Abstract
Population connectivity and spatial distribution are fundamentally related to ecology, evolution and behaviour. Here, we combined powerful genetic analysis with simulations of particle dispersal in a high-resolution ocean circulation model to investigate the distribution of green turtles foraging at the remote Palmyra Atoll National Wildlife Refuge, central Pacific. We analysed mitochondrial sequences from turtles (n = 349) collected there over 5 years (2008–2012). Genetic analysis assigned natal origins almost exclusively (approx. 97%) to the West Central and South Central Pacific combined Regional Management Units. Further, our modelling results indicated that turtles could potentially drift from rookeries to Palmyra Atoll via surface currents along a near-Equatorial swathe traversing the Pacific. Comparing findings from genetics and modelling highlighted the complex impacts of ocean currents and behaviour on natal origins. Although the Palmyra feeding ground was highly differentiated genetically from others in the Indo-Pacific, there was no significant differentiation among years, sexes or stage-classes at the Refuge. Understanding the distribution of this foraging population advances knowledge of green turtles and contributes to effective conservation planning for this threatened species.
Keywords: feeding ground, marine turtle, Chelonia mydas, mixed stock analysis, ocean currents, control region
1. Introduction
Movements that vary among stages shape the life histories of diverse taxa. Marine connectivity is thought to be greatly influenced by source population size and ocean circulation processes [1]. However, recent work has revealed increasingly complex scenarios with other factors, such as swimming behaviour [2,3], mortality [4,5] or intermittent climatic events like storms [6] playing key roles in determining the distributions of marine organisms. In numerous animal species with life cycles characterized by ontogenetic shifts in habitat utilization, population distribution remains insufficiently understood owing to cryptic stages and poorly defined linkages among stages [7]. Deficiencies in basic information on the distribution of such species impede the development of scientifically sound management recommendations and hinder understanding of population biology [8].
In marine turtles, after hatchlings emerge from nests on sandy beaches, they enter the ocean, where they are thought to spend their ‘lost years’. This stage is thus termed, because turtle location is largely unknown [9], although a testable hypothesis for green turtles (Chelonia mydas) has recently been proposed [7]. In this oceanic stage, turtles primarily drift with currents before settling into neritic habitats as juveniles [9]. After recruitment at approximately 3–5 years of age [10], green turtles are among species that generally forage in mixed aggregations drawn from various rookeries [7,8]. These feeding grounds (FGs) may contain a mix of stage-classes including transitory reproductive individuals moving through on their breeding migrations [11], or single stages as found in juvenile developmental habitats or adult FGs [12,13]. Adults undertake breeding migrations between FGs and nesting beaches that may be widely geographically separated [14–16]. Mating occurs offshore of the rookery and/or during reproductive migrations [17,18]. Many females return to nest in the area of their birth, a behaviour known as natal homing [9]. Less is known about males as their cryptic marine habitat makes them more difficult to research than females coming ashore to nest. However, recent studies have shown that adult males can migrate to disparate FGs like females, although males may return to breed more frequently than females [19–21]. Highly migratory green turtles are thus important elements of diverse and often-distant ecosystems. Effective conservation of these threatened [22,23] species requires understanding these connections [24–26].
Marine turtle researchers have primarily used genetic markers in combination with satellite telemetry and mark–recapture work to infer movement patterns and population distribution. The mixed stock analysis (MSA) method was developed to trace the natal origins of individuals at FGs [27,28]. In this approach, molecular markers are used to determine contributions of genetically differentiated source populations to FGs. Owing to natal homing, various Indo-Pacific green turtle rookery stocks are sufficiently differentiated at maternally inherited mitochondrial loci to allow for MSA [12,18,29–35]. Genetic analysis showed that Hawaiian FGs, for example, are composed almost entirely of individuals from the Hawaiian French Frigate Shoals rookery [30]. For management purposes, the Hawaiian Archipelago thus represents the Pacific North Central Regional Management Unit (RMU) [36]. By contrast, other regional FGs are mixed stocks with natal origins spanning local to distant rookeries and RMUs, that range from being undifferentiated to significantly different from other FGs [12,33,35].
