SUMMARY
Upon transit to colonization sites, bacteria often experience critical priming that prepares them for subsequent, specific interactions with the host; however, the underlying mechanisms are poorly described. During initiation of the symbiosis between the bacterium Vibrio fischeri and its squid host, which can be observed directly and in real time, ~5 V. fischeri cells aggregate along the mucociliary membranes of a superficial epithelium prior to entering host tissues. Here we show that these few early host-associated symbionts specifically induce robust changes in host gene expression that are critical to subsequent colonization steps. This exquisitely sensitive response to its specific symbiotic partner includes the upregulation of a host endochitinase, whose activity hydrolyzes polymeric chitin in the mucus into chitobiose, thereby priming the symbiont and also producing a chemoattractant gradient that promotes V. fischeri migration into host tissues. Thus, the host responds transcriptionally upon initial symbiont contact, which facilitates subsequent colonization.
INTRODUCTION
Bacterial partners, whether beneficial or pathogenic, often travel great distances from the initial site of contact to their eventual location of sustained colonization within the animal host. For example, the journey of a bacterium through the mammalian digestive tract to its target tissues in the hindgut is equivalent to the host animal migrating hundreds of kilometers. During such a journey, the microbes necessarily pass through a series of chemically and physically distinct environments that, in the case of some pathogens, primes them for colonization of a specific tissue (Krukonis and DiRita, 2003; Alvarez-Ordóñez et al., 2011). For example, exposure and subsequent response to acid stress of the stomach render Vibrio cholerae more effective at colonizing the intestine (Merrell et al., 2002); similarly, intracellular passage through a protist in the rumen enhances the pathogenicity of some Salmonella enterica strains (Rasmussen et al., 2005; Carlson et al., 2007). However, as symbiotic initiation is difficult to observe under natural conditions and many models bypass the normal inoculation route, questions about in vivo site-specific colonization by a microbial species have remained largely unexplored, such as: how does the passage through various host environments affect their ability to colonize a specific site; and, what is the molecular nature of the host-symbiont dialogue underlying the process?
The acquisition of the bacterial symbiont Vibrio fischeri by its squid host Euprymna scolopes offers the rare opportunity to probe colonization by specific bacteria directly and in real time in an intact, natural symbiosis using imaging technology. Although the process occurs across a distance of only ~100 μm, it is highly complex. In the established association, in which the symbiont population resides along the apical surfaces of microvillous epithelia within the crypts of the light organ, V. fischeri is the exclusive partner, i.e., in its absence, no other bacteria can colonize the organ.
Confocal microscopy has revealed that during passage to this site, V. fischeri cells interact first with a pair of juvenile-specific mucociliary epithelial surfaces on the light organ’s exterior (Figure 1A), which are concomitantly exposed to thousands of other environmental bacteria. A series of events in the first 3 h following hatching results in the specific enrichment of the symbiont along these surfaces. First, at environmentally relevant concentrations of V. fischeri (~5000 cells/mL of seawater) only 3–5 V. fischeri cells eventually attach to the cilia (Altura et al., 2013) and aggregate above the pores on the organ’s surface (Yip et al., 2006). Once aggregated, this population migrates to and through the pores, and then down the ducts to a physical bottleneck where one or two bacteria squeeze into each crypt and proliferate, filling the spaces. Only V. fischeri can negotiate this journey beyond the pores (McFall-Ngai and Ruby, 1991).
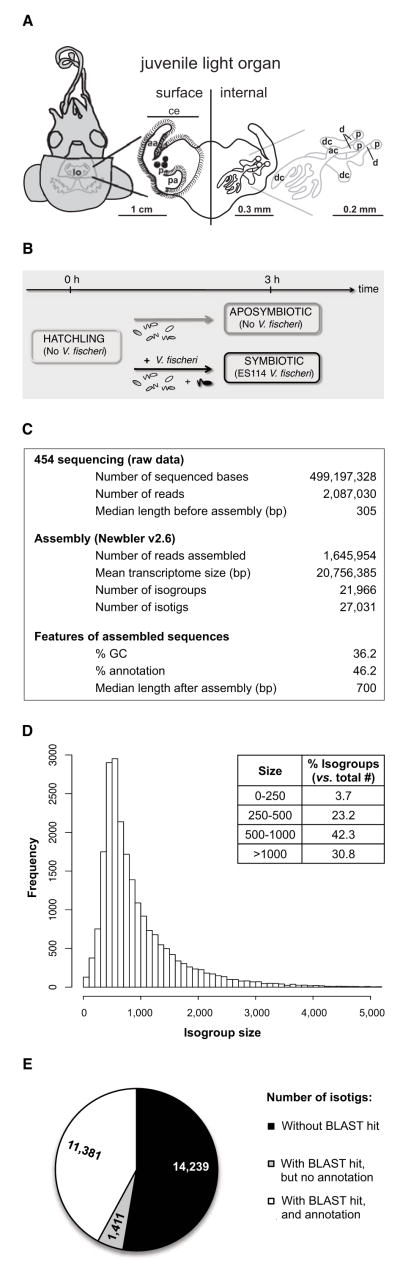
Figure 1. Symbiotic organ structure, experimental set-up, and generation of the squid light-organ transcriptome.
(A) (Left) Diagram of the ventral surface showing the light organ (lo) suspended in the body cavity, visible in this drawing through the translucent mantle. (Center) Enlargement of the light organ showing the surface in contact with the seawater (left), and the internal structure through which symbionts migrate (right). (Right) Detailed anatomy of the three crypts. ce: ciliated epithelium, p: pore, aa: anterior appendage, pa: posterior appendage, dc: deep crypt, ac: antechamber, d: duct. (B) Experimental set-up for the collection of squid used for 454 sequencing. Squid were incubated for 3h in seawater containing ~106 environmental bacteria ± 5000 V. fischeri cells/mL. (C) General statistics on 454 sequencing and assembly. (D) Size distribution of isogroups after assembly. (E) Summary of the gene ontology (GO) annotation results, using the score function of the Blast2GO software. This process consists of a BLAST annotation against databases and a mapping step for the assignation of GO terms. See also Figure S1.
Studies of the system indicate that the chemistry of the mucus is an important determinant of the events leading to colonization. During aggregation, other bacterial species are excluded as V. fischeri cells become primed for their colonization process. Briefly, within seconds of hatching from the egg, the animal begins ventilating seawater through its body cavity, and shedding mucus from the epithelial surface of the nascent light organ as a non-specific response to cell-wall (peptidoglycan) derivatives released by environmental bacteria (Nyholm et al., 2002). The mucus matrix contains antimicrobial biomolecules (Troll et al., 2010) and vesicles containing nitric oxide (NO) (Davidson et al., 2004). As in other host-symbiont interactions, including the highly complex mammalian-microbiota alliances (Duerkop et al., 2009), these antimicrobials provisioned into the mucus may create a selective ‘cocktail’ that functions in specificity and stability of the associations.
V. fischeri responds to this specific environment presented by the host during aggregation; e.g., the low concentration of NO present in the mucus up-regulates NO-detoxification mechanisms (Wang et al., 2010), which protect the symbiont against the more acute NO stress subsequently encountered in the ducts. Additionally, chemotaxis by V. fischeri mediates migration into the host tissues in response to signals like the dimeric breakdown product of chitin, chitobiose (di-N-acetyl-glucosamine) (Mandel et al., 2012). Thus, bacterial symbionts respond both transcriptionally and behaviorally to the host-derived conditions they encounter during initiation.
