Abstract
Hepatocellular carcinoma (HCC) is a primary malignancy of the liver of global importance. Recent studies of the expression and role of microRNA (miRNA) in HCC are providing new insights into disease pathogenesis. In addition, therapeutic efforts targeting specific miRNAs are being evaluated in animal models of HCC. The potential of miRNAs as biomarkers of disease or prognostic markers is being explored. Herein, we review studies of miRNA expression in human HCC, and discuss recent advances in knowledge about the involvement and role of selected miRNAs in disease pathogenesis, as biomarkers, or as therapeutic targets for HCC.
Hepatocellular cancer (HCC) is a primary malignancy arising within the liver. This disease is the third most common cause of cancer deaths in men worldwide. The incidence and mortality of HCC are increasing in many countries. These concerning trends are the opposite of those for several other cancers.1 The data underscore the unmet need for effective treatments for HCC. HCC often arises within a diseased liver and represents a lethal complication of cirrhosis. The poor prognosis of HCC patients reflects the lack of understanding of liver carcinogenesis and the failure to develop interventions that are aimed at blocking or reversing the steps of malignant transformation. Molecular targeted therapies focused on critical pathways are needed. Indeed, global gene expression profile studies have revealed the activation of several pathways that drive uncontrolled growth and aberrant survival signaling. Several key pathways have been identified. Recently, the mixed kinase inhibitor sorafenib has been approved for use in HCC. However, no novel gene markers or molecular prognostic factors have entered into clinical practice, emphasizing the need for new markers for diagnosis and prognosis of HCC patients.
microRNA EXPRESSION IN LIVER CANCERS
MicroRNAs (miRNAs) are a family of genes encoding small RNA molecules that play key roles in controlling gene expression. Mature miRNAs are the result of sequential processing of primary miRNA transcripts (pri- miRNA), mediated by two RNA III enzymes that act either in the nucleus (Drosha) or in the cytoplasm (Dicer).2 Mature miRNAs can negatively regulate protein expression of specific mRNA by either translational inhibition or mRNA degradation. These mechanisms have been extensively reviewed elsewhere.3 Widespread alterations of miRNAs occur across the human genome in a broad array of human cancers, and miRNA expression has been implicated in cancer genesis and progression. In fact, miRNAs may function either as tumor-suppressor genes or as oncogenes, by targeting and silencing mRNAs involved in carcinogenesis. Recent studies show that miRNA expression can be more useful than mRNA based profiling for identifying tissue type of tumor origin.
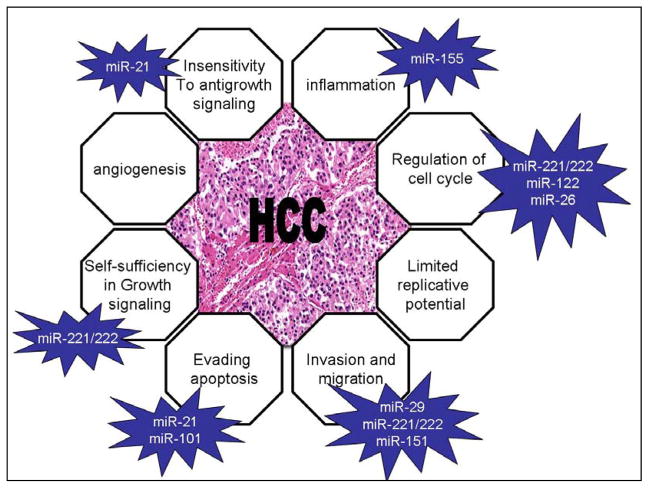
miRNAs might contribute to tumor progression, by modulating the expression of gene products involved in phenotypic characteristics of malignant cells, such as invasion and uncontrolled growth (Figure 1). Murakami et al observed that some miRNAs are associated with the degree of differentiation in hepatocellular carcinoma. MiR-222, miR-106a, miR-92, miR-17–5p, miR-20, and miR-18 are increased in poorly versus moderately versus well-differentiated hepatomas.4 The finding of alterations in miRNA and target gene expression in HCC and other cancers highlights potential mechanisms of tumor progression caused by a loss of ability to control proliferation or by gain of ability to survive in hostile environments related to these genetic changes in tumor cells. A comparison between nonmetastatic versus metastatic HCCs performed by Budhu et al5 identified a few miRNAs that are upregulated in metastatic tumors (miR-219 –1, miR-207, miR-38, miR-185) but whose targets are still unknown. Conversely, a long list of downregulated miRNAs has been identified in metastatic liver cancers. Budhu et al identified a 20- miRNAs signature that seems to be a significant independent predictor of patients’ survival and recurrence. In particular, high miR-219 and miR-207 and low miR-124a and miR-30c correlate with venous metastasis and poor survival. Consistently, miR-30 expression was found to be higher in HCC patients with tumor metastasis than in those without.6 Jiang et al7 identified a
Figure 1.
Several miRNAs act synergistically to promote HCC through the modulation of multiple cell phenotypes.
19-miRNA signature, including miR-26, miR-29, and miR-221, that correlates with survival risk. Recently, miR-26 was identified as an indicator of survival in the largest study of miRNA expression in HCC.8 These findings are extremely important in view of their relevance in clinical activity. HCC has a dismal outcome, mainly due to a high relapse rate after surgery. The possibility of using new miRNA-based predictive and prognostic factors might help to select patients who need adjuvant and/or more aggressive therapy.
