Abstract
Plant-associated bacteria fulfill important functions for plant growth and health. However, our knowledge about the impact of bacterial treatments on the host's microbiome and physiology is limited. The present study was conducted to assess the impact of bacterial inoculants on the microbiome of chamomile plants Chamomilla recutita (L.) Rauschert grown in a field under organic management in Egypt. Chamomile seedlings were inoculated with three indigenous Gram-positive strains (Streptomyces subrutilus Wbn2-11, Bacillus subtilis Co1-6, Paenibacillus polymyxa Mc5Re-14) from Egypt and three European Gram-negative strains (Pseudomonas fluorescens L13-6-12, Stenotrophomonas rhizophila P69, Serratia plymuthica 3Re4-18) already known for their beneficial plant-microbe interaction. Molecular fingerprints of 16S rRNA gene as well as real-time PCR analyses did not show statistically significant differences for all applied bacterial antagonists compared to the control. In contrast, a pyrosequencing analysis of the 16S rRNA gene libraries revealed significant differences in the community structure of bacteria between the treatments. These differences could be clearly shown by a shift within the community structure and corresponding beta-diversity indices. Moreover, B. subtilis Co1-6 and P. polymyxa Mc5Re-14 showed an enhancement of the bioactive secondary metabolite apigenin-7-O-glucoside. This indicates a possible new function of bacterial inoculants: to interact with the plant microbiome as well as to influence the plant metabolome.
Keywords: bioactive secondary metabolites, biological control agents, chamomile, microbial communities, soil-borne pathogens
Introduction
Whereas large-scale efforts have rapidly advanced the understanding of plant genomes, the impact and importance of the plant's microbiome is largely unexplored. Comparable to the human microbiome, millions of microbes inhabit plants, forming a complex ecological community that influences plant growth and health through its collective metabolic activities and host interactions (Berg, 2009; Lugtenberg and Kamilova, 2009). Currently, the many studies on plant-associated microorganisms reflect the full effect of ongoing research and the enormous interest in this topic (Mendes et al., 2011, 2013; Berendsen et al., 2012; Bakker et al., 2013). Viewing the microbiota from an ecological perspective could provide insight into how to promote health and stress tolerance of their hosts or how to adapt to a changing climate by targeting this microbial community. Furthermore, new functions of the plant microbiome can be detected. Several studies revealed that rhizobacteria have an effect on the aroma of fruits, e.g., strawberry and grapes (Pirlak and Köse, 2009; Verginer et al., 2010). Moreover, induced systemic resistance has been triggered in several crops by plant growth promoting bacteria (Murphy et al., 2003; Kloepper et al., 2004; Ryu et al., 2004a,b). Interestingly, certain plant growth-promoting rhizobacteria (PGPR) elicit induced systemic resistance and plant growth promotion in the absence of physical contact with plants via volatile organic compound (VOC) emissions (Farag et al., 2013). In addition, selected PGPR strains have been shown to reduce disease in plant parts through the induction of defense compounds, and especially members of the endomicrobiome have been shown to be involved in the production of bioactive compounds (Pimentel et al., 2011; Gutierrez et al., 2012). For several medicinal plants, bacteria and fungi were detected as producers of their active ingredients. Paclitaxel and maytansine, known as important anticancer lead molecules, were detected to be produced by endophytes (Chandra, 2012; Wings et al., 2013). Traditional Chinese Medicine is using indigenous medicinal plants integrated in an ancient healing system originating almost 4500 ago. For several of those plants it was shown that they are active due to its microbial endophytes (Miller et al., 2012; Zhao et al., 2012). However, these discoveries are only the beginning of understanding the complex interactions between plants and microbes, and new omics technologies will promote this.
Promotion of plant health or biological control is one of the well-studied functions of the plant microbiome. It is based on naturally occurring antagonists and offers sustainable solutions for plant protection (Weller, 2007; Berg, 2009; Raaijmakers et al., 2009). Gram-negative bacteria, especially those from the genus Pseudomonas, were identified as the dominant members of the indigenous antagonistic communities under humid conditions (Berg et al., 2005a; Haas and Defago, 2005; Costa et al., 2006; Zachow et al., 2008). In addition, these strains were also identified as a major group of disease-suppressive bacteria through pyrosequencing (Mendes et al., 2011). In contrast, under arid conditions, we found mainly Gram-positive bacteria as antagonistic counterparts (Köberl et al., 2011). To verify this finding as well as to find out which bacterial strains—indigenous Gram-positive strains in comparison with allochthonous Gram-negative strains—support plant growth under arid conditions in desert farming, different bacterial inoculants were developed and tested under field conditions.
The overall aim of this study was to evaluate the impact of several bacterial inoculants on the indigenous microbial community of chamomile plants and the production of bioactive secondary metabolites. We selected three Gram-positive strains isolated in Egypt and three European Gram-negative strains already known for their beneficial plant-microbe interactions and used as biocontrol agents (Lottmann and Berg, 2001; Zachow et al., 2010). Bacterial inoculants were applied to chamomile plants [Chamomilla recutita (L.) Rauschert] grown under field conditions with organic (biodynamic) management on Sekem farms in Egypt, and the impact of the treatment on the indigenous microbial communities was monitored. This is important to understand the potential risk of biocontrol but also to understand the mode of action of bacterial inoculants, which can be mediated by the plant microbiome as well (Scherwinski et al., 2008).
Results
Chemical analysis of chamomile secondary metabolites
HPLC-MS experiments of the flower extracts yielded slightly different contents of apigenin-7-O-glucoside and apigenin between the different treatments (Figure 1). For apigenin-7-O-glucoside, statistically significant higher contents than for the non-inoculated control plants (0.86 ± 0.04) were observed for the treatments with the indigenous Gram-positive strains Bacillus subtilis Co1-6 (1.06% ± 0.07) and Paenibacillus polymyxa Mc5Re-14 (1.04% ± 0.06). The autochthonous Streptomyces subrutilus Wb2n-11 (0.87% ± 0.09) and all three allochthonous Gram-negative strains Pseudomonas fluorescens L13-6-12 (0.79% ± 0.06), Stenotrophomonas rhizophila P69 (0.78% ± 0.15), and Serratia plymuthica 3Re4-18 (0.82% ± 0.06) showed no elevation of apigenin-7-O-glucoside content (Table S1). Highest contents of apigenin were obtained for treatments with the Gram-positive strains B. subtilis Co1-6 (0.95% ± 0.14) and P. polymyxa Mc5Re-14 (0.95% ± 0.10) as well, however without significant difference to the control (0.94% ± 0.10). Also P. fluorescens L13-6-12 (0.84% ± 0.07), S. rhizophila P69 (0.77% ± 0.12), S. plymuthica 3Re4-18 (0.91% ± 0.10), and S. subrutilus Wb2n-11 (0.78% ± 0.08) did not show an alteration (Table S2). Investigation of peaks which arose in the treatments with the two autochthonous Bacillales strains (m/z = 327 and 329 for [M-H]−) resulted in the possible identification of [C18H32O5 – H+]− (Rt: 13.15 min) and [C18H34O5 – H+]− (Rt: 14.8 min), respectively.
