Abstract
Progress in oncology drug development has been hampered by a lack of preclinical models that reliably predict clinical activity of novel compounds in cancer patients. In an effort to address these shortcomings, there has been a recent increase in the use of patient-derived tumour xenografts (PDTX) engrafted into immune-compromised rodents such as athymic nude or NOD/SCID mice for preclinical modelling. Numerous tumour-specific PDTX models have been established and, importantly, they are biologically stable when passaged in mice in terms of global gene-expression patterns, mutational status, metastatic potential, drug responsiveness and tumour architecture. These characteristics might provide significant improvements over standard cell-line xenograft models. This Review will discuss specific PDTX disease examples illustrating an overview of the opportunities and limitations of these models in cancer drug development, and describe concepts regarding predictive biomarker development and future applications.
Introduction
One of the most frequently cited reasons for the high failure rate of new agents in oncology is the lack of pre-clinical models that recapitulate the heterogeneity of tumours in patients.1 Although the advent of cancer cell-line culture techniques fuelled an acceleration and expansion of cancer biology discovery that continues to this day, the harsh reality is that our ability to translate these findings to clinical practice has been hampered by the very models that yielded such valuable insights. Numerous explanations have been suggested for this inconsistency, including the fact that cell lines, even when propagated in vivo, are derived from cancer cells that have adapted to growth outside a natural tumour microenvironment, resulting in genetic changes that are distinct from the genetic stress imposed on tumours in patients.2 Likewise, there is strong evidence that a greater genetic divergence exists between a primary tumour and the corresponding cell line derived from that tumour, versus a direct xenograft, even after several generations.2 Thus, although the ability to successfully engraft surgically-derived tumours from cancer patients has been established for decades, these preclinical models are just now being consistently characterized and applied towards drug development in oncology (Table 1).3–9 In this Review, we will present the opportunities and challenges of these models in oncology drug development, provide specific disease examples, and describe concepts regarding predictive biomarker development and future applications.
Table 1.
Preclinical drug screening in patient-derived tumour xenograft models
| Tumour model | Approved agent tested | Investigational agent tested |
|---|---|---|
| Pancreatic ductal adenocarcinoma |
Gemcitabine,11,20 erlotinib11,104 | Temsirolimus,11,35 saracatinib,106 bosutinib,105 MK-1775,112 IPI-504113 |
| NSCLC | Etoposide,13,21,114 carboplatin,13,21,114 gemcitabine,13,21,114 paclitaxel,13,21,114 vinorelbine,13,21,114 cetuximab,13,21,114 erlotinib,13,21,114 docetaxel,49 docetaxel–vinorelbine,49 docetaxel–gemcitabine,49 docetaxel–cisplatin,49 cisplatin64 |
Sagopilone,115 diaziquone,49 pazelliptine,49 retelliptine49 |
| Melanoma | Actinomycin-D,51 carmustine,51 doxorubicin,51,52 bleomycin,51 cisplatin,51,52 melphalan,51 mitomycin-C,51,52 vinblastine,51 cyclophosphamide,52 ifosfamide,52 lomustine,52 5-FU,52 methotrexate,52 etoposide,52 paclitaxel,52 vindesine,52 temozolomide54 |
NA |
| RCC | Sorafenib,89 sunitinib90 | NA |
| Breast cancer | Doxorubicin,74,76 cyclophosphamide,74,76 docetaxel,76 trastuzumab,76 ifosfamide,74 cisplatin,74 capecitabine74 |
Degarelix76 |
| HNSCC | Cisplatin,64,65 cetuximab27 | Diaziquone,64 pazelliptine,64 retelliptine64 |
| GBM | Bevacizumab92 | NA |
| Prostate cancer | Bicalutamide81 | NA |
| Ovarian cancer | 5-FU,116 cyclophosphamide,116 doxorubicin,116 methotrexate,116 hexamethylmelamine,116 cisplatin116 |
NA |
| HCC | 5-FU,117 oxaliplatin,117 doxorubicin,117 cisplatin,117 estradiol,117 progesterone,117 dihydrotestosterone117 |
Gefitinib,117 seocalcitol,117 brivanib118 |
Abbreviations: 5-FU, 5-fluorouracil; GBM, glioblastoma multiforme; HCC, hepatocellular carcinoma; HNSCC, head and neck squamous-cell carcinoma; NA, not applicable; NSCLC, non-small-cell lung cancer; RCC, renal cell carcinoma.
Methodology
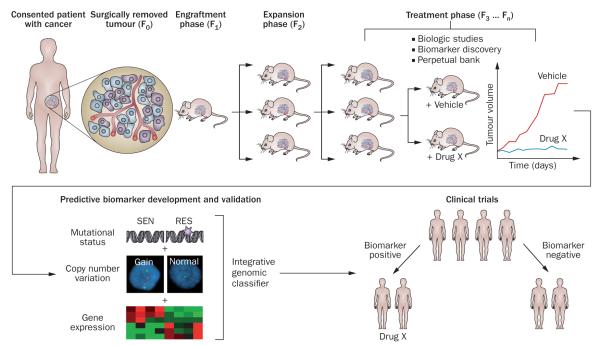
The methodology of initiation and propagation of patient-derived tumour xenografts (PDTX; Figure 1) has been covered in previous reviews by multiple groups.7–11 The approach is very straightforward, consisting of obtaining fresh surgical tissue, sectioning it into ~3 mm3 pieces, followed by subcutaneous or orthotopic implantation into the flank of an immunodeficient mouse or rat. The generation harbouring the patient-derived material is termed F0, with subsequent generations numbered consecutively (F1, F2, F3 and so on), although some groups have named these G0, G1 and so on (Figure 1).10 The amount of time required for tumour ‘take’ is variable among tumour types, location of implantation, and recipient strain, but in general this is between 2 months and 4 months, although failure of engraftment should not be ascertained until at least 6 months.7 In general, the third generation (F3 or G3) can be expanded for drug treatment, and most groups use early passages for such studies. However, the main determinant should be the extent to which the PDTX has diverged from the patient’s tumour in terms of genetics and histology (rather than an arbitrary passage number), two factors that are rarely presented when reporting results of therapeutic studies.
Figure 1.
Establishment and testing of PDTX models. Excess tumour specimens not needed for clinical diagnosis are obtained from the consented patients (F0). Non-necrotic areas of these tumours are sectioned into ~3 mm3 pieces and, after processing, implanted subcutaneously into anaesthetized 5-week to 6-week-old female athymic nude mice. During the engraftment phase, tumours are allowed to establish and grow and then are harvested upon reaching a size of 1,500 mm3 (F1). Similar protocols are employed for subsequent expansion cohort (F2) and treatment cohort (F3 … Fn). Typically, biological assays are performed on tumours in early generations (≤F5); these biological assays include drug efficacy studies, rational combination studies and the development of predictive biomarkers for novel targeted therapies. If the developed biomarkers achieved accurate prediction in a validation set of PDTX models (or ‘xenopatients’), they might be translated into early phase clinical trials as tools for patient selection strategies. Abbreviations: PDTX, patient-derived tumour xenografts; RES, resistant; SEN, sensitive.
