Abstract
During the past 6 years, focused virus hunting has led to the discovery of nine new human polyomaviruses, including Merkel cell polyomavirus, which has been linked to Merkel cell carcinoma, a lethal skin cell cancer. The discovery of so many new and highly divergent human polyomaviruses raises key questions regarding their evolution, tropism, latency, reactivation, immune evasion and contribution to disease. This Review describes the similarities and differences among the new human polyomaviruses and discusses how these viruses might interact with their human host.
The recent discovery of nine new human polyoma-viruses has re-invigorated the study of this group of DNA tumour viruses and their potential contributions to disease (TABLE 1). Several polyomaviruses have also been recently discovered in other primates, including chimpanzees, orangutans and gorillas1–4. This sudden burst of discovery has revealed that polyomaviruses represent a large genus with a high degree of divergence. Although all nine of the new human polyomaviruses have a genomic structure and gene organization similar to that of the canonical polyomavirus simian virus 40 (SV40) (FIG. 1a), there are significant differences in sequence that might have an impact on the life cycle and host cell tropism of the new human viruses. The increasing number of human and animal polyomaviruses suggests that there are many more to be discovered and raises key questions regarding their tropism, spread and disease-causing potential.
Table 1.
Human polyomaviruses
| Full name | Abbreviation | Alternative names |
Year of discovery |
NCBI RefSeq accession |
Associated disease | Source of isolation |
Seroprevalence (%) |
|---|---|---|---|---|---|---|---|
| BK polyomavirus6 | BKPyV | BK, BKV | 1971 | NC_001538.1 | Polyomavirus-associated nephropathy (PVAN), haemorrhagic cystitis | Urine | 82 (REF. 8), 92 (REF. 157) |
| JC polyomavirus7 | JCPyV | JC, JCV | 1971 | NC_001699.1 | Progressive multifocal leukoencephalopathy | Urine, brain | 40–55 (REFS 8,13,157,158) |
| Karolinska Institute polyomavirus16 | KIPyV | KI | 2007 | NC_009238.1 | None | Nasopharyngeal tissue | 55 (REF. 8), 70 (REF. 33), 90 (REF. 157) |
| Washington University polyomavirus17 | WUPyV | WU | 2007 | NC_009539.1 | None | Nasopharyngeal tissue | 70 (REF. 8), 80 (REF. 33), 90 (REF. 157) |
| Merkel cell polyomavirus22 | MCPyV | MCV | 2008 | NC_010277.1 | Merkel cell carcinoma | Lesion | 60 (REF. 157), 65 (REF. 159) |
| Human polyomavirus 6 (REF. 23) | HPyV6 | None | 2010 | NC_014406.1 | None | Skin | 70 (REF. 23) |
| Human polyomavirus 7 (REF. 23) | HPyV7 | None | 2010 | NC_014407.1 | None | Skin | 35 (REF. 23) |
| Trichodysplasia spinulosa-associated polyomavirus21 | TSPyV | None | 2010 | NC_014361.1 | Trichodysplasia spinulosa, pilomatrix dysplasia | Lesion | 70–80 (REF. 160) |
| Human polyomavirus 9 (REFS 24,27) | HPyV9 | Lymphotropic virus | 2011 | NC_015150.1 | None | Skin, blood, urine | 25–50 (REFS 161,162) |
| Malawi polyomavirus18,28,29 | MWPyV | MXPyV, HPyV10 | 2012 | NC_018102.1 | None | Stool, wart | Unknown |
| St Louis polyomavirus19 | STLPyV | None | 2012 | NC_020106.1 | None | Stool | Unknown |
Figure 1. Genomic organization and structure of polyomaviruses.
a | The prototypical circular double-stranded DNA genome has three main regions: a non-coding control region containing the early and late promoters, their transcription start sites and the origin of replication; an early region encoding large T antigen (LT) and small T antigen (ST) and an alternatively spliced LT (LT′); and a late region encoding the viral coat proteins VP1, VP2, VP3 and VP4. The reading frames for VP2, VP3 and VP4 are identical, but translation starts at successive initiating AUG codons to generate the different proteins. VP4 has been confirmed only in the primate species simian virus 40 (SV40). Agnoprotein (Agno) is encoded by a late transcript from JC polyomavirus and BK polyomavirus, but has yet to be confirmed in the new human polyomaviruses. b | Electron micrograph of purified recombinant VP1 pentamers (from murine polyomavirus), which have a diameter of ~9 nm (scale bar represents 100 nm). c | Electron micrograph of purified murine polyomavirus virions, which have a diameter of ~50 nm (scale bar represents 200 nm).
More than 50 years of research on SV40 and murine polyomavirus (MPyV) have provided crucial insights into the cellular and molecular biology of these viruses and established a strong foundation for the study of the many newly discovered polyomaviruses5. The first two human polyomavirus species to be characterized, JC polyomavirus (JCPyV) and BK polyomavirus (BKPyV), were identified in 1971 and named after the index case patients6,7. Both are benign infectious agents in most individuals, but are capable of causing severe pathological manifestations in individuals who are immunosuppressed. Epidemiological and clinical studies revealed that infection with JCPyV and BKPyV typically occurs at a young age, and the viruses establish a life-long persistent infection in sanctuary sites such as the proximal renal tubule8–13. However, in patients with HIV/AIDS and, more recently, in patients with multiple sclerosis who have been treated with natalizumab, JCPyV is the causative agent of progressive multifocal leukoencephalopathy (PML)14, a frequently fatal disease of the central nervous system. Uncontrolled BKPyV infection contributes to polyomavirus-associated nephropathy (PVAN) in patients who have received a renal transplant and to haemorrhagic cystitis in patients who have received a haematopoietic stem cell transplant15.
Since 2007, nine human polyomavirus species have been identified. These new viruses are named after the institution where the discovery was made (Karolinska Institute polyomavirus (KIPyV) and Washington University polyomavirus (WUPyV))16,17; after the source of the original virus isolate (Malawi polyomavirus (MWPyV) and St Louis polyomavirus (STLPyV))18,19; after the disease association (Merkel cell polyomavirus (MCPyV) and trichodysplasia spinulosa-associated polyomavirus (TSPyV))20,21,22; or in the temporal order of discovery (human polyomavirus 6 (HPyV6), HPyV7 and HPyV9 (REFS 23,24))) The new viruses were identified using a variety of techniques. Most of the discovery protocols involved virion enrichment from samples by treatment with DNase to eliminate un-encapsidated DNA, followed by protease treatment to disrupt the virions, and then DNA sequencing. These techniques are now widely used in mining microbiome niches25. WUPyV and KIPyV were found by high-throughput sequencing of DNA from nasopharyngeal samples, followed by comparison with known human and pathogen sequences16,17. Electron microscopy evidence of a hair follicle infected with a 38 nm diameter polyomavirus-like particle in a patient with trichodysplasia spinulosa anticipated the discovery of a virus20; TSPyV was subsequently identified from trichodysplasia spinulosa lesions by a strategy that exploited the circular nature of polyomavirus genomes by using rolling-circle amplification by highly processive DNA polymerases followed by digestion with restriction enzymes that typically cut only once in polyomavirus genomes21,26. HPyV6 and HPyV9 were also identified using the rolling-circle amplification method23,27. HPyV7 was identified from DNA isolated from skin using degenerate PCR primers corresponding to WUPyV and HPyV6 (REF. 23). MWPyV was first isolated from stool specimens by enriching for potential virus-like particles (VLPs)18. Two other groups also identified MWPyV, from stool (originally called MX polyomavirus (MXPyV))28 and from a patient with WHIM (warts, hypogammaglobulinaemia, infections and myelokathexis) syndrome (originally called HPyV10)29. The most recent polyomavirus to be discovered, STLPyV19, was isolated from stool specimens obtained from both the Gambia and the United States, and its close relationship to MWPyV supports its tentative assignment as a new human polyomavirus.
MCPyV was identified using digital transcriptome subtraction, in which cDNA libraries prepared from Merkel cell carcinoma (MCC) specimens were sequenced using pyrosequencing. The human sequences were subtracted to identify novel sequences, which were then aligned with known viral sequences by a BLAST homology search to detect polyomavirus-like genes22. Importantly, the initial discovery of MCPyV in these MCC specimens revealed that the viral genome was clonally integrated into the tumour cell DNA. Although MCPyV is a typical polyomavirus with an episomal circular double-stranded DNA (dsDNA) genome, this genome becomes stably integrated into a chromosome in MCC cells. Examination of the MCPyV genomes identified in MCC specimens revealed that they contain mutations and deletions that render the viral DNA incapable of supporting viral DNA replication and generating viable virions.
With the discovery of so many new viruses, the nomenclature of polyomaviruses has continued to evolve30. The former family designation Papovaviridae included both the polyomaviruses and the papillomaviruses, but the new designation Polyomaviridae includes Polyomavirus as the only genus. The Polyomaviridae Study Group of the International Committee on Taxonomy of Viruses (ICTV) proposed several criteria for polyomavirus classification, including dividing the polyomaviruses into three genera, with Orthopolyomavirus and Wukipolyomavirus containing all the mammalian species and Avipolyomavirus restricted to bird polyomaviruses30. In addition, the committee proposed that for a polyomavirus to be designated a unique species within the family, the whole-genome nucleotide sequence must have less than 81% nucleotide identity to the sequence of known polyomavirus species.
HPyV9 has been isolated from blood and urine, which are normally sterile, and this strongly supports the candidacy of this virus as a human polyomavirus. However, as some of the human polyomavirus sequences (HPyV6, HPyV7, MWPyV and STLPyV) have been isolated from human stool, respiratory secretions or skin swab specimens, they could represent environmental contaminants and be considered ‘potential’ human viruses. Evidence supporting their classification as human polyomaviruses includes isolation from an infected cell and detection of seropositivity, as has been described for HPyV6 and HPyV7 (REF. 23).