Figure 1.
Content (%) of apigenin-7-O-glucoside (A) and apigenin (B) in Chamomilla recutita (L.) Rauschert samples. Averages of individual HPLC-MS measurements and confidences are shown. Significant differences (p < 0.05) of bacterial treatments (Streptomyces subrutilus Wb2n-11, Bacillus subtilis Co1-6, Paenibacillus polymyxa Mc5Re-14, Pseudomonas fluorescens L13-6-12, Stenotrophomonas rhizophila P69, Serratia plymuthica 3Re4-18) to the control are indicated by asterisks.
Molecular fingerprinting of the chamomile-associated bacterial communities
In the molecular fingerprinting approach, universal primers were used to get a first overview about the whole bacterial community associated with Chamomilla recutita (L.) Rauschert as well as Pseudomonas- and Firmicutes-specific primers. Community composition was determined by image analysis of the band profiles generated by single-stranded conformational polymorphism (SSCP) analysis, and differences in the bacterial community composition based on these band patterns were calculated using the Pearson's correlation index. Cluster analysis of the microbial profiles resulted in grouping of rhizosphere samples at different sampling times at about 10% similarity (Figure 2). These differences between the two sampling times were confirmed by a principal component analysis (Figure 3). Within each cluster, only several of the samples from the different treatments were grouped together. Generally, samples from different treatments were found to be more similar to each other than the samples of the five replicates of each specific treatment, suggesting no significant differences arising from the bacterial inoculants. Moreover, no cluster group was found containing exclusively SSCP patterns of one specific treatment.
Figure 2.
Cluster analysis of eubacterial community fingerprints. Similarities between SSCP fingerprints were calculated using the curve-based Pearson correlation coefficient and grouped according to their similarity using the hierarchical UPGMA. Treatments (Streptomyces subrutilus Wb2n-11, Bacillus subtilis Co1-6, Paenibacillus polymyxa Mc5Re-14, Pseudomonas fluorescens L13-6-12, Stenotrophomonas rhizophila P69, Serratia plymuthica 3Re4-18, and water control) and sampling times (1st) after 4 weeks and (2nd) after 8 weeks are indicated. Numbers 1–5 mark the five independent replicates per treatment.
Figure 3.

Principal component analysis (PCA) of eubacterial (A), Pseudomonas (B) and Firmicutes (C) community fingerprints. Treatments (Streptomyces subrutilus Wb2n-11, Bacillus subtilis Co1-6, Paenibacillus polymyxa Mc5Re-14, Pseudomonas fluorescens L13-6-12, Stenotrophomonas rhizophila P69, Serratia plymuthica 3Re4-18, and water control) are indicated by colors. Sampling time (1st) after 4 weeks is shown on the left side and (2nd) after 8 weeks is shown on the right side. PCA was calculated based on relative positions and intensity of DNA bands.
The bacterial community associated with C. recutita (L.) Rauschert showed a high abundance of Bacillus spp. (Table S3). In the Pseudomonas community two dominant bands could be detected, which were abundant in both sampling times. These two bands were identified by partial 16S rRNA gene sequence analysis as Pseudomonas sp. (closest database match Pseudomonas sp. MOC14, 100% similarity to JX122114.1) (Table S4). In the Firmicutes community Bacillus spp. were found in all samples, whereas Bacillus sp. BMR7, 99–100% similarity to JX434152.1 and Bacillus sp. DV9-6, 99% similarity to GQ407151.1, represented the most abundant species (Table S5).
Pyrosequencing-based 16S rRNA profiling of chamomile-associated bacterial communities
To deeply investigate the diversity and the composition of the bacterial communities associated with C. recutita (L.) Rauschert, a pyrosequencing approach was employed. Rarefaction analysis was performed to an extent of diversity coverage (Figure S1). Assessment of richness revealed that pyrosequencing effort attained 35.8-46.5% of estimated richness at a genetic similarity of 97% (Table 1). At the genetic similarity levels of 95% and 80%, amplicon libraries covered 41.7–49.7% and 56.6–88.8% of estimated richness, respectively (Table 1). Taxonomic composition of bacterial communities was similar at phylum level, comprising Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria as the most dominant phyla (Figure 4). However, the phylum Verrucomicrobia was only present in the sample from the treatment with S. rhizophila P69, considering only taxa covering more than 1% of quality sequences. Acidobacteria were observed in samples treated with B. subtilis Co1-6, S. rhizophila P69, and S. plymuthica 3Re4-18. At genus level, Rhizobium (phylum Proteobacteria), Pseudoxanthomonas (phylum Proteobacteria), Pseudomonas (phylum Proteobacteria), Flavobacterium (phylum Bacteroidetes), and Arthrobacter (phylum Actinobacteria) represented the most abundant genera, showing a different composition according to the different treatments. Alpha-diversity of the amplicon libraries was characterized by Shannon index (H') for 97, 95, and 80% similarity levels. Slight differences between treatments where revealed by the comparison of the index values (Table 1). Jackknifed weighted UniFrac two-dimensional (Figure 5) and three-dimensional (Figure 6) principal coordinates analysis (PCoA) biplots were constructed in order to visualize relationships among samples based on differences in taxonomic diversity. Weighted biplots showed that the samples were clearly separated, implying a difference in bacterial community composition according to the treatments.
Table 1.
Richness estimates and diversity indices for amplicon libraries of rhizosphere samplesa.
| Genetic similarityb | Samplec | No. of OTUs | Chao1 | Coverage (%) | H'd |
|---|---|---|---|---|---|
| 97% | Wb2n-11 | 557 | 1443 | 38.6 | 5.14 |
| Co1-6 | 596 | 1280 | 46.5 | 5.32 | |
| Mc5Re-14 | 729 | 1928 | 37.8 | 5.66 | |
| P69 | 559 | 1209 | 46.2 | 5.23 | |
| 3Re4-18 | 546 | 1525 | 35.8 | 5.37 | |
| 95% | Wb2n-11 | 433 | 871 | 49.7 | 4.80 |
| Co1-6 | 477 | 998 | 47.8 | 5.00 | |
| Mc5Re-14 | 560 | 1342 | 41.7 | 5.26 | |
| P69 | 435 | 930 | 46.8 | 4.90 | |
| 3Re4-18 | 419 | 973 | 43.1 | 5.01 | |
| 80% | Wb2n-11 | 81 | 143 | 56.6 | 2.85 |
| Co1-6 | 76 | 86 | 88.8 | 3.05 | |
| Mc5Re-14 | 90 | 134 | 67.4 | 2.87 | |
| P69 | 92 | 120 | 76.9 | 3.11 | |
| 3Re4-18 | 89 | 100 | 88.6 | 3.20 |
The number of sequences of each sample was normalized to 1858.
Genetic similarities represent the taxonomic levels of species (97%), genera (95%), and phyla (80%).