Unfortunately, there has not been a comprehensive comparison among recipient strains or hosts, such as athymic nude mice, rats, or NOD/SCID mice, with regards to time-to-engraftment, take rate, genetics, or histology. The majority of investigators report take rates of >75% using athymic nu/nu mice, and NOD/SCID mice are more often used exclusively in F1 or in instances where engraftment is being assessed as a primary end point (such as early stage or adjuvant studies).12,13 The development of the NOD/SCID/IL2Rγnull mice has allowed for even greater take rates (approaching 95–100%) for tumours that are particularly difficult to engraft, as it further inhibits innate immunity by blocking the maturation of natural killer (NK) T cells.14 In addition to improved engraftment efficiency, this model has also been reported to maintain human tumour-associated leukocytes such as effector memory T cells for up to 9 weeks after implantation, providing an improved model of tumour–stromal interactions.15
Multilayered biological assays can be performed on early-passage (≤F5) PDTX to characterize these models for predictive biomarker development (Figure 1).16 One of the main advantages often noted with PDTX models is maintenance of the original tumour architecture and histological characteristics, although there has been controversy over how long and to what extent the human-derived microvasculature is maintained.17,18 For example, Gray et al.18 demonstrated that prostate cancer tumours implanted in nude mice maintained vessels lined with human endothelial cells and increased mean vessel density over time; however, a similar study with renal cell carcinoma revealed a decrease in human-derived vasculature over time. An interesting approach to circumvent this issue used tissue microarrays generated from 150 PDTX samples, which were assessed for VEGF-A, integrin β1, cathepsin B, proteinase-activated receptor 1, and MMP1, revealing profiles of an ‘angiogenic’ phenotype that could be selected for therapies targeting the tumour microenvironment.19 Such approaches, including gene-set enrichment analysis of angiogenic and metastatic pathways, might bypass concerns regarding the ability to completely recapitulate the human microenvironment in PDTX models.20
Genomic comparisons of PDTX models
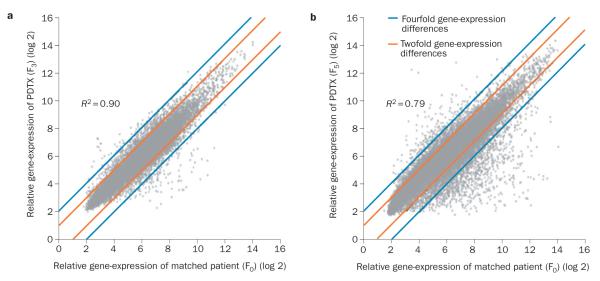
An important question regarding PDTX model stability is whether the process of engraftment and expansion changes the genetic features of the tumours. Comprehensive genome-wide gene-expression analysis studies have demonstrated that PDTX maintain the majority of the key genes and global pathway activity in primary tumours.2,21 For example, in non-small-cell lung cancer (NSCLC) PDTX models,21 unsupervised hierarchical clustering of genome-wide gene-expression profiles revealed that nine of the 17 primary tumours clustered directly with the derived PDTX models, with correlation coefficients ranging from 0.78 to 0.95. Importantly, 10 of the 17 primary–PDTX tumour pairs exhibited correlation coefficients >0.90 indicating a high degree of similarity between the primary cancer and the corresponding PDTX model.21 Similarly, in pancreatic cancer PDTX models, 10 out of 12 primary–PDTX (F0 versus F3) tumour pairs were found to be concordant for KRAS mutational and SMAD4 expression status.11 Interestingly, some of the pancreatic cancer PDTX models have been used to enrich the tumour DNA content for the pancreatic cancer genome sequencing project.22 In a comparative study using small-cell lung cancer (SCLC) PDTX models, Daniel et al.2 generated PDTX models from chemotherapy-naive patients with SCLC and compared them to cell lines derived from each PDTX and to subsequent (after 6 months in culture) cell-line derived PDTX using an Affymetrix® platform.2 The direct comparisons among the samples analysed (PDTX and the two cell line types) revealed three sets of differentially expressed genes: 395 were significantly different when comparing PDTX to their matched initial cell line, 152 were different when comparing PDTX to their derivative secondary cell line, whereas only 26 genes were differentially expressed when comparing the initial cell line to those derived from PDTX after 6 months in vitro. These results suggest that global gene expression can change when cell lines are derived in vitro, and that the expression of a significant number of such genes is not restored when the derivative cell line is returned to growth in vivo, supporting the notion that ‘never in culture’ PDTX models may more closely recapitulate the genetic characteristics of tumours in patients.2 Figure 2 depicts two comparative examples of matched patient–PDTX models using genome-wide gene expression of colorectal cancer (CRC) and pancreatic ductal adenocarcinoma (PDA). The F3 CRC PDTX model and the F5 PDA PDTX model demonstrated high correlation of global gene expression with their matched primary tumours (F0).
Figure 2.
Comparison of genome-wide gene-expression profiles between primary patient tumours and PDTX tumours. a | Matched patient primary CRC tumour (F0) and PDTX (F3). Genome-wide gene-expression profiles of a patient with CRC and their matched PDTX were profiled with Affymetrix® HuGene 1.0 ST arrays. b | Matched patient primary PDA tumour (F0) and PDTX (F5). Genome-wide gene-expression profiles of a patient with PDA and their matched PDTX were profiled with Affymetrix® HG-U133 Plus 2.0 arrays. High correlations were observed in both PDTX models and their matched primary tumours. Abbreviations: CRC, colorectal cancer; PDA, pancreatic ductal adenocarcinoma; PDTX, patient-derived tumour xenografts.
Colorectal cancer
In CRC, there is a long history of success in the establishment, maintenance, and study of PDTX models.23–26 CRC PDTX models are quite easy to establish with take rates of over 75%, they can be successfully cryopreserved prior to implantation, and closely recapitulate the genetic alterations and histology of the fresh tumour.23–26 Indeed, even in early stage CRC tumours exhibiting chromosomal instability, establishment and propagation as PDTX retains the intratumoural clonal heterogeneity, chromosomal instability, and histology of the parent tumour for up to 14 passages, providing a unique opportunity to study new agents for adjuvant therapy targeted towards specific molecular subtypes.25
An important component in the validation of disease-specific PDTX is determining the response to standard agents and correlating responses of the xenograft to the response of the patient. In one study, 15 CRC PDTX models were established and treated with 5-fluorouracil, oxaliplatin, or irinotecan with reasonable concordance between known response rates to these drugs and between the patient and corresponding xenograft.26 For example, five out of 15 xenografts were treated with cytostatic chemotherapeutics and all five of these exhibited similar responses to their corresponding patients.26 In addition, these models retained the histological features of the parent tumour, although there was an increase in epithelial (EpCAM) and tumour markers (CEA) with passage.26
With the advent of biological agents for the treatment of CRC, there has been a recent focus on the use of PDTX models to further refine the mechanisms of resistance to these agents and develop rational combination strategies. In one such study, 23 CRC PDTX models were treated with the EGFR inhibitor cetuximab, profiled for KRAS, NRAS, BRAF, and assessed for epiregulin, amphiregulin, as well as total and activated EGFR, MET, AKT, and HER3, to derive a ‘cetuximab response score’.27 The cetuximab response score comprised positive points associated with high levels of EGFR, epiregulin and/or amphiregulin, and negative points for KRAS, NRAS and/or BRAF mutations or high levels of activated MET, HER3, AKT, or undetectable EGFR. Although not independently validated in another set of PDTX models, the cetuximab response score was 90% accurate in predicting the responsiveness of the CRC explants, indicating the utility of these models in generating clinically relevant hypotheses that can be subsequently tested in other PDTX and, ultimately, in patients.27
PDTX have been used to functionally cross validate predictive biomarkers obtained retrospectively; KRAS, NRAS and BRAF mutations predicted non-responsiveness to cetuximab in both patients and the corresponding PDTX, which led to the identification of HER2 amplification in cetuximab-resistant tumours that were wild type for KRAS, NRAS, BRAF and PI3K.28 This study illustrates the ability to conduct ‘xenopatient’ trials where the rational combination of cetuximab with pertuzumab or lapatinib was assessed in the subset of tumours with resistance to cetuximab and HER2 amplification.28 These studies, and others, emphasize the potential impact of combining genomics data and the PDTX models with a closely associated clinical translation to accelerate drug development and predictive biomarker identification and validation in CRC and other diseases.16,29,30
Often overlooked, owing to the greater emphasis placed on in vivo models, the establishment of ‘new’ patient-derived cell lines is also important to update the current library of CRC cell lines in which initial drug screening and functional studies can be performed. PDTX models can also be used for establishing new cell lines; however, available data suggest, not surprisingly, that there is greater genetic divergence from the parental tumour when enzymatically dispersed and cultured in vitro, and that successful establishment requires initial passage as xenografts.24 Finally, in CRC, PDTX models have been used to establish and characterize a stem-cell compartment in CRC, which should facilitate future drug development in this area.31
Pancreatic cancer
Recent advances in molecular and genetic techniques have resulted in a significant increase in the scientific understanding of the complex genetics of PDA. Whole-exome sequence analysis of primary PDA tumours elucidated a core set of genetic pathways altered in this disease in association with an average of 63 genetic aberrations occurring within an individual tumour.22 Unfortunately, the acquisition of increased genetic information has not yet translated into improved clinical outcomes for this patient population.
Historically, pancreatic cancer cell lines have been used both in vitro and in vivo for preclinical analyses of therapeutic interventions. Cell line-based xenografts grow primarily as homogeneous masses of cancer cells with minimal stromal infiltration; therefore, they might not recapitulate the human PDA tumour architecture and interactions between stromal components and PDA cells, although studies directly comparing these aspects have yet to be performed. It is also becoming increasingly appreciated that the desmoplastic reaction of PDA decreases intratumoural perfusion, which in turn may alter the intratumoural pharmacokinetics of chemotherapeutic agents.32 As a result, cell line xenograft models may overestimate the antitumour effects of a given therapeutic strategy. PDTX pancreatic models employ the implantation of primary human PDA specimens as either heterotopic or orthotopic models.33,34 These PDTX models are characterized by the maintenance of the original tumour architecture, although the human stroma is replaced by murine stroma with sequential passage. Orthotopic PDTX models retain a greater proportion of stromal components and develop locoregional and distant metastases.33,34 In PDA, initial use of PDTX focused on optimizing the platform for the development of predictive and pharmacodynamic end points for molecularly targeted therapies.11 These studies were complicated by significant intratumoural heterogeneity of gene and protein expression. In the case of mTOR inhibitors, this model predicted that patients with high baseline expression of phosphorylated p70 S6 kinase would preferentially respond to mTOR inhibition, a result which unfortunately did not translate to patients.35 Failure of translation could be because of the low stringency in defining a ‘response’ in the PDTX model, or the multiple feedback loops in the mTOR pathway. Additional attempts to optimize the system included the development of a tumour biopsy strategy followed by ex vivo therapeutic treatment and pharmacodynamic end point analysis.36 This strategy demonstrated that a polo-like kinase inhibitor induced growth inhibition, particularly in gemcitabine-refractory PDTX models and that cyclin B1 was a biomarker of response.37 Another approach that has been used in PDA is the empirical treatment of a patient-specific PDTX with a panel of drugs whilst the patient is receiving first-line therapy.38 At the time of disease progression, the patient then receives the therapy demonstrating the most activity in their PDTX. This strategy combined with whole-exome sequencing was used to demonstrate the activity of mitomycin C and cisplatin in a patient harbouring a PALB2 mutation.39,40 Caveats to this approach include delayed or unsuccessful engraftment of the PDTX and the requirement for considerable resources. A modification would be to perform complete genomic analysis of a patient’s tumour upon engraftment to serve as a basis for molecular therapy selection of the PDTX, with results used to direct treatment of the patient at the time of disease progression.