MSAs indicate that FG genetic composition may be related to rookery size, geographical distance, ocean currents, severe weather and/or juvenile natal homing. In the latter process, young post-pelagic sea turtles move among FGs towards their natal region to feed [37]. However, MSA limitations can include broad confidence intervals (CIs), incomplete sampling, widely shared haplotypes and confounding effects of recent population history [38]. Fine-scale variation must be assessed prior to carrying out MSAs, for example to avoid error from non-resident males or females transiting through the FG [11,39]. Further, temporal variation has been investigated infrequently in Indo-Pacific green turtles, and research is needed into this issue that can impact MSAs assuming temporal constancy [40].
In a complementary approach, simulating hatchling dispersal within an ocean circulation model allows researchers to generate spatially explicit predictions based on trajectories for thousands of virtual particles. These predictions of transport can be compared with genetic MSA results to investigate effects of ocean currents on FG distribution and natal origins. Recent studies tracking particles released at Atlantic rookeries revealed significant impacts of ocean currents on genetic connectivity [7,24,41]. In this study, thousands of virtual particles were released into an ocean circulation model in the vicinity of our focal FG and tracked backwards through time to identify possible oceanic pathways available to turtles for reaching that FG—the Palmyra Atoll National Wildlife Refuge (PANWR).
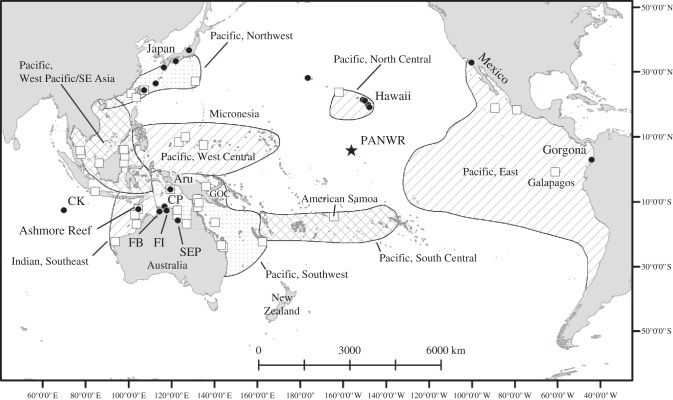
The PANWR is located within the Pacific Remote Islands Marine National Monument, about halfway between Hawaii and American Samoa, and is one of the least impacted coral reef systems in the central Pacific (figure 1). Palmyra Atoll is not currently inhabited except for limited occupation by management and research personnel. The Refuge contains a mixed stage-class green turtle FG that includes post-pelagic juveniles, subadults and adults. Also found there were a few turtles exhibiting the tapered carapace, light skin and darker carapace coloration described for eastern Pacific green turtle populations [42] and turtles caught in Central North Pacific fisheries [43]. Hawksbill turtles (Eretmochelys imbricata) were also present though less common, and turtle nesting is rare [44,45]. Green turtles at the PANWR were reported to lack the fibropapilloma tumours [45] prevalent in other populations such as Hawaii [46]. Surveys revealed an uneven distribution of green turtles around the 12 km2 atoll, with abundance hot spots off the reef flats to the north, south, west and east [45]. The PANWR FG is likely connected to other regional areas for breeding and possibly foraging, and turtles leaving these protected waters may be subjected to threats such as habitat loss, harvest and fishery interactions [23], underscoring the need to reveal their unknown connectivity.
Figure 1.
Location of the PANWR (star) with respect to other C. mydas rookeries (white squares), RMUs (references in table 1) and FGs (black dots) previously subject to genetic analysis. References and/or abbreviations for FGs are as follows—Hawaii [30]; Australasia [33], CK, Cocos Keeling; FB, Fog Bay; FI, Field Island; CP, Cobourg Peninsula; SEP, Sir Edward Pellews Island; GOC, Gulf of Carpentaria; Gorgona, Colombia: [12]; Japan [35].