Here we show that, although exposed to the myriad of other bacteria in the seawater, host tissues respond to the few attaching V. fischeri cells with significant changes in gene expression, most notably in genes encoding proteins that alter the environment to favor symbiont colonization. To explore the impact of symbiont-induced host transcription on the environment presented to the symbiont, we examined in depth one up-regulated gene and its encoded protein, a chitotriosidase. We provide evidence that symbiont-induced up-regulation of the host chitotriosidase gene results in two downstream events: a priming of V. fischeri by shaping of the biochemistry where they aggregate before entering host tissues, an activity that primes the symbiont for responses to chitin polymers; and the production of a gradient of chitobiose that mediates effective symbiont migration into host tissues. These studies provide a detailed window into the first minutes to hours of interaction between the tissues of a host animal and its symbiotic partner.
RESULTS
The transcriptome of the nascent organ is diverse
To determine whether changes in gene expression accompany early symbiotic events, we first constructed a reference transcriptome (Figure 1). Libraries built from extracted organs included: (i) ‘hatchling’ – <15 min after hatching from the egg; (ii) ‘aposymbiotic’ – 3 h after hatching and incubation in Hawaiian offshore seawater (HOSW), which contains environmental bacteria (~106/mL), but is devoid of V. fischeri, and (iii) ‘symbiotic’ – the ‘aposymbiotic’ condition, but with wild-type V. fischeri added at a typical, field-relevant concentration of ~5000 cells/mL (Figure 1B).
This database of early-stage symbiosis increased the number of known transcripts for the E. scolopes light-organ transcriptome by ~40% over a previously constructed Expressed Sequence Tags database (Chun et al., 2006). Pyrosequencing yielded about 2 × 106 reads, from which 76% were assembled with an inferred read error of 1.5–2.4% (Figures 1C and S1C). Assembly yielded >20,000 isogroups, i.e., unique sequence assemblies potentially representing gene loci, with a median length of 700 bp (Figures 1C and 1D), and mean coverage of 12x. These isogroups were constituted on average by 1.23 isotigs, alternative transcripts associated with a single locus. About 46% of the isotigs could be annotated by Blast2GO software (Figure 1E) with relatively high similarity and low e-values (Figure S1D–E). These annotations were inferred principally from UniProtKB by electronic annotation against several animal species (Figure S1F–H). Each annotated isotig was assigned an average of 4.8 Gene Ontology (GO) terms (Figures 1E and S1I–K). Assignment to Cluster of Orthologous Groups (COG) revealed enrichment of clusters related to cellular processes and signaling (39%) compared with information storage and processing (21%), and metabolism (19%); 21% were poorly characterized (Figure S1L).
Only a few V. fischeri cells are needed to induce specific changes in host gene expression
We compared the transcriptome of symbiotic organs during the aggregation of the ~5 V. fischeri cells with that of aposymbiotic organs. It is important to note that these aggregating V. fischeri cells were in contact with only 2–3 host cells, although the libraries were generated with entire juvenile organs, which have approximately 104 eukaryotic cells each. As such, any changes in the inductive signal from V. fischeri must be either highly up-regulated in the 2–3 interacting host cells or spread across the entire organ. Recent studies also indicate that, at low bacterial inoculum, V. fischeri products that likely induce host responses would be in concentrations too low to cause these widespread effects (Altura et al., 2013).
We first analyzed a subset of the 3-h symbiotic animals, obtained concurrently with tissue harvested for the production of the reference transcriptome, to confirm that an average of 5.2 ± 0.8 V. fischeri cells directly interact with squid tissues after a 3-h incubation (Altura et al., 2013; Figures 2A and S1A–B). Examination of the 454-sequencing libraries identified 84 isotigs (0.37% of the total) differentially represented between the apo- and symbiotic conditions, of which 72% had annotations (Figure 2B, Table S1). Functional-enrichment analysis of these differentially expressed isotigs revealed two major predicted molecular signatures of first contact between symbiotic partners: protease and hydrolase activities directed at sugar bonds, especially those associated with chitin metabolism (Figure 2C). None of these transcripts was differentially regulated in the hatching compared to the aposymbiotic condition (Table S1). Taken together, the data demonstrate that the changes in the transcription of the 3-h symbiotic light organ are due to host-tissue discrimination of only a few associating V. fischeri cells.
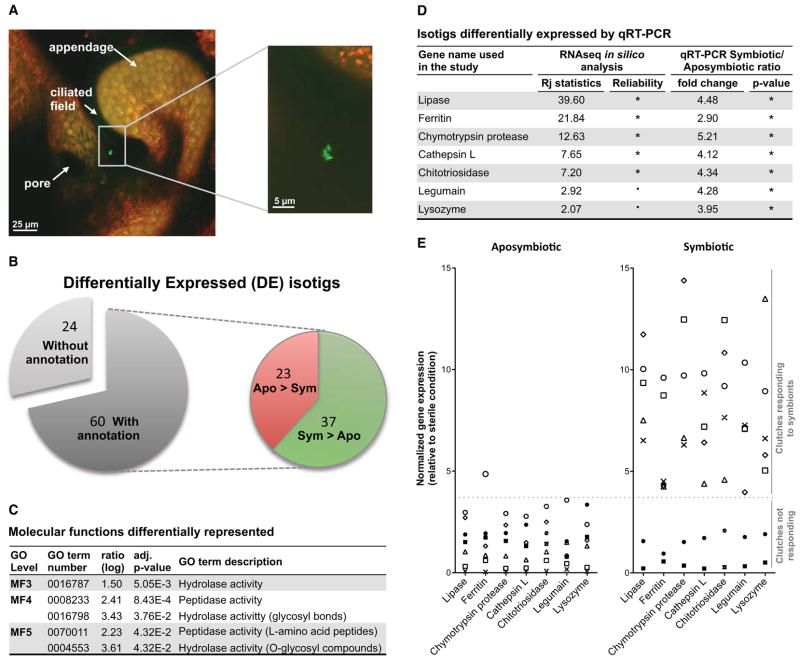
Figure 2. Effect of contact with V. fischeri on the expression of host genes.
(A) Confocal images showing an aggregate of 4 V. fischeri cells (GFP-ES114, green) in association with host tissues (CellTracker Orange). (B) Distribution of the differentially expressed (DE) isotigs between aposymbiotic (Apo) and symbiotic (Sym) light-organ libraries. (C) Functional enrichment analysis based on molecular functions (MF) (FatiGO software). Ratios correspond to the percentage of a particular GO term in the list containing DE isotigs divided by its percentage in the reference list (aposymbiotic condition). MF1, 2, 6, 7, 8 and 9 did not exhibit any significant GO terms. (D) Summary of differential expression of candidate genes based on in silico database analysis (Rj statistics from Stekel et al., 2000; *, Rj > 3; ., Rj > 2) and quantitative RT-PCR (n = 7 clutches, data detailed in panel (E); *, p < 0.05 with paired t-test between aposymbiotic and symbiotic conditions followed by FDR adjustment). (E) Differential expression of candidate genes by qRT-PCR (n = 7 clutches from different females, each represented by a different symbol). Expression of candidate genes was first normalized to the expression of three housekeeping genes. The expression data from aposymbiotic and symbiotic light organs, originating from squid incubated for 3h in HOSW, were then normalized to the expression data from aposymbiotic light organs originating from squid incubated for 3h in FSIO (the sterile condition). See also Table S1.
To reveal changes in gene expression that would result from exposure to non-specific environmental microbes and/or post-embryonic development over the first 3-h posthatch, we also compared the transcriptomes of hatchling light organs to those of the aposymbiotic condition. Based on functional enrichment analysis, we detected a significant differential representation of transcripts only in GO terms associated with the ribosomal machinery (Table S1). This differential expression is also detectable between hatchling and symbiotic light organs (Table S1).