The stromal components of the desmoplastic reaction associated with PDA might represent a novel treatment strategy for this disease; the importance of the stroma in PDA is exemplified by the observation that PDTX engraftment is associated with the expression of stromal gene pathways and decreased patient survival.20 In PDA, both genetically engineered and PDTX models have demonstrated that stromal modulation may increase intratumoural gemcitabine concentrations to improve therapy efficacy.32,41 Likewise, hedgehog pathway inhibition denudes the stroma, possibly owing to the induction of apoptosis in pancreas stellate cells, resulting in increased vascular patency.42 Interestingly, treatment of pancreas PDTX models with gemcitabine and nabpaclitaxel preferentially decreased the intratumoural desmoplastic reaction resulting in increased intratumoural gemcitabine concentrations and growth inhibition in comparison to each individual drug.41 These observations were tested in a phase I–II clinical trial of patients with advanced-stage PDA in which 44 patients were treated at the defined maximum tolerated dose and a median survival of 12.2 months was observed,41 suggesting that PDTX models can be used to derive strategies that target tumour stroma in PDA.
Lung cancer
Since the approval of the ALK inhibitor crizotinib and the EGFR inhibitor erlotinib for the treatment of NSCLC, there has been a focus on the use of preclinical models and fresh tissue biopsies from patients to probe mechanisms of responsiveness and resistance to these agents.43–47 These PDTX models have been assessed for their similarity to the parent tumour in terms of histology and genetics, as well as responsiveness to commonly used agents in lung cancer.21,48,49 Owing to the initial poor engraftment rate of these models, some groups have used the technique of implanting fresh patient-derived tumours in the subrenal capsule location in NOD/SCID mice, with the intent of capitalizing on the greater perfusion of blood vessels in this area and more-rapid development of a supporting tumour microvasculature.50 In one cohort of 14 PDTX derived from lung cancer patients with a range of stages and histologies, there was a >95% engraftment rate and the histological features of the parent tumour were largely retained.50 Although there were genetic changes that occurred with subsequent passage of the PDTX, genetic characteristics between the parent tumour and early passage PDTX were similar by comparative genomic hybridization and fluorescence in situ hybridization (FISH), and later changes were reflective of known genetic aberrations in lung cancer. Interestingly, in this study, the presence of infiltrating mouse stromal cells was documented in a lung carcinosarcoma PDTX, using species-specific pan-centromeric FISH probes.50
Clearly, an area of interest in lung cancer is the characterization of PDTX models using standard chemotherapeutic agents and assessment of the correlation of putative predictive biomarkers with clinical outcome. Examples of this include two studies, one that assessed docetaxel as a single agent or in a ‘doublet’ combination, and the other that assessed a wide range of standard agents alone; the studies had the intent of determining whether drug resistance21 or DNA repair markers49 could predict response to therapy. Although both studies are limited by the small number of PDTX models, seven and 24, respectively, the combined results indicate that these models recapitulate the magnitude and variability of response to therapy that is observed in lung cancer patients, and that standard biomarkers of drug resistance or DNA repair do not facilitate patient selection.21,49 In another cohort of 25 PDTX derived from patients with primarily non-metastatic lung cancer, the histology as well as markers for anti-EpCAM and Ki-67 were retained, although less heterogeneity was observed in the first few passages of the PDTX, thought to be due to the replacement of human accessory cells with murine stromal tissue.13 Using gene profiling, nine out of 17 NSCLC PDTX tumours clustered with their parent tumour using unsupervised hierarchical clustering, while of the eight that did not, five had <10% tumour tissue.21 These results indicate that although there can be a high degree of similarity between primary tumour and PDTX, this similarity should not be assumed. Therefore, studies that rely on a patient’s own PDTX to make treatment decisions should be thoroughly characterized with regards to histology and gene expression at a minimum, if not by the use of next-generation sequencing technology. Interestingly, the majority of gene-expression changes (193 probe sets) between the primary and the PDTX were genes that were downregulated, while the pathways represented by the unique genes (134) in these probe sets represented those involved in cell adhesion and immune response.21
An interesting and potentially clinically relevant application of these PDTX models in NSCLC would be the ability to predict, based on PDTX engraftment success, those patients who are more likely to relapse after curative surgery, with the added benefit of having an available ‘xenopatient’ for therapeutic testing prior to actual relapse in the patient. In one such study, 63 of 157 specimens from curative resections in NSCLC engrafted into the mice were compared to patient outcomes, and demonstrated a statistically significant shorter disease-free survival in a multivariate analysis.12 Factors associated with engraftment included large tumour size, squamous histology, and poor differentiation.12 Another study engrafted 32 curative resection specimens from patients with early stage NSCLC and subjected them to one to three typical chemotherapeutic regimens.48 Two observations of interest include the fact that even responding PDTX had nests of viable cells, indicating intratumoural heterogeneity of drug response, and PDTX from six of seven patients who had recurrence or clinical relapse did not respond to ex vivo chemotherapy, supporting the use of these models in early stage disease to develop more-effective (and patient-directed) adjuvant chemotherapy.
Melanoma
Until recently, patients with advanced-stage melanoma had limited therapeutic options, but with the approval of the BRAF inhibitor vemurafenib and the CTLA-4 inhibitor ipilimumab the therapeutic field has been reinvigorated and thus PDTX models will likely be used to define resistance pathways and rational combination strategies in this disease; however, these models are limited to non-immunotherapeutic approaches. Although the feasibility of establishing PDTX models of melanoma was demonstrated decades ago,51,52 studies characterizing large numbers of models derived from a broad patient base have been lacking and many centres have focused on the development of patient-derived cell lines. Early studies using a melanoma PDTX derived from a primary tumour and a metastatic lesion from the same patient sought to compare responses to anticancer agents between cell lines derived from the tumours versus PDTX. Although most of the responses were comparable, perhaps not surprisingly, unpredictable discordant sensitivity to certain agents was also observed.51 More recently, gene-array data from a larger panel of 22 melanoma PDTX tumours was used to develop a predictive gene signature to 11 standard cytotoxic agents.52 The data generated from these studies exhibited acceptable predictive value; however, no further clinical assessment has been reported.
PDTX models have also been used to identify melanoma tumour-initiating cells. After serial passage in nude mice, isolation and re-implantation of ABCB5+ cells from PDTX were capable of regenerating tumour heterogeneity and selective targeting of this subpopulation resulted in tumour growth inhibition.53 A report described the establishment and characterization of a human uveal melanoma PDTX model in SCID mice with an engraftment rate of 28% (25 of 90) and concordance between the primary and corresponding PDTX in terms of histology, genetic profiles, and tumour antigen expression.54 Further, when the engrafted mice were treated with temozolomide, a standard chemotherapeutic agent for uveal melanoma, the response was reflective of the clinical outcome observed in the patients.54
Head and neck cancer
Head and neck squamous-cell carcinoma (HNSCC) is a particularly challenging disease owing to the rarity of activating oncogene mutations and prevalence of mutations in tumour-suppressor genes, such as TP53 and Notch1 with 47% and 19% mutation rates, respectively.55,56 Cetuximab is the only approved targeted drug for HNSCC,57,58 but to date, and despite its quite low clinical efficacy, there are no validated patient selection strategies. To address these shortcomings, efforts have been undertaken to develop PDTX models of HNSCC. There have been several reports showing that the engraftment rate of patient-derived HNSCC biopsies in mice is between 29% and 44%.59–63 Of note, no difference was observed between patients whose tumours engrafted and those who did not with respect to tumour biology or clinical findings, including survival.61 In another report, no differences were observed in engraftment rates between NOD/SCID and Rag2DKO mice.60 However, relatively few efforts in HNSCC PDTX have been directed towards drug development. As part of a collaborative programme sponsored by the European Organisation for Research and Treatment of Cancer to evaluate the use of several PDTX models to predict phase II clinical drug activity, Langdon et al.,64 tested cisplatin, diaziquone and two investigational agents, pazelliptine and retelliptine.64 For the HNSCC models, no response to cisplatin was observed in five tumours tested; four out of five tumours responded to diaziquone, as measured by tumour growth inhibition >50%. Thus, these models seem to have underpredicted the clinical response to cisplatin and overpredicted the response to diaziquone.64 Another group used HNSCC PDTX models to study cellular mechanisms behind resistance to cisplatin with respect to DNA repair, apoptosis and cell-cycle regulation.65 They found that xenografts with mutations in TP53 or amplification of CCND1 were significantly more resistant to this agent (P <0.001). These results are consistent with previous findings demonstrating a correlation between TP53 mutations and a poor prognosis in patients with HNSCC and to neoadjuvant chemotherapy resistance.66–68 A recent study tested the response to cetuximab against a large panel of PDTX cancer models including HNSCC;27 of the eight HNSCC xenografts tested, five were responsive (63%), which was the highest response rate of any cancers tested. However, unlike the case for CRC and NSCLC, there was no correlation of response with mutations in KRAS, BRAF or NRAS, nor to amplification of receptors such as HER3 or MET.27 A large colony of HNSCC and skin squamous-cell carcinoma is being generated to aid in further drug development (J. J. Tentler et al., personal communication); clinical hypotheses based on these HNSCC PDTX studies have been translated into three currently active clinical trials (NCT01255800,69 NCT01252628,70 NCT0120409971), highlighting the translational impact of these models.