MECHANISMS OF miRNA ALTERATION IN HCC
Genetic and Epigenetic Alterations
The causes of the widespread miRNA mis-expression in cancers are not clearly understood; however, the origins of such abnormalities seem to be multiple. Calin et al9 investigated the association between various cytogenetic and molecular abnormalities and the location of miRNA genes and found that more than half of miRNA genes were located in cancer-associated genomic regions. Sixty-five miRNAs were located exactly in minimal regions of loss of heterozygosity (LOH) and 15 miRNAs in minimal regions of amplification, described in a variety of tumors, including lung, breast, ovarian, colon, gastric, and hepatocellular carcinomas, as well as leukemias and lymphomas. In addition, looking at 113 fragile sites (FRA) scattered in the human karyotype, they found that 61 miRNAs were located in the same cytogenetic positions with FRA.9 The first suggestion of a relation between miRNA location and chromosomal alterations came from studying hematologic malignancies where translocations are frequent events. MiR-15a and miR-16 –1, which are downregulated in approximately 68% of chronic lymphocytic leukemia (CLL) cases, were found to reside in 13q14.3, a region that is frequently deleted in CLL.10 Based on this evidence we might speculate that miRNA deregulation could be related to chromosomal alterations in HCC. The region where miR-122a is encoded (18q21.31) is frequently lost in HCC, suggesting that LOH might be responsible for the downregulation of miR-122 in liver cancer. miR-151 is located in a site (8q24.3) that is frequently amplified in HCC. Indeed, miR-151 expression is correlated with intrahepatic metastasis of HCC and increases HCC cell migration and invasion by directly targeting RhoGDIA.11 The dimensions of miRNAs are quite small in respect to the cDNAs of protein-coding genes, and therefore mutations in miRNA transcripts are likely to be rare events. A combination of LOH–mutation was reported to inactivate “two-hit” events in two cases of CLL.12 Yang et al13 analyzed sequence variations of 59 miRNAs in HCC and reported that mutation of miRNA is a sporadic event in HCC and thus may not represent a main mechanism underlying hepatocarcinogenesis.
Single-nucleotide polymorphisms (SNPs) in miRNA- encoded regions have been associated with susceptibility to lung cancer, breast cancer, and congenital heart disease.14 Recent studies in Chinese populations showed that miR-196A2 polymorphism was associated with susceptibility to hepatitis B virus (HBV)-related HCC, but these data were observed only in males.15 Conversely, the combination of nonrandom chromosomal abnormalities and other types of epigenetic events could contribute to downregulation or overexpression of miRNAs. It was proved that DNA demethylation and histone deacetylase inhibition can activate expression of miR-127, which may act as a tumor suppressor by targeting the B-cell leukemia/lymphoma 6 proto-oncogene.16 We showed that the pro-inflammatory cytokine interleukin-6 (IL-6) may potentially regulate miR-370 expression in cholangiocarcinoma cells, by altering the methylation status of its promoter.17 Analogously, promoter methylation was shown to be responsible for the downregulation of miR-1 in HCC.18 We also observed that the overexpression of miR-34a in HCC might be due to a general status of hypomethylation.19 Thus, modulation of the methylation of miR promoters may be responsible for miR mis-expression in HCC and prompts the clinical evaluation of epigenetics drugs in liver cancer.
Viral Disease
Chronic infection with HBV predisposes to the development of HCC. A salient feature of these chronic infections is the integration of subgenomic HBV-DNA fragments into many different locations within the host DNA. HBV integration is considered one of the events involved in hepatocarcinogenesis. Several critical genes involved in control of cell growth and adhesion, such as cyclin A1, retinoic acid receptor, calmodulin 1, MLL2, SERCA-1, and p53, have been identified adjacent to integrated HBV-DNA. However, because HBV-DNA integrates into random positions of the human genome, all HBV- DNA integration does not directly transform hepatocytes. Recently, the discovery of noncoding RNAs suggested the hypothesis that HBV-DNA integration can exert an oncogenetic role not only by promoting genetic instability with alteration of protein encoding genes but also by modulating the expression of regulatory genes such as miRNAs.
HBV integration is associated with genetic instability at different locus sited near FRA.20 For example, HBV- DNA integrates at 1p36, near FRA1A. Liver dysplastic nodules reportedly have gains in 1p and 1q,21 but during tumor progression there is LOH in these regions,22 suggesting that the consequences of HBV integration and associated genetic instability are often dynamic during carcinogenesis. MiR-34a is encoded on chromosome 1p36 and is frequently upregulated in HCC (Table 1). The mechanism by which this miRNA is upregulated is still under investigation. p53, which seems to positively regulate miR-34 in other tumor types, is often silenced in HCC and thus the relationship between miR-34a and p53 in HCC is not fully understood. HBV-DNA integration near the miR-34a site might provide a further reasonable explanation for the upregulation of this miRNA in HCC. Likely, HBV integration acts synergistically on different regulatory genes that have an additional effect on cell proliferation and transformation. In fact, integration at FRA1A may also silence expression of miRNA-200a, which is known to be decreased in HCC compared to non-tumor tissue (Table 1).
Table 1.