Focusing on molecular signatures highlighted by functional analysis in the symbiotic condition, we confirmed the RNAseq results by determining expression of isotigs encoding proteins associated with protease activity (chymotrypsin protease, cathepsin L, legumain) and chitin metabolism (chitotriosidase) by quantitative RT-PCR (Figures 2D and 2E). Additionally, we tested differentially expressed isotigs encoding proteins involved in immune responses to bacteria (ferritin and lysozyme). Squid light organs from multiple egg clutches (7 individual clutches laid by different captive females, thus different genetic backgrounds) were assayed to control for genetic variability that may influence development and responsiveness to a symbiotic infection. Five of the seven clutches responded to contact with V. fischeri by up-regulating a set of several isotigs encoding, in order of highest to lowest fold change: chymotrypsin protease, lipase, chitotriosidase, legumain, cathepsin L, lysozyme, and ferritin. The overall within-clutch patterns were consistent, i.e., all of these genes were either up-regulated or not regulated in the organs of a given clutch. Despite this biological variability, these data demonstrate that the ciliated epithelial cells are able to sense only a few associating V. fischeri cells, in a background of 106 non-symbiotic bacterial cells, and induce specific transcriptional changes in the juvenile light organ.
A catalytically active chitotriosidase localizes to the apical surfaces of epithelial cells, pores and ducts of the organ surface, and is secreted into the associated acidic mucus matrix
Because chitin and its breakdown products play major roles in both the establishment and maintenance of the symbiosis (Wier et al., 2010; Heath-Heckman and McFall-Ngai, 2011; Miyashiro et al., 2011; Mandel et al., 2012), as well as in the pathogenesis of other Vibrio species (Meibom et al., 2005; Blokesch and Schoolnik, 2007), we chose to focus on an in-depth characterization of the symbiont-induced chitotriosidase gene and its encoded protein.
Analysis of the derived amino-acid sequence of this putative chitotriosidase supported its role in chitin degradation. Based on the full-length sequence of EsChitotriosidase obtained by Rapid Amplification of cDNA Ends (RACE), the encoded protein was similar in sequence (Figure S2) and tertiary structure (Figure 3A) to a highly conserved animal endochitinase. Chitotriosidase catalyzes the random hydrolytic cleavage of β-1,4 glycosidic bonds of the chitin polymer; as an endochitinase, it releases chitobiose and larger multimers, but not the monomer, GlcNAc. It contains a signal peptide for secretion, a glycosyl-hydrolase, GH18-type catalytic domain with the critical DxxDxDxE residues essential for activity (Fusetti et al., 2002) (swissmodel Z-score = −2.53 against human chitotriosidase complexed with chitobiose; Benkert et al., 2011), and two chitin-binding domains with conserved cysteine residues critical for substrate binding. As such, it is predicted to be a secreted, catalytically active protein. We confirmed its catalytic activity using affinity-purified EsChitotriosidase (Figures 3B and 3C).
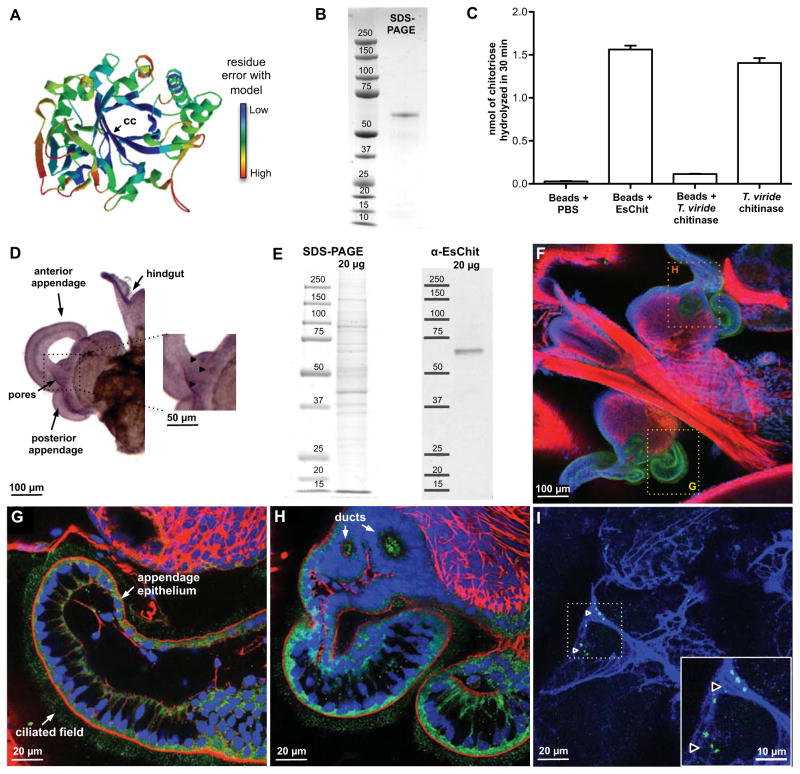
Figure 3. Localization of chitotriosidase (EsChit) in the squid light organ and protein characterization.
(A) Predicted 3D-structure of the catalytic EsChit glycosyl hydrolase 18 domain. Swissprot model: crystal structure of human chitotriosidase in complex with chitobiose (1LG1; Fusetti et al., 2002). Although the protein sequences are only 46% identical, the structure of EsChit is predicted to be similar to that of human chitotriosidase (Z-score = −2.53), especially in the proximity of the chitobiose-binding site (cc, catalytic center). Gradient from cold to warm colors corresponds to residue error compared to the human chitotriosidase structure. (B) SDS-PAGE gel of purified EsChit (SYPRO Ruby staining). (C) Chitotriosidase activity of EsChit purified using chitin-magnetic beads (mean ± SE; measurements done in triplicate). The activity was measured in assay buffer (Sigma). Chitinase (100 ng) from the fungus Trichoderma viride (T. viride; Sigma) was used as a positive control for the effect of binding to beads on chitinase activity. (D) In situ hybridization of EsChit in a symbiotic light organ (anti-sense probe). Purple staining, corresponding to the RNA expression of EsChit, is particularly high in the proximity of the pores (triangles, inset), in the epithelium of the appendages, and in the hindgut. (E) Coomassie staining of an SDS-PAGE gel of squid-tissue soluble-protein extract (left) and its associated western blot against EsChit (right, expected size: 59.2 kDa without signal peptide). (F–I) Immunocytochemistry of EsChit in the light organ (F). The antigen labeling is particularly bright in the ciliated field and the cytosol of the appendage epithelium (G), and in the pores/duct region (H). Colored boxes in (F) show representative locations of pictures zoomed in panels (G) and (H). Green, α-EsChit; red, rhodamine phalloidin (f-actin); blue, TOTO-3 (nuclei). (I) Immunocytochemistry of EsChit in the mucus coating the appendage epithelium. Triangles indicate foci of the protein in the mucus. The inset is a higher magnification of the region highlighted by the box. Green, α-EsChit; blue, Alexa633-WGA (mucus-binding lectin wheat-germ agglutinin). Negative controls for ISH (sense probe) and ICC, as well as the RACE sequence of EsChit and its predicted domains / functional residues, are presented in Figure S2.
