Abstract
Rheumatoid arthritis (RA) is a multifactorial disease and requires interaction between genetic and environmental factors for predisposition. The presence of bacterial DNA of the gut residing commensals in synovium as well as dysbiosis of certain commensal bacteria in faecal samples of RA patients as compared to controls suggest a significant role of the gut flora in pathogenesis of RA. The gut commensals are involved in host immune development and function suggesting they might be critical epigenetic factors modifying autoimmune diseases like RA. This raises the question if gut-derived commensal can be exploited to generate a biomarker profile along with genetic factors to define individuals at risk. Genomic wide association studies have confirmed the HLA (human leukocyte antigen) class II genes as the strongest risk factor for predisposition to RA. HLA-DQ8 and DRB1*0401 molecules predispose to develop arthritis while DRB1*0402 provides protection. Interaction between host genetic factors like major histocompatibility complex (MHC) and gut microbiota and its impact on the development of RA is difficult to study in humans due to high variability in the genetic factors and diet. Animal models provide a means to study the molecular basis of pathogenesis thereby providing a basis for developing therapeutic strategies. Using transgenic mice expressing RA-associated and resistant HLA genes, we have developed a collagen-induced arthritis (CIA) model that shares similarities with human disease in sex-bias, autoantibody profile and phenotype. Studies in transgenic mice suggest that arthritis-susceptibility may be associated with dysbiosis in the gut microbiome. Studies in animal models underscore the impact of the gut flora in extra-intestinal diseases. Exploring the role of gut microbes will significantly advance our understanding of RA pathogenesis and may further help develop strategies for mucosal modulation of RA.
Keywords: Autoimmunity, CIA model, gut, microbiome, microbiota, rheumatoid arthritis, type 1 diabetes
Rheumatoid arthritis (RA) is an autoimmune disease that is characterized by destruction of joints causing disability. Though the aetiology of RA is unknown, epidemiological studies suggest that it results from the complex interactions between genes, environmental factors and the immune system1. Longitudinal studies show that the autoimmune aspects may begin many years before the clinical manifestation of RA2. The role of environmental factors such as commensal intestinal microbiota, microbial infections and their immunological consequences in genetically susceptible individuals are not yet properly understood.
Among the known genetic factors, certain major histocompatibility complex (MHC) genes provide the strongest association with the development of RA. Epidemiology studies in various populations have shown an increased presence of DRB1*0401 and alleles that share the 3rd hypervariable region with DRB1*0401 gene, known as the ‘shared epitope’ hypothesis in RA patients3. In contrast, DRB1*0402 confers resistance to the development of arthritis. Recent genome wide association studies (GWAS) have confirmed *0401 association with RA and also shown some non-MHC gene involvement in RA4. However, genetic factors account for only 50 per cent of the risk factors for RA. Interaction between the genetic and environmental factors is required for the onset of disease. While many environmental factors have been suggested to contribute to the pathogenesis, smoking has been the one that is studied extensively. For many decades, numerous infectious agents that include Epstein Barr, rubella virus and parvovirus among others have been implicated in pathogenesis of RA. Support for an infectious aetiology for RA comes from (i) studies in animal models where arthritis can be induced with bacterial products, (ii) presence of certain oral and gut commensal bacterial antigens in the synovial fluids of patients, and (iii) anti-microbial activity of certain disease-modifying drugs2,5,6. Recent studies have implicated commensals occurring in oral cavity such as Porphyromonas gingivalis, a Gram-negative anaerobe, in pathogenesis of periodontitis-associated RA7.
Commensals and immune system
The human intestine is colonized by a large number of microorganisms (around 1014 bacteria) that exceeds the number of cells in the human body. The intestinal microbial colonization begins at birth and it continues to change depending on the environment during the various maturation phases of life that support a variety of physiological functions. Thus each individual harbours a unique intestinal microbiota which is influenced by various factors including food, geographical location, climate and personal hygiene. Turnbagh and coworkers8 suggested that a set of core microbiome is present in humans living in a certain habitat. Variability among individuals could arise due to the host lifestyle, diet, health, immune system and environment. However, the factors that influence the major deviation from the core microbiome are still unknown. Our gut is the primary port of entrance for various environmental antigens that can be in the form of food or infectious agents. The intestinal microflora forms an immunological barrier between the environment and the intestine and helps to maintain a healthy gastro-intestinal tract.
Nearly two decades ago, scientists put forth a concept called the ‘hygiene hypothesis. According to this hypothesis, an improvement in personal hygiene as observed in the developed countries has led to an increase in the risk of allergic and autoimmune disease9. Increase in incidences of various inflammatory and autoimmune diseases like inflammatory bowel disease (IBD), asthma, type 1 diabetes (T1D), and rheumatoid arthritis in the developed countries support this concept. An essential part of mucosal system for mounting protective immune responses is the fact that the mucosal immune system should be able to distinguish between ‘dangerous’ and ‘nondangerous’ agents10. While the skin surface is protected by several layers of epithelial cells, the mucosal surface including that of the digestive, urogenital tract, respiratory, eye and ducts of exocrine glands is covered mostly with a single-layered epithelium11. Mucosal surfaces, therefore, require more effective protection that can efficiently dispose the majority of external agents. The mucosal surfaces have a strongly developed and highly specialized immune system, the mucosa-associated lymphoid tissue (MALT) that harbours the majority of immunologically active cells in the body. The preferential induction of inhibition of the responses to non-dangerous antigens (mucosal tolerance) is a characteristic feature of mucosal immunity distinguishing it from systemic immunity11,12,13,14.
Immunomodulation by commensal bacteria/probiotics
The microbiota in the gut is the largest source of microbes that can exert beneficial as well as pathogenic effect on human health. The intestinal immune system is unique in its properties as the gut normally maintains a tolerant state in the face of all the antigens it encounters. Recent research has shown that gut microbiota is essential for maintaining homeostasis and development of the gut immune system. To eliminate pathogenic bacteria from the gut, the intestinal mucosa needs to generate a controlled response so that the beneficial commensals can be maintained15,16,17,18,19,20,21,22. The regulatory processes in the intestinal immune system lead to generation of an immune response while disposing microorganisms that are pathogenic but are still able to maintain a tolerance towards the commensals and food antigens.