miRNA expression profiling studies in human liver tissues
| Reference | Human tissue | differentially expressed miRNA | ||
|---|---|---|---|---|
| Author | HCC | control liver (adjacent or normal) | UP in HCC | DOWN in HCC |
| Murakami | 24 | 22 | mir-18, miR-224, miR-221/222, miR-20, miR-106 | mir-199a, miR-195, miR-200a, miR-125 |
| Pineau | 104 | 111 | mir-106b, miR-21, miR-210, miR-221/222, miR-34a, miR-425, miR-519, miR-93, miR-96 | let-7c |
| Gramantieri | 17 | 21 | mir-221/222, miR_21 | let-7, miR-122, miR-199a, miR- 200a, miR-223, miR-155 |
| Li-Fu | 20 | 20 | mir-21, miR-182, miR-200c | mir-101, miR-148a, miR-422, miR-41 |
| Jiang | 43 | 43 | mir-21, miR-18, miR-33, miR-130b, miR_135, miR-221/222, miR-301 | mir-101, miR-199a, miR-139, miR-150, miR-200b, miR-214, miR-224 |
| Li | 78 | 88 | mir-181a, miR-221/222, miR-25, miR-34a, miR-125 | mir-101, miR-148a, miR-214, miR-29c, miR-424, miR-224 |
| Su | 5 | 3 | mir-18, miR-25, miR-93, miR-127-3p, miR-210, miR-216, miR-222, miR-224, miR-326-5p, miR-382, miR-491-5p, miR- 527 | mir-29c, miR-99a, miR-100, miR-101, miR-125b, miR-199a, miR-215, miR-223, miR-424 |
| Wang | 19 | 19 | mir-224, miR-182, miR-183, miR-96, miR-9, miR-221/222, miR-216, miR-21, miR-186, miR-301, miR-155, miR-25, miR-324, miR-137, miR-364, miR-151 | mir-214, miR-145, miR-139 |
| Connolly | 4 | 4 | mir-20, miR-93, miR-18a, miR-148a, miR-106b, miR-324, miR-19b, miR-17, miR- 27, miR-21, miR-15a, miR-92 | mir-22, miR-99a, miR-130a, miR-126, let-7a, miR-122 |
Interestingly, other mechanisms involving miRNA may contribute to interactions between HBV and hepatocytes. Jin et al23 have recently shown that HBV encodes a miRNA in the precore– core region of its genome. Although it was speculated that this viral (v)- miRNA could affect the expression pattern of host cell genes, only the viral polymerase gene was experimentally found as its target. Therefore, conversely to what happens to v-miRNAs encoded by other viruses, the HBV-miRNA does not seem to participate in the hepatitis-induced transformation of hepatocytes. However, it might influence the capability of the virus to proliferate and integrate into the host genome, modulating indirectly the expression of cellular miRNAs as described above.
HBV-related HCCs have higher levels of miR-143 expression, whose induction seems to be caused by nuclear factor kappaB (NF-KB) activation. miR-143 was shown to promote HCC metastasis in an animal model by targeting the fibronectin type III domain containing the 3B gene, which regulates cell motility.24
In the Western world, most cases of HCC are related to hepatitis C virus (HCV) infection. Remarkable effort has been put into assessing the effects of miRNAs on HCV replication (see below). However, additional evidence shows that miRNA expression may be modulated by HCV. Several miRNA expression profiling studies show that HCV-related HCCs have a different miRNA expression signature when compared to HCV- negative HCCs. Ura et al25 observed that some miRNAs, including miR-26 and miR-29, were commonly repressed in HBV- or HCV-related HCC in comparison to normal liver tissues. Conversely, the expression of other miRNAs could differentiate HBV- from HCV-related HCCs, and thereby suggest differences in the mechanisms by which these viruses may modulate miRNA expression.25 Studies in HCV-infected HCC cell lines confirmed the HCV-dependent modulation of miRNA expression.26–28 Less clear is the mechanism by which HCV modulates miRNA expression. We have shown that overexpression of HCV-proteins can modulate miRNA expression. Upregulation of miR-193b in HCV-positive cells may regulate apoptosis by modulating the expression of the anti-apoptotic protein Mcl-1.28
miR-155 was found to be overexpressed in HCV- replicating cells.26 Interestingly a correlation between pre-miR-155 (BIC) and serum HCV RNA has been recently described in HCV-infected patients. The lowest expression of BIC was found in patients that eliminated HCV RNA from both serum and macrophages.29 Even though additional studies are warranted to evaluate the interaction between HCV and miR-155, these data are promising for the development of novel miRNA-based molecular markers in the assessment of HCV-positive patients.
SELECTED miRNAs AND ROLE IN PATHOGENESIS OF LIVER CANCERS
miR-221/-222
miR-221 and miR-222 are encoded in tandem from a gene cluster located on chromosome X (Xp11.3) and have identical 5= regions that enable them to target the same genes.30 They behave as oncogenes in several malignancies. In HCC miR-221 and miR-222 were found to be significantly upregulated when compared with adjacent liver tissues (Table 1). Murakami et al4 compared miRNA expression in tumors with different degrees of differentiation and found miR-222 as one of those miRNAs that significantly increases in poorly differentiated HCC. Recently, miR-221 and miR-222 were found to be consistently dysregulated through each step going from human normal liver to cirrhosis to full blown HCC.31 These miRNA enhance tumorigenesis in vitro and in vivo by regulating several targets. HCC has a disrupted expression of p27Kip1, a member of the Cip/Kip family of cyclin-dependent kinase (CDK) inhibitors that function to negatively control cell cycle progression.32 Recent data strongly support the hypothesis that miR-221/-222 regulate p27Kip1 expression at a post-transcriptional level.31,33 Cell cycle progression is also affected by miR-221 through the modulation of CDKN1C/p57, which was shown to correlate with a higher biological aggressiveness, advanced stage, poor differentiation, larger size, portal invasion, and high proliferative activity in HCC. Furthermore, a low CDKN1C/p57 labeling index is associated with worse outcomes and lower disease-free survival after surgery, suggesting that miR-221/-222– dependent downregulation of CDKN1C/p57 might contribute to the progression of HCC.34
In addition to the modulation of cell cycle, miR-221 contributes to the progression of HCC through the inhibition of apoptosis. Bmf is a pro-apoptotic member of theBcl-2 family that triggers caspase activation in HCC. Upregulation of miR-221 is responsible for the lack of Bmf expression in human HCC tissues.35 How- ever, the multiple targets of miR-221 may explain why HCCs with higher miR-221 levels were associated with a shorter time to recurrence, whereas low Bmf levels were associated with a trend toward shorter time to recurrence; this difference did not achieve statistical significance.35 Furthermore, miR-221/-222 can affect the expression of DNA damage inducible transcript 4 which is a modulator of the mammalian target of rapamycin (mTOR) pathway.31 Wong et al36 studied gene expression profiling in HCC cells treated with anti– miR-222 and observed that Akt signaling was the major pathway implicated in the miR-222–affected biological networks. They further proved protein phosphatase 2A subunit B to be a target of miR-222.36 Garofalo et al37 showed the miR-221/-222 target PTEN and TIMP3 tumor suppressors, and enhance cellular migration through the activation of the AKT pathway and metallopeptidases. Thus, it is likely that miR-221/-222 can act synergistically on several targets, which results in the modulation of the PI3K-akt-mTOR pathway.