The serology for BKPyV and JCPyV indicates that infection typically occurs at an early age, and persistent, sustained antibody titres throughout life indicate a persistent infection. For many of the human polyomaviruses surveyed, the seroprevalence is high in the general population8,31–33. Serological assays have evolved (BOX 1), and the use of recombinant viral VP1 capsid proteins (FIG. 1b) specific for each species has enabled the development of high-throughput screens and sensitive competition assays to distinguish between infections with the various polyomaviruses. The seroprevalence of the new polyomaviruses is as high as 90% of individuals (TABLE 1), with the possible exceptions of HPyV7 and HPyV9, although more data are needed. The seroprevalence of JCPyV also appears to be lower than this, in the 40–60% range; this incidence rate has important risk stratification consequences for patients receiving natalizumab and related immunosuppressive drugs34.
Box 1 | Serological methods for studying polyomavirus infection.
The serological analysis of polyomavirus infection has been facilitated by the recombinant production of the viral protein VP1. Expressing VP1 in eukaryotic cells — for example, in the baculovirus expression system — results in the formation of assembled virus-like particles (VLPs) that can be isolated and bound directly to ELISA (enzyme-linked immunosorbent assay) plates. This assay can be further refined by subjecting borderline samples to a secondary screen using pre-incubation of the serum sample with VP1 VLPs in solution. In this competition step, specificity for the particular VP1 is confirmed. The production and purification of VLPs can be difficult, and expression of glutathione-S-transferase (GST)-conjugated VP1 in Escherichia coli has been used as a convenient alternative150. The GST–VP1 proteins are purified by affinity chromatography as pentameric capsomeres (rather than assembled VLPs) and then bound to ELISA wells that have been coated with casein-conjugated glutathione8. This method also allows the use of competition assays by first pre-incubating the sera with non-conjugated VP1 pentamers of the same or different polyomavirus. Using such competition assays, the human antibody response has been generally found to be specific to each polyomavirus species, although cross-reactivity has been detected151,152. This technique has been adapted to incorporate Luminex bead technology, in which the GST–VP1 proteins from different polyomaviruses are bound to glutathione-conjugated beads of varying colours, allowing for the simultaneous assay of up to 100 different species in a single serum sample (of microlitre quantity) for use in high-throughput screens153. Most recently, pseudovirus assays, similar to those developed for measuring papillomavirus serology, have been used to assess polyomavirus neutralizing antibody titres31. Pseudoviruses are generated by transfecting 293TT cells (engineered to overexpress SV40 large T antigen) with expression plasmids for the VP1, VP2 and VP3 genes of the particular polyomavirus along with a reporter plasmid. VLPs are formed, and the reporter plasmid is packaged in a subset of particles. Sera incubated with the purified pseudoviruses before ‘infection’ of permissive target cells then give a semi-quantitative measure of the neutralizing antibody titre as measured by the decrease in colorimetric signal from the reporter.
Given the increasing use of immunosuppressive therapy after renal and haematopoietic stem cell transplantation and for multiple sclerosis, rheumatoid arthritis and other autoimmune diseases, and the ability of some polyomaviruses to cause disease in some individuals receiving this therapy, it is likely that the frequency of diseases resulting from these viruses will increase. Although many questions remain unanswered, extrapolation from the known biology of JCPyV and BKPyV allows a predictive framework for the study of the new viruses. In this article, we compare the similarities and differences among the new human polyomaviruses with respect to their evolution, possible disease relationships and host interactions. For additional reviews addressing some of these issues, see REFS 14,15,35–37.
Genomic organization and phylogeny
Polyomaviruses are small viruses (~45 nm in diameter) containing a single circular dsDNA genome of ~5.2 kb (FIG. 1a,c). There are two distinct transcriptional units within the genome; the early region encodes the alternatively spliced large tumour (T) and small T antigens, named for their corresponding protein size, and the late region encodes the structural viral proteins VP1, VP2 and VP3, which form the viral capsid38. Alternatively spliced variants of large T antigen have been characterized for several polyomaviruses39 (an example, denoted LT′, is shown in FIG. 1a). STLPyV may express 229T, a third early transcript that shares the first 190 residues of small T antigen with an additional 39 residues from an alternative reading frame of large T antigen19. None of the human polyomaviruses appears to encode a middle T antigen, which is present in MPyV40. The late region transcript uses alternative translational start sites to encode VP1, VP2 and VP3. The late region of JCPyV and BKPyV, as well as that of several monkey polyomaviruses, including SV40, also encodes agnoprotein, a small protein of unknown function. The early and late regions are separated by a non-coding regulatory region containing the early and late promoters (with their corresponding transcriptional start sites) and the origin of viral DNA replication38.
Polyomavirus virions are non-enveloped, and the capsid comprises 72 pentamers (capsomeres) of the major structural protein VP1, each of which is associated with a single molecule of VP2 or VP3. SV40 also encodes VP4, which has been shown to act as a viroporin41. Because infectious viruses and permissive host cells have not yet been identified for the new human polyomaviruses, the prototypical gene products described in FIG. 1a have been for the most part deduced from their nucleotide sequences and have not yet been validated for each virus.
Phylogeny
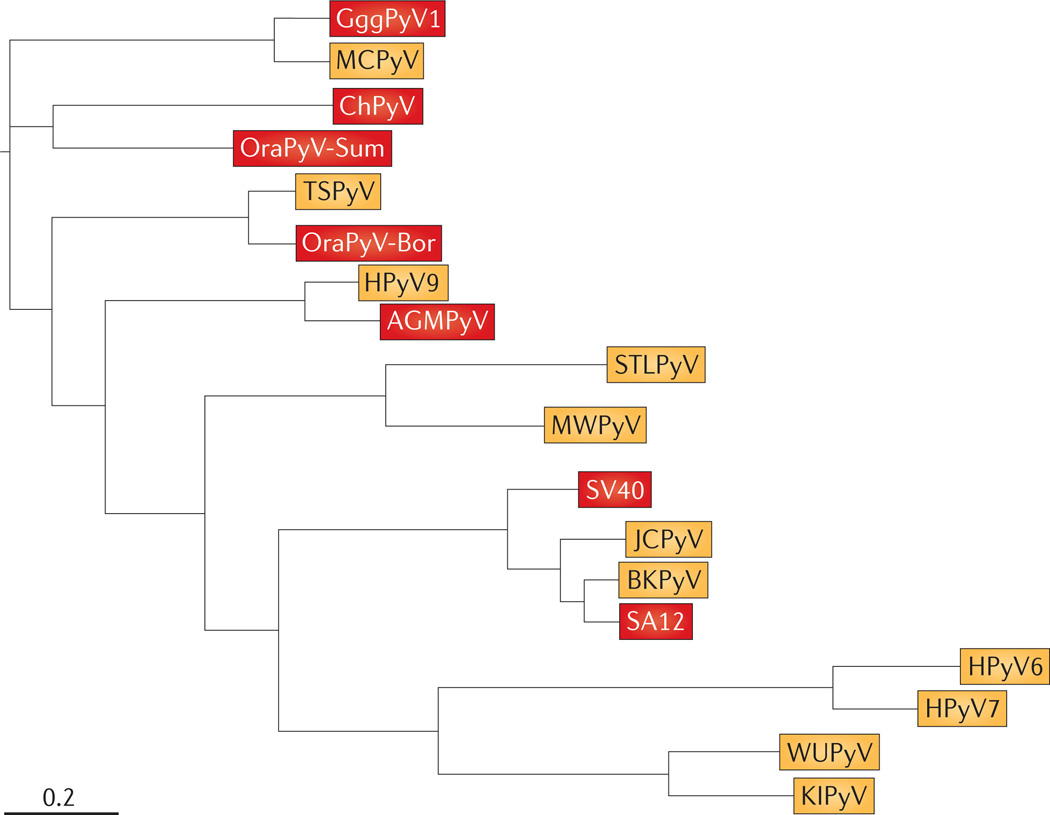
As more new polyomavirus sequences have been isolated, it has become clear, on the basis of sequence homologies, that there are distinct clades or species of polyomaviruses, and that within a species there are several sequence variants. It is currently unclear what the biological consequences of these variants are or whether these variants represent different serotypes. The amino acid relationships of the human and primate polyomavirus large T antigen and VP1 proteins of each species are shown in the simplified phylogenetic tree in FIG. 2. The groupings are perhaps not surprising, as several related species were isolated from the same tissue or sample types. For example, WUPyV and KIPyV, HPyV6 and HPyV7, and MWPyV and STLPyV are seen paired in their own distinct branches, mirroring their isolation from nasopharyngeal, skin and stool sources, respectively. The close protein homology between these pairs might reflect distinct viral protein–host protein interactions that support their tissue tropism42. Many human polyomaviruses appear to be closely related to primate relatives: BKPyV and JCPyV most closely resemble SV40 and simian agent 12 (SA12), a baboon virus; HPyV9 is very similar to African green monkey polyomavirus (AGMPyV; also known as B lymphotropic polyomavirus (LPyV)); MCPyV resembles Gorilla gorilla gorilla polyomavirus 1, as well as several isolates from chimpanzees4; and TSPyV resembles Bornean orangutan polyomavirus. It is difficult to assign the evolutionary derivation of these primate-related human virus species. Both co-evolution from a common ancestral polyomavirus and adaptation to a new host after zoonotic transmission of each primate virus have been considered43–45. Data obtained from polyomaviruses found in other species might inform these evolutionary hypotheses.
Figure 2. Phylogenetic tree relating polyomaviruses from human and primate isolates.