The immunomodulatory effects of the commensal bacteria as well as the probiotics rely on contact between bacteria and the host's intestinal epithelial cells23,24,25. Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD) isoforms expressed in the epithelial cells trigger innate responses by recognizing conserved microbial structures22. Because commensal bacteria differ in their ability to stimulate TLRs and other intracellular innate immunity receptors such as NOD like receptors, the pattern of released chemical mediators varies significantly determining pro-inflammatory and anti-inflammatory responses. Lipopolysaccharide (LPS) from Gram-negative bacteria binds TLR-4, while peptidoglycan (PGN) and other cell wall components from Gram-positive bacteria signal through TLR-2 pathway to generate immune response26,27.
The immune responses generated by the effector T cells are regulated by immune-suppressive regulatory T-cell (Treg) subsets. Dendritic cells (DCs) play an important role in instructing naive CD4+ T-cells to differentiate into T-cells producing Th1, Th17 or Th2 cytokines after encounter with an antigen. However, due to the unique gastrointestinal environment, mucosal DCs have been shown to favour conversion of T-cells into regulatory T-cells (Tregs), a process that is dependent on the presence of commensal bacteria28,29,30. Recent studies have shown that commensal bacteria such as Lactobacillus and Bifidobacterium infantis exert their anti-inflammatory effect through induction of CD4+CD25+FoxP3+ regulatory T-cells31,32. The commensal bacteria/probiotics can induce an anti-inflammatory action on the intestinal mucosal as well as on the peripheral immune system by suppressing T-cell proliferation33,34,35,36. The gut commensal B. infantis has been shown to inhibit nuclear factor- kappa B (NF-κB) activation in the gut resulting in the induction of an anti-inflammatory response by suppressing T-cell proliferation and production of interleukin (IL)-10 and Th2 cytokines systemically as well as in the gut37,38,39.
How commensals/probiotics suppress T-cell proliferation is not well understood and further studies are needed to more precisely determine their effects on the suppression of T-cell response in inflammatory diseases. Given the fact that DCs are instrumental in generation of the immune response, it has been hypothesized that commensals influence differentiation and function of DCs, thereby modulating the immune response40. Thus treatment with commensals/probiotics may provide benefits by modulating DC function and thus are relevant for treating inflammatory diseases including autoimmune diseases such as RA. This suggests that the gut microbiome may dictate a pro- or anti-inflammatory environment that can have a significant impact on the adaptive immune response away from gut41.
There is accumulating evidence suggesting that the gut microbiota plays a major role in determining the phenotype of the host. This is supported by the studies showing that gut commensals are critical for a variety of physiological and metabolic processes in the host42. Studies in gnotobiotic mice support this notion43. Recent studies in mice have suggested that host genotype is a stronger determinant of gut microbiota than sex44. We propose that gut microbiome, sex and genetic factors may be able to predict susceptibility to develop autoimmune disease like RA. Treatment with a potential anti-inflammatory commensal might help in stopping progression of the disease and reducing severity by changing gut permeability and immune environment.
Commensals in rheumatoid arthritis
Although the aetiology of RA is unknown, it is a polygenic disease that requires both, genetic and environmental factors for its onset. One of the most attractive explanations for this autoimmune phenomenon has centered on the exposure to various environmental factors such as infections that are capable of initiating disease in genetically predisposed individuals45,46,47. Synovial fluid of patients with RA shows the presence of bacterial DNA and their products arising from naturally occurring commensals in the gut and other mucosal surfaces. One explanation for the presence of gut commensals in the joints of RA patients could be a leaky gut or loss of intestinal integrity that facilitates the migration of gut commensals or their products to the peripheral organs. The bacterial products released in the joints may result in local and systemic immune stimulation.
Mucosal surfaces like skin and the gut are the most common portals of contact with microorganisms and other environmental factors. The hygiene hypothesis has been suggested as one reason for the increasing incidence of autoimmunity. This hypothesis is supported by the findings that interaction between the intestinal microbes and the innate immune system may be a critical epigenetic factor modifying inflammatory and autoimmune diseases such as T1D and rheumatoid arthritis49. Although progress has been made in understanding the composition and some of the functions of the mucosal immune system in the induction of protective immunity, the basic mechanisms and gut composition of various microbes involved in the development of immune responses remain largely unknown. It has long been thought that the gut microbiome contributes to the nutrition, maturation of the intestine, defense from pathogens, and helps maintain the integrity and the function of the gut50. As such, it has rich potential of manipulation of not only the gut but also the systemic immune system. The environment that is developed within the intestine is the product of the interaction between the host and the gut microbial ecology, ultimately for the benefit of both the host and micro community. Alterations in gut microbiota and their function have been associated with many inflammatory diseases like inflammatory bowel disease, type 1 diabetes, and RA in humans.
Studies have shown that specific intestinal commensals or their specific molecular patterns may induce production of pro- or anti-inflammatory cytokines thereby modulating the integrity of the intestinal mucosal barrier51,52. Thus, alterations of a normal gut microbiome can affect mucosal immunity. These changes may have an extended effect on non-intestinal diseases like diabetes and RA53. This premise is supported by a study where analysis of the faecal microbiome of patients with RA revealed significantly fewer Bifidobacterium and bacteria of the Bacteroides-Porphyromonas-Prevotella group, B. fragilis subgroup, and the Eubacterium rectale–Clostridium coccoides group than the faecal microbiota of patients with non-inflammatory fibromyalgia54. Because these bacterial species are known to belong to common taxa in the human faecal microbiome55,56,57, their absence in RA patients might suggest an altered gut microbiome. However, data in humans on the role of gut microbiome are limited at present.
There are no studies in humans on the role of commensals in arthritis. The only probiotics tried as treatment for RA are the lactobacilli. Their therapeutic application did not significantly improve the American College of Rheumatology (ACR) Scores in one study, but showed benefit in another58,59. Interaction between host genetic factors like MHC and gut microbiota and its impact on the development of RA is difficult to study in humans due to many confounding factors: (i) high variability in genetic factors, (ii) diets that can influence gut microbiome, and (iii) it is difficult to determine if the changes are causal association or the effect of diseases, as disease may have started by the time patients are seen in the clinics. Animal models provide means to study the molecular basis of pathogenesis60 as well as the basis for developing therapeutic strategies.