The MET proto-oncogene encodes the tyrosine kinase receptor for hepatocyte growth factor (HGF), a potent mitogen and motogen for epithelial cells. MET is overexpressed in HCC. Patients with high MET HCC have a significantly shorter 5-year survival than patients with low MET HCC.38 Recent evidence suggests that MET induces miR-221/-222 through AP-1 transcription factor, providing the molecular basis for the prognostic significance of miR-221/-222.37
miR-122
One of the most abundant miRNAs in the liver is miR-122a. It is expressed in normal hepatocytes but is downregulated in up to 70% of human HCCs (Table 1). MiR-122 is described as a liver-specific miRNA. The expression of miR-122 can be used as a marker of liver development and it undergoes liver-specific expression during embryonic development. A role for miR-122 in hepatocarcinogenesis is suggested by the differential expression of miR-122a in HCC versus non-tumor cirrhotic hepatic tissue, and by the studies by Kutay et al39 of miRNA expression in murine HCC. In these studies, miR-122a was decreased by 50% between 36 and 54 weeks of a folate-methyl– deficient diet, when neoplastic transformation also occurred. In fact, none of these animals that were switched to a folate-methyl–adequate diet after 36 weeks developed HCC or had aberrant expression of miR-122a in the liver.
Deregulation of cell cycle progression can contribute to hepatocarcinogenesis. Cyclin G1 promotes cell cycle progression and it may be associated with genomic instability. In HCC cells, an inverse correlation has been observed between miR122a and cyclin G1, which is a validated target of this miRNA. In experimental hepatocarcinogenesis, loss of cyclin G1 is associated with a significantly lower tumor incidence after carcinogenic challenges.40 Thus, deregulated expression of cyclin G1, in response to altered miR122a expression, could contribute to the pathogenesis of liver cancer.
While the adult liver is the major site for miR-122, CAT-1 (cationic amino acid transporter-1) is universally expressed in all adult mammalian tissues except the liver. During mouse embryonic development, miR-122 becomes expressed in the liver as CAT-1 mRNA begins to decrease, suggesting that CAT-1 mRNA could be an in vivo target for degradation directed by miR-122.41
CAT-1 is a carrier protein required in the regenerating liver for the transport of cationic amino acids and polyamines in the late G1 phase, a process that is essential for liver cells to enter mitosis.41 Given these data, modulation of expression of CAT-1 may be another mechanism through which this miRNA regulates cell cycle in normal and transformed hepatocytes. Additionaly studies by Bai et al42 identified several other targets of miR-122, including ADAM10, SRF, and IGFR1. All of these proteins are upregulated in HCC and are involved in the modulation of cell adhesion, motility, and growth, providing additional evidence for the contribution of miR-122 to hepatocarcinogenesis.
These observations acquire clinical relevance in view of the correlation between overall survival and miR-122 expression levels in human HCCs.43 Interestingly, miR-122 was found to be repressed in HCCs that harbor an hepatoblast signature, which is specific for poor differentiation, or c-met and late transforming growth factor-13 (TGF-13) signatures, which are associated with poor prognosis. In support of previous findings, miR-122 loss was also associated with high proliferation and low apoptotic features.43 Together, these data suggest that loss of function of miR-122 can pre- dispose hepatocytes to neoplastic transformation and contribute to disease progression and worsening of patients’ outcomes. Therapeutic strategies aimed at re- expressing miR-122 have resulted in inhibition of tumor growth, and, moreover, enforced expression of miR-122 in HCC cells has been shown to enhance sorafenib-induced apoptosis, providing a rationale for future studies to test the therapeutic potential of a combination of a miR-122 mimetic and sorafenib.42
miR-122 may also facilitate HCV replication, promoting the development of cirrhosis and liver cancer. A binding site for miR-122 was identified in the 5″ non- coding region of HCV RNA and is highly conserved in all six HCV genotypes. Studies with replication-defective viral RNAs demonstrated that miR-122 affected RNA abundance by positively modulating RNA replication.27,44–46 Conversely, functional inactivation of miR-122a led to 80% reduction of HCV RNA replication, suggesting that loss of miR-122a in HCC may increase resistance of cancer cells to HCV replication. This also explains why HCV RNA can replicate only in miR-122– positive hepatoma cells, such as Huh7 cells, and not in HepG2 cells, which lack miR-122 expression.45 Recently, it was shown that the antiviral effect of interferon-13 against HCV is mediated by some induced miRNAs, including miR-196 and miR-448, which might act synergistically with the downregulated miR-122 to prevent replication of the HCV genome.47
These studies are interesting not only because they highlight a link between miRNA expression and HCV replication, but also because for the first time miRNAs are shown to target the 5″ -end region of target RNA and to increase its expression rather than suppress it. Although a common presumption is that miRNAs can behave as oncogenes or oncosuppressors depending on their target genes, enhancing target gene expression may be important for selective targets.