With microscopy, we localized the encoding transcript and the protein in the light organ. Whole-mount in situ hybridization revealed abundant transcript in the superficial ciliated epithelium and the cells surrounding the light-organ pores (Figure 3D). Confocal immunocytochemistry localized the protein to intracellular regions along the apical surfaces of the cells of the ciliated field, pores and ducts of the organ (Figure 3E–H). In addition, the antibody recognized the secreted protein in the mucus matrix overlying the light-organ ciliated surface (Figure 3I). These data demonstrate that the gene is transcribed and the protein produced and secreted in locations where V. fischeri aggregates and establishes dominance over other seawater bacteria.
The EsChitotriosidase occurs in sites with an optimal pH for its activity
Using a chitinase activity assay, we determined that the pH optimum for in vitro endochitinase activity (i.e., the conversion of chitotriose to chitobiose) of the EsChitotriosidase is between pH 5.0 and 7.5 (Figure 4A), typical of chitotriosidases (e.g., Xu and Zhang, 2012). Because the pH of the seawater surrounding the squid in nature (~8.2) is well above this range, we hypothesized that the environment created by the host in the location where V. fischeri aggregates is more acidic. To determine the pH of the ciliary-mucus environment, we constructed a pH-sensitive probe coupled to wheat-germ agglutinin (WGA), a lectin known to bind the mucus (Nyholm et al., 2002). The pH in the mucus along the organ’s ciliated appendages was approximately 6.4, and 5.9 in the region of the pores (Figure 4B and 4C). Thus, the mucus matrix has a pH range different from that of the seawater passing over these tissues, but compatible with the optimal pH for EsChitotriosidase activity. No difference in pH was observed in these regions over the first 3 h following hatching under any condition, i.e., interaction of host tissues with V. fischeri cells did not influence the pH.
Figure 4. Activity of EsChitotriosidase and the effect of pH of the light-organ surface.
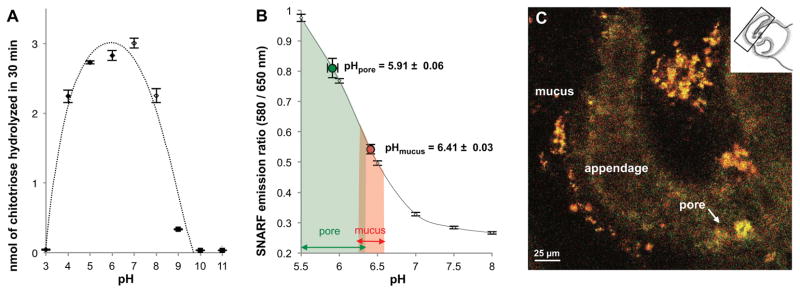
(A) Effect of pH on EsChitotriosidase activity (mean ± SE; measurements done in triplicate; black dotted line, polynomial regression). Activity in 2.5% NaCl was measured: (◆), at pH 3, 4, 5, and 6 in 100 mM sodium acetate; (◇), at pH 7 and 8 in 100 mM Tris; (◆), at pH 9, 10, and 11 in 100 mM sodium carbonate. (B, C) pH was estimated using WGA coupled to a pH-sensitive fluorescent probe (SNARF). This probe is excited at 488 nm and selectively emits at two different wavelengths (580 nm and 650 nm). The emission ratio is characteristic of a specific pH, estimated by a calibration curve. (B) Determination of pH; mean ± SE; n = 10 squid (10 measurements/squid). The red and green areas indicate the range of pH values observed in the mucus and around the pores, respectively. Calibration curve (grey) was generated in pH-adjusted mPBS; mean ± SE; n = 20 measurements. (C) Example of WGA-SNARF staining in the mucus coating the appendage and in the pore. Gradient colors from yellow/red (in the mucus) to green/yellow (close to the pore) match with mild-acidic to acidic pHs, respectively. The inset orientates the imaged tissue within the light organ.
Chitin derivatives are presented in the mucus and ducts where the EsChitotriosidase occurs
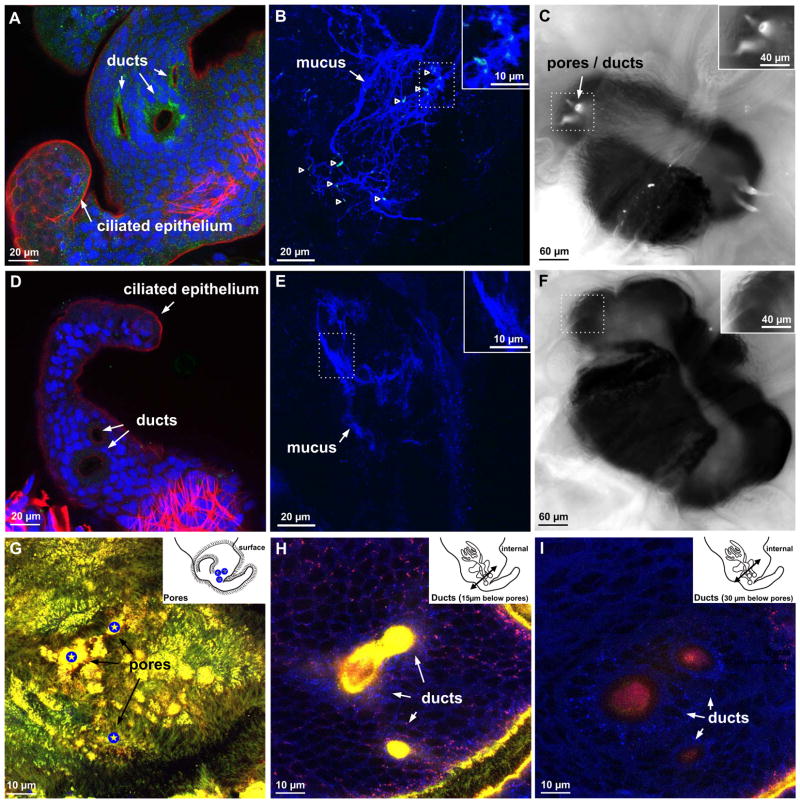
To present V. fischeri with chitobiose for either priming or the creation of a chemoattractant gradient, the host chitotriosidase must have the substrate chitin, the production of which has been shown to be dependent on chitin synthase in other systems. A chitin synthase localized mainly to the apical surfaces of the ciliated epithelia and to regions of the pores and ducts (Figure 5A). For chitin, we used three different labeling methods. A previous study with a chitin-binding protein (CBP; New England Biolabs) that detects polymers larger than hexamers showed no labeling at the pores or in the ducts (Heath-Heckman and McFall-Ngai, 2010); this study did not examine the mucus. Using a fix to preserve the mucus, the CBP labeled chitin polymers in the mucus matrix (Figure 5B). In animal tissues, calcofluor white binds to polysaccharides. This reagent labeled strongly in regions of the pores and ducts (Figure 5C). To characterize the type of polysaccharides present in these regions, we performed a complementary staining method able to differentiate GlcNAc and small chitin breakdown products from sialic acid. GlcNAc residues were present on the organ surface and to ~10–12 μm into the ducts; in contrast, only sialic acid residues were present further down in the ducts (Figure 5G–I). These results suggest a relative enrichment of small polymers of chitin surrounding the pore and superficial duct regions, where V. fischeri senses a chitobiose gradient.
Figure 5. Expression of chitin synthase and presence of chitin derivatives in the squid light organ.