HLA transgenic mice as model for rheumatoid arthritis
Associations between various factors and disease in humans are based on epidemiological studies making it difficult to define the mechanism of association. Animal models help delineate the effects of a specific gene as experiments can be conducted in controlled conditions. Collagen-induced arthritis (CIA) is an animal model that shares many characteristics of inflammatory arthritis in humans and has been used as a model for RA. Immunization of certain strains of mice with type II collagen (CII) leads to the development of CIA. It is a CD4+ T and B cell dependent autoreactive response to CII. Similar to RA, susceptibility to develop CIA is determined by polymorphism in the MHC class II loci, H2A61.
The H2A locus of mouse is homologous to human leukocyte antigen (HLA)-DQ loci while H2E is homologous to HLA-DR loci in humans. The model generated with mice has advanced our understanding of pathogenesis of RA but has limitations such as (i) response is restricted by mouse class II molecules making it irrelevant to human RA, (ii) disease incidence is similar for sexes, (iii) arthritic mice did not produce rheumatoid factor, and (iv) activated mouse T cells do not express class II alleles thus do not participate in immune response locally in the joint62.
Mice expressing human HLA genes but lacking all four classical murine chains, Aα, Aβ, Eα, Eβ chains has greatly enhanced our capability to overcome the limitations of the mouse model63. Using transgenic mice expressing RA-associated DRB1*0401 and RA-resistant *0402 genes (*0401.AE-/- and *0402.AE-/-, respectively), we have developed a CIA model that mimics human disease64,65. Our studies with *0401 mice showed that they developed CIA with sex-bias (female to male ratio 3:1), produce rheumatoid factor (RF) and anti-citrullinated peptide antibodies (ACPAs) and phenotype. On the other hand, *0402 mice were resistant to CIA (Fig. 1). Observations in *0402 mice suggest that the protection from arthritis could be due to (i) negative selection of autoreactive cells in the thymus, (ii) higher number of regulatory cells, (iii) increased AICD, and (iv) low T cell proliferation and production of Th1 cytokines and tumour necrosis factor-α (TNF)-α. Transgenic mice express class II molecules on activated T cells, as do humans but unlike wild mice. Because human class II molecules shape the T-cell repertoire in these humanized mice, they show the same HLA restrictions in an immune response as humans65,66. Our data with mice expressing the RA-susceptible and resistant haplotype, *0401/DQ8 and *0402/DQ8, respectively, suggest that DRB1 polymorphism modulates DQ8-mediated CIA. Our data are in confirmation with genome wide association67 suggesting HLA genes as major risk factors for predisposition to RA. Thus HLA transgenic mice provide a good model to study the role of triggering environmental factors and their interaction with genetic risk factors that may influence pathogenesis in RA.
Fig. 1.
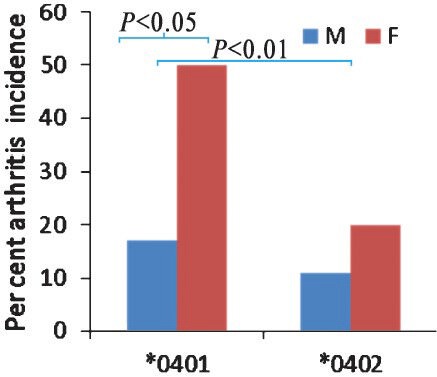
Incidence of collagen-induced arthritis in HLA transgenic mice. Transgenic mice expressing HLA-DRB1 *0401 and DRB1*0402 genes on endogenous class II knockout (AE-/-) background (*0401.AE-/- and *0402.AE-/-, respectively) were immunized with type II collagen and followed for onset and severity of arthritis. Incidence of arthritis in female DRB1*0401 mice was three times compared to males. DRB1*0402 mice are generally resistant to develop arthritis. P<0.05 DRB1*0401 male versus female, P<0.01 *0401 versus *0402 mice. Source: Ref. 64.
Gut and arthritis in mice
Host genotype affects the gut microbial composition in humans and mice68,69. There is accumulating evidence suggesting a role of gut microbiota in pathogenesis as well as protection in murine models of various diseases. Based on the experiments in germ free facility, a dysbiosis in the gut flora has been suggested as the basis of disease phenotype53. The abundance of certain bacterial groups has been reported in autoimmunity; segmented filamentous bacteria (SFB) related to Clostridium, have been linked to an increase in pro-inflammatory responses in arthritis and diabetes driven by an increase in Th-17 cells70,71,72. Similarly, Lactobacillus can induce arthritis in IL-1ra-/- mice. However, L. casei potentiates antigen-specific oral tolerance and suppresses Th1-type immune responses suppressing arthritis in a mouse model73,74. A normal intestinal microbiota that lives in symbiosis with its host can positively influence immune responses and may be able to protect against the development of inflammatory diseases in various inflammatory models20,49,75. Conversely, studies with germ free and pathogen free mice have shown that disruptions in gut microbiota may induce production of pro-inflammatory cytokine and interlukin-17 producing Th-17 cells at increased levels, even in tissues away from the gut76. It is under debate whether gut microbiota is controlled by immune system or vice versa52,71,77. These studies have enhanced our understanding of the gut microbiome and its influence in host functions. The limitations of these studies are that the host genotype is endogenous MHC molecules and arthritis does not mimic human disease. Moreover, germ free mice constituted with a single commensal do not provide its role in a community.
Recent advances in technology have made it possible to study the gut microbiome composition and structure and its influence on diseases. However, it is difficult to study interactions between host genetic factors like HLA genes and gut microbiota, and their impact on the development of RA due to a high variability in genetic factors in humans. Also, it is difficult to establish a causal relationship between RA and the gut microbiota as a variable diet and geographical location can influence the gut microbiome composition. Thus, HLA transgenic mice can be used to understand the interaction of the gut microbiota and immune system and their role in causation of RA. Since HLA transgenic mice mimic human disease in sex-bias, they also provide a tool to understand the basic differences in the gut immune system of male and female mice. We have used the arthritis-susceptible *0401 and arthritis-resistant *0402 mice to understand the influence of the host genotype on the gut microbiome. The advantage of using this model is that (i) the mice are kept in identical conditions, (ii) the diet is controlled, and (iii) the only difference in the genotype of mice is the HLA transgene that differs by three amino acids and thus this model can provide information about how genotype can influence gut microbiota and also define a profile that can be used for diagnostic purpose.