Breast cancer
Over the past decade, major advances in our understanding of breast cancer biology at the molecular and genetic level have revealed a diverse and complex disease consisting of distinct molecular subtypes, each with a unique gene-expression profile, which will likely influence responses to standard and/or novel therapies.72 These molecular classifications: luminal A, luminal B, triple-negative, basal-like and HER2-positive, present a unique opportunity to individualize therapies tailored specifically to a patient’s defined tumour subtype. Attempts to develop PDTX models of hormone-driven cancers such as breast and prostate cancer have met with limited success. However, recent larger scale efforts involving orthotopic implantation of human breast tumour tissue into cleared mammary fat pads of NOD/SCID or athymic nude mice, with oestrogen supplementation of oestrogen receptor (ER)-positive tumours, have resulted in improved engraftment efficiency such that enough PDTX models have now been generated to fully represent the diverse subtypes observed in clinical breast cancer.73–76 For example, DeRose et al.75 implanted 49 primary and metastatic breast cancer biopsies into NOD/SCID mice, from which 18 engrafted (37%) and 12 were successfully maintained in successive passages as PDTX (24% of total). Representative phenotypes included ER-positive and progesterone receptor (PR)-positive, ER-negative and PR-negative, and HER2-positive tumours. Interestingly, neither the source of the tumour (primary or metastatic) nor the ER, PR or HER2 status was significantly associated with engraftment success. However, tumours that were ER-negative, PR-negative and HER2-negative (triple negative) grew at the fastest rate, consistent with the aggressive clinical manifestation of this disease.75 The rate of engraftment was a prognostic factor for patient survival time even in individuals with newly diagnosed disease who did not have detectable metastases at time of surgery. Thus, the ability of a tumour to engraft in mice could potentially be used as a surrogate prognostic indicator of aggressiveness and risk of disease progression. Importantly, in several other aspects, these breast cancer PDTX models recapitulated the biology and disease outcomes of the patient tumours from which they were derived. As in other models, tumour architecture was maintained through several passages in mice, including its supportive stroma; however, staining with antibodies specific to mouse CD45 revealed that this stroma was primarily comprised of infiltrating mouse leukocytes. The tumours also maintained their oestrogen dependence and responsiveness through serial passages in mice. Another important aspect of these PDTX was their ability to metastasize to sites similar to those observed in the original subjects.75
Loss of expression of the DNA repair-related BRCA2 gene is responsible for a large percentage of hereditary breast and ovarian cancers and studies of BRCA2 function in tumour biology in vivo have been impeded by a lack of relevant models. One such PDTX model of BRCA2-mutant breast cancer has been described in which the basal-like morphology and tumour architecture including stroma were conserved in the xenograft, as were gene-expression patterns over successive generations.74 This PDTX model exhibited differential responses to standard chemotherapies, including pronounced sensitivity to anthracyline-based therapies as well as to a cisplatin and ifosfamide combination, but resistance to agents targeting microtubules, such as taxanes.
PDTX models of breast cancer have also been used to study metabolomic profiles and their role in breast tumours, which to date is poorly understood.77 Concentrations of choline-derived metabolites were measured in xenografts of primary breast biopsies representing basal-like and luminal-like subtypes. Differences in choline metabolite concentrations observed between these two models in mice correlated well with similar profiles and gene-expression patterns observed in material from patients with ER-positive and PR-positive breast cancer, and triple-negative breast cancer, suggesting that these PDTX xenografts are relevant models for studying choline metabolism in these subtypes.77
Prostate cancer
Historically, there has been a lack of reliable preclinical models for studying prostate cancer biology and therapy due to the difficulty in establishing prostate cancer cell lines in vitro. Several groups have established prostate cancer PDTX models that have allowed the preclinical study of a wide variety of prostate cancer-specific biological processes, such as chromosomal aberrations,78 angiogenesis,79 identification of pluripotent stem cells,80 and response to androgen ablation therapies.81 Comparative studies have been undertaken to examine the efficiency and histopathological consequences of engraftment of human prostate cancer tissue to subcutaneous, subcapsular renal and prostatic orthotopic sites.82 The subcutaneous grafts demonstrated the lowest take rates and also the lowest level of histodifferentiation. The most efficient engraftment was renal subcapsular (>90%), which also demonstrated differentiation and continued expression of the androgen receptor and prostate-specific antigen (PSA). However, the orthotopic models have consistently exhibited the best degree of differentiation and expression of androgen receptor and PSA.82
Renal cell carcinoma
Cytokine therapy with interleukin-2 or interferon-α was the standard of care for the first-line treatment of metastatic renal cell carcinoma (RCC). However, in the past 5 years, the arrival of molecularly targeted agents has revolutionized the management of metastatic RCC.83 PDTX models of RCC have been used for decades and may provide several advantages over cell line xenografts.80–85 RCC cells continuously cultured in vitro are known to acquire many genetic alterations not found in the original tumour.86 In addition, the limited number of RCC cell lines available means that they lack the heterogeneity that characterizes kidney cancer in the human population.18,87 Several studies have demonstrated that RCC PDTX models are similar to parental tumours as assessed by histology, karyotype, DNA-ploidy, and molecular cytogenetic analyses.80–85,88 Grisanzio et al.80 have also created orthotopic PDTX that exhibit high levels of histological, immunophenotypical and genetic concordance between the PDTX and the corresponding primary tumour. Another advantage of an RCC PDTX model is the ability to evaluate tumour angiogenesis, and studies have demonstrated that the vasculature of RCC PDTX are predominantly comprised of human endothelial cells, up to 35 days after implantation.17,18 Currently, RCC PDTX are being used to evaluate the effects of targeted agents. Yuen et al.89 evaluated the effects of the tyrosine kinase inhibitor sorafenib on different RCC subtypes, demonstrating inhibition of both angiogenic and non-angiogenic targets. In another study, PDTX of RCC tumours that initially responded to another tyrosine kinase inhibitor (sunitinib) and eventually progressed, were used to interrogate resistance mechanisms to sunitinib.90 This study demonstrated that de novo onset of an epithelial–mesenchymal transition-like phenotype was associated with acquired resistance to sunitinib, and reversion to an epithelial histology in a PDTX model was associated with a return of responsiveness to sunitinib. As the knowledge of mechanisms of action and resistance to targeted agents increases in RCC, new potential combinations may arise, as exemplified in a study where investigators demonstrated that the combination of vascular disrupting agents and everolimus resulted in enhanced reduction of blood volume in orthotopic RCC xenograft tumours in mice.91
Glioblastoma multiforme
Glioblastoma multiforme (GBM) are aggressive and malignant primary brain tumours that are highly resistant to therapy. Recent GBM research has focused on the identification of aberrant genetic events and signalling pathways, and has provided insight into the molecular mechanisms of the disease as well as the clinical evaluation of new investigational compounds that target these pathways. A clinically relevant orthotopic model of GBM has been developed that uses human GBM biopsies.92,93 Unlike many other PDTX models that directly engraft tumour tissue into mice immediately after surgery, GBM biopsies were cut to 0.3 mm samples and cultured in agar in vitro for the development of spheroids and subsequently implanted into the brain of athymic nude rats. In this model, the average culture time for spheroid formation was 18 days and the take rate was 96% (28 out of 29 tumours).93 More importantly, this model seemed to recapitulate the disease course of GBM in humans, as evidenced by high vascularity, glioma tumour cell invasion and necrosis.93 This model was also shown to be genetically stable.92 Comparison of patient GBM to the PDTX by comparative genomic hybridization exhibited great similarity. As there is a high degree of vascularity in GBM tumors, a GBM PDTX orthotopic model was used to assess the efficacy of the antiangiogenic drug bevacizumab.92 Although anti-VEGF therapy resulted in a significant decrease in tumour volume and vascularity, the hypoxic microenvironment that resulted induced a more-invasive phenotype that seemed to be dependent on upregulation of the PI3K/Akt signalling pathway. These results suggest that combinational strategies involving bevacizumab and a molecularly targeted compound such as a PI3K inhibitor may be more effective. Therefore, the use of such GBM PDTX models might further facilitate the development of novel combination strategies that will hopefully improve outcomes in this patient population.