(A, D) Immunocytochemical localization of E. scolopes chitin synthase (EsCS). (A) Expression of EsCS in duct cells. (D) The negative control using IgY instead of the antibody, because the EsCS antibody was generated in chicken. Green, α-EsCS (A) or α-IgY (D); red, rhodamine phalloidin (f-actin); blue, TOTO-3 (nuclei). (B, E) Localization of chitin in the mucus surrounding the ciliated appendages. (B) Chitin residues in the mucus labeled with CBP (chitin-binding protein). (E) Negative control, without added CBP. Inset: magnification of the boxed region. Green, FITC-CBP (chitin); blue, WGA (mucus). (C, F) Localization of polysaccharides in the light organ. (C) Calcofluor staining in the pores and ducts. (F) Negative control, no calcofluor added. Insets: higher magnification of the pore/duct region. (G–I) Confocal images of WGA labeling at different levels, from the pores (blue star) to the ducts. Inset: illustration showing the location of the section (arrow) along the light organ. Red, Alexa633-WGA (GlcNAc and sialic-acid residues); green, FITC-succinylated WGA (GlcNAc residues only); blue, CellTracker Orange (animal cells).
Degradation of chitin promotes V. fischeri chemoattraction and efficient host colonization
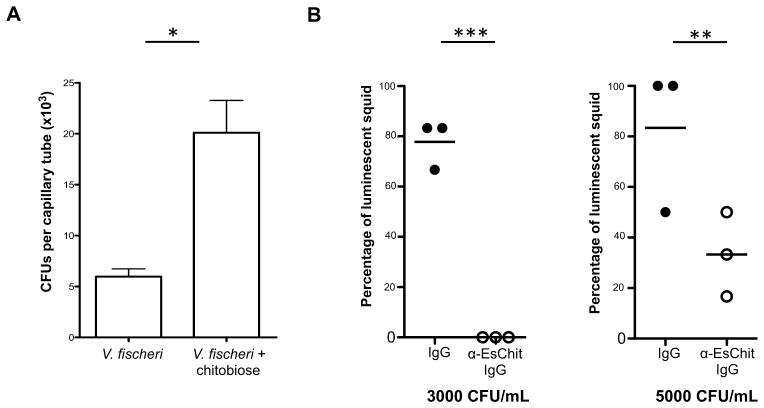
While V. fischeri cells exhibit chemotaxis towards chitobiose during early symbiotic initiation, laboratory studies directly assaying this behavior suggest that, when grown under the conditions used for squid colonization experiments, these bacteria exhibit minimal chemotaxis towards chitobiose (Figure S3). As EsChitotriosidase is localized not only close to the pores, but also in the mucus along the ciliated field (Figure 3I), we hypothesized that: (i) the EsChitotriosidase activity would produce low levels of chitin breakdown products in the mucosal environment; and, (ii) during aggregation, V. fischeri cells would be presented with these products, priming these cells to subsequently sense the chitin-based chemoattractant gradient that mediates migration into the host ducts. To test whether pre-exposure to chitin breakdown products does, in fact, increase chemotactic recognition of chitobiose by V. fischeri, we pre-incubated cells in culture medium either with or without added chitobiose, and measured their subsequent chemoattraction towards chitobiose using capillary assays (Figure 6A). V. fischeri cells that were previously exposed to chitobiose showed increased migration towards chitobiose. These data suggest that exposure of V. fischeri to chitin in the aggregate primes the symbionts to later respond to a chitobiose gradient during transit through the pores.
Figure 6. Role of EsChitotriosidase in early colonization events.
(A) Importance of priming of V. fischeri for chemotaxis towards 10 mM chitobiose, tested using capillary assays (mean ± SE; 3 biological replicates with 2 technical replicates each). Pretreatment was performed with 1 mM chitobiose in SWT. Wilcoxon’s test; *, p < 0.05. See also Figure S3. (B) Effect of co-incubation with α-EsChit antibody on colonization of the light organ by V. fischeri using different inocula (n = 3 replicates of 6 squid/inoculation/condition). Fisher’s Exact Test; ***, p < 0.001; **, p < 0.01. The luminescence values reflect differences in CFU colonization levels.
To determine whether EsChitotriosidase played a role in chitobiose priming for symbiont chemotaxis, we measured the effect of the chitotriosidase inhibition on colonization efficiency by adding the anti-chitotriosidase antibody (20 μg/mL) to the surrounding seawater as symbionts were initiating colonization. Antibody binding significantly decreased chitotriosidase activity in vitro (ANOVA, treatment: F = 21.7, p = 0.007; replicate: F = 18.4, p = 0.009; comparison α-Eschit IgG/IgG: padj = 0.01). In vivo efficiency of symbiosis initiation, as measured by the proportion of V. fischeri-exposed animals that had been successfully colonized one day after inoculation, was statistically lower in animals that had been exposed to the antibody during initiation (Figure 6B), suggesting that antibody binding of the EsChitotriosidase compromises the ability of the symbiont to colonize normally.
DISCUSSION
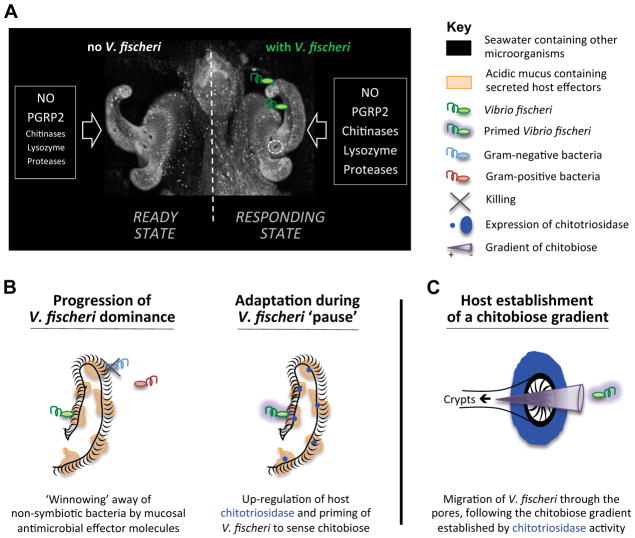
In this study, we demonstrate that, during the initiation phase of an animal-bacterial association, partner interactions create an environment that facilitates the species-specific colonization of distant host tissues. In the squid-vibrio symbiosis, the initiation of colonization occurs as a progression of events that cover a distance of only a few tens of microns during the first ~3 h after the host hatches from its egg. These discrete events have allowed a fine-scale spatiotemporal resolution of the colonization process, leading to the construction of a model describing the host’s progression from a state of readiness to one of selective responsiveness (Figure 7). This model illustrates two major findings of our study: (i) the remarkable sensitivity of host gene expression to the presence of just 3–5 cells of the specific symbiont, and (ii) the link between these changes in expression and the symbionts’ successful migration into distant tissues.
Figure 7. Model for early colonization.
(A) The initial contact of V. fischeri with host tissues induces the expression of several genes (e.g., proteases, chitinases such as EsChitotriosidase, lysozyme) that, when supplemented with components already present in the mucus (NO, EsPGRP2), affect the chemistry of the mucus matrix, shaping the specificity and preparing for future colonization events. (B) Course of events that allow selective colonization by V. fischeri. (left) Antimicrobial compounds (e.g., lysozyme, PGRP2) are activated by acidic proteases in the low pH environment and participate in the selective exclusion of non-symbiotic bacteria. (right) While V. fischeri cells are ‘pausing’ in the aggregate, the up-regulation of EsChitotriosidase in the ciliated field of the light organ hydrolyzes chitin into chitobiose, which prepares V. fischeri to sense and be attracted towards chitobiose. (C) EsChitotriosidase, which is highly expressed close to the pores and optimally active at low pH, degrades chitin produced by the host into chitobiose, thereby establishing a chitobiose gradient extending out of the pores. Primed V. fischeri cells are attracted by the chitobiose gradient, and migrate through the pores (Mandel et al., 2012).