Can gut microbiome be used to predict susceptibility to arthritis?
The need to high-throughput DNA sequencing of the intestinal microbiota has been highlighted in recent studies that could identify patients at high risk41,78. We addressed the question if gut microbiota can be used to predict susceptibility to arthritis by deep rDNA pyrosequencing of the hypervariable V3-V5 region of the16S ribosomal RNA (rRNA) gene of the bacterial community combined with phylogenetic analysis of faecal samples from the naïve *0401 and *0402 mice79. The gut flora of transgenic mice showed that the gut microbial composition of transgenic mice shares similarities with human mucosal microbiome in presence of different genera (Fig. 2). Non-metric multidimensional scaling (NMMDS) and analysis of similarities (ANOSIM) based on Bray-Curtis similarity indexes were used to assess how the microbial communities differed among subjects. We tested if susceptibility to develop arthritis may be related to the presence or absence of specific bacteria by characterizing abundance of specific operational taxonomic units (OTUs). Taxonomic classification of each OTU (clustered at 97% sequence similarity) was obtained by Blast alignment to the NCBI reference sequence database80. The OTUs were mostly classified in the phyla Bacteroidetes (46% of all reads), followed by Firmicutes (36%), Actinobacteria (14%) and Verrucomicrobia (4%)41.
Fig. 2.
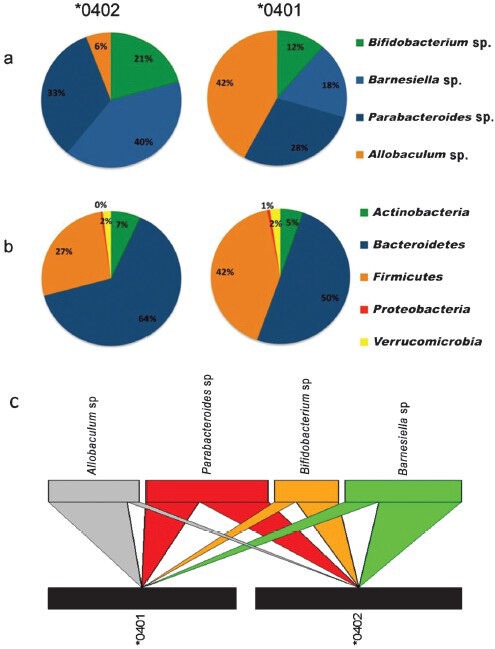
Gut microbiome of arthritis-susceptible and –resistant mice is different. (A) Genera distribution of the main Operational Taxonomic Units (OTUs) driving the differences in *0402 and *0401 mice. Allobaculum occurred with abundance in *0401 mice while Barnesiella sp. was most abundant in *0402 mice, (B) Phyla distribution of the total OTUs detected in *0402 and *0401 mice according to the SIMPER analysis. Bacteroidetes: Firmicutes ratios are more even in *0401 mice compared to *0402 mice. The genera distributions of the OTUs driving the differences follow a similar pattern to the phyla distributions in all the detected OTUs, and (C) Bipartite interaction matrix plot showing the relationships between the main OTUs detected in each of the mice strains. The position, width and direction of the upper multicolor blocks and wedges show how abundant each OTU is in each strain. Source: Ref. 79.
A comparison of the microbial community in faecal samples showed minor differences between the two strains79. However, abundance and richness of various taxa showed a different composition of gut microbial community for the susceptible and resistant strains, suggesting a dysbiosis in arthritis-susceptible mice. A percentage-species contribution analysis (SIMPER) and taxonomic search suggested that Bacteroidetes and Firmicutes were present evenly in *0401 mice while *0402 mice harboured Bacteriodetes and Firmicutes at a ratio of 2:1 suggesting a role of Bacteriodetes in the maintenance of a homeostatic GI tract. The *0401 mice showed an abundance of Allobaculum sp. (87% identity with an unclassified member of the Clostridials) while *0402 mice showed significant abundance of Bifidobacterium sp. (Fig. 2). As these mice were naïve and healthy, it suggested that the host genotype might have a strong influence on the gut microbiome. How HLA genes influence selection of commensals is not clear. We have shown that T cell repertoire selection by *0401 and *0402 differs in transgenic mice81. One can speculate that positively selected T cells may have a role in selection of gut bacterial communities. This would imply a genetic predisposition to develop RA may result from an interaction between the genes and certain commensals in certain conditions where an abnormal immune response is generated. Alternatively, a disruption of gut microbial community may trigger an inflammatory response in the gut that is carried out to tissues distant from it; suggesting that gut bacteria can control adaptive immune response and influence autoimmunity.
Our observations may imply that in the microbial ecology of the gut, the most abundant OTUs are the ones driving the most important processes in the community including immune regulation. These data suggest that *0401 may regulate the initial gut microbiome. However, over time with exposure to various environmental factors and infections, there may be perturbations in the microbiome that influences the strong regulatory environment maintained by a stable host microbiome, thus changing gut permeability. This may lead to transport of luminal contents to the periphery, generating an immune response and a break in tolerance in genetically predisposed mice82.
Arthritis-resistant mice show dynamic age and sex dependent gut microbiome changes
A dynamic change in gut microbiota composition and functionality occurs as we age. As early as in 1990, it was shown that Bifidobacterium declined with age while Clostridium species increased, suggesting that a change in gut flora may be associated with healthy ageing83. However, it is unknown if ageing gut flora maintains a symbiotic relation with the host. Probiotics to modulate gut flora may be one way of healthy ageing, however, other factors that may be involved in impacting ageing gut flora need to be defined. It is possible that gut flora during ageing in combination with genetic factors may have a role in predisposition to age related and inflammatory diseases. If this is the case, the gut microbiome profile along with genetic factors may provide a biomarker profile for diagnostic purposes. Also, using animal models, it may be possible to generate a profile that can predict susceptibility to various diseases.