Paediatric cancers
Within the paediatric oncology community, PDTX has been used to establish most of the current models, and these have been of value in developing multiple effective therapies, most notably both topotecan and irinotecan for solid tumours, and combinations of these agents with both standard cytotoxic and experimental agents.94–96 The predictive value of these PDTX xenografts led to the National Cancer Institute supporting the Pediatric Preclinical Testing Program (PPTP).97 Although paediatric cancer represents only a small percentage of all cancers, this clinical community realized in the early 1980s the deficits of cell-line derived models, demonstrated that greater genetic drift occurred in culture than for mouse-passaged tumours, and showed the maintenance of both expression profiles and genomic characteristics in PDTX.97 One example is ependymoma, which is the third most common malignant brain tumour of children with limited treatment options. Even after total resection of the tumour followed by radiation treatment, recurrence and metastasis occurs in about half of patients.98–100 Preclinical studies to identify new therapies for this cancer have been difficult owing to a lack of good animal models. Yu et al.101 successfully developed an orthotopic ependymoma PDTX mouse model that demonstrated similar histological and invasive growth characteristics between the xenograft tumours and the original tumour in the patient. Not only did they show that by gene-array analysis the expression profiles were maintained, they demonstrated the CD133+ cancer stem cells were preserved in the tumour and when cell lines were made from this xenograft, the differentiation capabilities were also maintained.
Predictive biomarker discovery
Gene-expression profiling and other genome-wide high-throughput technologies have advanced our understanding of cancer biology, enabling the discovery of biomarkers for classifying tumour responsiveness to treatment using PDTX models (Box 1). In one study, cDNA microarrays were employed to interrogate the expression of 23,040 genes in 85 PDTX models of various solid tumours for genes associated with chemosensitivity to nine anticancer drugs.102 Of the 1,578 genes whose expression levels correlated significantly with chemosensitivity, 333 genes showed significant correlation with more than two drugs, and 32 correlated with six or seven drugs. This study represents one of the earliest using microarray analysis on PDTX models to develop predictive biomarkers for standard anticancer agents.
Box 1. Strategy for the use of PDTX models for predictive biomarker discovery.
Step 1—training set
▪ Determine drug responses for a panel of PDTX mice of specific disease type
▪ Analyse and annotate molecular features for the PDTX model from fresh-frozen or FFPE tumour samples (for example, genome sequencing, transcriptome profiling, proteomics, metabolomics)
▪ Employ computational and statistical methods to derive predictive classifiers by integrating these molecular features
Step 2—assay development
▪ Determine the feasibility of developing an assay from the biomarkers identified from the training set (for example, mutation analysis by sequencing, gene expression by reverse transcription PCR, protein expression by immunohistochemistry, copy number changes by fluorescence in situ hybridization)
▪ Calibrate the biomarkers on archival FFPE samples or fresh-frozen tumours
▪ Determine the parameters and thresholds for the predictive biomarkers of the assay by testing on the training set
Step 3—preclinical trial design
▪ Design biomarker-driven clinical trials based on the prevalence of the identified biomarkers
▪ Determine appropriate sample size and power estimation for small-scale preclinical trial
Step 4—validation or test set
▪ Determine predictive biomarkers on the remaining PDTX models using the developed assay
▪ Select appropriate PDTX models with positive and negative predictive biomarkers
▪ Determine drug responses on the selected PDTX models
▪ Determine the accuracy of the predictive biomarkers
▪ Potential refinement of the predictive biomarkers by repeating steps one to three Abbreviations: PDTX, patient-derived tumour xenografts; FFPE, formalin-fixed, paraffin-embedded.
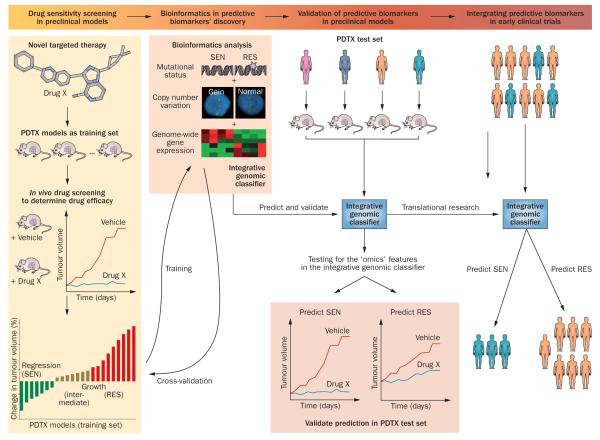
Our group has used PDTX models to develop predictive biomarkers for several targeted anticancer agents in CRC and PDA using gene-expression profiling and oncogene sequencing. We have employed a novel comparison-based method, k-TSP (k-disjoint Top Scoring Pairs) that seeks to identify pairs of genes whose expression levels typically invert from sensitive to resistant PDTX models.103 The gene-pair comparisons can be easily interpreted as ‘IF–ELSE’ decision rules, which can be measured by reverse transcription PCR and are thus well suited for clinical applications. Among the targeted therapies studied by our group using this methodology are inhibitors of EGFR,104 IGF-1R,16 Src,30,105,106 MEK,29 and mTOR.35 Two of the predictive biomarkers developed are currently being assessed in clinical trials (NCT00735917107 and NCT01016860108). Importantly, we continue to revise and refine the integration of k-TSP with other predictive biomarkers, to facilitate the development of integrated classifiers that may incorporate data from diverse platforms (Figure 3).16
Figure 3.
Predictive biomarker development strategy in PDTX models. A novel targeted therapy (drug X) will be screened in a cohort of tumour-specific PDTX models to determine efficacy and the PDTX will be classified into SEN, RES and intermediate groups. These models will be used as the training set to develop predictive biomarkers. Multiple layers of ‘omics’ technologies will be employed to characterize these PDTX training sets to derive an integrative genomic classifier to predict SEN and RES models in response to drug X. A cross-validation step can be used to refine the classifier, which could be developed into assays for predicting responsiveness to drug X in fresh frozen or FFPE tissue samples. The classifier would require validation by correct prediction in an independent cohort of PDTX models (test set). If the classifier achieved a high level of accuracy in the test set, it could potentially be translated into early clinical trials to select patients to be treated with drug X. Abbreviations: FFPE, formalin-fixed, paraffin-embedded; PDTX, patient-derived tumour xenograft; RES, resistant; SEN, sensitive.
Conclusions
It is becoming increasingly clear that novel preclinical models that more closely recapitulate the heterogeneity of human tumours are needed for more-efficient oncology drug development. PDTX models in many ways represent a major advancement in that direction, but need to be viewed within the context of their inherent challenges and potential opportunities. Challenges include the immense resources that are needed to establish and maintain such ‘live’ tumour banks, often with limited funding sources; the lack of current standardized criteria for assessing and reporting histological, stromal, and genetic drift from the primary (F0) tumour; low stringency in ‘response’ criteria that may not translate to benefit in cancer patients; and the numerous biological challenges of using immunocompromised rodent models as hosts for human tumours. The drift of stromal components of PDTX tumours from primarily human to primarily mouse is likely to present a relevant drawback of these models, given the importance of the tumour microenvironment in numerous aspects of cancer biology. This limitation is especially apparent in studies involving species-specific anticancer compounds that target the microenvironment, such as bevacizumab, or agents that target the immune system, such as ipilimumab. In addition, the response of a human tumour to a particular anticancer agent may be determined by systemic exposure in the murine background. Thus, when drugs are tolerated at high doses there may be an overprediction of response, and likewise, where mice are less tolerant it may produce false-negative results. Some of the challenges could be ameliorated by the development of standard criteria for reporting the extent of pheno typic and genetic drift from the F0 generation when presenting therapeutic results, by increasing the stringency of the response criteria to require regression or at least 80% tumour growth inhibition, and by ensuring that ‘xeno patient’ trials are large enough to lead to clinically meaningful results. Along these lines, one could envision national and international PDTX cooperative groups conducting collaborative studies within molecularly defined tumour subsets that would result in hypothesis-driven personalized medicine strategies for cancer patients. Future directions for these models are numerous and include the development of more-sophisticated orthotopic models, models in which the patient’s own bone marrow stem cells are engrafted along with an orthotopic tumour, in addition to advancements in transfection technology to facilitate in vivo functional short-hairpin RNA genomic screens. Clearly, PDTX models need to be viewed as complementary to other preclinical models, such as genetically engineered mouse models, which are well suited for studying drug responses within a well-defined genetic background.109 Although the concept and initial establishment of PDTX models has been in existence for decades, their value in oncology drug development is just becoming realized as individualized therapy approaches transform cancer therapy.