miR-26
Growing evidence suggests a role for miR-26 as a mediator of liver cancer progression. Selective activatio of the oncogene MYC in the liver results in the development of hepatocarcinomas. miRNA expression profiling studies in tumor tissues from transgenic animals with selective hepatic MYC activation showed a remarkable repression of miR-26a compared to the expression in the liver of control mice. Tumor-suppressor properties of this miRNA are supported by the induction of cell cycle arrest in HCC cells with enforced expression of miR-26. These effects are mediated by the direct modulation of cyclin E2 and cyclin D2. In vivo delivery of miR-26 reduced cellular proliferation and increased apoptosis in mouse liver tumors, suggesting a promising role for a miR-26 mimetic-based therapeutics.48 In support of a role of miR-26 in liver carcinogenesis is the observed difference in miR-26 expression between sexes. Males are significanlty more at risk of developing HCC than females. Recent evidence linked this discrepancy to the hormonal-dependent different levels of IL-6.49 Profiling studies in a cohort of 224 HCC patients revealed significantly higher levels of miR-26 expression in the non-tumoral liver tissue of women compared to men. Although this difference has not been observed in tumor tissues, miR-26 expression is reduced in HCC compared to non-tumoral liver regardless of sex. Moreover, tumors with lower levels of miR-26 expression have shorter overall survival.8 A multivariate analysis validated miR-26 as a prognostic marker for HCC and confirmed previous studies of gene expression also in the liver tissue surrounding HCC.50 In accordance with previous data, tumors with low miR-26 expression showed a distinct mRNA expression compared with tumors with high miR-26. Enrichment of IL-6 and NF-KB genes was noted, highlighting an important role for this cytokine in HCC progression (Figure 2). Along with IL-6, many other immune-related genes (such as those encoding for other inflammatory cytokines) are activated in low miR-26 tumors, providing an interesting link between miR-26 and immune response. This is in line with the correlation found between miR-26 expression and the response to interferon-a in HBV-related HCC.8 Thus, miR-26 might represent a promising prognostic marker for HCC, able to distinguish those tumors that are characterized by a poorer outcome but a favorable response to adjuvant therapy with interferon-a.
Figure 2.
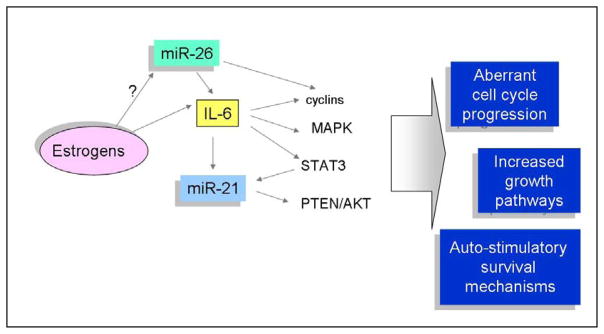
The inflammatory cytokine Interleukin 6 (IL-6) plays an important role in hepatocarcinogenesis through the modulation of STAT3 pathway, MAPK route and cell cycle progression. Growing evidence suggests a role for miRNAs in the regulation of IL-6. miR-26 was shown to be inversely related to IL-6 expression in a cohort of Chinese HBV-related HCC where low expression of miR-26 correlated with dismal outcome. Liver expression of miR-26 is higher in women than men. Consistently, estrogen-dependent IL-6 over-expression in female rats was shown to be responsible for the lower incidence of HCC in female. The oncogenic miR-21 was shown to be induced by IL-6 in a STAT-3-dependent mechanism, suggesting an auto-stimulatoruy loop.
miR-21
miR-21 is consistently identified as being significantly overexpressed in HCC cells when compared to normal liver and cirrhotic cells both in animals and in humans. An important role for miR-21 in growth regulation is suggested by studies in which it was shown to regulate cell cycle progression and apoptosis.51,52 The role of miR-21 has been studied in several different pathologic conditions and processes that are characterized by an increased cell proliferation. However, the role of aberrant expression of miR-21 has been studied most widely in tumors. The contribution of this miR-21 to the pathogenesis of cancer is suggested by studies showing deregulated expression in several neoplasms. Aberrant expression has been observed in cervical, head and neck, pancreatic, and breast cancers, as well as in glioblastoma, cholangiocarcinoma, CLL, diffuse large B-cell lymphoma, and other tumors.53 In HCC, miR-21 is highly overexpressed (Table 1) and can contribute to tumor growth and spread by modulating the expression of gene products involved in phenotypic characteristics of cancer cells such as cell growth, migration, and invasion.51 miR-21 was found to be increased in the early stages of liver regeneration, suggesting a contribution of this miRNA to hepatocyte proliferation.54 miR-21 was shown to target PTEN in HCC cells. PTEN is a tumor-suppressor gene that is frequently decreased in liver cancer and correlates with poor prognosis. Overexpression of miR-21 was found to silence PTEN, leading to phosphorylation of FAK and metalloproteinases, which resulted in promotion of cell migration and invasion.51 Other targets of miR-21 include the programmed cell death 4 (PDCD4) gene, which is reduced in HCC; peli-1, which regulates NF-KB signaling; and tropomyosin 1, a tumor-suppressor protein reduced in HCV-associated HCC.52,55 The anti-apoptotic activity of miR-21 is also supported by the increased expression of this miRNA following cytotoxic drugs. In fact, we have previously shown that miR-21 modulates gemcitabine-induced apoptosis by PTEN-dependent activation of PI3-kinase signaling in cholangiocarcinoma cells.56 Rossi et al57 have shown that 5-fluorouracil (5-FU) upregulates miR-21 in colorectal cancer cells and suggested that the drug- induced miRNA dysregulation could be the expression of cellular response to the toxic effects of 5-FU. In view of these results, miR-21 might be further studied as a potential target of therapeutic strategies. The combination of anti–miR-21 therapy with more classical chemo- therapy agents might help to overcome chemoresistance, promote cell death, and limit liver cancer growth and metastasis. HCC represents a classical case of inflammation- linked cancer. IL-6 isa multifunctional cytokine that mediates the hepatic response to systemic inflammation and likely contributes to hepatocarcinogenesis.58
High serum level of IL-6 seems to be a risk factor for the development of HCC.49,59 Recent data that postulate a potential role for miR-21 in the regulation of inflammatory cytokine production and the evidence of an IL-6 – dependent induction of miR-21 in myeloma cells60 suggest a link between overexpression of miR-21 and cytokines in the development and maintenance of liver cancer.