In humans it is difficult to differentiate if certain gut flora may have changed due to ageing process or due to disease onset. Mean age of onset for arthritis is around 50 years. Difference in gut flora of RA patients compared to controls observed in a study could be due to the ageing process as well as disease. In healthy individuals, a deficiency of Bifidobacteria in the gut during ageing may contribute to the pathogenic immune response in RA. We took advantage of transgenic mice to determine if there is a correlation between ageing and any changes in the gut microbial community that may be associated with disease susceptibility. Our observations suggested that the gut flora of arthritis-resistant mice showed a dynamic change as they aged while in comparison arthritis-susceptible mice harboured a gut microbiome that was characterized by an abundance and/or lack of specific commensals that did not show significant difference at any age79.
According to ANOSIM test there were significant differences in the bacterial community structures between male and female *0402 mice that were driven by specific bacteria. The OTUs distributions at the genus and phylum levels confirmed that the higher abundance of Actinobacteria (including Bifidobacterium) and Bacteroidetes, and lower levels of Firmicutes (including Allobaculum) in *0402 females drive the observed differences between sexes in *0402 mice. In contrast, the ANOSIM test for *0401 mice did not show differences in faecal microbiota sex-based suggesting arthritis-susceptible mice had lost the microbial dynamism required for immune regulation in mice. However, *0401 males showed a significantly higher abundance of B. pseudolongum compared to females79 (Fig. 3). These data suggest that a loss of the gut microbial dynamism in females may, in combination with the genetic factors, contribute to disease susceptibility. These studies also point out that sex hormones may have a significant role in modulation of the gut microbiome. In humans, men with RA do have higher levels of estrogen84, suggesting that host gut-hormonal axis may modulate immune response in favour of susceptibility in genetically susceptible individuals.
Fig. 3.
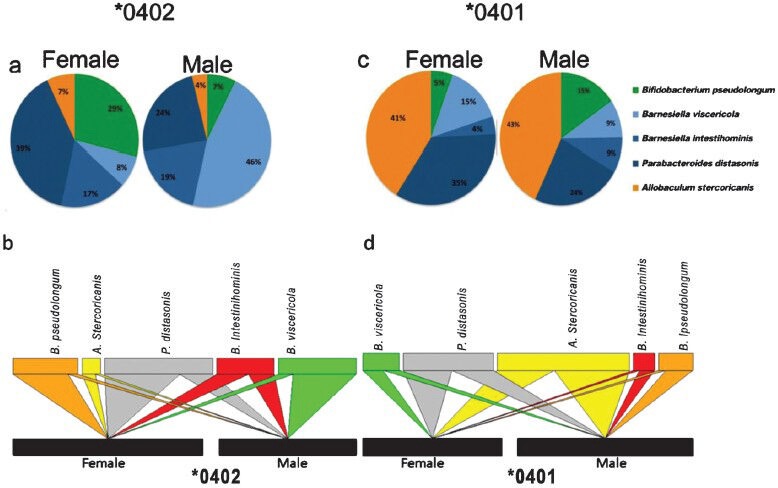
Relative abundance of OTUs in the faecal microbiomes of male and female *0401 and *0402 mice. (A) *0402 females show significantly higher relative abundances of Bifidobacterium-Parabacteroides OTUs while males harbour high levels of Barnesiella viscericola. (B) Bipartite interaction matrix plot showing the relationships between the main OTUs detected in male and female *0402 mice. (C) Male *0401 mice gut microbiome has higher frequency of Bifidobacteria compared to females. (D) Bipartite interaction matrix plot showing the relationships between the main OTUs detected and sex. The position, width and direction of the upper multicolor blocks and wedges show how abundant each OTU is in males or females. Source: Ref. 79.
Gut microbiome is related to mucosal immune function
Absence of intestinal microbiota in animals has been associated with significant impairment in cellular and humoral immune responses, suggesting a pivotal role of the gut microbiota in the development of effector immune responses85. While in humans such clear associations are not feasible, an association between the gut microbes and immune response is supported by change in systemic immune response, specially improvement in phagocytic activity following probiotics supplementation83. This suggests that a normal gut microbial community is necessary for intestinal health, whereby an altered or abnormal gut flora, a dysbiosis may contribute to the alterations in mucosal immune system generating a pathogenic response.
We investigated if dysbiosis in arthritis-susceptible transgenic mice was associated with an altered immune system in the gut when compared to the arthritis-resistant mice. Jejuna of naïve mice were tested for cytokine transcripts involved in the Th17 network by real time PCR. Our data showed a differential expression of Th17 cytokines in guts of both strains79. The *0401 mice showed an increase in transcripts of proinflammatory cytokines like IL-17, IL-2, IFN-γ and CXCL5 while negative regulators like IL-21, CCL22 were observed with significantly lower levels. CCL22 has been shown to recruit T regulatory cells and IL-21 has immunomodulatory activities for T as well as B cell function. Comparison of male and female *0401 mice showed that females had much higher levels of proinflammatory cytokines like IL-17 and IL-23. However, chemokines CCl20 and CCl22, known to regulate immune response, were much lower in females compared to males79.
Several studies have suggested that certain bacteria like segmented filamentous bacteria (SFB) produce Th17 cytokines and lead to the development of arthritis72,86. We performed correlation analysis to determine if specific gut bacteria were associated with certain cytokines in transgenic mice. Our observations79 suggested that the presence of Allobaculum species in *0401 mice was negatively correlated with IL-21, a cytokine known to regulate CD4 T cell differentiation to Th17, and determine B cell survival and differentiation to plasma cells. The other species found in much lower levels in arthritis-susceptible mice, Bifidobacterium, also showed a negative correlation with IL-17. These data suggest a role of gut bacteria in determining the mucosal immune system that may migrate to the periphery via different means resulting in modulation of adaptive immune system. The gut microbiome provides a link between the innate and adaptive immune system that can be targeted for therapy.