Viruses identified from human isolates are shown in yellow, and viruses identified from primate isolates are shown in red. This simplified tree was constructed by concatenating the amino acid sequences for the capsid protein VP1 and large T antigen from 19 separate isolates. Sequences were then aligned using MUSCLE 3.8.31 with default parameters154. Alignment positions containing more than 90% gaps were filtered using filter_alignment.py from QIIME155. The phylogenetic tree was then constructed using FastTree 2.1.4 with default parameters156. Branch length is in substitutions per site. Note that Washington University polyomavirus (WUPyV), Karolinska Institute polyomavirus (KIPyV), human polyomavirus 6 (HPyV6) and HPyV7 all belong to the genus Wukipolyomavirus, and all other isolates shown belong to the genus Orthopolyomavirus30. The RefSeq accession numbers for the human viruses are listed in TABLE 1. The primate viruses shown, and their GenBank or RefSeq accession numbers, are: Gorilla gorilla gorilla polyomavirus 1 (GggPyV1; HQ385752), chimpanzee polyomavirus (ChPyV; NC_014743), Sumatran orangutan polyomavirus (OraPyV-Sum; FN356901), Borneo OraPyV (OraPyV-Bor; NC_013439), African green monkey polyomavirus (AGMPyV; NC_004763), simian virus 40 (SV40; NC_001669) and simian agent 12 (SA12; NC_012122). Merkel cell polyomavirus (MCPyV) is also related to a number of chimpanzee isolates (not shown)4. Tree constructed by D. McDonald, University of Colorado at Boulder, USA. BKPyV, BK polyomavirus; JCPyV, JC polyomavirus; MWPyV, Malawi polyomavirus; STLPyV, St Louis polyomavirus; TSPyV, trichodysplasia spinulosa-associated polyomavirus.
Polyomavirus proteins
The polyomaviruses encode a limited number of proteins, all of which are dependent on specific interactions with host cell proteins to promote viral replication and sustain viability. The history of polyomavirus research has repeatedly revealed that the identification of specific viral protein–host protein interactions leads to key insights into eukaryotic cell biology and mammalian tumour biology.
T antigens
All human polyomaviruses express distinct early or late mRNAs. The early mRNAs are expressed soon after infection and are alternatively spliced to yield large and small T antigens that share the amino-terminal 75–80 residues encoded by the first exon of the large T antigen gene (FIG. 3a–c). The small T antigen mRNA reads through the first splice donor site of the large T antigen gene to encode a unique region that is not shared with large T antigen. The small T antigen unique region, stop codon and small T-specific splice donor site all reside within the intron of the large T antigen gene and use the splice acceptor of the large T antigen gene to make the complete small T antigen mRNA. Although all polyomaviruses produce an early mRNA that encodes the full-length large T antigen, several (including MCPyV, BKPyV and JCPyV) produce alternatively spliced mRNAs in which regions encoding the central or carboxy-terminal domains of large T antigen have been deleted39 (such as the LT′ mRNA in FIG. 1a). The MCPyV small T antigen mRNA can also undergo this alternative splicing, but this does not affect the small T antigen-coding region39.
Figure 3. The functional domains of polyomavirus large and small T antigens.
a | The amino-terminal DnaJ (J) domain is shared between small and large T antigens. Large T antigen also contains a retinoblastoma-associated protein (RB)-binding LXCXE motif, a threonine–proline–proline–lysine (TPPK) motif, a nuclear-localization sequence (NLS), a DNA-binding domain (DBD), a helicase domain and a host range and adenovirus helper function (HR–AH) domain. Small T antigen contains an amino-terminal J domain followed by a unique region that is not shared with large T antigen and contains two zinc-fingers. b | Large T antigen binds to many cellular proteins. The J domain recruits heat shock cognate 71 kDa protein (HSC70) homologues. The region between the J domain and LXCXE motif has been shown to bind independently to cullin 7 (CUL7), BUB1 and insulin receptor substrate 1 (IRS1). Merkel cell polyomavirus (MCPyV) large T antigen binds to the endosomal protein VPS39. The LXCXE domain binds to retinoblastoma-associated protein (RB) and related proteins (p107 and p130). Phosphorylation of the threonine residue in the TPPK motif is required for large T antigen-mediated viral DNA replication. The nuclear localization signal (NLS) binds specifically to KPNA family importin homologues. The DBD and helicase domains are required for viral replication and recruit cellular DNA replication factors (DNA polymerase-α catalytic subunit (POLA), the replication protein A complex (RPA) and the DNA primase complex (PRIM) for DBD; and EP300, CREBBP, p53 and DNA topoisomerase 1 (TOP1) for the helicase domain) to the replicating simian virus 40 (SV40) genome. The carboxyl termini of SV40, JC polyomavirus (JCPyV) and BK polyomavirus (BKPyV) large T antigens each contain a threonine residue (threonine 701 in SV40, threonine 691 in BKPyV and threonine 684 in JCPyV) that, when phosphorylated, competes with phosphorylated G1–S-specific cyclin E1 and MYC for binding to the cullin RING ligase substrate adaptor FBXW7. FAM111A has recently been implicated in the SV40 host range restriction and adenovirus helper function. c | Small T antigen binds specifically to the A and C subunits of protein phosphatase 2A (PP2A). d | Polyomavirus genome replication. Replication begins when the large T antigen DBD binds to the origin of replication. Two hexamers of large T antigen form in a head-to-head orientation at the origin. The helicase domains of the large T antigen hexamers initiate unwinding of viral DNA followed by bidirectional replication.
Large T antigen contains several intrinsic biochemical activities and binds specifically to cellular proteins required for polyomavirus replication (FIG. 3b). The N terminus of all polyomavirus T antigens contains a DnaJ domain, or J domain, which contributes to efficient viral DNA replication, although it is not clear how this is accomplished46. In addition to the J domain, all human polyomavirus large T antigens contain an LXCXE motif47. This motif binds directly to the tumour suppressor retinoblastoma-associated protein (RB) and to the RB-related proteins p130 (also known as RBL2) and p107 (also known as RBL1). The J domain cooperates with the LXCXE motif to disrupt the interaction between RB and the E2F family transcription factors in order to promote cell cycle entry and progression48. In addition to RB, MCPyV large T antigen binds the endosomal protein VPS39, although the biological consequences of this interaction are not clear49. Following the LXCXE motif, all large T antigens contain a threonine–proline–proline–lysine (TPPK) motif, a nuclear localization sequence (NLS), a DNA-binding domain (DBD) and a helicase domain. Polyomavirus DNA replication is initiated when two hexamers of large T antigen form in a head-to-head orientation on the origin of replication. These double hexamers twist in opposite orientations to each other, unwinding the viral DNA, and recruit cellular DNA polymerase–primase to initiate DNA replication50. Following initiation, the double hexamers separate from each other and move in opposite directions to replicate viral DNA in a bidirectional manner51(FIG. 3d).
The outside surface of the helicase domain of SV40 large T antigen binds to the tumour suppressor protein p53 (REF. 52). Large T antigen binding to p53 blocks p53-dependent gene expression in response to DNA damage signals. Some human polyomavirus large T antigens, including those from JCPyV and BKPyV, bind to p53 in a similar manner to the SV40 protein. A recent report also provides evidence for a complex association between p53 and large T antigen from WUPyV, HPyV6 and HPyV7 (REF. 42); however, it is unclear whether MCPyV large T antigen binds p53. Furthermore, in Merkel cell carcinoma samples, the large T antigen gene has undergone mutations that result in expression of a truncated molecule which lacks the DBD and helicase domain, thereby rendering the virus unable to replicate the integrated viral origin of replication. This truncation deletes the region homologous to the p53-binding region of SV40 large T antigen and the resultant protein must therefore ne unable to bind p53 even if the wild-type, full-length version could bind39.
After the helicase domain, JCPyV and BKPyV large T antigen contains a C-terminal region that bears some homology to the SV40 C-terminal domain (FIG. 3b). This region of homology includes a threonine residue (threonine 701 in SV40) that, when phosphorylated in SV40, serves as a phospho-degron decoy for the cullin RING ligase substrate adaptor FBXW7. Large T antigen phosphorylated on threonine 701 competes with phosphorylated cyclin E and MYC for binding to FBXW7. This competition interferes with the degradation of the G1–S-specific cyclin E1 and MYC, thereby increasing their overall levels and contributing to cellular growth and proliferation53. Recently, the C-terminal region of SV40 large T antigen was shown to bind to FAM111A, a previously uncharacterized protein that contributes to viral gene expression, host range restriction and adenovirus replication in monkey cells54(FIG. 3b). It is not known whether FAM111A binds to any of the human polyomavirus large T antigens. All the human polyomavirus large T antigens except those of JCPyV and BKPyV contain much shorter C-terminal regions with little homology to each other or to that of SV40.
All human polyomavirus small T antigens contain an N-terminal J domain followed by a unique region that contains two zinc-fingers which participate in binding to cellular protein phosphatase 2A (PP2A). Normally, PP2A contains three subunits: the regulatory A subunit, the substrate adaptor B subunit and the catalytic C subunit. Small T antigen binds the A and C subunits of PP2A and displaces the B subunit, thereby altering the substrate specificity or inhibiting the enzymatic activity of PP2A55,56. In addition to the highly conserved J domain and zinc-fingers, small T antigen contains a highly divergent region of 30–40 residues that links these two regions. It is highly likely that this uncharacterized small T antigen region can bind to additional cellular proteins, as it is oriented away from the PP2A-binding regions. Recently, it has been shown that MCPyV small T antigen can affect the phosphorylation state and activity of eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) in a manner that is independent of PP2A binding57.
Several human polyomaviruses encode a microRNA (miRNA). Evidence for an miRNA expressed by JCPyV, BKPyV and MCPyV has been obtained from infected tissues as well as from in vitro reporter constructs58–60. The miRNA is encoded by the late transcript and contains sequences that are complementary to the early region mRNA encoding large T antigen and small T antigen (FIG. 1a). The miRNA from JCPyV, BKPyV and MCPyV targets the early mRNA for cleavage, resulting in reduced levels of large T antigen expression. As expression of large T antigen promotes the expression of the late mRNA and therefore of the miRNA, the miRNA is part of a negative feedback loop that modulates the expression levels of large T antigen, thereby reducing the host’s immune response61. It has also been suggested that the miRNAs from JCPyV, BKPyV and MCPyV target cellular genes involved in the immune response60,62. However, it is unclear whether this plays a part in latency or disease.
VP1 structures and sialic acid binding
VP1 capsomeres are the major structural subunit of the polyomavirus capsid (FIG. 1b). Each of the 72 capsomeres in the capsid shell is associated with one copy of either VP2 or VP3. Although recombinant VP1 capsomeres can self-assemble in vitro or in vivo into VLPs that resemble the viral capsid63,64, the minor capsid proteins are required for the formation of infectious virions but are not required for haemagglutination65. VP4 is not part of the virion but functions as a viroporin that can perforate membranes to promote virion release from the cell66.
