–2934. doi: 10.1182/blood-2011-07-367755. [DOI] [PubMed] [Google Scholar]
- 31.Kumari U., Ya Jun W., Huat Bay B., Lyakhovich A. Evidence of mitochondrial dysfunction and impaired ROS detoxifying machinery in Fanconi Anemia cells. Oncogene. 2014;33:165–172. doi: 10.1038/onc.2012.583. [DOI] [PubMed] [Google Scholar]
- 32.Kraemer K.H., Patronas N.J., Schiffmann R., Brooks B.P., Tamura D., DiGiovanna J.J. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi M. Roles of oxidative stress in xeroderma pigmentosum. Adv. Exp. Med. Biol. 2008;637:120–127. doi: 10.1007/978-0-387-09599-8_13. [DOI] [PubMed] [Google Scholar]
- 34.Pereboeva L., Westin E., Patel T., Flaniken I., Lamb L., Klingelhutz A., Goldman F. DNA damage responses and oxidative stress in dyskeratosis congenita. PLoS ONE. 2013;8:e76473. doi: 10.1371/journal.pone.0076473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirwan M., Beswick R., Walne A.J., Hossain U., Casimir C., Vulliamy T., Dokal I. Dyskeratosis congenita and the DNA damage response. Br. J. Haematol. 2011;153:634–643. doi: 10.1111/j.1365-2141.2011.08679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks P.J. Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst.) 2013;12:656–671. doi: 10.1016/j.dnarep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kashiyama K., Nakazawa Y., Pilz D.T., Guo C., Shimada M., Sasaki K., Fawcett H., Wing J.F., Lewin S.O., Carr L. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am. J. Hum. Genet. 2013;92:807–819. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dokal I. Dyskeratosis congenita. Hematology (Am Soc Hematol Educ Program) 2011;2011:480–486. doi: 10.1182/asheducation-2011.1.480. [DOI] [PubMed] [Google Scholar]
- 39.Shtessel L., Ahmed S. Telomere dysfunction in human bone marrow failure syndromes. Nucleus. 2011;2:24–29. doi: 10.4161/nucl.2.1.13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savage S.A., Bertuch A.A. The genetics and clinical manifestations of telomere biology disorders. Genet. Med. 2010;12:753–764. doi: 10.1097/GIM.0b013e3181f415b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cawthon R.M. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