Conclusions
Most inflammatory and autoimmune diseases are multifactorial in nature, requiring genetic and environmental factors (Fig. 4). While the genetic factors have unequivocally shown HLA genes as the strongest, the role of environmental factors has been difficult to delineate in humans. Transgenic mice carrying HLA genes provide a means to understand the role of environmental factors and their interaction with genes. We have shown that transgenic mice carrying HLA-DRB1*0401 and *0402 genes develop arthritis that mimics human disease. Our data in naïve mice suggest that the difference of only three amino acids in *0401 and *0402 mice could lead to dysbiosis in the arthritis-susceptible mice with a significant difference in the gut microbiota and resulting immune system. The gut microbiota is crucial to the development of postnatal immune system. Mucosal modulation of arthritis may be an effective target for therapy. If dysbiosis in an individual can be corrected via manipulation of gut commensals, it may lead to benefits without side effects. Our studies are a step towards individualized medicine; however, more detailed studies are needed to determine the mechanism and action of specific commensals on mucosal and systemic immunity.
Fig. 4.
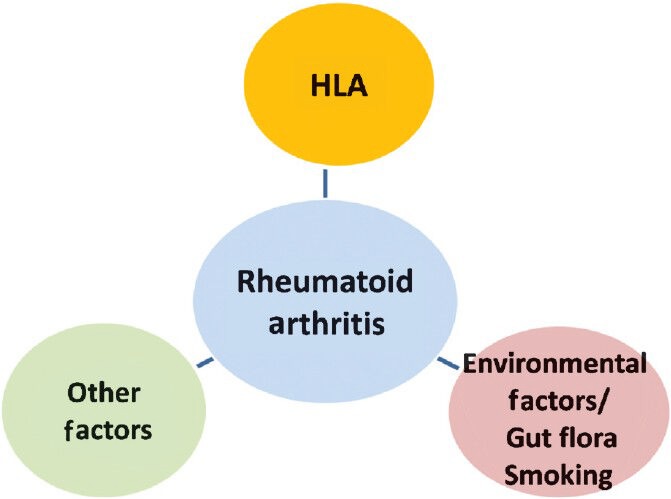
Multifactorial nature of autoimmune disease like rheumatoid arthritis. Among genetic factors, human leukocyte antigen (HLA) class II genes provide the strongest risk to develop autoimmunity. Interaction between HLA and environmental factors is required for the onset of disease. The environmental factors may include cigarette smoking, infectious agents and microbial flora of the gut. The core gut microbiome is associated with genotype though it is modulated by diet and other environmental factors. The gut flora can influence immune system locally and systemically. Thus a dysbiosis in the gut microbiome in a genetically susceptible individual can lead to a proinflammatory response systemically generating a pathogenic response resulting in the onset of disease. Genetic factors other than HLA class II molecules are also known to be associated with RA.
Acknowledgment
Authors thank Michele Smart and Julie Hanson for generation and characterization of transgenic mice. The work is supported by AI075262, Department of Defense grant to VT and funds from Mayo Clinic/UIUC alliance.
References
- 1.Svendsen AJ, Kyvik KO, Houen G, Junker P, Christensen K, Christiansen L, et al. On the origin of rheumatoid arthritis: the impact of environment and genes - a population based twin study. PLoS One. 2013;8:e57304. doi: 10.1371/journal.pone.0057304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klareskog L, Padyukov L, Lorentzen J, Alfredsson L. Mechanisms of disease: Genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat Clin Pract. 2006;2:425–33. doi: 10.1038/ncprheum0249. [DOI] [PubMed] [Google Scholar]
- 3.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 4.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moen K, Brun JG, Valen M, Skartveit L, Eribe EK, Olsen I, et al. Synovial inflammation in active rheumatoid arthritis and psoriatic arthritis facilitates trapping of a variety of oral bacterial DNAs. Clin Exp Rheumatol. 2006;24:656–63. [PubMed] [Google Scholar]
- 6.Kempsell KE, Cox CJ, Hurle M, Wong A, Wilkie S, Zanders ED, et al. Reverse transcriptase-PCR analysis of bacterial rRNA for detection and characterization of bacterial species in arthritis synovial tissue. Infect Immun. 2000;68:6012–26. doi: 10.1128/iai.68.10.6012-6026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hitchon CA, El-Gabalawy HS. Infection and rheumatoid arthritis: still an open question. Curr Opin Rheumatol. 2011;23:352–7. doi: 10.1097/BOR.0b013e3283477b7b. [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Rook GA, Brunet LR. Microbes, immunoregulation, and the gut. Gut. 2005;54:317–20. doi: 10.1136/gut.2004.053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzinger P. An innate sense of danger. Semin Immunol. 1998;10:399–415. doi: 10.1006/smim.1998.0143. [DOI] [PubMed] [Google Scholar]
- 11.Tlaskalova-Hogenova H, Tuckova L, Lodinova-Zadnikova R, Stepankova R, Cukrowska B, Funda DP, et al. Mucosal immunity: its role in defense and allergy. Int Arch Allergy Immunol. 2002;128:77–89. doi: 10.1159/000059397. [DOI] [PubMed] [Google Scholar]
- 12.Mestecky J, Russell MW, Jackson S, Michalek SM, Tlaskalova-Hogenova H, Sterzl J. New York: Plenum Press; 1995. Advances in mucosal immunology. [Google Scholar]
- 13.Ogra PL, Mestecky J, Lamm ME, Strober W, McGhee J, Bienenstock J. New York: Academic Press; 1999. Handbook of muocosal immunology. [Google Scholar]
- 14.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–64. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 15.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 16.Bocker U, Yezerskyy O, Feick P, Manigold T, Panja A, Kalina U, et al. Responsiveness of intestinal epithelial cell lines to lipopolysaccharide is correlated with Toll-like receptor 4 but not Toll-like receptor 2 or CD14 expression. Int J Colorectal Dis. 2003;18:25–32. doi: 10.1007/s00384-002-0415-6. [DOI] [PubMed] [Google Scholar]
- 17.Bourlioux P, Koletzko B, Guarner F, Braesco V. The intestine and its microflora are partners for the protection of the host: Report on the Danone Symposium “The Intelligent Intestine”, held in Paris. Am J Clin Nutr. 2002;78:675–83. doi: 10.1093/ajcn/78.4.675. [DOI] [PubMed] [Google Scholar]
- 18.Fedorak RN. Understanding why probiotic therapies can be effective in treating IBD. J Clin Gastroenterol. 2008;42(Suppl 3):S111–5. doi: 10.1097/MCG.0b013e31816d922c. [DOI] [PubMed] [Google Scholar]