Much as in the newborn mammal, the juvenile squid hatches from an essentially sterile environment that surrounds the embryo; however, its first postembryonic ventilations bring myriad environmental microbes into the juvenile’s body cavity. Perhaps one of the most remarkable observations in the present study is that as few as 3–5 V. fischeri cells are sufficient to induce a specific transcriptional expression pattern in light-organ tissues, suggesting that the animal hatches from the egg in a state of readiness, but is highly selective in its response (Figure 7A). In an analogous manner, although transcriptomic studies of the initial hours of host-microbe interaction have not been conducted in mammals, normal embryonic development and communication across the placenta result in the production of specific cells (e.g., RORγt+ cells; Eberl, 2012) and biomolecules (e.g., maternal antibodies; Blümer et al., 2007) that ready the host’s immune system for subsequent early postembryonic exposure to microbes.
The mucociliary membranes of the nascent light organ, where the first steps of colonization occur, have a similar structure, activity, and biochemistry as epithelial surfaces of the airway, excretory, and reproductive systems of mammals, as well as of the peritrophic membranes that line the midgut of insects (Dinglasan et al., 2009; Hegedus et al., 2009). For example, in both the squid (Davidson et al., 2004; Troll et al., 2010) and mammalian model systems (Duerkop et al., 2009), antimicrobial compounds incorporated into the mucus influence the composition of the native microbiota as well as the successful colonization by potential pathogens. Such symbiont-partner selection has also been explored at the cellular and molecular levels in basal metazoans. Specifically, in certain cnidarian species, the secretion of mucus containing antimicrobial peptides along the apical surfaces of the superficial epithelia is critical for the establishment of the normal community composition of the native microbiota (Fraune et al., 2010). Our data suggest that the mucus matrix of the host squid changes as the nascent tissues modulate their gene expression in response to interactions with V. fischeri. It is tempting to speculate that the regulation of squid genes, such as those encoding potentially antimicrobial acidic proteases, iron-sequestering proteins, and chitinases, which all have homologs associated with general inflammatory responses (Cho et al., 2002; Conus and Simon, 2010; Lee et al. 2011), is a mechanism through which proper symbionts are selected. Interestingly, many of these proteins are regulated in response to Microbe-Associated Molecular Patterns (MAMPs) in other systems (Ong et al. 2005; Badariotti et al., 2007; Liu et al. 2009), and have activities with optima at low pH, like that found in the light-organ mucus (Figure 4). Thus, it can be postulated that the matrix on the surface of epithelia is responsible for a type of ‘ecosystems management’, where fluid dynamics (e.g., ciliary activity, peristalsis) function together with the biochemistry of the mucus to control microbial associations.
In what ways might such changes in host gene expression also control symbiont specificity and colonization efficiency? In this study, we considered in depth one such V. fischeri-induced host gene, encoding EsChitotriosidase, and tested the hypothesis that its degradation of chitin to chitobiose might increase the efficiency of symbiont colonization. Between aggregation and migration, V. fischeri cells ‘pause’, a behavior that has been associated with preparation for the upcoming oxidative environment of the duct (Wang et al., 2010) and that may also similarly prime them for other conditions, like a host-derived chemotactic gradient. Many species of the Vibrionaceae, including V. fischeri, are chemotactic towards chitooligosaccharides (Bassler et al., 1991; Hirano et al., 2011), and chitobiose specifically induces a subset of V. fischeri genes, including several encoding certain chemotaxis proteins (A. Schaefer, personal communication). In addition, a recent study (Mandel et al., 2012) showed that V. fischeri uses chemotaxis toward chitobiose to navigate into host tissues; however, when and how this chemoattractant gradient is produced was not addressed. We showed that the initial host-symbiont contact results in an up-regulation of EsChitotriosidase, whose catalytic function releases chitobiose. The data suggest that this activity establishes a chitobiose gradient in the vicinity of the pores and primes the symbionts to induce chitin-responsive gene transcription in the aggregates. As a result, pre-exposure to chitin breakdown products enhances the ability of symbiotic V. fischeri cells to subsequently sense a chemoattractant gradient toward chitobiose (Figure 6). Taken together, these results suggest that symbiont induction of host EsChitotriosidase shapes the local chemical environment in a way that favors both the eventual migration of the symbionts through the pores, and thus efficient establishment of a symbiotic colonization. Chitobiose may not be the only chemoattractant in the system; other chemoattractants may be responsible for bringing the bacteria to their final residence, the crypt spaces. This priming behavior is not unlike that of the human symbiont Bacteroides thetaiotaomicron, which responds to host-derived fucose residues by inducing enterocyte glycosylation (Bry et al. 1996). Interestingly, a pathogenic Escherichia coli strain has recently been shown to also sense Bacteroides-released fucose and, as a result, use it as a cue to induce expression of virulence genes (Pacheco et al., 2012). Whether other hosts produce (oligo)saccharide signals that are used by bacteria to establish a persistent association remains to be determined, but the existing examples already point to the role of such signaling in inter-domain communication between coevolved partners (Oldroyd, 2013; McFall-Ngai et al., 2013).
Surprisingly, our results suggest the possibility that interaction with only a few symbiont cells results in the propagation of a transcriptional signal for EsChitotriosidase across the ~10,000 cells of the entire organ; i.e., the pattern revealed by in situ hybridization provides evidence that the induction of gene expression is not confined to the few host cells directly interacting with the symbiont aggregate (Figure 2A), but is shared by much of the ciliated surface epithelium. These results contrast with a previous study of whole light organs in which in situ hybridization indicated that a symbiont-induced increase in expression of the C8 subunit of the proteasome was confined to one cell type in the ciliated fields (Kimbell et al., 2006). The results of the present study suggest that localized signaling by V. fischeri cells not only promotes specificity on the ciliated surface, and migration into host tissues, but also may prepare distant tissues (e.g., the crypt epithelium) for the eventual colonization by the symbiont. More refined techniques, such as laser-capture microdissection (Hooper, 2004), will be required to determine the types and amounts of transcripts that change in those host cells that directly bind symbionts, compared to adjacent epithelial cells. Also, further studies will determine whether the observed changes in gene expression are either transient, and thus specific to these early events, or part of a longer-term or permanent change in response to the symbiont. Sequential waves of transcriptomic changes have been detected following induction of different immune responses in influenza-infected lungs (Pommerenke et al., 2012), a kinetic pattern that may also occur during the subsequent 3–5 h transit of V. fischeri cells from the surface to the deep crypts.
In summary, the squid-vibrio system provides an ideal candidate for testing recent models of epithelial selection, which predict parameters that favor particular bacterial species and stabilize a host-bacteria mutualism (Schluter and Foster, 2012). While this study has afforded a window into initial animal host responses to a bacterial partner, rapid development of new sequencing technologies promise to provide robust methods for the study of single-cell transcriptomics for bacteria such as V. fischeri. These horizons will present the opportunity to eavesdrop into the very first conversations of an animal host with its coevolved partner.
EXPERIMENTAL PROCEDURES
Adult E. scolopes were collected in Oahu (Hawaii) and bred in the laboratory. All experiments conform to the relevant regulatory standards established by UW-Madison.