Abbreviations correspond to the treatments with Streptomyces subrutilus (Wb2n-11), Bacillus subtilis (Co1-6), Paenibacillus polymyxa (Mc5Re-14), Stenotrophomonas rhizophila (P69), and Serratia plymuthica (3Re4-18).
Shannon diversity indices.
Figure 4.
Classification of bacterial communities associated with Chamomilla recutita (L.) Rauschert. Pyrosequencing reads were classified at phylum (A) and genus (B) level against RDP core set within QIIME pipeline with an 80% confidence threshold. Taxa below 1% of relative abundance are included in “Other.” Multi-colored charts at the legend are shown for each sample correspondingly.
Figure 5.

Comparison of the microbial communities of Chamomilla recutita (L.) Rauschert rhizosphere by jackknifed principal coordinate analysis. The 2D-plot illustrates the compositional similarity between samples based on weighted UniFrac. The positions of the points are the averages for the jackknifed replicates generated by QIIME and are shown with ellipses representing the interquartile range (IQR) in each axis. Larger ellipses represent more diverse communities. Colors correspond to the different treatments.
Figure 6.
Comparison of the microbial communities of Chamomilla recutita (L.) Rauschert rhizosphere by jackknifed principal coordinate analysis. The biplot illustrates the compositional similarity between samples based on weighted UniFrac. Taxa coordinates are indicated by gray orbs with size, as a function of relative abundance. To confine the biplot, the number of the displayed taxa was restricted to 5. The positions of the points are the averages for the jackknifed replicates generated by QIIME and are shown with ellipses representing the interquartile range (IQR) in each axis. Larger ellipses represent more diverse communities. Colors correspond to the different treatments.
Quantitative analysis of bacterial abundances
The bacterial abundances of total bacteria and Firmicutes were determined by quantitative PCR. For total bacteria, abundances ranged from 7.45 log10 copies per g root fresh weight (fw) for the treatment with S. subrutilus Wb2n-11 to 8.03 log10 copies per g fw for the treatment with P. polymyxa Mc5Re-14 (Figure S2). Abundances for Firmicutes ranged from 6.97 log10 copies per g fw for the treatment with S. subrutilus Wb2n-11 to 7.54 log10 copies per g fw for the treatment with P. polymyxa Mc5Re-14 (Figure S2). However, no statistically significant differences between the treatments could be detected.
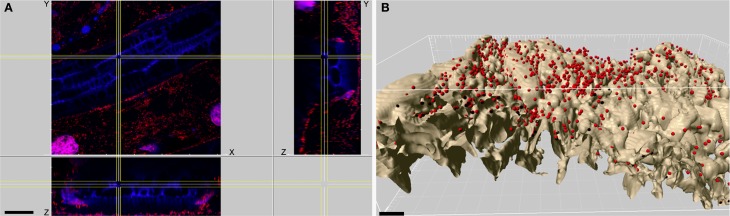
The colonization pattern of the labeled bacterial (DsRed) strain S. plymuthica 3Re4-18 was monitored with confocal laser scanning microscopy. For chamomile seedlings cultivated in a gnotobiotic system, cells of S. plymuthica 3Re4-18 were able to colonize the rhizosphere. (Figure 7A). Two different colonization patterns could be observed: single cells, covering the root surface and forming a dense network. Furthermore, cells were often found as surrounding clouds (Figure 7B), where they were loosely arranged around the root surface.
Figure 7.
Root epidermis of a chamomile seedling 14 days after inoculation. Visualization of DsRed marked bacteria with confocal laser scanning microscopy. (A) xy, xz, and yz maximum projections showing a colonizing of small colonies and single cells of Serratia plymuthica 3Re4-18 of the surrounding area of the root. Red: bacterial cells, blue: root, scale bar: 30 μm. (B) Surface model of (A) shows the root-surface localization of Serratia plymuthica 3Re4-18 in the three-dimensional space. Red: bacterial cells, brown: root, scale bar: 10 μm.
Discussion
In this study, we examined the impact of six bacterial inoculants on the native bacterial community of C. recutita (L.) Rauschert as well as on the production of flavonoid compounds. We found an impact of the treatments on both, host's microbiome and physiology.
The impact of bacterial inoculants was analyzed by three methods. No impact was found at quantitative level: all abundances were highly similar and showed no statistically significant differences. Quantitative insight into the microbial communities in the rhizosphere showed that total bacteria (up to 8.03 log10 copies per g root fw for the treatment with P. polymyxa Mc5Re-14) as well as Firmicutes (up to 7.54 log10 copies per g fw for the treatment with P. polymyxa Mc5Re-14) resulted in a high abundance in the rhizosphere. The phenomenon that the number of microorganisms in the rhizosphere is enhanced as a result of exudation of compounds by the root was described as the rhizosphere effect by Lynch (1990). However, no clear influence on the abundance of microorganisms by bacterial inoculants could be detected, even though results give a hint that Gram-positive bacteria are more dominant in the rhizosphere than introduced Gram-negative strains. The high abundance of Gram-positive bacteria in bulk soil was already described by Smalla et al. (2001).
At qualitative level, the applied methods resulted in different conclusions. All molecular fingerprints obtained with universal and group-specific primers revealed high similarity of microbial community composition and were strongly related to the sampling times, showing about 10% similarity. In both sampling times, the bacterial rhizosphere community did not differ between the treatments and the control. The rhizosphere of the young chamomile plant was shown to be colonized mainly by Bacillus spp. and Pseudomonas spp. The high abundance of Bacillus species in the field soil at Sekem farms was already described by Köberl et al. (2011). Pseudomonads represent important members of the rhizosphere microbial community due to their aggressive colonization (Scherwinski et al., 2006). Their plant growth promoting ability was described in several studies (Bloemberg and Lugtenberg, 2001; Weller, 2007). Generally, the species composition differed slightly between the two sampling times. However, due to the limited number of identified strains, these results should be interpreted carefully. The SSCP analysis showed that there was no clear influence by the bacterial inoculants on the diversity of the naturally occurring bacterial populations in the rhizosphere of C. recutita (L.) Rauschert. Next-generation sequencing allows a deeper insight into microbial community composition, and therefore answers to the following questions can be provided: (i) how are microbial communities composed on different taxonomic levels? (ii) how does the structure of communities look like between different samples? (iii) how do communities change across treatments? To address these questions, samples from different treatments with bacterial inoculants were investigated and compared among each other. At first, the pyrosequencing approach showed similar patterns of bacterial diversity between the different treatments, whereas Proteobacteria, Actinobacteria, Acidobacteria, Chloroflexi, Verrucomicrobia, Bacteroidetes, Planctomycetes, Gemmatimonadetes, and Firmicutes represented the most abundant phyla in all analyzed samples (>1% of all sequences). Janssen (2006) already described members of these identified phyla as the most abundant soil bacteria. The phylum Verrucomicrobia was only present in the sample from the treatment with S. rhizophila P69 (=DSM14405T). In other ad planta studies, this strain was found to have an indirect positive interaction with their host plants by altering fungal communities (Schmidt et al., 2012). When bacteria were analyzed at the genus level, the relative abundances of different genera belonging to phyla Proteobacteria, Bacteroidetes, and Actinobacteria varied among the samples under different treatments. These results showed that it is of crucial importance to be aware of using the appropriate levels of taxonomy for different investigations. Therefore, this study suggests that for the detection of differences within microbial communities deriving from bacterial inoculants the use of genus or even lower levels of taxonomy yield the greatest benefits. Unlike alpha diversity measurements, such as species richness and rarefaction curves, beta diversity measures the degree of similarity, e.g., phylogenetic relatedness between pairs of communities (Caporaso et al., 2010). Therefore, phylogenetic beta diversity metrics were used to investigate the structure of communities between the samples. PCoA plots showed that samples differed according to the treatments. Despite the SSCP results, pyrosequencing results suggest that the introduction of antagonistic bacterial strains leads to changes in the native bacterial community structure.