miR-155
niR-155 was found within the BIC gene on chromosome 21 in humans. The genomic structure of human BIC consists of three exons, However, it lacks a large open reading frame and therefore its sole function may be to give rise to miR-155 encoded within exon 3. MiR-155 is involved in regulating the innate immune response. It is induced by inflammatory stimuli such as the bacterial endotoxin61 and it has been identified as a target of synthetic viral intermediates and of interferon-13, the host antiviral response cytokine.62 These data indicate that immune cells, such as macrophages, respond to viral cues by strongly upregulating miR-155 and suggest that miR-155 is a component of the primary macrophage response to different types of inflammatory mediators. It appears that miR-155 exerts control of the mammalian immune system, at least in part, by regulating cytokine production.63 miR-155 is up- regulated in diffuse large B-cell lymphoma, CLL, Hodgkin disease, and pancreatic and colorectal cancers, showing that it can behave as an oncogene in several different malignancies.64, HCC frequently develops in a background of chronic inflammatory disease of the liver. As a component of the inflammatory response, miR-155 links inflammation and cancer. miR-155 was upregulated in mice treated with a choline-deficient, low-methionine amino acid (CDAA) diet, which induces steatohepatitis at early stages and leads to the formation of pre-neoplastic nodules after 65 weeks and hepatocellular adenomas and carcinomas after 84 weeks.65 miR-155 expression level correlated positively with the number of miR-155–positive cells, as well as the extent of inflammation in these mice, suggesting that miR-155 could play a causal role in CDAA diet–induced pathogenesis. In support of this hypothesis, miR-155 was found to be increased in human HCC, while its tumor-suppressor target C/EBPB was reduced.65 In addition, miR-155 was significantly upregulated in chronic liver disease compared to normal liver and high miR-155 expression in non-tumoral tissues was associated with poor overall survival in HCC patients.66
miR-199a* (miR-199a-3p)
The miR-199a gene is located on chromosomes 1 and 19. miR-199a-1 is located in an intron of the Dynamin 2 (DNM2) gene on chromosome 19p13.2. miRNA genes may be expressed as either the mature strand or the miRNA* strand. Although the miRNA strand is generally expressed at higher levels than the miRNA* strand, this is not the case for the miR-199a gene in hepatic tissues, where miR-199a* expression is 100- to 300-fold higher than miR-199a expression.7 Three independent studies showed that miR-199a and miR-199a* are reduced by approximately twofold in HCC compared to adjacent benign and nondiseased liver.4,7,40 We have assayed miR-199a and miR-199a* by quantitative polymerase chain reaction (qPCR) in a variety of HCC cell lines (Skep-1, SNU449, SNU182, HepG2, Hep38, Huh7, and PLC/PRF5) and have shown that neither mature miR-199a nor miR-199a* is detectable in any of these HCC cell lines nor was any found in the 37 cell lines assayed from a variety of tumor types including breast, lung, colorectal, prostate, head and neck, pancreas cancers, and hematologic malignancies (unpublished data). Therefore, miR-199a* is reduced in HCC tumor specimens, albeit at modest levels of reduction, and is undetectable by qPCR in a variety of HCC cells lines. Deregulation of miR-199a* has been shown to occur in liver tissue under conditions that generally predispose the liver to HCC. Steatohepatitis frequently precedes liver fibrosis, liver failure, and often HCC.67,68 Using gene expression profiling, Dolganiuc et al demonstrated that miR-199a* (along with miR-182 and-183) was downregulated in a murine model of alcohol-induced steatohepatitis (ASH), while three miRNAs were upregulated in the murine model for nonalco-holic steatohepatitis (NASH).69 It is intriguing that all three miRNAs were regulated to the same but opposite degree in ASH versus NASH. Additional studies are warranted to determine what, if any, role miR-199a* plays in HCC patients who are inflicted with NASH. HCV infection is a known cause of chronic hepatitis, liver cirrhosis, and HCC.70 It has been demonstrated that certain miRNAs play important roles in regulating viral replication and proliferation.45 Murakami et al discovered that miR-199a* has similar sequences to the HCV genome that is conserved among all HCV genotypes. Their study demonstrated that miR-199a* regulates HCV replication by serving as an inhibitor to cell function. They propose that this may serve as an avenue for new HCV-targeted therapy and hence a preventative therapy for HCC.71
Other possibilities exist in which miR-199a* regulates cell function and also may contribute to the development of HCC. miR-199a* has been shown to target the MET proto-oncogene.72 MET produces the transmembrane tyrosine kinase receptor for hepatocyte growth factor receptor. The c-MET protein promotes invasive growth by inducing cell motility, invasion, and protection from apoptosis. Increased c-MET activation has been associated with poor survival in HCC patients.73 miR-199a* mimetic reversed the MET- induced invasive growth in a variety of cancer cell lines. In tumors that were actively proliferating, the miR-199a/a* locus was heavily methylated and the only cells expressing these miRNAs were the fibroblasts within the tumor. When miR-199a* was introduced back into the tumor cells, apoptosis was induced.72 Fornari et al demonstrated that the addition of miR-199a* to HCC tissues led to a G1-phase cell cycle arrest and reduced invasion through its interactions with c- MET production.74 The mTOR protein inversely correlated with miR-199a* in HCC cell lines. Increasing miR-199a* levels resulted in increased doxorubicin sensitivity, increased susceptibility to hypoxia, induced apoptosis, G1-phase cell cycle arrest, and reduced invasive capability. The levels of miR-199a* and mTOR were evaluated in HCC, cirrhotic, and normal human liver specimens; the levels of mTOR and miR-199a* were inversely correlated in the HCC tissues. A shorter time to reoccurrence also was seen in the patients whose tissue was low in miR-199a*.74 One final validated target of miR-199a* is ERK.72 While this is a downstream target of c-MET, it has been independently shown to be downregulated by miR-199a*. Therefore miR-199a* appears to have multiple levels of control over the c-MET signaling pathway and hence decreased levels of miR-199a* may have dramatic alterations in HCC.
Other miRNAs
Downregulation of miR-29 is a frequent event in HCC. An association between decreased miR-29b expression and an increased alpha-fetoprotein (AFP) level was observed. Lower miR-29b level is associated with shorter disease-free survival and is an independent predictor for shorter disease-free survival of HCC patients.75 In vitro evidence shows a pro-apoptotic effect of miR-29, which is mediated by the modulation of Mcl-1 and Bcl-2. These data implicate the potential application of miR-29 in prognosis prediction and cancer therapy for HCC patients.