Transcriptomic database using 454 pyrosequencing
Juvenile animals were collected within 15 min of hatching and randomly segregated into various experimental conditions in Hawaiian Offshore SeaWater (HOSW), which contains ~106 environmental bacteria/mL but undetectable V. fischeri: hatchling, aposymbiotic, symbiotic (with ~5000 cells/mL of ES114 strain added to the HOSW). Light organs were dissected and placed into RNAlater (Ambion) (n=400/condition for 454 sequencing) and frozen at −80°C until RNA extraction (RNeasy kit, Qiagen). cDNA was generated (SMART cDNA synthesis kit, Clontech Laboratories) and its quality checked using a Bioanalyzer 2100 (Agilent Technologies). Samples were prepared for the 454 sequencing and sequenced on a GS-FLX system with Titanium chemistry (Roche Life Sciences). Sequences were processed and assembled through Newbler, annotated for Blast results and Gene ontology by Blast2GO, and clustered in orthologous groups. Reads were quantified (RSEM package, R software), and their differential expression was estimated using the Stekel’s method (Rj > 3 were differentially expressed) and FatiGO software. For details and references, see Supplemental Experimental Procedures.
Expression of candidate genes
Squid were collected as for the 454 sequencing, with an additional ‘sterile’ condition in FSIO (filter-sterilized Instant Ocean). Quantitative RT-PCR procedures conform to MIQE guidelines (Bustin et al., 2009) for light-organ collection (20 squid/replicate), RNA preparation, cDNA preparation, qPCR amplification, expression normalization against 3 housekeeping genes, and data analysis.
Full-length sequence was confirmed by Rapid Amplification of cDNA Ends. In situ probes were synthetized from cDNA using a T7 polymerase (Promega) and primers containing the T7-binding site at the 5′ end. Whole mount in situ hybridizations were performed as described in Lee et al. (2009) with incubations at 60°C. For details, see Supplemental Experimental Procedures.
Immunocytochemistry (ICC) and protein biochemistry
Polyclonal antibodies against the squid chitotriosidase and chitin synthase were generated from synthetic peptides made to unique regions in the protein (GenScript), and were determined to be highly specific by Western blot when used against squid protein extracts. For ICC, the fixation, permeabilization, blocking, and washing procedures were modified from Troll et al., 2010 (see Supplementary Experimental Procedures). For ICC experiments involving mucus staining, the procedure was identical to classical ICCs, except that squid were fixed in Bouin’s solution for 3 h, permeabilized for 2 h, and the mucus was counterstained with 10 μg/mL Alexa633-conjugated WGA.
The chitotriosidase was purified by affinity chromatography: squid protein extract was incubated for 1 h at 4 °C with 50 μL of pre-washed chitin bound to magnetic beads (New Englands Biolabs). Unbound proteins were washed, and purified proteins were loaded (after beads boiled for 10 min in 1X TCEP-loading buffer) into a NuPAGE 4–12 % Bis-Tris gel and stained with SYPRO Ruby (Life Technologies). Chitotriosidase activity was tested using a chitinase assay kit (Sigma-Aldrich) on the bead-protein complex against 50 μg of 4-methylumbelliferyl-β-D-N,N′,N″-triacetylchitotriose, which fluoresces at 535 nm after cleavage by chitotriosidase (for details, see Supplementary Experimental Procedures).
Determination of mucus pH
Wheat germ agglutinin (WGA, Vector laboratories), which labels host mucus, was mixed with the dual emission pH probe SNARF-4F 5-(and -6) carboxylic acid (SNARF, Molecular Probes) at a 1:10 ratio for 15 min with agitation. SNARF was covalently linked to WGA by incubating EDAC at a 1:10 ratio with the protein overnight at 4°C, reaction that was quenched by adding 1 M glycine. Free SNARF was rinsed from the protein by filtration through Amicon Ultra-4 Centrifugal Filter Units (EMD Millipore) with mPBS. A calibration curve of SNARF, at a working concentration of 10 μM in mPBS at pH values from 5.5 to 8, was generated after excitation at 488 nm. Squid were incubated for 30 min in 10 μg/mL WGA-SNARF in FSIO, and washed 3 times in FSIO before anesthesia. The signal intensity in 100 μm2-squares was counted at 580 nm and 650 nm emissions.
Localization of chitin derivatives
The 3h-squid were incubated with a mix of 20 μg/mL of Alexa633-WGA, which stains GlcNAc residues and sialic acid, 20 μg/mL of FITC-succinylated WGA, which stains GlcNAc residues only, and 10 mM Cell Tracker orange in FSIO for 30 min. Squid were washed 3 times in FSIO and anesthetized before mounting. To localize chitin in the light organ, squid were incubated in 1% calcofluor white, which intercalates into chitin and cellulose, in FSIO for 5 min following the manufacturer’s instructions (Sigma-Aldrich). For FITC-conjugated chitin-binding protein (New England Biolabs) staining, squid were fixed and permeabilized as for the ICC protocol in mucus, and CBP was added at a dilution of 1:250 in mPBS for 3 days.
Capillary assays for chemoattraction
Chemotaxis toward 10 mM chitobiose was measured in 1 μL capillary tubes, after growth in SWT supplemented or not with 1 mM chitobiose ((GlcNAc)2). Non-primed cells showed chemotaxis toward GlcNAc (Figure S3), which indicated that they were both motile and chemotactic. To further control for any indirect effects during growth in the presence of a sugar, we measured chemotaxis towards chitobiose by V. fischeri cells that were pre-exposed to either 100 μM or 1 mM GlcNAc, and observed no response (fewer than 4000 CFUs per capillary, data not shown). For details, see Supplemental Experimental Procedures.
Squid colonization after chitotriosidase inhibition
Squid were pre-incubated in 12-wells plates for 2 h in 2 mL FSIO+IgG; i.e. FSIO containing either α-EsChitotriosidase antibody (20 μg/mL) to bind specific sites, or rabbit IgG (20 μg/mL, Genscript) as a control (6 squid/well, 3 wells/condition). Squid were then transferred for 3 h into FSIO+IgG containing either 3000 or 5000 V. fischeri per mL. Squid were quickly rinsed to stop colonization, transferred into FSIO+IgG, and incubated overnight. Luminescence was scored after 18 h in a luminometer (Turner Designs), and bacterial density was checked at the end of the experiment to ensure a similar inoculation for both conditions. Because colonization data follow a binomial distribution, statistics were calculated using the Fisher’s Exact Test (R software, version 2.14.1). Control with 20 μg/mL of bovine serum albumin (BSA) was also performed to rule out any potential effect of the IgG (Fisher’s Exact Test between IgG and BSA (3000 CFU/mL, p = 0.71; 5000 CFU/mL, p = 1.00). Reduction of chitotriosidase activity against 4-methylumbelliferyl-β-D-N,N′,N″-triacetylchitotriose was tested in vitro as described above, with no addition, addition of 40 μg of α-EsChitotriosidase antibody or rabbit IgG (n=3 replicates/condition).
Supplementary Material
HIGHLIGHTS.
Contact with a few V. fischeri induces specific gene expression changes in host squid
V. fischeri induced host genes include a catalytically active chitotriosidase
Chitotriosidase is secreted into the acidic mucus where chitin derivatives are present
Chitin degradation promotes V. fischeri chemoattraction and efficient host colonization
Acknowledgments
We thank Fabrice Vavre for helpful discussions, and Dorina Ölsner for assistance. This work was supported by NIH grants AI R01 50661 to MM-N and RR12294 to EGR and MM-N, NSF grant IOS 0817232 to MM-N and EGR, the DFG priority program 1399 and structural funds of the DFG Clusters of Excellence Inflammation Interfaces and Future Ocean to PR, and Marie Curie Actions FP7-PEOPLE-2010-IOF/272684/SymbiOx to NK.