- 19.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 21.Vanderpool C, Yan F, Polk DB. Mechanisms of probiotic action: Implications for therapeutic applications in inflammatory bowel diseases. Inflammatory Bowel Dis. 2008;14:1585–96. doi: 10.1002/ibd.20525. [DOI] [PubMed] [Google Scholar]
- 22.Kelly D, Conway S, Aminov R. Commensal gut bacteria: mechanisms of immune modulation. Trends Immunol. 2005;26:326–33. doi: 10.1016/j.it.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Boirivant M, Strober W. The mechanism of action of probiotics. Curr Opin Gastroenterol. 2007;23:679–92. doi: 10.1097/MOG.0b013e3282f0cffc. [DOI] [PubMed] [Google Scholar]
- 24.Kagnoff MF. Microbial-epithelial cell crosstalk during inflammation: the host response. Ann NY Acad Sci. 2006;1072:313–20. doi: 10.1196/annals.1326.038. [DOI] [PubMed] [Google Scholar]
- 25.Walker WA. Mechanisms of action of probiotics. Clin Infect Dis. 2008;46(Suppl 2):S87–91. doi: 10.1086/523335. [DOI] [PubMed] [Google Scholar]
- 26.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–7. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 27.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 28.Baba N, Samson S, Bourdet-Sicard R, Rubio M, Sarfati M. Commensal bacteria trigger a full dendritic cell maturation program that promotes the expansion of non-Tr1 suppressor T cells. J Leukocyte Biol. 2008;84:468–76. doi: 10.1189/jlb.0108017. [DOI] [PubMed] [Google Scholar]
- 29.Christensen HR, Frokiaer H, Pestka JJ. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J Immunol. 2002;168:171–8. doi: 10.4049/jimmunol.168.1.171. [DOI] [PubMed] [Google Scholar]
- 30.Kelsall BL, Leon F. Involvement of intestinal dendritic cells in oral tolerance, immunity to pathogens, and inflammatory bowel disease. Immunol Rev. 2005;206:132–48. doi: 10.1111/j.0105-2896.2005.00292.x. [DOI] [PubMed] [Google Scholar]
- 31.O’Mahony C, Scully P, O’Mahony D, Murphy S, O’Brien F, Lyons A, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathogens. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karimi K, Inman MD, Bienenstock J, Forsythe P. Lactobacillus reuteri-induced regulatory T cells protect against an allergic airway response in mice. Am J Respir Crit Care Med. 2009;179:186–93. doi: 10.1164/rccm.200806-951OC. [DOI] [PubMed] [Google Scholar]
- 33.Braat H, van den Brande J, van Tol E, Hommes D, Peppelenbosch M, van Deventer S. Lactobacillus rhamnosus induces peripheral hyporesponsiveness in stimulated CD4+ T cells via modulation of dendritic cell function. Am J Clin Nutr. 2004;80:1618–25. doi: 10.1093/ajcn/80.6.1618. [DOI] [PubMed] [Google Scholar]
- 34.Pessi T, Isolauri E, Sutas Y, Kankaanranta H, Moilanen E, Hurme M. Suppression of T-cell activation by Lactobacillus rhamnosus GG-degraded bovine casein. Int Immunopharmacol. 2001;1:211–8. doi: 10.1016/s1567-5769(00)00018-7. [DOI] [PubMed] [Google Scholar]
- 35.Pessi T, Sutas Y, Saxelin M, Kallioinen H, Isolauri E. Antiproliferative effects of homogenates derived from five strains of candidate probiotic bacteria. Appl Environ Microbiol. 1999;65:4725–8. doi: 10.1128/aem.65.11.4725-4728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz M, Linde HJ, Lehn N, Zimmermann K, Grossmann J, Falk W, et al. Immunomodulatory consequences of oral administration of Lactobacillus rhamnosus strain GG in healthy volunteers. J Dairy Res. 2003;70:165–73. doi: 10.1017/s0022029903006034. [DOI] [PubMed] [Google Scholar]
- 37.Liu YJ, Soumelis V, Watanabe N, Ito T, Wang YH, Malefyt Rde W, et al. TSLP: an epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol. 2007;25:193–219. doi: 10.1146/annurev.immunol.25.022106.141718. [DOI] [PubMed] [Google Scholar]
- 38.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 39.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–90. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies JM, Sheil B, Shanahan F. Bacterial signalling overrides cytokine signalling and modifies dendritic cell differentiation. Immunology. 2009;128:e805–15. doi: 10.1111/j.1365-2567.2009.03086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 44.Kovacs A, Ben-Jacob N, Tayem H, Halperin E, Iraqi FA, Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microbial Ecol. 2011;61:423–8. doi: 10.1007/s00248-010-9787-2. [DOI] [PubMed] [Google Scholar]
- 45.Toussirot E, Roudier J. Pathophysiological links between rheumatoid arthritis and the Epstein-Barr virus: an update. Joint Bone Spine. 2007;74:418–26. doi: 10.1016/j.jbspin.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Tarkowski A. Infection and musculoskeletal conditions: Infectious arthritis. Best Prac Res. 2006;20:1029–44. doi: 10.1016/j.berh.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Trombone AP, Claudino M, Colavite P, de Assis GF, Avila-Campos MJ, Silva JS, et al. Periodontitis and arthritis interaction in mice involves a shared hyper-inflammatory genotype and functional immunological interferences. Genes Immun. 2010;11:479–89. doi: 10.1038/gene.2010.13. [DOI] [PubMed] [Google Scholar]
- 48.Edwards CJ. Commensal gut bacteria and the etiopathogenesis of rheumatoid arthritis. J Rheumatol. 2008;35:1477–797. [PubMed] [Google Scholar]
- 49.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Round JL, O’Connell RM, Mazmanian SK. Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun. 2010;34:J220–5. doi: 10.1016/j.jaut.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–74. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Rheumatology. 2011;7:569–78. doi: 10.1038/nrrheum.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35:1500–5. [PubMed] [Google Scholar]