Key points.
▪ Many preclinical animal models fail to accurately predict the clinical efficacy of novel anticancer agents, largely due to their inability to reflect the complexity and heterogeneity of human tumours
▪ Patient-derived tumour xenograft models (PDTX), where surgically resected tumour samples are engrafted directly into immune-compromised mice, offer several advantages over standard cell-line xenograft models
▪ PDTX tumours maintain the molecular, genetic and histological heterogeneity typical of tumours of origin through serial passaging in mice
▪ The tumour histology of PDTX models provides an excellent in vivo preclinical platform to study cancer stem-cell biology and stromal–tumour interactions; novel cancer therapeutics can also be assessed
▪ Well-characterized PDTX models represent an information-rich preclinical resource for analysis of drug activity, including novel–novel drug combinations, as well as predictive biomarker discovery
▪ The PDTX approach to modelling of specific cancer types could potentially reduce non-informative animal studies while providing a more-relevant system to test clinically directed hypotheses
Review criteria.
Literature searches were performed using the PubMed database and the following search terms: “patient-derived xenografts”, “colorectal cancer”, “pancreatic cancer”, “melanoma”, “breast cancer”, “lung cancer”, “prostate cancer”, “head and neck cancer”, “glioblastoma”, “pediatric cancer”. Searches were performed over the time period of September 2011 to January 2012. Only articles published in English were considered. The development of head and neck squamous cell carcinoma patient-derived tumour xenograft models at the University of Colorado School of Medicine mentioned in this Review was conveyed via personal communication from A. Jimeno.
Footnotes
Author contributions All authors contributed equally to all aspects of the article, including researching data, discussion of content, writing, reviewing and editing the manuscript before submission.
Competing interests The authors declare no competing interests.
References
- 1.Johnson JI, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br. J. Cancer. 2001;84:1424–1431. doi: 10.1054/bjoc.2001.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel VC, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giovanella BC, et al. DNA topoisomerase I–targeted chemotherapy of human colon cancer in xenografts. Science. 1989;246:1046–1048. doi: 10.1126/science.2555920. [DOI] [PubMed] [Google Scholar]
- 4.Houghton JA, Maroda SJ, Jr, Phillips JO, Houghton PJ. Biochemical determinants of responsiveness to 5-fluorouracil and its derivatives in xenografts of human colorectal adenocarcinomas in mice. Cancer Res. 1981;41:144–149. [PubMed] [Google Scholar]
- 5.Houghton JA, Taylor DM. Growth characteristics of human colorectal tumours during serial passage in immune-deprived mice. Br. J. Cancer. 1978;37:213–223. doi: 10.1038/bjc.1978.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin K, et al. Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin. Transl. Oncol. 2010;12:473–480. doi: 10.1007/s12094-010-0540-6. [DOI] [PubMed] [Google Scholar]
- 7.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat. Protoc. 2007;2:247–250. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 8.Rubio-Viqueira B, Hidalgo M. Direct in vivo xenograft tumor model for predicting chemotherapeutic drug response in cancer patients. Clin. Pharmacol. Ther. 2009;85:217–221. doi: 10.1038/clpt.2008.200. [DOI] [PubMed] [Google Scholar]
- 9.Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006;66:3351–3354. doi: 10.1158/0008-5472.CAN-05-3627. [DOI] [PubMed] [Google Scholar]
- 10.Jin K, et al. Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin. Transl. Oncol. 2010;12:473–480. doi: 10.1007/s12094-010-0540-6. [DOI] [PubMed] [Google Scholar]
- 11.Rubio-Viqueira B, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin. Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 12.John T, et al. The ability to form primary tumor xenografts is predictive of increased risk of disease recurrence in early-stage non-small cell lung cancer. Clin. Cancer Res. 2011;17:134–141. doi: 10.1158/1078-0432.CCR-10-2224. [DOI] [PubMed] [Google Scholar]
- 13.Merk J, Rolff J, Becker M, Leschber G, Fichtner I. Patient-derived xenografts of non-small-cell lung cancer: a pre-clinical model to evaluate adjuvant chemotherapy? Eur. J. Cardiothorac. Surg. 2009;36:454–459. doi: 10.1016/j.ejcts.2009.03.054. [DOI] [PubMed] [Google Scholar]
- 14.Shultz LD, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2Rγnull mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 15.Simpson-Abelson MR, et al. Long-term engraftment and expansion of tumor-derived memory T cells following the implantation of non-disrupted pieces of human lung tumor into NOD-scid IL2Rγnull mice. J. Immunol. 2008;180:7009–7018. doi: 10.4049/jimmunol.180.10.7009. [DOI] [PubMed] [Google Scholar]
- 16.Pitts TM, et al. Development of an integrated genomic classifier for a novel agent in colorectal cancer: approach to individualized therapy in early development. Clin. Cancer Res. 2010;16:3193–3204. doi: 10.1158/1078-0432.CCR-09-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanz L, et al. Differential transplantability of human endothelial cells in colorectal cancer and renal cell carcinoma primary xenografts. Lab. Invest. 2009;89:91–97. doi: 10.1038/labinvest.2008.108. [DOI] [PubMed] [Google Scholar]
- 18.Gray DR, et al. Short-term human prostate primary xenografts: an in vivo model of human prostate cancer vasculature and angiogenesis. Cancer Res. 2004;64:1712–1721. doi: 10.1158/0008-5472.can-03-2700. [DOI] [PubMed] [Google Scholar]
- 19.Smith V, Wirth GJ, Fiebig HH, Burger AM. Tissue microarrays of human tumor xenografts: characterization of proteins involved in migration and angiogenesis for applications in the development of targeted anticancer agents. Cancer Genomics Proteomics. 2008;5:263–273. [PMC free article] [PubMed] [Google Scholar]
- 20.Garrido-Laguna I, et al. Tumor engraftment in nude mice and enrichment in stroma- related gene pathways predict poor survival and resistance to gemcitabine in patients with pancreatic cancer. Clin. Cancer Res. 2011;17:5793–5800. doi: 10.1158/1078-0432.CCR-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fichtner I, et al. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin. Cancer Res. 2008;14:6456–6468. doi: 10.1158/1078-0432.CCR-08-0138. [DOI] [PubMed] [Google Scholar]
- 22.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linnebacher M, et al. Cryopreservation of human colorectal carcinomas prior to xenografting. BMC Cancer. 2010;10:362. doi: 10.1186/1471-2407-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dangles-Marie V, et al. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: comparison of success rate and cell line features. Cancer Res. 2007;67:398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- 25.Guenot D, et al. Primary tumour genetic alterations and intra-tumoral heterogeneity are maintained in xenografts of human colon cancers showing chromosome instability. J. Pathol. 2006;208:643–652. doi: 10.1002/path.1936. [DOI] [PubMed] [Google Scholar]