TGF-13 has paradoxical roles in tumorigenesis by functioning both as a tumor-suppressor and as an oncogene based on the stage of tumor progression.76 The late TGF-13 signature showed an aggressive and invasive tumor phenotype relative to early TGF-13–responsive genes.43 Interestingly, TGF-13 was shown to modulate also miRNA expression. In particular, miR-181 is induced by TGF-13, likely as result of a late TGF-13 response (after 24 hours).77 Indeed, miR-181 has oncogenic properties that mediate liver carcinogenesis through the TIMP-3–mediated modulation of metalloproteinases and enhanced chemoresistance of HCC cells.77 Along with miR-181, TGF-13 can also modulate other miRNAs that may be involved in HCC, such as miR-23, miR-27, miR-24, miR-200, miR-21, miR-99, miR106, miR-125, miR-101, miR-222, and miR-193. In accordance with TGF-13’s effects on prognosis, miR-23a, miR-27a, and miR-24 closely correlated with intrahepatic metastasis of HCC. Similarly, HCC tissues with liver cirrhosis also had a higher expression of miR-23a and miR-27a compared with those without liver cirrhosis.78 miR-101 is a tumor-suppressor miRNA that modulates cell migration and cell growth by acting on several targets including FOS and Mcl-1.79,80 Members of the Let-7 family have been found to be reduced in HCC.81 In particular, Let-7g was shown to act as a tumor-suppressor gene in liver cancer cells, by modulating the ability of the cells to proliferate and to migrate.81,82 Indeed, Let-7g was reduced in human HCC with metastasis in comparison with those with no distant metastasis and was predictive of poor survival in HCC patients.81 These effects seem to be mediated by the targeting of several genes, including Bcl-xL, type 1 collagen alpha2, and likely c-Myc.81–83
miRNA IN DIAGNOSIS AND PROGNOSIS OF HCC
Several recent studies have been performed of miRNA profiling in human HCC. Similar to observations in other cancers, these studies have revealed altered expression of several miRNAs (Table 1). Further definition of the miRNAs that are aberrantly expressed in HCC and elucidation of their contribution to the pathophysiology of HCC is likely to be useful in establishing a role for miRNA. On the basis of these profiling studies, signatures that are associated with cancer or pre- cancerous changes can be identified. miRNA expression has been studied in HCC tissues in comparison with adjacent non-neoplastic liver tissues. Among the miRNAs that are aberrantly expressed there are several that are increased in expression, including miR-21, miR-221/-222, and miR-181, whereas others are significantly decreased in expression, such as miR-122, miR-101, miR-29, miR-200, miR-148a, and mir-199a (Table 1). Thus, expression profiling in the absence of a defined role may be useful for the diagnosis of HCC or in defining prognosis. Moreover, identification of a role of aberrantly expressed miRNAs in cellular processes associated with cancer such as proliferation, apoptosis, or deregulated intracellular signaling is likely to provide additional insight into the pathogenesis of HCC that may eventually yield newer approaches to therapy.
miRNA AS THERAPEUTIC TARGETS FOR HCC
A therapeutic approach for HCC that targets miRNA would be highly innovative. Such an approach would be promising given the evidence to date of the involvement of miRNA in HCC pathogenesis and biology. Systemic administration of antisense LNA (locked nucleic acid) oligonucleotides to miR-122 was recently shown to modulate miRNA and target gene expression in the liver and result in the loss of HCV with minimal toxicities in a non-human primate.84 This study showed the feasibility of this approach for targeting aberrantly overexpressed miRNA in the liver. In another recent study, the therapeutic value of a miRNA replacement strategy was described for miR-26a, a miRNA whose expression is frequently lost in HCC.48 miR-26a can target cyclins D2 and E2 and induce G1 arrest when expressed in human liver cancer cells. Using a mouse model of HCC, miR-26 replacement delivered using a self-complementary adeno-associated virus vector dramatically suppressed disease progression. Tumor-specific apoptosis was activated. These findings support the potential of targeting tumor-sup- pressing miRNAs or aberrantly expressed miRNA as a powerful and highly specific anticancer therapeutic modality for HCC.
Acknowledgments
This study was supported by Grant No. DK069370 from the National Institutes of Health (TP).
Footnotes
Financial disclosures: None of the authors has any financial disclosures.