Many polyomaviruses use a ganglioside modified with a glycan terminating in sialic acid as a primary cell attachment molecule67,68. The preference for sialic acid was initially suggested by the ability of MPyV, JCPyV and BKPyV to haemagglutinate erythrocytes and by the loss of cell infectivity after neuraminidase treatment. Studies using liposomes mixed with specific synthetic gangliosides in flotation assays, and the addition of specific gangliosides to cells to restore infectivity, further defined the precise glycan linkages for each virus69. More recently, recombinant VP1 capsomeres have been used as reagents in glycan array binding screens to identify candidate glycans for subsequent validation. Structural studies have confirmed the glycan recognition of many of the new polyomaviruses. In one such study, researchers initially crystallized the MPyV capsomere, solved its structure to atomic resolution and then soaked the crystals in sialyllactose, which led to the identification of the sugar-binding pocket on the VP1 capsomere70,71. This technique has allowed the subsequent crystallographic characterization of several new human polyomaviruses and their glycan receptors72,73 (FIG. 4). These structures also demonstrate both the remarkable conservation of the core eight-stranded antiparallel β-barrel of the capsomere and the extensive elaboration of surface loops that represent the most disparate aspects of the capsid structure between viruses.
Figure 4. Glycan receptors used by polyomaviruses.
Host glycans with different branching patterns are linked to lactosylceramide (Lac–Cer), which provides anchorage in the cell membrane. Although the affinity constants for a single glycan–VP1 interaction are low, the avidity of multiple interactions on the viral surface provides a stable attachment. Subtle modifications of the glycan are used by different viruses, as indicated, and several glycans have been co-crystallized in complex with the specific VP1 pentamer (see Functional Glycomics Gateway page for VP1). Glycan GT1b has been suggested as one glycan receptor for Merkel cell polyomavirus (MCPyV) on the basis of in vitro binding studies and X-ray crystallography69,73. It is also possible that receptor recognition changes for a particular virus, as demonstrated by the glycan preferences of ‘large plaque’ and ‘small plaque’ murine polyomavirus (MPyV)86.
There are exceptions to the general rule of VP1 binding to gangliosides and the ability of polyomaviruses to haemagglutinate. SV40 infectivity is insensitive to neuraminidase, and the virus does not haemagglutinate. These properties result from the narrow specificity of SV40 for the glycan GM1 (monosialotetrahexosylganglioside), which is not present on erythrocytes, and the inability of neuraminidase to cleave the sialic acid modification from GM1 (REF. 74). MCPyV was found to haemagglutinate erythrocytes and bind glycan GT1b in a liposome flotation assay69. However, it has been suggested that glycosaminoglycans are the primary cell attachment molecules for MCPyV and that sialylated-glycan binding occurs at a subsequent step75. JCPyV does not bind to a glycan-modified ganglioside, but rather binds to a linear pentasaccharide, LSTc (N-acetylneuraminic acid-α2,6-galactose-β1,4-N-acetylglucosamine-β1,3-galactose-β1,4-glucose), present on a glycoprotein68; however, JCPyV variants have been identified that have lost recognition of LSTc76. Thus, there are some caveats to the assays that have been used to identify putative binding molecules for the new polyomaviruses, and the results obtained from these assays should be validated with infectivity analyses.
Sequence variants
Complicating the search for pathological correlations is the occurrence of sequence variants within a species, some of which seem to arise in vivo during persistent infection. The number of reported variants has been increasing for the well-studied viruses JCPyV and BKPyV. In vivo, JCPyV seems to select for VP1 variants and agnogene deletions that might predispose to brain tropism and PML76–79. When isolated from diseased tissues, the JCPyV genome often contains rearrangements in the non-coding regulatory region that might favour higher expression levels of large T antigen80,81. BKPyV variants (both in the non-coding region and VP1) also exist, but the pathophysiological differences of these variants are unclear82. BKPyV has been categorized into four genotypes that can be distinguished by serology as well as by viral-neutralization assays83.
The ramifications of subtle changes in glycan recognition could be significant for polyomavirus pathogenesis. As first described for MPyV, a single amino acid substitution in the sialic acid-binding site can change the virus from having a ‘small plaque’ phenotype to having a ‘large plaque’ phenotype84. This phenotype seems to result from an altered glycan recognition specificity that allows the virus to spread more rapidly by reducing the variety of glycans that can be bound85,86. Such behaviour can be anticipated in the human polyomaviruses. For example, JCPyV, which is latent in the kidney, can reactivate under conditions of immunosuppression and spread to the brain, causing PML. JCPyV isolated from PML lesions has been found to have mutations in VP1 that are thought to enable the virus to more effectively cross the blood–brain barrier and infect oligodendrocytes. Despite the fidelity of the host DNA polymerase involved in viral replication, over many years such mutant viruses could accumulate in the reservoir virus population in the host. In any case, given the appropriate selection pressure in culture, SV40 VP1 acquires mutations that alter receptor usage and cell tropism74. It is likely that additional factors other than glycan recognition will contribute to the cellular tropism adopted by the new human viruses as they fill unique niches, particularly in the skin, for which the interaction between papillomaviruses and heparan sulphate serves as an example87,88.
In vitro culture methods have not yet succeeded for any of the new human polyomaviruses, although stable, high-level expression of the corresponding large and small T antigens in cell lines might help support MCPyV virion production89,90. This method could be successful for other human polyomaviruses91. Alternatively, cell culture-adapted strains of the new human polyomaviruses might be identified, with rearrangements in the early promoter that support high levels of large T antigen expression similar to those found in SV40 and BKPyV.
Human polyomavirus infection and disease
The host cells and tissues that support infection by the new human polyomaviruses in vivo are unknown, and their identification is one of the most pressing research questions. On the basis of the sites where the polyomavirus DNA has been isolated, suspected sites of infection include the nasopharynx and lung for WUPyV and KIPyV, the skin for MCPyV, HPyV6, HPyV7, TSPyV and HPyV9, and the gastrointestinal tract for MWPyV and STLPyV. We anticipate, by extrapolation, that the new human polyomaviruses are transmitted horizontally by direct contact or by aerosol or faecal–oral routes. However, the site of infection, the original site of polyomavirus identification and the distant organs in which the virus can be detected might be distinct and difficult to relate to any pathophysiology92. Although several of the new human polyomaviruses have been isolated from nasopharyngeal or stool samples, correlations between infection and respiratory symptoms or diarrhoeal illness remain circumstantial93–99. The data remain confounded by the presence of numerous viral agents in the samples studied, such that no single entity has been singled out as the causative agent100–103.
The clinical symptoms of primary infection with BKPyV and JCPyV have never been conclusively identified. Thus, it is perhaps not surprising that the new human polyomaviruses have not been associated with any clinical symptoms either. For example, in a recent study, serial collections of serum from a cohort of men with HIV/AIDS were examined for signs of recent MCPyV infection. Despite careful surveillance and detailed questionnaires, none of the newly MCPyV-infected individuals developed any signs or symptoms of a primary MCPyV infection104.
Attempts to link the new polyomaviruses to clinical disease have focused on older individuals, in whom immunity is waning, or in the immunosuppressed100,101,103. In particular, immunocompromised individuals have been studied with respect to opportunistic diseases, but no specific correlations with human polyomaviruses have been identified as yet. JCPyV and BKPyV can cause a lytic infection that results in the destruction of tissue. JCPyV infection of oligodendrocytes in the central nervous system (that is, PML) and BKPyV infection of renal tubule cells (that is, PVAN) result in loss of these cells, similar to the well-described cytopathic effect of SV40. However, except for MCPyV and TSPyV, there has been no obvious association of the new human polyomaviruses with a clinical syndrome105,106.
Merkel cell carcinoma
The key insight that prompted the hunt for a pathogen in MCC was the recognition that the incidence of MCC is higher in immunosuppressed recipients of organ transplants and in patients with HIV/AIDS than in non-immunocompromised individuals. MCC was first described in 1972, followed by several reports that this rare cancer can occur in patients with a weakened immune status107. Patients with haematological malignancies, especially chronic lymphocytic leukaemia, are at an increased risk of developing MCC108. There have been several reports of MCC developing in patients with autoimmune diseases such as psoriatic and rheumatoid arthritis, particularly when treated with immunesuppressive therapy109. Additional risk factors for MCC include an age of greater than 60 years, and most MCCs occur in sun-exposed areas of the skin110. In addition, it has been reported that pharmacological use of statins and environmental exposure to arsenic increase the risk of developing MCC, although the mechanisms involved are unclear111,112.
Clonal integration of the MCPyV genome has been nearly universally identified in MCC cells22,113,114. In addition to integration of the viral genome, the large T antigen gene is mutated, resulting in expression of a truncated molecule that lacks the DBD and the helicase domain, thereby inactivating its ability to replicate the integrated viral origin of replication39,114 (FIG. 5a,b). Thus, similar to cervical lesions, which often contain integrated human papillomavirus genomes, MCC tumours arise from the selection of cells containing integrated polyomavirus genomes with mutations that eliminate viral replication while preserving expression of the oncogenic viral proteins39. A few integration sites have been mapped for MCPyV; it seems that the virus integrates near fragile sites in locations similar to those where integrated human papillomavirus genomes have been found in cervical cancer22,115,116.
Figure 5. Merkel cell polyomavirus and Merkel cell carcinoma.
a | In Merkel cell carcinoma, the Merkel cell polyomavirus (MCPyV) DNA undergoes mutations, leaving small T antigen and the J domain and LXCXE motif of large T antigen intact, but typically deleting the DNA-binding domain and helicase domain of large T antigen. b | An MCPyV genome (black) undergoes random integration into the host cell DNA (blue). If wild-type large T antigen is present in the cell, then repeated rounds of bidirectional viral DNA replication result in numerous copies of viral DNA, appearing as onion skinning, within the host chromosome. c | If a wild-type genome is integrated into the host and then obtains a mutation that inactivates large T antigen, the viral genome cannot be replicated bidirectionally and instead undergoes amplification in a manner analogous to amplification of cellular oncogenes such as MYC. d | An alternative model to explain the high copy number of integrated viral DNA is that rolling-circle amplification of an episomal MCPyV genome containing a mutation in the large T antigen gene results in a concatemer of mutant viral genomes. This concatemer then becomes randomly integrated into the host cell chromosome, resulting in a Merkel cell carcinoma containing a high copy number of viral genomes per cell.