- 55.Finegold SM, Mathisen SVGE. Normal indigenous intestinal flora. In: Hentges DJ, editor. Human intestinal microflora in health and disease. New York: Academic Press; 1983. pp. 3–31. [Google Scholar]
- 56.Langendijk PS, Schut F, Jansen GJ, Raangs GC, Kamphuis GR, Wilkinson MH, et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–75. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ventura M, Turroni F, Canchaya C, Vaughan EE, O’Toole PW, van Sinderen D. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci. 2009;14:3214–21. doi: 10.2741/3445. [DOI] [PubMed] [Google Scholar]
- 58.Hatakka K, Martio J, Korpela M, Herranen M, Poussa T, Laasanen T, et al. Effects of probiotic therapy on the activity and activation of mild rheumatoid arthritis - a pilot study. Scand J Rheumatol. 2003;32:211–5. doi: 10.1080/03009740310003695. [DOI] [PubMed] [Google Scholar]
- 59.Nenonen MT, Helve TA, Rauma AL, Hanninen OO. Uncooked, lactobacilli-rich, vegan food and rheumatoid arthritis. Br J Rheumatol. 1998;37:274–81. doi: 10.1093/rheumatology/37.3.274. [DOI] [PubMed] [Google Scholar]
- 60.Taneja V, David CS. Lessons from animal models for human autoimmune diseases. Nature Immunology. 2001;2:781–4. doi: 10.1038/ni0901-781. [DOI] [PubMed] [Google Scholar]
- 61.Wooley PH, Luthra HS, Stuart JM, David CS. Type II collagen-induced arthritis in mice. I. Major histocompatibility complex (I region) linkage and antibody correlates. J Exp Med. 1981;154:688–700. doi: 10.1084/jem.154.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taneja V, David CS. Role of HLA class II genes in susceptibility/resistance to inflammatory arthritis: studies with humanized mice. Immunol Rev. 2010;233:62–78. doi: 10.1111/j.0105-2896.2009.00858.x. [DOI] [PubMed] [Google Scholar]
- 63.Taneja V, David CS. HLA transgenic mice as humanized mouse models of disease and immunity. J Clin Invest. 1998;101:921–6. doi: 10.1172/JCI2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taneja V, Behrens M, Basal E, Sparks J, Griffiths MM, Luthra H, et al. Delineating the role of the HLA-DR4 “shared epitope” in susceptibility versus resistance to develop arthritis. J Immunol. 2008;181:2869–77. doi: 10.4049/jimmunol.181.4.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taneja V, Behrens M, Mangalam A, Griffiths MM, Luthra HS, David CS. New humanized HLA-DR4-transgenic mice that mimic the sex bias of rheumatoid arthritis. Arthritis Rheum. 2007;56:69–78. doi: 10.1002/art.22213. [DOI] [PubMed] [Google Scholar]
- 66.Geluk A, Taneja V, van Meijgaarden KE, Zanelli E, Abou-Zeid C, Thole JE, et al. Identification of HLA class II-restricted determinants of Mycobacterium tuberculosis-derived proteins by using HLA-transgenic, class II-deficient mice. Proc Natl Acad Sci USA. 1998;95:10797–802. doi: 10.1073/pnas.95.18.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaahtovuo J, Toivanen P, Eerola E. Bacterial composition of murine fecal microflora is indigenous and genetically guided. FEMS Microbiol Ecol. 2003;44:131–6. doi: 10.1016/S0168-6496(02)00460-9. [DOI] [PubMed] [Google Scholar]
- 69.De Palma G, Capilla A, Nadal I, Nova E, Pozo T, Varea V, et al. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Curr Issues Mol Biol. 2009;12:1–10. [PubMed] [Google Scholar]
- 70.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–73. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanov II, Littman DR. Segmented filamentous bacteria take the stage. Mucosal Immunol. 2010;3:209–12. doi: 10.1038/mi.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.So JS, Kwon HK, Lee CG, Yi HJ, Park JA, Lim SY, et al. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol Immunol. 2008;45:2690–9. doi: 10.1016/j.molimm.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 74.So JS, Lee CG, Kwon HK, Yi HJ, Chae CS, Park JA, et al. Lactobacillus casei potentiates induction of oral tolerance in experimental arthritis. Mol Immunol. 2008;46:172–80. doi: 10.1016/j.molimm.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 75.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615–22. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chervonsky AV. Influence of microbial environment on autoimmunity. Nature Immunol. 2010;11:28–35. doi: 10.1038/ni.1801. [DOI] [PubMed] [Google Scholar]
- 78.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–41. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, J. Murray JA, et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One. 2012;7:e36095. doi: 10.1371/journal.pone.0036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 81.Taneja V, Taneja N, Behrens M, Pan S, Trejo T, Griffiths M, et al. HLA-DRB1*0402 (DW10) transgene protects collagen-induced arthritis-susceptible H2Aq and DRB1FNx010401 (DW4) transgenic mice from arthritis. J Immunol. 2003;171:4431–8. doi: 10.4049/jimmunol.171.8.4431. [DOI] [PubMed] [Google Scholar]
- 82.Arrieta MC, Bistritz L, Meddings JB. Alterations in intestinal permeability. Gut. 2006;55:1512–20. doi: 10.1136/gut.2005.085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tiihonen K, Ouwehand AC, Rautonen N. Human intestinal microbiota and healthy ageing. Ageing Res Rev. 2010;9:107–16. doi: 10.1016/j.arr.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 84.Tengstrand B, Carlstrom K, Fellander-Tsai L, Hafstrom I. Abnormal levels of serum dehydroepiandrosterone, estrone, and estradiol in men with rheumatoid arthritis: high correlation between serum estradiol and current degree of inflammation. J Rheumatol. 2003;30:2338–43. [PubMed] [Google Scholar]
- 85.Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–51S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 86.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]