The impact on the physiology of the host was found for the treatments with the two Gram-positive strains B. subtilis Co1-6 and P. polymyxa Mc5Re-14, yielding a significantly higher content of apigenin-7-O-glucoside compared to the treatments with Gram-negative strains. Apigenin was only found in traces and may have been produced postharvest by hydrolysis from apigenin-7-O-glucoside (Bauer and Wagner, 1991; Maier et al., 1993). Likewise, previous studies have isolated and identified the presence of both mono- and di-acetylated apigenin-7-O-glucoside, which are known to undergo rapid ester hydrolysis leading to the formation of apigenin-7-O-glucoside (Redaelli et al., 1981; Svehlikova et al., 2004). Moreover, apigenin-7-O-glucoside is highly susceptible for the hydrolysis to its corresponding aglycone in presence of acid or hydrolytic enzymes. As a result, the formation or degradation of apigenin-7-O-glucoside may lead to a falsification of quantification. To date, only few studies have considered the influence of rhizobacteria on the secondary metabolites of plants (Zuanazzi et al., 1998; Singh et al., 2002; Orhan et al., 2006). Therefore, little is known about the mechanisms behind positive influence of rhizobacterial secondary metabolites on the plants secondary metabolite production. Plants have the ability to acquire enhanced level of resistance to pathogenic microorganisms, a mechanism first described by Van Peer et al. (1991) and Wei et al. (1991) as induced systemic resistance (ISR). However, ISR can also be induced by various non-pathogenic microorganisms that may activate inducible defense mechanisms in the plant in a similar way to pathogenic microorganisms (Loon, 2007). Activation of defense mechanisms by plants suggests that even a beneficial rhizobacterium may be recognized as a potential threat, leading to the production of resistance compounds (Kloepper et al., 2004; Ongena et al., 2005; Schuhegger et al., 2006; Arkhipova et al., 2007; Vleesschauwer and Höfte, 2007; Lehr et al., 2008; Choudhary and Johri, 2009; Lal and Tabacchioni, 2009).
Next-generation sequencing allows a deeper insight into plant-associated microbial communities. This was already shown for suppressive soils (Mendes et al., 2011) and for the analysis of core microbiomes (Lundberg et al., 2012). In our study we show that they also improve our understanding of the mode of interaction of bacterial inoculants. The latter are promising for a sustainable agriculture and the challenges for crop production in the context of climate change. To understand the effects and interactions of biocontrol agents will allow the improvement of biocontrol products.
Materials and methods
The bacterial strains used in this study are summarized in Table 2. For the preparation of inoculums, several colonies of each bacterial strain were inoculated in 500 ml liquid LB medium (Roth, Karlsruhe, Germany) and grown at 30°C and 150 rpm for 24 h (Co1-6, L13-6-12, P69, and 3Re4-18) and for 48 h (Wb2n-11 and Mc5Re-14). To harvest the cells, bacterial suspensions were centrifuged at 13,500 g for 20 min. Pellets were dissolved in 2 ml sucrose solution (1%), serving as cryoprotectant agent, and frozen to −70°C for 5 h. Tubes containing the frozen bacterial suspension were put into ampules and connected to a freeze-dryer (Labconco FreeZone 4.5 Liter Benchtop, USA) for 12 h under vacuum at <0.1 Pa.
Table 2.
Bacterial strains used in this study.
| Bacterial strain | Origin | References |
|---|---|---|
| Streptomyces subrutilus Wb2n-11 | Desert soil in Sinai, Gram-positive | Köberl et al., 2011 |
| Bacillus subtilis subsp. subtilis Co1-6 | Rhizosphere of Calendula officinalis (L.), Gram-positive | Köberl et al., 2011 |
| Paenibacillus polymyxa Mc5Re-14 | Endorhiza of Chamomilla recutita (L.) Rauschert, Gram-positive | Köberl et al., 2011 |
| Pseudomonas fluorescens L13-6-12 | Rhizosphere of Solanum tuberosum (L.), Gram-negative | Lottmann and Berg, 2001 |
| Stenotrophomonas rhizophila P69 (= DSM14405T) | Rhizosphere of Brassica napus (L.), Gram-negative | Wolf et al., 2002 |
| Serratia plymuthica 3Re4-18 | Endorhiza of Solanum tuberosum (L.), Gram-negative | Berg et al., 2005b; Grosch et al., 2005 |
Field experiment and sampling procedure
Prior to planting, one-month old field-grown C. recutita (L.) Rauschert seedlings were root-dipped in suspensions of the bacteria for 15 min. The non-treated control seedlings were dipped in tap water and planted. Five replicates of each treatment were performed in a randomized block design (1 × 2 m per plot) at a field at Adleya farm/Sekem (30°22′88″N; 31°39′41″E). During the growth stage, the field was irrigated with water (2607 l m−3 on average per year) coming from the Nile or from local groundwater drillings—drip irrigation systems were used. Composite soil samples were collected from each plot at a surface depth of 0–5 cm, and the soil-texture was classified as sandy silt with a pH of 8.48, 254.75 μ S/m electrical conductivity, 1.44% organic matter, 0.82% organic carbon, 0.15% total nitrogen, 0.01% total phosphorous, and 0.07% total potassium. Rhizosphere samples were taken at 4 and 8 weeks after planting, each with five replicates per treatment. A sample consisted of 5 g roots with adhering soil. In the end of the growing season (March 2012), chamomile flowers were harvested, each with five replicates per treatment.