The number of copies of the MCPyV viral genome found integrated in MCC can range from a single copy to several thousand copies113,117,118. Two distinct models could explain the sequence of events that transforms an infected cell containing a wild-type episomal MCPyV genome into a tumour cell containing multiple copies of tandemly arrayed integrated mutant viral genomes. The mutation that truncates large T antigen, as well as the amplification of the viral genome copy number, could occur either before or after integration into the host genome. If random integration of a wild-type genome occurs first, then this must be followed by mutation of the large T antigen gene to disable the ability of the encoded protein to initiate replication of the integrated genome39(FIG. 5b,c). The integrated mutant genome would subsequently undergo amplification in a process that is perhaps similar to that whereby a cellular oncogene such as MYC undergoes copy number amplification. Alternatively, mutation of the large T antigen gene could occur in the episomal viral genome before integration. This mutant genome would subsequently undergo rolling-circle amplification instead of the normal polyomavirus bidirectional genome replication (FIG. 5d). Rolling-circle amplification could occur when the viral genome contains a single-stranded DNA break; in this situation, one large T antigen hexamer would continue to replicate the intact strand, whereas the other hexamer would be unable to advance (FIG. 5d). The extended, head-to-tail, mutated genome multimer would subsequently be integrated intact into the host genome. In both models, selection occurs for cancer cells that express the truncated large T antigen and intact small T antigen.
Expression of the MCPyV T antigens in MCC tumour cells probably contributes to disease initiation and maintenance. The MCPyV truncated large T antigen expressed in MCC typically preserves the J domain and LXCXE RB-binding motif49,113,114 (FIG. 5a). Expression of the truncated large T antigen that binds RB is required for continued growth of several MCPyV-positive MCC cell lines119. Recently, it has been shown that the RB-binding motif of MCPyV large T antigen contributes to high levels of survivin (also known as BIRC5) expression and that treatment with a small-molecule inhibitor of survivin reduces the growth rate of MCC cell line xenografts120. In addition to large T antigen, small T antigen is required for cellular transformation57. Although MCPyV small T antigen can bind to PP2A, this activity might not be sufficient for the transforming functions of the virus. MCPyV small T antigen mutants with single amino acid substitutions that reduce PP2A binding retain some transforming functions in vitro57. MCPyV small T antigen promotes hyperphosphorylation and inactivation of the translation inhibitor 4EBP1 and increases host gene expression57. Small T antigens from other polyomaviruses might bind to PP2A, as well as to additional cellular proteins that contribute to the viral life cycle.
Studies using immunohistochemistry with antibodies specific for MCPyV T antigens have indicated that T antigen expression occurs in up to 97% of all MCCs57,113,121–124. MCC tumours exhibit a range of expression levels of the viral T antigens, from barely detectable to extremely high levels57,113,118. This raises the question of whether MCCs with low or negligible levels of MCPyV T antigen expression have specific mutations in cellular oncogenes or tumour suppressor genes that contribute to cancer formation and maintenance. Mutations in TP53 (encoding p53) and PIK3CA (encoding phosphoinositide-3-kinase catalytic subunit α-isoform) have been reported to occur in less than 10% of all MCC studied to date113,125–128. Notably, mutations in TP53 might be more frequent in MCCs with low levels or even a complete lack of MCPyV T antigen expression. Recently, two groups reported that expression of p63, a p53-related protein, is correlated with poor prognosis in MCC129–131; by contrast, two other groups did not find a significant correlation between p63 expression and prognosis132,133.
There has been some suggestion that high levels of MCPyV T antigen expression have a positive impact on the overall prognosis of MCC. The presence of serum antibodies against large T antigen correlates with disease status and might be useful as a biomarker134. A striking feature in some MCC tumours is the presence of an abundant T cell infiltration within the tumour itself135,136. A strong T cell infiltration of MCC is associated with better prognosis136.
Other human polyomaviruses and cancer
MCC is the only human cancer that contains clonally integrated polyomavirus DNA. MCPyV has been detected in other human cancers such as chronic lymphocytic leukaemia (CLL) and squamous cell skin carcinomas, although at low levels and not integrated into the genome108,137–139. This situation is similar to that of JCPyV and BKPyV, which have been detected in a variety of gastrointestinal and genitourinary cancers140–142. However, there has been no evidence that the tumours contain integrated JCPyV or BKPyV genomes. SV40 has been considered as a zoonotic agent involved in human oncogenesis, but the low copy number (<1/1,000 cells), the inconsistent results between laboratories and the near-undetectable seroprevalence of the virus has not supported this hypothesis8,143,144. The presence of episomal polyomavirus DNA is insufficient to support a causal role of these viruses in the pathogenesis of a cancer, because the tumour itself might be permissive for high levels of viral replication. Interestingly, however, a polyomavirus was isolated as an episome from high-grade brain cancers in ten wild raccoons exhibiting abnormal behaviour145. In these brain tumours, the raccoon polyomavirus expressed large T antigen, and this was associated with high levels of host cell p53.
Conclusions
Although the cornucopia of recently discovered human polyomaviruses might have unique cellular tropisms, it is likely that they will share certain characteristics with the well-studied species MPyV, SV40, BKPyV and JCPyV. For example, we expect to find a ubiquitous sero-epidemiological prevalence for these viruses, although the few viruses with limited prevalence could be of great interest. Of clinical importance are the 50% of JCPyV-negative humans, who might be at a muchreduced risk of developing PML after immunosuppression with natalizumab, owing to their lack of virus. The 50% of patients who are serologically negative for BKPyV subtype IV and receive a renal transplant are also speculated to be at greater risk of developing PVAN resulting from infection with a BKPyV subtype IV-positive kidney. Another likely common theme is that an asymptomatic primary infection occurs at a young age and is followed by life-long persistence. The host cell reservoir for persistence might differ among polyomavirus species, but these cell types will be the source of reactivated virus and potentially also disease sites.
Despite all these expectations, the real unknowns for polyomavirus researchers continue to be the mechanisms of persistence and reactivation, how the unique features of each viral protein contribute to these processes, and the precise cell types that serve as viral reservoirs. It would be useful to be able to define the latency state, even if it is simply a low level of continuous lytic infection. Some progress has been made in understanding the host immunological response to polyomavirus infection in the mouse146, but latency remains a puzzle. One must also wonder whether infection and persistence with these normally benign viruses confers some advantage to the host.
In addition to MCPyV, another polyomavirus with possible oncogenic manifestations is HPyV9. HPyV9 is related to AGMPyV, which can infect and immortalize human B cells27,24,147,148. If the newly discovered HPyV9 has the same tropism, it might have a causal relationship with B cell lymphomas. However, except for MCPyV, the most common pathological manifestation of infection with the new viruses identified so far is a lytic infection following reactivation in an immunosuppressed host. These lytic reactivations can be serious or even fatal, as suggested by PVAN and PML. For these diseases, antiviral therapies are desperately needed. Possible therapeutic strategies include small-molecule inhibitors of virus–glycan binding or of T antigen functions, booster immunization in anticipation of immunosuppressive regimens to increase antibody levels, or enhancement of cell-mediated immunity in elderly patients. The increasing use of immunosuppressive therapies might lead to more cases of polyomavirus-induced disease and could create a market for effective therapeutics that inhibit polyomavirus replication. In addition, new therapeutics that target the oncogenic activity of the MCPyV T antigens or boost the immune response would be extremely beneficial in MCC.
The evolution of polyomaviruses within and between species remains an active area of investigation. The close relationship with primate polyomaviruses also suggests that more human viruses might be discovered that are related to primate polyomaviruses which have yet to be associated with a human relative. The discovery of the skin-resident polyomaviruses HPyV6 and HPyV7, with receptor specificities that extend to glycosaminoglycans as well as glycans, indicates that the polyomaviruses might be able to invade the papillomavirus niche. These two types of virus were once classified in a single family, Papovaviridae, and have many related features, so it would not be surprising if their infectious patterns intersect. Indeed, two of the most interesting species in this intersection are bandicoot papillomatosis carcinomatosis viruses 1 and 2, which are recombinant viruses with the early region of a polyomavirus and the late genes of a papillomavirus, and which occupy a skin niche149.
The explosion in microbiome discovery will undoubtedly lead to the identification of more viruses and a growing family tree for the polyomaviruses. Much work lies ahead in filling in the biological and clinical data for each species and determining their relationship to human health
Acknowledgements
This work was supported in part by US Public Health Service grants P01CA050661, RO1CA93804 and R01CA63113 to J.A.D. and RO1CA37667 to R.L.G. The authors thank D. McDonald for assistance with the bioinformatics.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
James A. Decaprio’s homepage: http://dms.hms.harvard.edu/BBS/fac/DeCaprio.php
Robert L. Garcea’s homepage: http://garcealab.colorado.edu
Functional Glycomics Gateway page for VP1: http://www.functionalglycomics.org/CFGparadigms/index.php/Polyomavirus_capsid_protein_(VP1)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Contributor Information
James A. DeCaprio, Email: james_decaprio@dfci.harvard.edu.
Robert L. Garcea, Email: robert.garcea@colorado.edu.