Footnotes
ACCESSION NUMBERS
Sequences were deposited in the Sequence Read Archive, under the biological project PRJNA205147. The Genbank accession number of EsChitotriosidase is KF015222.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altura MA, Heath-Heckman EAC, Gillette A, Kremer N, Krachler AM, Brennan CA, Ruby EG, Orth K, McFall-Ngai MJ. First engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol. 2013 doi: 10.1111/1462-2920.12179. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ordóñez A, Begley M, Prieto M, Messens W, López M, Bernardo A, Hill C. Salmonella spp. survival strategies within the host gastrointestinal tract. Microbiology. 2011;157:3268–81. doi: 10.1099/mic.0.050351-0. [DOI] [PubMed] [Google Scholar]
- Badariotti F, Thuau R, Lelong C, Dubos MP, Favrel P. Characterization of an atypical family 18 chitinase from the oyster Crassostrea gigas: evidence for a role in early development and immunity. Dev Comp Immunol. 2007;31:559–70. doi: 10.1016/j.dci.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Gibbons PJ, Yu C, Roseman S. Chitin utilization by marine bacteria. Chemotaxis to chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24268–75. [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–50. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M, Schoolnik GK. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 2007;3:e81. doi: 10.1371/journal.ppat.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümer N, Pfefferle PI, Renz H. Development of mucosal immune function in the intrauterine and early postnatal environment. Curr Opin Gastroenterol. 2007;23:655–60. doi: 10.1097/MOG.0b013e3282eeb428. [DOI] [PubMed] [Google Scholar]
- Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–3. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carlson SA, Sharma VK, McCuddin ZP, Rasmussen MA, Franklin SK. Involvement of a Salmonella genomic island 1 gene in the rumen protozoan-mediated enhancement of invasion for multiple-antibiotic-resistant Salmonella enterica serovar Typhimurium. Infect Immun. 2007;75:792–800. doi: 10.1128/IAI.00679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Park IY, Kim HS, Lee WT, Kim MS, Kim SC. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 2002;16:429–31. doi: 10.1096/fj.01-0736fje. [DOI] [PubMed] [Google Scholar]
- Chun CK, Scheetz TE, Bonaldo MdF, Brown B, Clemens A, Crookes-Goodson WJ, Crouch K, DeMartini T, Eyestone M, Goodson MS, et al. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics. 2006;7:154. doi: 10.1186/1471-2164-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conus S, Simon HU. Cathepsins and their involvement in immune responses. Swiss Med Wkly. 2010;140:w13042. doi: 10.4414/smw.2010.13042. [DOI] [PubMed] [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–51. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Dinglasan RR, Devenport M, Florens L, Johnson JR, McHugh CA, Donnelly-Doman M, Carucci DJ, Yates JR, 3rd, Jacobs-Lorena M. The Anopheles gambiae adult midgut peritrophic matrix proteome. Insect Biochem Mol Biol. 2009;39:125–34. doi: 10.1016/j.ibmb.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–76. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Eberl G. Development and evolution of RORγt+ cells in a microbe’s world. Immunol Rev. 2012;245:177–88. doi: 10.1111/j.1600-065X.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, Samoilovich MP, Bosch TCG. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci U S A. 2010;107:18067–72. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusetti F, von Moeller H, Houston D, Rozeboom HJ, Dijkstra BW, Boot RG, Aerts JMFG, van Aalten DMF. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. J Biol Chem. 2002;277:25537–44. doi: 10.1074/jbc.M201636200. [DOI] [PubMed] [Google Scholar]
- Heath-Heckman EAC, McFall-Ngai MJ. The occurrence of chitin in the hemocytes of invertebrates. Zoology (Jena) 2011;114:191–8. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegedus D, Erlandson M, Gillott C, Toprak U. New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol. 2009;54:285–302. doi: 10.1146/annurev.ento.54.110807.090559. [DOI] [PubMed] [Google Scholar]
- Hirano T, Aoki M, Kadokura K, Kumaki Y, Hakamata W, Oku T, Nishio T. Heterodisaccharide 4-O-(N-acetyl-β-D-glucosaminyl)-D-glucosamine is an effective chemotactic attractant for Vibrio bacteria that produce chitin oligosaccharide deacetylase. Lett Appl Microbiol. 2011;53:161–6. doi: 10.1111/j.1472-765X.2011.03083.x. [DOI] [PubMed] [Google Scholar]
- Hooper LV. Laser microdissection: exploring host-bacterial encounters at the front lines. Curr Opin Microbiol. 2004;7:290–5. doi: 10.1016/j.mib.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Kimbell JR, Koropatnick TA, McFall-Ngai MJ. Evidence for the participation of the proteasome in symbiont-induced tissue morphogenesis. Biol Bull. 2006;211:1–6. doi: 10.2307/4134572. [DOI] [PubMed] [Google Scholar]
- Krukonis ES, DiRita VJ. From motility to virulence: Sensing and responding to environmental signals in Vibrio cholerae. Curr Opin Microbiol. 2003;6:186–90. doi: 10.1016/s1369-5274(03)00032-8. [DOI] [PubMed] [Google Scholar]
- Lee PN, McFall-Ngai MJ, Callaerts P, de Couet HG. Whole-mount in situ hybridization of Hawaiian bobtail squid (Euprymna scolopes) embryos with DIG-labeled riboprobes: II. Embryo preparation, hybridization, washes, and immunohistochemistry. Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.prot5322. pdb.prot5322. [DOI] [PubMed] [Google Scholar]
- Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, He CH, Takyar S, Elias JA. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 2011;73:479–501. doi: 10.1146/annurev-physiol-012110-142250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li-Ling J, Hou L, Li Q, Ma F. Identification and characterization of a chitinase-coding gene from Lamprey (Lampetra japonica) with a role in gonadal development and innate immunity. Dev Comp Immunol. 2009;33:257–63. doi: 10.1016/j.dci.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EAC, Deloney-Marino CR, McFall-Ngai MJ, Ruby EG. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol. 2012;78:4620–4626. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Hadfield MG, Bosch TCG, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110:3229–36. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–4. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310:1824–7. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- Merrell DS, Hava DL, Camilli A. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol. 2002;43:1471–91. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG. The N-acetyl-D-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol Microbiol. 2011;82:894–903. doi: 10.1111/j.1365-2958.2011.07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;68:5113–22. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- Ong DST, Wang L, Zhu Y, Ho B, Ding JL. The response of ferritin to LPS and acute phase of Pseudomonas infection. J Endotoxin Res. 2005;11:267–80. doi: 10.1179/096805105X58698. [DOI] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–7. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommerenke C, Wilk E, Srivastava B, Schulze A, Novoselova N, Geffers R, Schughart K. Global transcriptome analysis in influenza-infected mouse lungs reveals the kinetics of innate and adaptive host immune responses. PLoS One. 2012;7:e41169. doi: 10.1371/journal.pone.0041169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen MA, Carlson SA, Franklin SK, McCuddin ZP, Wu MT, Sharma VK. Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect Immun. 2005;73:4668–75. doi: 10.1128/IAI.73.8.4668-4675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biol. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–61. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, McFall-Ngai MJ. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. 2010;12:2190–2203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-vibrio symbiosis. Mol Microbiol. 2010;78:903–15. doi: 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A. 2010;107:2259–64. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Zhang S. Identification, expression and bioactivity of a chitotriosidase-like homolog in amphioxus: dependence of enzymatic and antifungal activities on the chitin-binding domain. Mol Immunol. 2012;51:57–65. doi: 10.1016/j.molimm.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol. 2006;62:1586–600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.