Determination of flavonoids
Fresh chamomile flowers were dried in a forced-convection oven at 40°C and milled to a fine powder using a universal IKA M 20 grinding mill (IKA-Werke, Staufen, Germany). The extraction of chamomile flower heads was performed using an accelerated solvent extractor, ASE 200 (Dionex Corporation, Sunnyvale, CA, USA). Extraction was carried out at a temperature of 68°C, with a constant pressure of 69 bar and a static time of 5 min using 100% methanol (v/v) as extraction solvent. Based on preliminary experiments, conditions of ASE 200 were set as follows: no preheating period, heating time of 5 min, flush volume at 30% of the extraction cell volume, three extraction cycles, nitrogen purge time of 60 s. The extracts were filtered (Whatman No. 42) and stored at 4°C in darkness until the chromatographic analysis. HPLC analysis was performed on an UltiMate 3000 RS chromatographic system (Dionex, Sunnyvale, CA, USA). A LiChrospher 100 RP-18 (125x4 mm, 5 μm) (Merck, Darmstadt, Germany) was employed for the separation. The binary mobile phase consisted of solvents A (water) and B (acetonitrile) according to the following profile: 0–17 min, 15–40% B; 17–18 min, 40–75% B; 18–29 min, 75% B. The flow rate was 1.0 ml/min and the injection volume was 5 μ l. Chromatograms were recorded at a wavelength of 340 nm. For the mass analysis, a linear ion trap quadrupole (LTQ) XL mass spectrometer (Thermo Scientific, San Jose, CA, USA) with an electrospray interface (ESI) was used. ESI negative ion mode conditions were set as follows: source voltage 3.0 kV, sheath gas flow rate 50 au, auxiliary gas flow rate 10 au, source current 100.0 μ A, capillary voltage −45.0 kV, and capillary temperature 330°C. The screening was performed in full scan, covering the range from m/z 50 up to 2000. Calibration curves of apigenin-7-O-glucoside and apigenin were obtained by the external standard method. Apigenin-7-O-glucoside and apigenin were quantified using Xcalibur Quan Browser (Version 2.0, Thermo Fisher, San Jose, CA, USA). Chromatograms of each treatment were overlaid with the control using MZmine (Version 2.8, Katajamaa et al., 2006) in order to identify new peaks arising from the treatments. Identification of new peaks was performed using a Dionex Ultimate 3000 UHPLC focused LC system coupled via a heated electrospray source HESI2 to a Q-Exactive mass spectrometer (Thermo Fisher Scientific). HPLC conditions were set as described above. HESI conditions were set as follows: spray voltage: −3 kV, capillary temp: 450°C, sheath and auxiliary gas: N2, flow: 75 & 20 instrument units, gas temp heater: 350°C. The MS instrument was operated in negative mode, externally mass calibrated, and with the resolving power sat to 140000 (FWHM), scanned between m/z 133-2000. With an exclusion time of 10 s, the five most intense ions were selected for collision induced fragmentation. The selection window was 0.4 Dalton, and the fragmentation energy NCE sat to 30 ± 20% instrument units. MS2 spectra were obtained with the resolving power sat to 35000. Putative compounds were identified using the online METLIN metabolite database (http://metlin.scripps.edu).
Rhizosphere sampling and total community DNA isolation
The bacterial fraction associated with C. recutita (L.) Rauschert was extracted using the protocol adapted from Opelt and Berg (2004). In brief, for each rhizosphere sample, 5 g of roots with adhering soil were mixed with 45 ml NaCl solution (0.85%) and vortexed for 5 min. A total volume of 4 ml of the suspension was centrifuged at 16,000 g at 4°C for 20 min, and the pellet was used for isolation of the total community DNA. For mechanical lysis, the cells were homogenized in a FastPrep FP120 Instrument (MP Biomedicals, Solon, OH, USA) for 40 s at speed 6.0. The obtained DNA was purified using the FastDNA SPIN Kit for Soil (MP Biomedicals) according to the manufacturer's protocol. Final aliquots of the total community DNA were further used for PCR-SSCP, 454 pyrosequencing, and qPCR.
Microbial fingerprinting by PCR-SSCP
Fingerprinting of microbial communities was carried out by PCR-based SSCP described by Schwieger and Tebbe (1998). Bacterial 16S rRNA gene sequences were amplified by PCR using the universal eubacterial primer pair Unibac-II-515f/Unibac-II-927rP (Zachow et al., 2008). The 60 μl reaction mixture contained 1×Taq-&Go Ready-to-use PCR Mix (MP Biomedicals), 3 mM MgCl2, 0.2 μM of each primer, and 1 μl DNA template (95°C, 5 min; 32 cycles of 95°C, 20 s; 54°C, 15 s; 72°C, 30 s; and elongation at 72°C, 10 min). For the analysis of the Pseudomonas community, a nested PCR was performed. In the first PCR, the Pseudomonas specific primer pair F311Ps/1459rPsP (Milling et al., 2005) was used in a 20 μl reaction mixture containing 1×Taq-&Go Ready-to-use PCR Mix, 3 mM MgCl2, 0.5 mg/ml BSA, 1.5% DMSO, 0.2 μM of each primer, and 1 μl DNA template (94°C, 7 min; 30 cycles of 94°C, 45 s; 56°C, 2 min; 72°C, 2 min; and elongation at 72°C, 10 min). Samples served as templates for the second PCR using the primer pair Unibac-II-515f/Unibac-II-927rP. For the analysis of the Firmicutes community, the universal eubacterial primer pair 27f/1492r (Lane, 1991) was used in a 20 μl reaction mixture containing 1×Taq-&Go Ready-to-use PCR Mix, 3 mM MgCl2, 0.2 μM of each primer, and 1 μl DNA template (95°C, 5 min; 30 cycles of 95°C, 30 s; 57°C, 30 s; 72°C, 90 s; and elongation at 72°C, 5 min). In the second PCR, the Firmicutes specific primer pair BLS342f/BACr833rP (Blackwood et al., 2005) was used in a 60 μl reaction mixture containing 1×Taq-&Go Ready-to-use PCR Mix, 0.2 μM of each primer, and 3 μl of the product from the first PCR (95°C, 5 min; 30 cycles of 95°C, 45 s; 57°C, 60 s; 72°C, 45 s; and elongation at 72°C, 10 min). The obtained amplicons were separated using the INGENY phorU system (INGENY International BV, Goes, Netherlands) at 400 V and 26°C followed by silver staining. Dominant bands were excised from SSCP gels as described by Schwieger and Tebbe (1998). Extracted DNA fragments were re-amplified by PCR and sequenced. For phylogenetic analysis and identification of related sequences, the obtained sequences were aligned with reference gene sequences from GenBank using BLAST algorithm.
Amplicon sequencing of bacterial communities
To characterize the rhizosphere bacterial communities associated with C. recutita (L.) Rauschert, the V4-V5 hypervariable region of the bacterial 16S rRNA gene (Escherichia coli positions 515 to 927) was chosen for the amplification and subsequent pyrosequencing of the PCR products. Due to lack of funds, only samples from the treatments Wb2n-11, Co1-6, Mc5Re-14, P69, and 3Re4-18 from the first sampling time were sequenced. The V4-V5 region was amplified using the primer pair Unibac-II-515f/Unibac-II-927r. The 20 μl reaction mixture contained 1×Taq-&Go Ready-to-use PCR Mix, 3 mM MgCl2, 0.5 μM of each primer, and 1 μl of template DNA (95°C, 2 min; 34 cycles of 95°C, 20 s; 65°C, 15 s; 72°C, 29 s; and elongation at 72°C, 10 min). PCR products from four samples of the same treatment were purified with Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). Amplicons of each treatment were pooled together in an equimolar ratio and subjected to pyrosequencing using a Roche 454 GS-FLX+ Titanium platform executed by Eurofins MWG (Ebersberg, Germany).