References
- 1.Johne R, Enderlein D, Nieper H, Muller H. Novel polyomavirus detected in the feces of a chimpanzee by nested broad-spectrum PCR. J. Virol. 2005;79:3883–3887. doi: 10.1128/JVI.79.6.3883-3887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groenewoud MJ, et al. Characterization of novel polyomaviruses from Bornean and Sumatran orangutans. J. Gen. Virol. 2010;91:653–658. doi: 10.1099/vir.0.017673-0. [DOI] [PubMed] [Google Scholar]
- 3.Deuzing I, et al. Detection and characterization of two chimpanzee polyomavirus genotypes from different subspecies. Virol. J. 2010;7:347. doi: 10.1186/1743-422X-7-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leendertz FH, et al. African great apes are naturally infected with polyomaviruses closely related to Merkel cell polyomavirus. J. Virol. 2011;85:916–924. doi: 10.1128/JVI.01585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howley PM, Livingston DM. Small DNA tumor viruses: large contributors to biomedical sciences. Virology. 2009;384:256–259. doi: 10.1016/j.virol.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (BK) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 7.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 8. Kean JM, Rao S, Wang M, Garcea RL. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5:e1000363. doi: 10.1371/journal.ppat.1000363. The largest seroepidemiological study of the new viruses.
- 9.Walker DL, Padgett BL. The epidemiology of human polyomaviruses. Prog. Clin. Biol. Res. 1983;105:99–106. [PubMed] [Google Scholar]
- 10.Stolt A, Sasnauskas K, Koskela P, Lehtinen M, Dillner J. Seroepidemiology of the human polyomaviruses. J. Gen. Virol. 2003;84:1499–1504. doi: 10.1099/vir.0.18842-0. [DOI] [PubMed] [Google Scholar]
- 11.Knowles WA. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV) Adv. Exp. Med. Biol. 2006;577:19–45. doi: 10.1007/0-387-32957-9_2. [DOI] [PubMed] [Google Scholar]
- 12.Knowles WA, et al. Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J. Med. Virol. 2003;71:115–123. doi: 10.1002/jmv.10450. [DOI] [PubMed] [Google Scholar]
- 13.Egli A, et al. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J. Infect. Dis. 2009;199:837–846. doi: 10.1086/597126. [DOI] [PubMed] [Google Scholar]
- 14.Ferenczy MW, et al. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin. Microbiol. Rev. 2012;25:471–506. doi: 10.1128/CMR.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuypers DR. Management of polyomavirus-associated nephropathy in renal transplant recipients. Nature Rev. Nephrol. 2012;8:390–402. doi: 10.1038/nrneph.2012.64. [DOI] [PubMed] [Google Scholar]
- 16. Allander T, et al. Identification of a third human polyomavirus. J. Virol. 2007;81:4130–4136. doi: 10.1128/JVI.00028-07. The discovery of KIPyV, the first new human polyomavirus to be discovered in 36 years.
- 17. Gaynor AM, et al. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007;3:e64. doi: 10.1371/journal.ppat.0030064. The discovery of WUPyV by next-generation sequencing.
- 18.Siebrasse EA, et al. Identification of MW polyomavirus, a novel polyomavirus in human stool. J. Virol. 2012;86:10321–10326. doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim ES, et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology. 2012;436:295–303. doi: 10.1016/j.virol.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haycox CL, et al. Trichodysplasia spinulosa–a newly described folliculocentric viral infection in an immunocompromised host. J. Investig. Dermatol. Symp. Proc. 1999;4:268–271. doi: 10.1038/sj.jidsp.5640227. [DOI] [PubMed] [Google Scholar]
- 21. van der Meijden E, et al. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6:e1001024. doi: 10.1371/journal.ppat.1001024. The discovery of TSPyV using rolling-circle amplification to identify a virus that had been detected by electron microscopy.
- 22. Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. The discovery of MCPyV as a result of the recognition that MCC occurs more frequently in immunocompromised patients than in non-immunocompromised individuals. Southern blotting also demonstrates the clonal integration of MCPyV in tumour cells.
- 23.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scuda N, et al. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 2011;85:4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nature Rev. Microbiol. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rector A, Tachezy R, Van Ranst M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004;78:4993–4998. doi: 10.1128/JVI.78.10.4993-4998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauvage V, et al. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg. Infect. Dis. 2011;17:1364–1370. doi: 10.3201/eid1708.110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G, et al. Discovery of a novel polyomavirus in acute diarrheal samples from children. PLoS ONE. 2012;7:e49449. doi: 10.1371/journal.pone.0049449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buck CB, et al. Complete genome sequence of a tenth human polyomavirus. J. Virol. 2012;86:10887. doi: 10.1128/JVI.01690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johne R, et al. Taxonomical developments in the family Polyomaviridae. Arch. Virol. 2011;156:1627–1634. doi: 10.1007/s00705-011-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastrana DV, et al. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 2009;5:e1000578. doi: 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viscidi RP, et al. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clin. Vaccine Immunol. 2011;18:1737–1743. doi: 10.1128/CVI.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen NL, Le BM, Wang D. Serologic evidence of frequent human infection with WU and KI polyomaviruses. Emerg. Infect. Dis. 2009;15:1199–1205. doi: 10.3201/eid1508.090270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gorelik L, Goelz S, Sandrock AW. Asymptomatic reactivation of JC virus in patients treated with natalizumab. N. Engl. J. Med. 2009;361:2487–2488. doi: 10.1056/NEJMc0909622. author reply 2489–2490. The worrisome reactivation of JCPyV in patients treated with immunomodulatory therapies.
- 35.Feltkamp MC, Kazem S, van der Meijden E, Lauber C, Gorbalenya AE. From Stockholm to Malawi: recent developments in studying human polyomaviruses. J. Gen. Virol. 2013;94:482–496. doi: 10.1099/vir.0.048462-0. [DOI] [PubMed] [Google Scholar]
- 36.Dalianis T, Garcea RL. Welcome to the Polyomaviridae. Semin. Cancer Biol. 2009;19:209–210. doi: 10.1016/j.semcancer.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Jiang M, Abend JR, Johnson SF, Imperiale MJ. The role of polyomaviruses in human disease. Virology. 2009;384:266–273. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tooze J, Acheson NH. DNA Tumor Viruses. Cold Spring Harbor Laboratory; 1981. [Google Scholar]
- 39. Shuda M, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl Acad. Sci. USA. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. Mutations in MCPyV large T antigen, found in MCC, disable the ability of the virus to replicate viral origin-containing DNA.
- 40.Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. Cellular transformation by Simian Virus 40 and Murine Polyoma Virus T antigens. Semin. Cancer Biol. 2009;19:218–228. doi: 10.1016/j.semcancer.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raghava S, Giorda KM, Romano FB, Heuck AP, Hebert DN. The SV40 late protein VP4 is a viroporin that forms pores to disrupt membranes for viral release. PLoS Pathog. 2011;7:e1002116. doi: 10.1371/journal.ppat.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rozenblatt-Rosen O, et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature. 2012;487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krumbholz A, Bininda-Emonds OR, Wutzler P, Zell R. Phylogenetics, evolution, and medical importance of polyomaviruses. Infect. Genet. Evol. 2009;9:784–799. doi: 10.1016/j.meegid.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Losada M, et al. Comparing phylogenetic codivergence between polyomaviruses and their hosts. J. Virol. 2006;80:5663–5669. doi: 10.1128/JVI.00056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warden CD, Lacey SF. Updated phylogenetic analysis of polyomavirus-host co-evolution. J. Bioinfo. Res. 2012;1:46–49. [Google Scholar]
- 46.Campbell KS, et al. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 47.DeCaprio JA, et al. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 48.Stubdal H, et al. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, et al. Merkel cell polyomavirus large T antigen disrupts lysosome clustering by translocating human Vam6p from the cytoplasm to the nucleus. J. Biol. Chem. 2011;286:17079–17090. doi: 10.1074/jbc.M110.192856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sowd GA, Fanning E. A wolf in sheep’s clothing: SV40 co-opts host genome maintenance proteins to replicate viral DNA. PLoS Pathog. 2012;8:e1002994. doi: 10.1371/journal.ppat.1002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yardimci H, et al. Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012;492:205–209. doi: 10.1038/nature11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 2006;20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welcker M, Clurman BE. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J. Biol. Chem. 2005;280:7654–7658. doi: 10.1074/jbc.M413377200. [DOI] [PubMed] [Google Scholar]
- 54.Fine DA, et al. Identification of FAM111A as an SV40 host range restriction and adenovirus helper factor. PLoS Pathog. 2012;8:e1002949. doi: 10.1371/journal.ppat.1002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pallas DC, et al. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 56.Pallas DC, et al. The third subunit of protein phosphatase 2A (PP2A), a 55-kilodalton protein which is apparently substituted for by T antigens in complexes with the 36- and 63-kilodalton PP2A subunits, bears little resemblance to T antigens. J. Virol. 1992;66:886–893. doi: 10.1128/jvi.66.2.886-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. The finding that MCPyV small T antigen is expressed in most MCCs, is oncogenic and can promote phosphorylation of 4EBP1.
- 58.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J. Virol. 2008;82:9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Lee S, et al. Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. J. Clin. Virol. 2011;52:272–275. doi: 10.1016/j.jcv.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 62.Bauman Y, et al. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9:93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 63.Salunke DM, Caspar DLD, Garcea RL. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- 64.Montross L, et al. Nuclear assembly of polyomavirus capsids in insect cells expressing the major capsid protein VP1. J. Virol. 1991;65:4991–4998. doi: 10.1128/jvi.65.9.4991-4998.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cole CN, Landers T, Goff SP, Manteuil-Brutlag S, Berg P. Physical and genetic characterization of deletion mutants of simian virus 40 constructed in vitro. J. Virol. 1977;24:277–294. doi: 10.1128/jvi.24.1.277-294.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giorda KM, Raghava S, Hebert DN. The Simian virus 40 late viral protein VP4 disrupts the nuclear envelope for viral release. J. Virol. 2012;86:3180–3192. doi: 10.1128/JVI.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Neu U, Stehle T, Atwood WJ. The Polyomaviridae contributions of virus structure to our understanding of virus receptors and infectious entry. Virology. 2009;384:389–399. doi: 10.1016/j.virol.2008.12.021. An excellent review of the polyomavirus receptors.