- 26.Fichtner I, et al. Anticancer drug response and expression of molecular markers in early-passage xenotransplanted colon carcinomas. Eur. J. Cancer. 2004;40:298–307. doi: 10.1016/j.ejca.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Krumbach R, et al. Primary resistance to cetuximab in a panel of patient-derived tumour xenograft models: activation of MET as one mechanism for drug resistance. Eur. J. Cancer. 2011;47:1231–1243. doi: 10.1016/j.ejca.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Bertotti A, et al. A molecularly annotated platform of patient-derived xenografts (‘xenopatients’) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 29.Tentler JJ, et al. Identification of predictive markers of response to the MEK1/2 inhibitor selumetinib (AZD6244) in K-ras-mutated colorectal cancer. Mol. Cancer Ther. 2010;9:3351–3362. doi: 10.1158/1535-7163.MCT-10-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arcaroli JJ, et al. Gene array and fluorescence in situ hybridization biomarkers of activity of saracatinib (AZD0530), a Src inhibitor, in a preclinical model of colorectal cancer. Clin. Cancer Res. 2010;16:4165–4177. doi: 10.1158/1078-0432.CCR-10-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl Acad. Sci. USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MP, et al. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat. Protoc. 2009;4:1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu X, Guadagni F, Hoffman RM. A metastatic nude-mouse model of human pancreatic cancer constructed orthotopically with histologically intact patient specimens. Proc. Natl Acad. Sci. USA. 1992;89:5645–5649. doi: 10.1073/pnas.89.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrido-Laguna I, et al. Integrated preclinical and clinical development of mTOR inhibitors in pancreatic cancer. Br. J. Cancer. 2010;103:649–655. doi: 10.1038/sj.bjc.6605819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubio-Viqueira B, et al. Optimizing the development of targeted agents in pancreatic cancer: tumor fine-needle aspiration biopsy as a platform for novel prospective ex vivo drug sensitivity assays. Mol. Cancer Ther. 2007;6:515–523. doi: 10.1158/1535-7163.MCT-06-0388. [DOI] [PubMed] [Google Scholar]
- 37.Jimeno A, et al. A fine-needle aspirate-based vulnerability assay identifies polo-like kinase 1 as a mediator of gemcitabine resistance in pancreatic cancer. Mol. Cancer Ther. 2010;9:311–318. doi: 10.1158/1535-7163.MCT-09-0693. [DOI] [PubMed] [Google Scholar]
- 38.Hidalgo M, et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol. Cancer Ther. 2011;10:1311–1316. doi: 10.1158/1535-7163.MCT-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villarroel MC, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol. Cancer Ther. 2011;10:3–8. doi: 10.1158/1535-7163.MCT-10-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Von Hoff DD, et al. Gemcitabine plus nabpaclitaxel is an active regimen in patients with advanced pancreatic Cancer: a phase I/II trial. J. Clin. Oncol. 2011;29:4548–4554. doi: 10.1200/JCO.2011.36.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey JM, et al. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin. Cancer Res. 2008;14:5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamazaki S, et al. Pharmacokineticpharmacodynamic modeling of crizotinib for anaplastic lymphoma kinase inhibition and anti-tumor efficacy in human tumor xenograft mouse models. J. Pharmacol. Exp. Ther. 2012;340:549–557. doi: 10.1124/jpet.111.188870. [DOI] [PubMed] [Google Scholar]
- 44.Christensen JG. Proof of principle for crizotinib in anaplastic lymphoma kinase-positive malignancies was achieved in ALK-positive nonclinical models. Mol. Cancer Ther. 2011;10:2024. doi: 10.1158/1535-7163.MCT-11-0721. [DOI] [PubMed] [Google Scholar]
- 45.Sasaki T, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–6060. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ercan D, et al. Amplification of EGFR T790M causes resistance to an irreversible EGFR inhibitor. Oncogene. 2010;29:2346–2356. doi: 10.1038/onc.2009.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida T, et al. Effects of Src inhibitors on cell growth and epidermal growth factor receptor and MET signaling in gefitinib-resistant non-small cell lung cancer cells with acquired MET amplification. Cancer Sci. 2010;101:167–172. doi: 10.1111/j.1349-7006.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong X, et al. Patient-derived first generation xenografts of non-small cell lung cancers: promising tools for predicting drug responses for personalized chemotherapy. Clin. Cancer Res. 2010;16:1442–1451. doi: 10.1158/1078-0432.CCR-09-2878. [DOI] [PubMed] [Google Scholar]
- 49.Nemati F, et al. Preclinical assessment of cisplatin-based therapy versus docetaxel-based therapy on a panel of human non-small-cell lung cancer xenografts. Anticancer Drugs. 2009;20:932–940. doi: 10.1097/CAD.0b013e32833009cc. [DOI] [PubMed] [Google Scholar]
- 50.Cutz JC, et al. Establishment in severe combined immunodeficiency mice of subrenal capsule xenografts and transplantable tumor lines from a variety of primary human lung cancers: potential models for studying tumor progression-related changes. Clin. Cancer Res. 2006;12:4043–4054. doi: 10.1158/1078-0432.CCR-06-0252. [DOI] [PubMed] [Google Scholar]
- 51.Taetle R, et al. Use of nude mouse xenografts as preclinical screens. Characterization of xenograft-derived melanoma cell lines. Cancer. 1987;60:1836–1841. doi: 10.1002/1097-0142(19871015)60:8<1836::aid-cncr2820600827>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 52.Fiebig HH, et al. Gene signatures developed from patient tumor explants grown in nude mice to predict tumor response to 11 cytotoxic drugs. Cancer Genomics Proteomics. 2007;4:197–209. [PubMed] [Google Scholar]
- 53.Schatton T, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nemati F, et al. Establishment and characterization of a panel of human uveal melanoma xenografts derived from primary and/or metastatic tumors. Clin. Cancer Res. 2010;16:2352–2362. doi: 10.1158/1078-0432.CCR-09-3066. [DOI] [PubMed] [Google Scholar]
- 55.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vermorken JB, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 58.Bonner JA, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 59.Hennessey PT, et al. Promoter methylation in head and neck squamous cell carcinoma cell lines is significantly different than methylation in primary tumors and xenografts. PLoS ONE. 2011;6:e20584. doi: 10.1371/journal.pone.0020584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prince ME, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl Acad. Sci. USA. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen J, Milo GE, Shuler CF, Schuller DE. Xenograft growth and histodifferentiation of squamous cell carcinomas of the pharynx and larynx. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 1996;81:197–202. doi: 10.1016/s1079-2104(96)80415-x. [DOI] [PubMed] [Google Scholar]
- 62.Zatterstrom UK, et al. Growth of xenografted squamous cell carcinoma of the head and neck–possible correlation with patient survival. APMIS. 1992;100:976–980. doi: 10.1111/j.1699-0463.1992.tb04028.x. [DOI] [PubMed] [Google Scholar]
- 63.Wennerberg J, Trope C, Biorklund A. Heterotransplantation of human head and neck tumours into nude mice. Acta Otolaryngol. 1983;95:183–190. doi: 10.3109/00016488309130933. [DOI] [PubMed] [Google Scholar]
- 64.Langdon SP, et al. Preclinical phase II studies in human tumor xenografts: a European multicenter follow-up study. Ann. Oncol. 1994;5:415–422. doi: 10.1093/oxfordjournals.annonc.a058872. [DOI] [PubMed] [Google Scholar]
- 65.Henriksson E, et al. p53 mutation and cyclin D1 amplification correlate with cisplatin sensitivity in xenografted human squamous cell carcinomas from head and neck. Acta Oncol. 2006;45:300–305. doi: 10.1080/02841860600547380. [DOI] [PubMed] [Google Scholar]
- 66.Peltonen JK, et al. Specific TP53 mutations predict aggressive phenotype in head and neck squamous cell carcinoma: a retrospective archival study. Head Neck Oncol. 2011;3:20. doi: 10.1186/1758-3284-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cabelguenne A, et al. p53 alterations predict tumor response to neoadjuvant chemotherapy in head and neck squamous cell carcinoma: a prospective series. J. Clin. Oncol. 2000;18:1465–1473. doi: 10.1200/JCO.2000.18.7.1465. [DOI] [PubMed] [Google Scholar]
- 68.Koch WM, et al. p53 mutation and locoregional treatment failure in head and neck squamous cell carcinoma. J. Natl Cancer Inst. 1996;88:1580–1586. doi: 10.1093/jnci/88.21.1580. [DOI] [PubMed] [Google Scholar]
- 69.US National Library of Medicine ClinicalTrials.gov. 2011 [online], http://www.clinicaltrials.gov/ct2/show/NCT01255800.
- 70.US National Library of Medicine ClinicalTrials.gov. 2012 [online], http://www.clinicaltrials.gov/ct2/show/NCT01252628.
- 71.US National Library of Medicine ClinicalTrials.gov. 2012 [online], http://www.clinicaltrials.gov/ct2/show/NCT01204099.