Processing of pyrosequencing data
Raw sequencing reads were demultiplexed, quality and length filtered using ribosomal database project's (RDP) pyrosequencing pipeline (Cole et al., 2009). Primers were cropped and all sequence reads shorter than 150 bp—with a minimum average quality score <20 and with any ambiguous characters were discarded. Data were normalized to the same number of sequences using an in-house developed Perl script (10 times random re-samplings followed by subset formation (Bragina et al., 2013). A further downstream analysis of normalized data was achieved using the QIIME toolkit (Caporaso et al., 2010). Bacterial sequences were clustered into OTUs using 3, 5, and 20% dissimilarity thresholds with UCLAST (Edgar, 2010), and the most abundant sequence from each OTU was selected as a representative sequence for that OTU. Taxonomy was assigned by using a QIIME-based wrapper of the RDP classifier program (Wang et al., 2007) against the RDP core set (Cole et al., 2009) using an 80% confidence threshold for taxonomic assignment. Rarefaction analysis and estimation of alpha-diversity was performed using Chao1, Shannon, and observed OTU metrics at 3, 5, and 20% dissimilarity. Beta-diversity was examined using weighted UniFrac distances (Lozupone and Knight, 2005) between samples sub-sampled 20 times, with replacement, at a depth of 100 sequences per sample. This method takes phylogenetic relationships between community members in account, incorporating the abundances of phylotypes into the pairwise community comparisons (Eilers et al., 2010). The compositional similarity of all samples was visualized in a three-dimensional principal coordinate system (PCoA) based on previously calculated jackknifed principal coordinates. To reveal the most abundant taxa in different areas of the PCoA plot, taxonomic classification of 20% genetic distance was included.
Quantitative polymerase chain reaction (qPCR)
The same region of the 16S rRNA gene was amplified by quantitative PCR to determine the total bacterial as well as the Firmicutes abundances in the rhizosphere of C. recutita (L.) Rauschert. For the total bacteria, the universal eubacterial primer pair Unibac-II-515f/Unibac-II-927r was used, while the Firmicutes specific primer pair BLS342f/BACr833r was used for Firmicutes. To estimate bacterial gene abundances, standard curves were generated using 10-fold serial dilutions of plasmid DNA containing a full-length copy of either the P. polymyxa PB71 16S rRNA gene or the B. subtilis Sd3-12 16S rRNA gene (Köberl et al., 2011). For the total bacteria, the qPCR 10 μl reaction mixture contained 1x KAPA SYBR FAST qPCR MasterMix Universal (PEQLAB, Polling, Austria), 0.25 μM of each primer, and 1 μl of the standard and DNA template (95°C, 5 min; 35 cycles of 95°C, 20 s; 54°C, 15 s; 72°C, 30 s; and melt from 72 to 95°C). For Firmicutes, the 10 μl qPCR reaction mixture contained 1x KAPA SYBR FAST qPCR MasterMix Universal, 0.25 μM of each primer, and 1 μl standard and DNA template (95°C, 5 min; 30 cycles of 95°C, 45 s; 56°C, 60 s; 72°C, 45 s; and melt from 72 to 95°C). qPCR was performed in duplicate for each sample using the Rotor-Gene 6000 real-time rotary analyser (Corbett Research, Sydney, Australia). The melting curve analysis of the PCR products was performed immediately after the amplification. Bacterial copy numbers for each reaction were generated from the standard curves and calculated to copy number per g root fresh weight (fw). Each replicate was analyzed two times in two independent runs.
Confocal laser scanning microscopy (CLSM)
DsRed2-labeled bacteria (3Re4-18) were grown in 5 ml nutrient broth (NB) supplemented with tetracycline (40 μ g ml−1) at 30°C and 120 rpm for 24 h. Bacterial cells were collected by centrifugation at 13,500 g for 5 min and resuspended in fresh NB medium without addition of antibiotics. The cell suspension was adjusted to an optical density corresponding to a cell count of 109 cells ml−1. Seeds were mixed with 1 ml of the cell suspension in 1.5 ml Eppendorf tubes and incubated at room temperature for 15 min, followed by a washing step with sterile NaCl solution (0.85%). Seeds were placed on a filter paper in moist chambers which were kept at 22°C for 5 days (16/8 h day/night). Seeds incubated with sterile water served as control. Roots of chamomile were examined on a Leica TCS SPE confocal scanning microscope (Leica Microsystems GmbH, Wetzler, Germany).
Statistics
Computer-assisted comparisons of SSCP generated community profiles were performed using GelComparII (version 5.1, Applied Maths, Kortrijk, Belgium). For the cluster analysis, similarity matrices based on Pearson's correlation coefficients were constructed, and a dendrogram using the unweighted paired group means algorithm (UPGMA) was created. Relative positions and intensity of DNA bands were used for a principal component analysis (PCA). Significances in the difference between the treatments were calculated using PASW Statistics 18 (SPSS Inc., Chicago, IL, USA). First, data were checked for normal distribution and homogeneity of variance. Second, an one-way ANOVA analysis was performed with data that follow a normal distribution, and a post-hoc test was applied depending on the homogeneity of the variance index using the Tukey honestly significant difference (HSD) analysis for qPCR data and the Tamhane T2 test for HPLC-MS data.