- 68.Neu U, et al. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe. 2010;8:309–319. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Erickson KD, Garcea RL, Tsai B. Ganglioside GT1b is a putative host cell receptor for the Merkel cell polyomavirus. J. Virol. 2009;83:10275–10279. doi: 10.1128/JVI.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stehle T, Harrison SC. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 1997;16:5139–5148. doi: 10.1093/emboj/16.16.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stehle T, Yan Y, Benjamin TL, Harrison SC. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- 72.Neu U, Wang J, Macejak D, Garcea RL, Stehle T. Structures of the major capsid proteins of the human Karolinska Institutet and Washington University polyomaviruses. J. Virol. 2011;85:7384–7392. doi: 10.1128/JVI.00382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Neu U, et al. Structures of Merkel cell polyomavirus VP1 complexes define a sialic acid binding site required for infection. PLoS Pathog. 2012;8:e1002738. doi: 10.1371/journal.ppat.1002738. The latest data on structural differences in the VP1 proteins of the new human polyomaviruses.
- 74.Magaldi TG, et al. Mutations in the GM1 binding site of simian virus 40 VP1 alter receptor usage and cell tropism. J. Virol. 2012;86:7028–7042. doi: 10.1128/JVI.00371-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schowalter RM, Reinhold WC, Buck CB. Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS ONE. 2012;7:e42181. doi: 10.1371/journal.pone.0042181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gorelik L, et al. Progressive multifocal leukoencephalopathy (PML) development is associated with mutations in JC virus capsid protein VP1 that change its receptor specificity. J. Infect. Dis. 2011;204:103–114. doi: 10.1093/infdis/jir198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reid CE, et al. Sequencing and analysis of JC virus DNA from natalizumab-treated PML patients. J. Infect. Dis. 2011;204:237–244. doi: 10.1093/infdis/jir256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sunyaev SR, Lugovskoy A, Simon K, Gorelik L. Adaptive mutations in the JC virus protein capsid are associated with progressive multifocal leukoencephalopathy (PML) PLoS Genet. 2009;5:e1000368. doi: 10.1371/journal.pgen.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dang X, Wuthrich C, Gordon J, Sawa H, Koralnik IJ. JC virus encephalopathy is associated with a novel agnoprotein-deletion JCV variant. PLoS ONE. 2012;7:e35793. doi: 10.1371/journal.pone.0035793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Major EO, Amemiya K, Tornatore CS, Houff SA, Berger JR. Pathogenesis and molecular biology of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin. Microbiol. Rev. 1992;5:49–73. doi: 10.1128/cmr.5.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pfister LA, Letvin NL, Koralnik IJ. JC virus regulatory region tandem repeats in plasma and central nervous system isolates correlate with poor clinical outcome in patients with progressive multifocal leukoencephalopathy. J. Virol. 2001;75:5672–5676. doi: 10.1128/JVI.75.12.5672-5676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gosert R, et al. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J. Exp. Med. 2008;205:841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pastrana DV, et al. Neutralization serotyping of BK polyomavirus infection in kidney transplant recipients. PLoS Pathog. 2012;8:e1002650. doi: 10.1371/journal.ppat.1002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Freund R, Garcea RL, Sahli R, Benjamin TL. A single-amino-acid substitution in polyomavirus VP1 correlates with plaque size and hemagglutination behavior. J. Virol. 1991;65:350–355. doi: 10.1128/jvi.65.1.350-355.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bauer PH, et al. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J. Virol. 1995;69:7925–7931. doi: 10.1128/jvi.69.12.7925-7931.1995. The demonstration that MPyV spread in the host is dependent on receptor usage.
- 86.Bauer PH, et al. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J. Virol. 1999;73:5826–5832. doi: 10.1128/jvi.73.7.5826-5832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giroglou T, Florin L, Schafer F, Streeck RE, Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson KM, et al. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J. Virol. 2009;83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Generation of HPV pseudovirions using transfection and their use in neutralization assays. Methods Mol. Med. 2005;119:445–462. doi: 10.1385/1-59259-982-6:445. [DOI] [PubMed] [Google Scholar]
- 90.Schowalter RM, Pastrana DV, Buck CB. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011;7:e1002161. doi: 10.1371/journal.ppat.1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Broekema NM, Imperiale MJ. Efficient propagation of archetype BK and JC polyomaviruses. Virology. 2012;422:235–241. doi: 10.1016/j.virol.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dalianis T, Ramqvist T, Andreasson K, Kean JM, Garcea RLKI. WU and Merkel cell polyomaviruses: a new era for human polyomavirus research. Semin. Cancer Biol. 2009;19:270–275. doi: 10.1016/j.semcancer.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 93.Bialasiewicz S, et al. Presence of the newly discovered human polyomaviruses KI and WU in Australian patients with acute respiratory tract infection. J. Clin. Virol. 2008;41:63–68. doi: 10.1016/j.jcv.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bialasiewicz S, Whiley DM, Lambert SB, Nissen MD, Sloots TP. Detection of BK, JC, WU, or KI polyomaviruses in faecal, urine, blood, cerebrospinal fluid and respiratory samples. J. Clin. Virol. 2009;45:249–254. doi: 10.1016/j.jcv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 95.Han TH, Chung JY, Koo JW, Kim SW, Hwang ES. WU polyomavirus in children with acute lower respiratory tract infections, South Korea. Emerg. Infect. Dis. 2007;13:1766–1768. doi: 10.3201/eid1311.070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neske F, et al. WU polyomavirus infection in children, Germany. Emerg. Infect. Dis. 2008;14:680–681. doi: 10.3201/eid0104.071325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ren L, et al. WU polyomavirus in fecal specimens of children with acute gastroenteritis, China. Emerg. Infect. Dis. 2009;15:134–135. doi: 10.3201/eid1501.080693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Le BM, et al. Clinical and epidemiologic characterization of WU polyomavirus infection, St. Louis, Missouri. Emerg. Infect. Dis. 2007;13:1936–1938. doi: 10.3201/eid1312.070977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wattier RL, et al. Role of human polyomaviruses in respiratory tract disease in young children. Emerg. Infect. Dis. 2008;14:1766–1768. doi: 10.3201/eid1411.080394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rao S, Garcea RL, Robinson CC, Simoes EA. WU and KI polyomavirus infections in pediatric hematology/oncology patients with acute respiratory tract illness. J. Clin. Virol. 2011;52:28–32. doi: 10.1016/j.jcv.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Debiaggi M, et al. Molecular epidemiology of KI and WU polyomaviruses in infants with acute respiratory disease and in adult hematopoietic stem cell transplant recipients. J. Med. Virol. 2010;82:153–156. doi: 10.1002/jmv.21659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dang X, et al. Infrequent detection of KI, WU and MC polyomaviruses in immunosuppressed individuals with or without progressive multifocal leukoencephalopathy. PLoS ONE. 2011;6:e16736. doi: 10.1371/journal.pone.0016736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mourez T, et al. Polyomaviruses KI and WU in immunocompromised patients with respiratory disease. Emerg. Infect. Dis. 2009;15:107–109. doi: 10.3201/1501.080758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tolstov YL, et al. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg. Infect. Dis. 2011;17:1371–1380. doi: 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Norja P, Ubillos I, Templeton K, Simmonds P. No evidence for an association between infections with WU and KI polyomaviruses and respiratory disease. J. Clin. Virol. 2007;40:307–311. doi: 10.1016/j.jcv.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Payungporn S, et al. Prevalence and molecular characterization of WU/KI polyomaviruses isolated from pediatric patients with respiratory disease in Thailand. Virus Res. 2008;135:230–236. doi: 10.1016/j.virusres.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Toker C. Trabecular carcinoma of the skin. Arch. Dermatol. 1972;105:107–110. [PubMed] [Google Scholar]
- 108.Koljonen V, et al. Chronic lymphocytic leukaemia patients have a high risk of Merkel-cell polyomavirus DNA-positive Merkel-cell carcinoma. Br. J. Cancer. 2009;101:1444–1447. doi: 10.1038/sj.bjc.6605306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.de Giorgi V, Benemei S, Grazzini M, Lotti T, Geppetti P. Rapid growth of Merkel cell carcinoma during etanercept treatment of psoriatic arthritis: cause or coincidence? Acta Derm. Venereol. 2011;91:354–355. doi: 10.2340/00015555-1038. [DOI] [PubMed] [Google Scholar]
- 110.Paulson KG, Iyer JG, Nghiem P. Asymmetric lateral distribution of melanoma and Merkel cell carcinoma in the United States. J. Am. Acad. Dermatol. 2011;65:35–39. doi: 10.1016/j.jaad.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohnishi Y, et al. Merkel cell carcinoma and multiple Bowen’s disease: incidental association or possible relationship to inorganic arsenic exposure? J. Dermatol. 1997;24:310–316. doi: 10.1111/j.1346-8138.1997.tb02796.x. [DOI] [PubMed] [Google Scholar]
- 112.Sahi H, et al. Increased incidence of Merkel cell carcinoma among younger statin users. Cancer Epidemiol. 2012;36:421–424. doi: 10.1016/j.canep.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 113. Rodig SJ, et al. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J. Clin. Invest. 2012;122:4645–4653. doi: 10.1172/JCI64116. This study raises the possibility that all MCCs contain MCPyV.