- 72.Carey LA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 73.Beckhove P, et al. Efficient engraftment of human primary breast cancer transplants in nonconditioned NOD/Scid mice. Int. J. Cancer. 2003;105:444–453. doi: 10.1002/ijc.11125. [DOI] [PubMed] [Google Scholar]
- 74.de Plater L, et al. Establishment and characterisation of a new breast cancer xenograft obtained from a woman carrying a germline BRCA2 mutation. Br. J. Cancer. 2010;103:1192–1200. doi: 10.1038/sj.bjc.6605900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marangoni E, et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res. 2007;13:3989–3998. doi: 10.1158/1078-0432.CCR-07-0078. [DOI] [PubMed] [Google Scholar]
- 77.Moestue SA, et al. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer. 2010;10:433. doi: 10.1186/1471-2407-10-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laitinen S, Karhu R, Sawyers CL, Vessella RL, Visakorpi T. Chromosomal aberrations in prostate cancer xenografts detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2002;35:66–73. doi: 10.1002/gcc.10097. [DOI] [PubMed] [Google Scholar]
- 79.Gray DR, et al. Short-term human prostate primary xenografts: an in vivo model of human prostate cancer vasculature and angiogenesis. Cancer Res. 2004;64:1712–1721. doi: 10.1158/0008-5472.can-03-2700. [DOI] [PubMed] [Google Scholar]
- 80.Grisanzio C, et al. Orthotopic xenografts of RCC retain histological, immunophenotypic and genetic features of tumours in patients. J. Pathol. 2011;225:212–221. doi: 10.1002/path.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshida T, et al. Antiandrogen bicalutamide promotes tumor growth in a novel androgen-dependent prostate cancer xenograft model derived from a bicalutamide-treated patient. Cancer Res. 2005;65:9611–9616. doi: 10.1158/0008-5472.CAN-05-0817. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, et al. Development and characterization of efficient xenograft models for benign and malignant human prostate tissue. Prostate. 2005;64:149–159. doi: 10.1002/pros.20225. [DOI] [PubMed] [Google Scholar]
- 83.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): a Cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 84.Beniers AJ, et al. Establishment and characterization of five new human renal tumor xenografts. Am. J. Pathol. 1992;140:483–495. [PMC free article] [PubMed] [Google Scholar]
- 85.Kopper L, et al. Renal cell carcinoma–xenotransplantation into immuno-suppressed mice. Oncology. 1984;41:19–24. doi: 10.1159/000225784. [DOI] [PubMed] [Google Scholar]
- 86.Beroukhim R, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.An Z, Jiang P, Wang X, Moossa AR, Hoffman RM. Development of a high metastatic orthotopic model of human renal cell carcinoma in nude mice: benefits of fragment implantation compared to cell-suspension injection. Clin. Exp. Metastasis. 1999;17:265–270. doi: 10.1023/a:1006654600095. [DOI] [PubMed] [Google Scholar]
- 88.Angevin E, et al. Human renal cell carcinoma xenografts in SCID mice: tumorigenicity correlates with a poor clinical prognosis. Lab. Invest. 1999;79:879–888. [PubMed] [Google Scholar]
- 89.Yuen JS, et al. Inhibition of angiogenic and nonangiogenic targets by sorafenib in renal cell carcinoma (RCC) in a RCC xenograft model. Br. J. Cancer. 2011;104:941–947. doi: 10.1038/bjc.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hammers HJ, et al. Reversible epithelial to mesenchymal transition and acquired resistance to sunitinib in patients with renal cell carcinoma: evidence from a xenograft study. Mol. Cancer Ther. 2010;9:1525–1535. doi: 10.1158/1535-7163.MCT-09-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ellis L, et al. Vascular disruption in combination with mTOR inhibition in renal cell carcinoma. Mol. Cancer Ther. 2012;11:383–392. doi: 10.1158/1535-7163.MCT-11-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keunen O, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc. Natl Acad. Sci. USA. 2011;108:3749–3754. doi: 10.1073/pnas.1014480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J, et al. A reproducible brain tumour model established from human glioblastoma biopsies. BMC Cancer. 2009;9:465. doi: 10.1186/1471-2407-9-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carol H, et al. Initial testing of topotecan by the pediatric preclinical testing program. Pediatr. Blood Cancer. 2010;54:707–715. doi: 10.1002/pbc.22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Houghton PJ, et al. Efficacy of topoisomerase I inhibitors, topotecan and irinotecan, administered at low dose levels in protracted schedules to mice bearing xenografts of human tumors. Cancer Chemother. Pharmacol. 1995;36:393–403. doi: 10.1007/BF00686188. [DOI] [PubMed] [Google Scholar]
- 96.Vassal G, et al. Potent therapeutic activity of irinotecan (CPT-11) and its schedule dependency in medulloblastoma xenografts in nude mice. Int. J. Cancer. 1997;73:156–163. doi: 10.1002/(sici)1097-0215(19970926)73:1<156::aid-ijc24>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 97.Houghton PJ, et al. The pediatric preclinical testing program: description of models and early testing results. Pediatr. Blood Cancer. 2007;49:928–940. doi: 10.1002/pbc.21078. [DOI] [PubMed] [Google Scholar]
- 98.Foreman NK, Love S, Thorne R. Intracranial ependymomas: analysis of prognostic factors in a population-based series. Pediatr. Neurosurg. 1996;24:119–125. doi: 10.1159/000121027. [DOI] [PubMed] [Google Scholar]
- 99.Merchant TE, et al. Preliminary results from a phase II trial of conformal radiation therapy and evaluation of radiation-related CNS effects for pediatric patients with localized ependymoma. J. Clin. Oncol. 2004;22:3156–3162. doi: 10.1200/JCO.2004.11.142. [DOI] [PubMed] [Google Scholar]
- 100.Pollack IF, et al. Intracranial ependymomas of childhood: long-term outcome and prognostic factors. Neurosurgery. 1995;37:655–666. doi: 10.1227/00006123-199510000-00008. [DOI] [PubMed] [Google Scholar]
- 101.Yu L, et al. A clinically relevant orthotopic xenograft model of ependymoma that maintains the genomic signature of the primary tumor and preserves cancer stem cells in vivo. Neuro. Oncol. 2010;12:580–594. doi: 10.1093/neuonc/nop056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zembutsu H, et al. Genome-wide cDNA microarray screening to correlate gene expression profiles with sensitivity of 85 human cancer xenografts to anticancer drugs. Cancer Res. 2002;62:518–527. [PubMed] [Google Scholar]
- 103.Tan AC, Naiman DQ, Xu L, Winslow RL, Geman D. Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics. 2005;21:3896–3904. doi: 10.1093/bioinformatics/bti631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jimeno A, et al. Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res. 2008;68:2841–2849. doi: 10.1158/0008-5472.CAN-07-5200. [DOI] [PubMed] [Google Scholar]
- 105.Messersmith WA, et al. Efficacy and pharmacodynamic effects of bosutinib (SKI-606), a Src/Abl inhibitor, in freshly generated human pancreas cancer xenografts. Mol. Cancer Ther. 2009;8:1484–1493. doi: 10.1158/1535-7163.MCT-09-0075. [DOI] [PubMed] [Google Scholar]
- 106.Rajeshkumar NV, et al. Antitumor effects and biomarkers of activity of AZD0530, a Src inhibitor, in pancreatic cancer. Clin. Cancer Res. 2009;15:4138–4146. doi: 10.1158/1078-0432.CCR-08-3021. [DOI] [PubMed] [Google Scholar]
- 107.US National Library of Medicine ClinicalTrials.gov. 2010 [online], http://www.clinicaltrials.gov/ct2/show/NCT00735917.
- 108.US National Library of Medicine ClinicalTrials.gov. 2010 [online], http://www.clinicaltrials.gov/ct2/show/NCT01016860.
- 109.Singh M, et al. Assessing therapeutic responses in Kras mutant cancers using genetically engineered mouse models. Nat. Biotechnol. 2010;28:585–593. doi: 10.1038/nbt.1640. [DOI] [PubMed] [Google Scholar]
- 110.Giovanella BC, Stehlin JS, Jr, Shepard RC, Williams LJ., Jr. Correlation between response to chemotherapy of human tumors in patients and in nude mice. Cancer. 1983;52:1146–1152. doi: 10.1002/1097-0142(19831001)52:7<1146::aid-cncr2820520704>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 111.Tentler JJ, et al. Assessment of the in vivo antitumor effects of ENMD-2076, a novel multitargeted kinase inhibitor, against primary and cell line-derived human colorectal cancer xenograft models. Clin. Cancer Res. 2010;16:2989–2998. doi: 10.1158/1078-0432.CCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rajeshkumar NV, et al. MK-1775, a potent Wee1 inhibitor, synergizes with gemcitabine to achieve tumor regressions, selectively in p53-deficient pancreatic cancer xenografts. Clin. Cancer Res. 2011;17:2799–2806. doi: 10.1158/1078-0432.CCR-10-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Song D, et al. Antitumor activity and molecular effects of the novel heat shock protein 90 inhibitor, IPI-504, in pancreatic cancer. Mol. Cancer Ther. 2008;7:3275–3284. doi: 10.1158/1535-7163.MCT-08-0508. [DOI] [PubMed] [Google Scholar]
- 114.Merk J, Rolff J, Dorn C, Leschber G, Fichtner I. Chemoresistance in non-small-cell lung cancer: can multidrug resistance markers predict the response of xenograft lung cancer models to chemotherapy? Eur. J. Cardiothorac. Surg. 2011;40:e29–e33. doi: 10.1016/j.ejcts.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 115.Hammer S, et al. Comparative profiling of the novel epothilone, sagopilone, in xenografts derived from primary non-small cell lung cancer. Clin. Cancer Res. 2010;16:1452–1465. doi: 10.1158/1078-0432.CCR-09-2455. [DOI] [PubMed] [Google Scholar]
- 116.Kolfschoten GM, et al. Development of a panel of 15 human ovarian cancer xenografts for drug screening and determination of the role of the glutathione detoxification system. Gynecol. Oncol. 2000;76:362–368. doi: 10.1006/gyno.1999.5689. [DOI] [PubMed] [Google Scholar]
- 117.Huynh H, Soo KC, Chow PK, Panasci L, Tran E. Xenografts of human hepatocellular carcinoma: a useful model for testing drugs. Clin. Cancer Res. 2006;12:4306–4314. doi: 10.1158/1078-0432.CCR-05-2568. [DOI] [PubMed] [Google Scholar]
- 118.Huynh H, et al. Brivanib alaninate, a dual inhibitor of vascular endothelial growth factor receptor and fibroblast growth factor receptor tyrosine kinases, induces growth inhibition in mouse models of human hepatocellular carcinoma. Clin. Cancer Res. 2008;14:6146–6153. doi: 10.1158/1078-0432.CCR-08-0509. [DOI] [PubMed] [Google Scholar]