Author contributions
Conceived and designed the experiments: Ruth Schmidt, Martina Köberl, Marlene Monschein, Gabriele Berg, Rudolf Bauer, and Elshahat M. Ramadan. Performed the experiments: Ruth Schmidt, Amr Mostafa, Martina Köberl, and Marlene Monschein. Analyzed the data: Ruth Schmidt, Martina Köberl, Marlene Monschein, and Kenneth B. Jensen. Contributed reagents/materials/analysis tools: Gabriele Berg. Wrote the paper: Ruth Schmidt and Gabriele Berg.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Ibrahim Abouleish and his family for their generous hospitality in Sekem. Furthermore, we want to thank co-workers from the Sekem companies and the Heliopolis University, especially to Angela Hofmann for competent advices, Fekria Mohamed, Ibrahim El Yamany, Ahmed Abdel El Salam, and Hosssam Said for fieldwork, and Mahmoud Saber, Christophe Floride, and Hassan Abouessa for their support. For relevant theoretical and practical support we thank Massimiliano Cardinale and Henry Müller (Graz). This project was partly funded by the Unchain program (University Chair on Innovation)—funded by the European Commission—Tempus IV (JP-00241-2008).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00064/abstract
References
- Arkhipova T. N., Prinsen E., Veselov S. U., Martinenko E. V., Melentiev A. I., Kudoyarova G. R. (2007). Cytokinin producing bacteria enhance plant growth in drying soil. Plant Soil 292, 305–315 10.1007/s11104-007-9233-5 [DOI] [Google Scholar]
- Bakker P. A., Berendsen R. L., Doornbos R. F., Wintermans P. C., Pieterse C. M. (2013). The rhizosphere revisited: root microbiomics. Front. Plant Sci. 4:165 10.3389/fpls.2013.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer R., Wagner H. (1991). Äußere Einflüsse auf den Flavonoidgehalt von Kamillenblüten. Sci. Pharm. 59, 3 [Google Scholar]
- Berendsen R. L., Pieterse C. M., Bakker P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Berg G. (2009). Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18 10.1007/s00253-009-2092-7 [DOI] [PubMed] [Google Scholar]
- Berg G., Eberl L., Hartmann A. (2005a). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685 10.1111/j.1462-2920.2005.00891.x [DOI] [PubMed] [Google Scholar]
- Berg G., Krechel A., Ditz M., Sikora R. A., Ulrich A., Hallmann J. (2005b). Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 51, 215–229 10.1016/j.femsec.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Blackwood C. B., Oaks A., Buyer J. S. (2005). Phylum- and class-specific PCR primers for general microbial community analysis. Appl. Environ. Microbiol. 71, 6193–6198 10.1128/AEM.71.10.6193-6198.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloemberg G. V., Lugtenberg B. J. (2001). Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 4, 343–350 10.1016/S1369-5266(00)00183-7 [DOI] [PubMed] [Google Scholar]
- Bragina A., Berg C., Müller H., Moser D., Berg G. (2013). Insights into functional bacterial diversity and its effects on Alpine bog ecosystem functioning. Sci. Rep. 3, 1955 10.1038/srep01955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S. (2012). Endophytic fungi: novel sources of anticancer lead molecules. Appl. Microbiol. Biotechnol. 95, 47–59 10.1007/s00253-012-4128-7 [DOI] [PubMed] [Google Scholar]
- Choudhary D. K., Johri B. N. (2009). Interactions of Bacillus spp. and plants – with special reference to induced systemic resistance (ISR). Microbiol. Res. 164, 493–513 10.1016/j.micres.2008.08.007 [DOI] [PubMed] [Google Scholar]
- Cole J. R., Wang Q., Cardenas E., Fish J., Chai B., Farris R. J., et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–D145 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R., Gotz M., Mrotzek N., Lottmann J., Berg G., Smalla K. (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56, 236–249 10.1111/j.1574-6941.2005.00026.x [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Eilers K. G., Lauber C. L., Knight R., Fierer N. (2010). Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903 10.1016/j.soilbio.2010.02.003 [DOI] [Google Scholar]
- Farag M. A., Zhang H., Ryu C. M. (2013). Dynamic chemical communication between plants and bacteria through airborne signals: induced resistance by bacterial volatiles. J. Chem. Ecol. 39, 1007–1018 10.1007/s10886-013-0317-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosch R., Faltin F., Lottmann J., Kofoet A., Berg G. (2005). Effectiveness of 3 antagonistic bacterial isolates to control Rhizoctonia solani Kühn on lettuce and potato. Can. J. Microbiol. 51, 345–353 10.1139/w05-002 [DOI] [PubMed] [Google Scholar]
- Gutierrez R. M., Gonzalez A. M., Ramirez A. M. (2012). Compounds derived from endophytes: a review of phytochemistry and pharmacology. Curr. Med. Chem. 19, 2992–3030 10.2174/092986712800672111 [DOI] [PubMed] [Google Scholar]
- Haas D., Defago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319 10.1038/nrmicro1129 [DOI] [PubMed] [Google Scholar]
- Janssen P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728 10.1128/AEM.72.3.1719-1728.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajamaa M., Miettinen J., Oresic M. (2006). MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics 22, 634–636 10.1093/bioinformatics/btk039 [DOI] [PubMed] [Google Scholar]
- Kloepper J. W., Ryu C. M., Zhang S. (2004). Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94, 1259–1266 10.1094/PHYTO.2004.94.11.1259 [DOI] [PubMed] [Google Scholar]
- Köberl M., Müller H., Ramadan E. M., Berg G. (2011). Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PLoS ONE 6:e24452 10.1371/journal.pone.0024452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S., Tabacchioni S. (2009). Ecology and biotechnological potential of Paenibacillus polymyxa: a minireview. Indian J. Microbiol. 49, 2–10 10.1007/s12088-009-0008-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. (1991). 16S/23S rRNA sequencing, in Nucleic Acids Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (Chichester: John Wiley & Sons; ), 115–147 [Google Scholar]
- Lehr N. A., Schrey S. D., Hampp R., Tarkka M. T. (2008). Root inoculation with a forest soil streptomycete leads to locally and systemically increased resistance against phytopathogens in Norway spruce. New Phytol. 177, 965–976 10.1111/j.1469-8137.2007.02322.x [DOI] [PubMed] [Google Scholar]
- Loon L. C. (2007). Plant responses to plant growth-promoting rhizobacteria. Eur. J. Plant Pathol. 119, 243–254 10.1007/s10658-007-9165-1 [DOI] [Google Scholar]
- Lottmann J., Berg G. (2001). Phenotypic and genotypic characterization of antagonistic bacteria associated with roots of transgenic and non-transgenic potato plants. Microbiol. Res. 156, 75–82 10.1078/0944-5013-00086 [DOI] [PubMed] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 10.1146/annurev.micro.62.081307.162918 [DOI] [PubMed] [Google Scholar]
- Lundberg D. S., Lebeis S. L., Paredes S. H., Yourstone S., Gehring J., Malfatti S., et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. M. (1990). The Rhizosphere. Chichester: John Wiley & Sons [Google Scholar]
- Maier R., Carle R., Kreis W., Reinhard E. (1993). Purification and characterization of a flavone 7-O-glucoside-specific glucosidase from ligulate florets of Chamomilla recutita. Planta Med. 59, 436–441 10.1055/s-2006-959727 [DOI] [PubMed] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J. H., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 10.1126/science.1203980 [DOI] [PubMed] [Google Scholar]