- 114.Schmitt M, Wieland U, Kreuter A, Pawlita M. C-terminal deletions of Merkel cell polyomavirus large T-antigen, a highly specific surrogate marker for virally induced malignancy. Int. J. Cancer. 2012;131:2863–2868. doi: 10.1002/ijc.27607. [DOI] [PubMed] [Google Scholar]
- 115.Kraus I, et al. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 2008;68:2514–2522. doi: 10.1158/0008-5472.CAN-07-2776. [DOI] [PubMed] [Google Scholar]
- 116.Duncavage EJ, et al. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J. Mol. Diagn. 2011;13:325–333. doi: 10.1016/j.jmoldx.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shuda M, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int. J. Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bhatia K, Goedert JJ, Modali R, Preiss L, Ayers LW. Merkel cell carcinoma subgroups by Merkel cell polyomavirus DNA relative abundance and oncogene expression. Int. J. Cancer. 2010;126:2240–2246. doi: 10.1002/ijc.24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Houben R, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int. J. Cancer. 2012;130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 120.Arora R, et al. Survivin is a therapeutic target in Merkel cell carcinoma. Sci. Transl. Med. 2012;4:133ra56. doi: 10.1126/scitranslmed.3003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Busam KJ, et al. Merkel cell polyomavirus expression in merkel cell carcinomas and its absence in combined tumors and pulmonary neuroendocrine carcinomas. Am. J. Surg. Pathol. 2009;33:1378–1385. doi: 10.1097/PAS.0b013e3181aa30a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schrama D, et al. Merkel cell polyomavirus status is not associated with clinical course of Merkel cell carcinoma. J. Invest. Dermatol. 2011;131:1631–1638. doi: 10.1038/jid.2011.115. [DOI] [PubMed] [Google Scholar]
- 123.Paik JY, et al. Immunohistochemistry for Merkel cell polyomavirus is highly specific but not sensitive for the diagnosis of Merkel cell carcinoma in the Australian population. Hum. Pathol. 2011;42:1385–1390. doi: 10.1016/j.humpath.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 124.Sihto H, et al. Merkel cell polyomavirus infection, large T antigen, retinoblastoma protein and outcome in Merkel cell carcinoma. Clin. Cancer Res. 2011;17:4806–4813. doi: 10.1158/1078-0432.CCR-10-3363. [DOI] [PubMed] [Google Scholar]
- 125.Hafner C, et al. Activation of the PI3K/AKT pathway in Merkel cell carcinoma. PLoS ONE. 2012;7:e31255. doi: 10.1371/journal.pone.0031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nardi V, et al. Activation of PI3K signaling in Merkel cell carcinoma. Clin. Cancer Res. 2012;18:1227–1236. doi: 10.1158/1078-0432.CCR-11-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lill C, et al. P53 mutation is a rare event in Merkel cell carcinoma of the head and neck. Eur. Arch. Otorhinolaryngol. 2011;268:1639–1646. doi: 10.1007/s00405-011-1529-7. [DOI] [PubMed] [Google Scholar]
- 128.Waltari M, et al. Association of Merkel cell polyomavirus infection with tumor p53, KIT, stem cell factor, PDGFR-alpha and survival in Merkel cell carcinoma. Int. J. Cancer. 2011;129:619–628. doi: 10.1002/ijc.25720. [DOI] [PubMed] [Google Scholar]
- 129.Asioli S, Righi A, Volante M, Eusebi V, Bussolati G. p63 expression as a new prognostic marker in Merkel cell carcinoma. Cancer. 2007;110:640–647. doi: 10.1002/cncr.22828. [DOI] [PubMed] [Google Scholar]
- 130.Asioli S, et al. Expression of p63 is the sole independent marker of aggressiveness in localised (stage I–II) Merkel cell carcinomas. Mod. Pathol. 2011;24:1451–1461. doi: 10.1038/modpathol.2011.100. [DOI] [PubMed] [Google Scholar]
- 131.Hall BJ, et al. Immunohistochemical prognostication of Merkel cell carcinoma: p63 expression but not polyomavirus status correlates with outcome. J. Cutan. Pathol. 2012;39:911–917. doi: 10.1111/j.1600-0560.2012.01964.x. [DOI] [PubMed] [Google Scholar]
- 132.Lim CS, et al. Increasing tumor thickness is associated with recurrence and poorer survival in patients with Merkel cell carcinoma. Ann. Surg. Oncol. 2012;19:3325–3334. doi: 10.1245/s10434-012-2509-x. [DOI] [PubMed] [Google Scholar]
- 133.Higaki-Mori H, et al. Association of Merkel cell polyomavirus infection with clinicopathological differences in Merkel cell carcinoma. Hum. Pathol. 2012;43:2282–2291. doi: 10.1016/j.humpath.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 134.Paulson KG, et al. Antibodies to merkel cell polyomavirus T antigen oncoproteins reflect tumor burden in merkel cell carcinoma patients. Cancer Res. 2010;70:8388–8397. doi: 10.1158/0008-5472.CAN-10-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Iyer JG, et al. Merkel cell polyomavirus-specific CD8+ and CD4+ T-cell responses identified in Merkel cell carcinomas and blood. Clin. Cancer Res. 2011;17:6671–6680. doi: 10.1158/1078-0432.CCR-11-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Paulson KG, et al. Transcriptome-wide studies of merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J. Clin. Oncol. 2011;29:1539–1546. doi: 10.1200/JCO.2010.30.6308. This study shows that patients with MCC who have a strong immune response have an improved prognosis compared with immunocompromised patients.
- 137.Tolstov YL, et al. Lack of evidence for direct involvement of Merkel cell polyomavirus (MCV) in chronic lymphocytic leukemia (CLL) Blood. 2010;115:4973–4974. doi: 10.1182/blood-2010-03-273177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reisinger DM, et al. Lack of evidence for basal or squamous cell carcinoma infection with Merkel cell polyomavirus in immunocompetent patients with Merkel cell carcinoma. J. Am. Acad. Dermatol. 2010;63:400–403. doi: 10.1016/j.jaad.2009.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rollison DE, et al. Case-control study of Merkel cell polyomavirus infection and cutaneous squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2012;21:74–81. doi: 10.1158/1055-9965.EPI-11-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Abend JR, Jiang M, Imperiale MJ. BK virus and human cancer: innocent until proven guilty. Semin. Cancer Biol. 2009;19:252–260. doi: 10.1016/j.semcancer.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pino L, et al. Bladder transitional cell carcinoma and BK virus in a young kidney transplant recipient. Transpl. Infect. Dis. 2012;15:e25–e27. doi: 10.1111/tid.12042. [DOI] [PubMed] [Google Scholar]
- 142.Vilkin A, et al. Presence of JC virus DNA in the tumor tissue and normal mucosa of patients with sporadic colorectal cancer (CRC) or with positive family history and Bethesda criteria. Dig. Dis. Sci. 2012;57:79–84. doi: 10.1007/s10620-011-1855-z. [DOI] [PubMed] [Google Scholar]
- 143.Garcea RL, Imperiale MJ. Simian virus 40 infection of humans. J. Virol. 2003;77:5039–5045. doi: 10.1128/JVI.77.9.5039-5045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Poulin DL, DeCaprio JA. Is there a role for SV40 in human cancer? J. Clin. Oncol. 2006;24:4356–4365. doi: 10.1200/JCO.2005.03.7101. [DOI] [PubMed] [Google Scholar]
- 145.Dela Cruz FN, Jr, et al. Novel polyomavirus associated with brain tumors in free-ranging Raccoons, Western United States. Emerg. Infect. Dis. 2013;19:77–84. doi: 10.3201/eid1901.121078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Swanson PA, Lukacher AE, Szomolanyi-Tsuda E. Immunity to polyomvirus infection: the polyomavirus–mouse model. Semin. Cancer Biol. 2009;19:244–251. doi: 10.1016/j.semcancer.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.zur Hausen H, Gissmann L. Lymphotropic papovaviruses isolated from African green monkey and human cells. Med. Microbiol. Immunol. 1979;167:137–153. doi: 10.1007/BF02121180. [DOI] [PubMed] [Google Scholar]
- 148.Kanda T, Takemoto KK. Monkey B-lymphotropic papovavirus mutant capable of replicating in T-lymphoblastoid cells. J. Virol. 1985;55:96–100. doi: 10.1128/jvi.55.1.96-100.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Woolford L, et al. A novel virus detected in papillomas and carcinomas of the endangered western barred bandicoot (Perameles bougainville) exhibits genomic features of both the Papillomaviridae and Polyomaviridae. J. Virol. 2007;81:13280–13290. doi: 10.1128/JVI.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sehr P, Zumbach K, Pawlita M. A generic capture ELISA for recombinant proteins fused to glutathione S-transferase: validation for HPV serology. J. Immunol. Methods. 2001;253:153–162. doi: 10.1016/s0022-1759(01)00376-3. [DOI] [PubMed] [Google Scholar]
- 151.Viscidi RP, Clayman B. Serological cross reactivity between polyomavirus capsids. Adv. Exp. Med. Biol. 2006;577:73–84. doi: 10.1007/0-387-32957-9_5. [DOI] [PubMed] [Google Scholar]
- 152.Randhawa P, et al. Identification of species-specific and cross-reactive epitopes in human polyomavirus capsids using monoclonal antibodies. J. Gen. Virol. 2009;90:634–639. doi: 10.1099/vir.0.008391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Waterboer T, et al. Multiplex human papillomavirus serology based on in situ purified glutathione S-transferase fusion proteins. Clin. Chem. 2005;51:1845–1853. doi: 10.1373/clinchem.2005.052381. [DOI] [PubMed] [Google Scholar]
- 154.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Carter JJ, et al. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J. Natl Cancer Inst. 2009;101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gorelik L, et al. Anti-JC virus antibodies: implications for PML risk stratification. Ann. Neurol. 2010;68:295–303. doi: 10.1002/ana.22128. [DOI] [PubMed] [Google Scholar]
- 159.Faust H, Pastrana DV, Buck CB, Dillner J, Ekstrom J. Antibodies to Merkel cell polyomavirus correlate to presence of viral DNA in the skin. J. Infect. Dis. 2011;203:1096–1100. doi: 10.1093/infdis/jiq173. [DOI] [PubMed] [Google Scholar]
- 160.van der Meijden E, et al. Seroprevalence of trichodysplasia spinulosa-associated polyomavirus. Emerg. Infect. Dis. 2011;17:1355–1363. doi: 10.3201/eid1708.110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nicol JT, et al. Seroprevalence and cross-reactivity of human polyomavirus 9. Emerg. Infect. Dis. 2012;18:1329–1332. doi: 10.3201/eid1808.111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Trusch F, et al. Seroprevalence of human polyomavirus 9 and cross-reactivity to African green monkey-derived lymphotropic polyomavirus. J. Gen. Virol. 2012;93:698–705. doi: 10.1099/vir.0.039156-0. [DOI] [PubMed] [Google Scholar]