References
- 1.Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116:544–73. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–5. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–45. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 5.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 6.Yao J, Liang L, Huang S, et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–56. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- 7.Jiang J, Gusev Y, Aderca I, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437– 47. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding J, Huang S, Wu S, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390–9. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793– 801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Zhou F, Xu T, et al. Analysis of sequence variations in 59 microRNAs in hepatocellular carcinomas. Mutat Res. 2008;638:205–9. doi: 10.1016/j.mrfmmm.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther. 2010;125:92–104. doi: 10.1016/j.pharmthera.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Qi P, Dou TH, Zhou FG, Gu X, Wang H, Gao CF. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol. 2010;71:621– 6. doi: 10.1016/j.humimm.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 16.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435– 43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 17.Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27:378– 86. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 18.Datta J, Kutay H, Nasser MW, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–58. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Meng FY, Braconi C, Swenson E, Khrapenko L, Patel T. Epigenetic activation of miR-34a contributes to an invasive phenotype in hepatocellular cancer. Hepatology. 2008;48:387A– 8A. [Google Scholar]
- 20.Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157–70. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Raidl M, Pirker C, Schulte-Hermann R, et al. Multiple chromosomal abnormalities in human liver (pre)neoplasia. J Hepatol. 2004;40:660– 8. doi: 10.1016/j.jhep.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Zondervan PE, Wink J, Alers JC, et al. Molecular cytogenetic evaluation of virus-associated and non-viral hepato-cellular carcinoma: analysis of 26 carcinomas and 12 concurrent dysplasias. J Pathol. 2000;192:207–15. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH690>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 23.Jin WB, Wu FL, Kong D, Guo AG. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31:124– 6. doi: 10.1016/j.compbiolchem.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–9. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 25.Ura S, Honda M, Yamashita T, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57– 67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 27.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci U S A. 2003;100:235– 40. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braconi C, Valeri N, Gasparini P, et al. Hepatitis C virus proteins modulate microRNA expression and chemosen- sitivity in malignant hepatocytes. Clin Cancer Res. 2010;16:957– 66. doi: 10.1158/1078-0432.CCR-09-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidorkiewicz M, Grek M, Jozwiak B, Majda-Stanislawska E, Piekarska A, Bartkowiak J. Expression of microRNA-155 precursor in peripheral blood mononuclear cells from hepatitis C patients after antiviral treatment. Acta Virol. 2010;54:75–8. doi: 10.4149/av_2010_01_75. [DOI] [PubMed] [Google Scholar]
- 30.le SC, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koff A. How to decrease p27Kip1 levels during tumor development. Cancer Cell. 2006;9:75– 6. doi: 10.1016/j.ccr.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 33.Visone R, Russo L, Pallante P, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14:791– 8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 34.Fornari F, Gramantieri L, Ferracin M, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651– 61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 35.Gramantieri L, Fornari F, Ferracin M, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15:5073– 81. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong QW, Ching AK, Chan AW, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867–75. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 37.Garofalo M, Di LG, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Ueki T, Fujimoto J, Suzuki T, Yamamoto H, Okamoto E. Expression of hepatocyte growth factor and its receptor c-met proto-oncogene in hepatocellular carcinoma. Hepatology. 1997;25:862– 6. doi: 10.1002/hep.510250413. [DOI] [PubMed] [Google Scholar]
- 39.Kutay H, Bai S, Datta J, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671– 8. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–9. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 41.Chang J, Nicolas E, Marks D, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 42.Bai S, Nasser MW, Wang B, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–27. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–36. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henke JI, Goergen D, Zheng J, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–10. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol. 2006;71:369–76. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- 46.Zhang R, Wang L, Yu GR, Zhang X, Yao LB, Yang AG. MicroRNA-122 might be a double-edged sword in hepatocellular carcinoma. Hepatology. 2009;50:1322–3. doi: 10.1002/hep.23108. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen IM, Cheng G, Wieland S, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–22. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naugler WE, Sakurai T, Kim S, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121– 4. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 50.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 51.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H, Ozaki I, Mizuta T, et al. Involvement of programmed cell deat h 4 in transforming growth factor- beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–12. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 53.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–25. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 54.Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol. 2010;298:G535– 41. doi: 10.1152/ajpgi.00338.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 56.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 57.Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56:248–53. doi: 10.1016/j.phrs.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 58.Moran DM, Mattocks MA, Cahill PA, Koniaris LG, McKillop IH. Interleukin-6 mediates G(0)/G(1) growth arrest in hepatocellular carcinoma through a STAT 3-dependent pathway. J Surg Res. 2008;147:23–33. doi: 10.1016/j.jss.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soresi M, Giannitrapani L, D’Antona F, et al. Interleukin-6 and its soluble receptor in patients with liver cirrhosis and hepatocellular carcinoma. World J Gastroenterol. 2006;12:2563–8. doi: 10.3748/wjg.v12.i16.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loffler D, Brocke-Heidrich K, Pfeifer G, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 61.Tili E, Michaille JJ, Cimino A, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/ TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 62.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macro- phage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 64.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 65.Wang B, Majumder S, Nuovo G, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152– 61. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Budhu A, Yu Z, Forgues M, Tang ZY, Croce C, Wang XW. Immune cell-related MICRORNA-155 is associated with human liver cirrhosis and hepatocellular carcinoma. ILCA Annual Conference (abstract); Milan, Italy. Sept 4–6, 2009. [Google Scholar]
- 67.Bugianesi E, Leone N, Vanni E, et al. Expanding the natural history of nonalcoholic steatohepatitis: from cryptogenic cirrhosis to hepatocellular carcinoma. Gastroenterology. 2002;123:134– 40. doi: 10.1053/gast.2002.34168. [DOI] [PubMed] [Google Scholar]
- 68.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87–96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 69.Dolganiuc A, Petrasek J, Kodys K, et al. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–10. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wasley A, Alter MJ. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin Liver Dis. 2000;20:1–16. doi: 10.1055/s-2000-9506. [DOI] [PubMed] [Google Scholar]
- 71.Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453– 60. doi: 10.1016/j.jhep.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 72.Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158– 66. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 73.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–95. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fornari F, Milazzo M, Chieco P, et al. MiR-199a–3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–93. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 75.Xiong Y, Fang JH, Yun JP, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836– 45. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 76.Massague J. A very private TGF-beta receptor embrace. Mol Cell. 2008;29:149–50. doi: 10.1016/j.molcel.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 77.Wang B, Hsu SH, Majumder S, et al. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787–97. doi: 10.1038/onc.2009.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang S, He X, Ding J, et al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;123:972– 8. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- 79.Li S, Fu H, Wang Y, et al. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194–202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- 80.Su H, Yang JR, Xu T, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135– 42. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 81.Ji J, Zhao L, Budhu A, et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52:690–7. doi: 10.1016/j.jhep.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lan FF, Wang H, Chen YC, et al. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and up-regulation of p16(INK4A) Int J Cancer. 2011;128:319–31. doi: 10.1002/ijc.25336. [DOI] [PubMed] [Google Scholar]
- 83.Shimizu S, Takehara T, Hikita H, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 84.Lanford RE, Hildebrandt-Eriksen ES, Petri A, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li W, Xie L, He X, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–22. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–15. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 87.Connolly E, Melegari M, Landgraf P, et al. Elevated expression of the miR-17–92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856–64. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]