- Miller K. I., Qing C., Sze D. M., Neilan B. A. (2012). Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS ONE 7:e35953 10.1371/journal.pone.0035953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milling A., Smalla K., Maidl F., Schloter M., Munch J. (2005). Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266, 23–39 10.1007/s11104-005-4906-4 [DOI] [Google Scholar]
- Murphy J. F., Reddy M. S., Ryu C. M., Kloepper J. W., Li R. (2003). Rhizobacteria-mediated growth promotion of tomato leads to protection against cucumber mosaic virus. Phytopathology 93, 1301–1307 10.1094/PHYTO.2003.93.10.1301 [DOI] [PubMed] [Google Scholar]
- Ongena M., Jacques P., Toure Y., Destain J., Jabrane A., Thonart P. (2005). Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl. Microbiol. Biotechnol. 69, 29–38 10.1007/s00253-005-1940-3 [DOI] [PubMed] [Google Scholar]
- Opelt K., Berg G. (2004). Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the Baltic Sea Coast. Appl. Environ. Microbiol. 70, 6569–6579 10.1128/AEM.70.11.6569-6579.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan E., Esitken A., Ercisli S., Turan M., Sahin F. (2006). Effects of plant growth promoting rhizobacteria (PGPR) on yield, growth and nutrient contents in organically growing raspberry. Sci. Hortic. 111, 38–43 10.1016/j.scienta.2006.09.002 [DOI] [Google Scholar]
- Pimentel M. R., Molina G., Dionísio A. P., Maróstica Junior M. R., Pastore G. M. (2011). The use of endophytes to obtain bioactive compounds and their application in biotransformation process. Biotechnol. Res. Int. 2011, 576286 10.4061/2011/576286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirlak L., Köse M. (2009). Effects of plant growth promoting rhizobacteria on yield and some fruit properties of strawberry. J. Plant Nutr. 32, 1173–1184 10.1080/01904160902943197 [DOI] [Google Scholar]
- Raaijmakers J., Paulitz T., Steinberg C., Alabouvette C., Moënne-Loccoz Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361 10.1007/s11104-008-9568-6 [DOI] [Google Scholar]
- Redaelli C., Formentini L., Santaniello E. (1981). Reversed-phase high-performance liquid chromatography analysis of apigenin and its glucosides in flowers of Matricaria chamomilla and chamomile extracts. Planta Med. 42, 288–292 10.1055/s-2007-971643 [DOI] [PubMed] [Google Scholar]
- Ryu C. M., Farag M. A., Hu C. H., Reddy M. S., Kloepper J. W., Paré P. W. (2004a). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026 10.1104/pp.103.026583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C. M., Murphy J. F., Mysore K. S., Kloepper J. W. (2004b). Plant growth-promoting rhizobacteria systemically protect Arabidopsis thaliana against cucumber mosaic virus by a salicylic acid and NPR1-independent and jasmonic acid-dependent signaling pathway. Plant J. 39, 381–392 10.1111/j.1365-313X.2004.02142.x [DOI] [PubMed] [Google Scholar]
- Scherwinski K., Grosch R., Berg G. (2008). Effect of bacterial antagonists on lettuce: active biocontrol of Rhizoctonia solani and negligible, short-term effects on nontarget microorganisms. FEMS Microbiol. Ecol. 64, 106–116 10.1111/j.1574-6941.2007.00421.x [DOI] [PubMed] [Google Scholar]
- Scherwinski K., Wolf A., Berg G. (2006). Assessing the risk of biological control agents on the indigenous microbial communities: Serratia plymuthica HRO-C48 and Streptomyces sp. HRO-71 as model bacteria. Biocontrol 52, 87–112 10.1007/s10526-006-9006-8 [DOI] [Google Scholar]
- Schmidt C., Alavi M., Cardinale M., Müller H., Berg G. (2012). Stenotrophomonas rhizophila DSM14405T promotes plant growth probably by altering fungal communities in the rhizosphere. Biol. Fertil. Soils 48, 947–960 10.1007/s00374-012-0688-z [DOI] [Google Scholar]
- Schuhegger R., Ihring A., Gantner S., Bahnweg G., Knappe C., Vogg G., et al. (2006). Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918 10.1111/j.1365-3040.2005.01471.x [DOI] [PubMed] [Google Scholar]
- Schwieger F., Tebbe C. C. (1998). A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64, 4870–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U. P., Sarma B. K., Singh D. P., Bahadur A. (2002). Plant growth-promoting rhizobacteria-mediated induction of phenolics in pea (Pisum sativum) after infection with Erysiphe pisi. Curr. Microbiol. 44, 396–400 10.1007/s00284-001-0007-7 [DOI] [PubMed] [Google Scholar]
- Smalla K., Wieland G., Buchner A., Zock A., Parzy J., Kaiser S., et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67, 4742–4751 10.1128/AEM.67.10.4742-4751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svehlikova V., Bennett R. N., Mellon F. A., Needs P. W., Piacente S., Kroon P. A., et al. (2004). Isolation, identification and stability of acylated derivatives of apigenin 7-O-glucoside from chamomile (Chamomilla recutita [L.] Rauschert). Phytochemistry 65, 2323–2332 10.1016/j.phytochem.2004.07.011 [DOI] [PubMed] [Google Scholar]
- Van Peer R., Niemann G. J., Schippers B. (1991). Induced resistance and phytoalexin accumulation in biological control of fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Phytopathology 81, 728–734 10.1094/Phyto-81-728 [DOI] [Google Scholar]
- Verginer M., Leitner E., Berg G. (2010). Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 58, 8344–8350 10.1021/jf100393w [DOI] [PubMed] [Google Scholar]
- Vleesschauwer D. D., Höfte M. (2007). Using Serratia plymuthica to control fungal pathogens of plants. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Res. 2:46 10.1079/PAVSNNR2007204621947430 [DOI] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Kloepper J. W., Tuzun S. (1991). Iduction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology 81, 1508–1512 10.1094/Phyto-81-1508 [DOI] [Google Scholar]
- Weller D. M. (2007). Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97, 250–256 10.1094/PHYTO-97-2-0250 [DOI] [PubMed] [Google Scholar]
- Wings S., Müller H., Berg G., Lamshöft M., Leistner E. (2013). A study of the bacterial community in the root system of the maytansine containing plant Putterlickia verrucosa. Phytochemistry 91, 158–164 10.1016/j.phytochem.2012.06.016 [DOI] [PubMed] [Google Scholar]
- Wolf A., Fritze A., Hagemann M., Berg G. (2002). Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int. J. Syst. Evol. Microbiol. 52, 1937–1944 10.1099/ijs.0.02135-0 [DOI] [PubMed] [Google Scholar]
- Zachow C., Fatehi J., Cardinale M., Tilcher R., Berg G. (2010). Strain-specific colonization pattern of Rhizoctonia antagonists in the root system of sugar beet. FEMS Microbiol. Ecol. 74, 124–135 10.1111/j.1574-6941.2010.00930.x [DOI] [PubMed] [Google Scholar]
- Zachow C., Tilcher R., Berg G. (2008). Sugar beet-associated bacterial and fungal communities show a high indigenous antagonistic potential against plant pathogens. Microb. Ecol. 55, 119–129 10.1007/s00248-007-9257-7 [DOI] [PubMed] [Google Scholar]
- Zhao L. X., Xu L. H., Jiang C. L. (2012). Methods for the study of endophytic microorganisms from traditional Chinese medicine plants. Methods Enzymol. 517, 3–21 10.1016/B978-0-12-404634-4.00001-2 [DOI] [PubMed] [Google Scholar]
- Zuanazzi J. A., Clergeot P. H., Quirion J. C., Husson H. P., Kondorosi A., Ratet P. (1998). Production of Sinorhizobium meliloti nod gene activator and repressor flavonoids from Medicago sativa roots. Mol. Plant Microbe Interact. 11, 784–794 10.1094/MPMI.1998.11.8.784 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.