Here, we combined genetic analysis and dispersal modelling to investigate the population distribution of green turtles foraging at the PANWR. Our goals were to: (i) characterize the genetic composition of the foraging population with mtDNA control region sequences; (ii) investigate fine-scale genetic variation on the atoll, including among years, stage-classes and sexes; (iii) assess genetic differentiation between the PANWR and other FGs; (iv) elucidate the natal origins of turtles foraging at the PANWR using MSA; (v) compare our dispersal modelling results to genetic MSA estimates; and (vi) consider effects of population size, geographical distance, natal homing and ocean currents on FG composition. Resolving the genetic structure of this foraging population will add to overall knowledge of marine connectivity, with a focus on Indo-Pacific green turtles, and improve our ability to form effective conservation plans. This is especially important given the variety of threats sea turtles face along their migratory pathways.
2. Material and methods
2.1. Genetic analysis
2.1.1. Sampling and laboratory procedures
Tissue or blood samples were collected from green turtles captured at the PANWR between 2008 and 2012 (n = 349) using standard and previously used protocols [39,47]. All turtles were examined, measured, tagged for individual identification and released. Samples were collected from 4–24 August 2008, 14 August to 10 September 2009, 14–28 July 2010, 20 July to 18 August 2011 and 30 June to 3 August 2012. The curved carapace length (CCL) of sequenced turtles ranged from 40.3 to 113.6 cm (average = 69.6 cm), which was virtually the same as the entire sampled population [45]. DNA extractions were performed using a DNeasy kit following the manufacturer's instructions (Qiagen Inc.). Primers LCM15382 and H950 were used to amplify an approximately 857 bp fragment of the mtDNA control region and two tRNAs [48]. Standard conditions and negative controls were used, and sequencing was carried out in both directions [39,47]. Sequences were aligned using Sequencher v. 4.6 (Gene Codes Corporation) or Geneious v. 6.1 (Biomatters Inc.) and named according to the standardized Southwest Fisheries Science Center (SWFSC) designations.
2.1.2. Genetic diversity and differentiation
In regional analyses, sequences were truncated to approximately 384 bp for comparison with previous studies (figure 1 and tables 1 and 2). All statistical analyses of Palmyra sequences were conducted using these truncated segments as well as the longer sequences (approx. 857 bp) for comparison. Geneious was used to construct a neighbour-joining tree of Palmyra subhaplotypes derived from these longer sequences using the Tamura–Nei model. Arlequin v. 3.11 [50] was used to calculate the number of haplotypes (a) as well as haplotype (h) and nucleotide (π) diversities [51]. Arlequin was also used to carry out pairwise and global exact tests of population differentiation [52] as well as pairwise tests and analysis of molecular variance (AMOVA) using F-statistics based on haplotype frequencies only [53]. In temporal analyses, samples were compared among years to assess whether the MSA assumption of temporal constancy was met.
Table 1.
Green sea turtle control region haplotype relative frequencies detected at Palmyra and the RMUs, with respect to total sample size (n). Also shown is number of nesting females per RMU, with references.
| FG |
Regional Management Units |
|||||||
|---|---|---|---|---|---|---|---|---|
| Indian |
Pacific |
|||||||
| Palmyra | Southeast | West Pacific/SE Asia | West Central and South Central | Southwest | Northwest | North Central | East | |
| haplotype | ||||||||
| CMP1 | 0.009 | 0.681 | 0.003 | |||||
| CMP2 | 0.148 | |||||||
| CMP3 | 0.170 | 0.035 | ||||||
| CMP4 | 0.020 | 0.643 | ||||||
| CMP5 | 0.109 | |||||||
| CMP6 | 0.161 | |||||||
| CMP7 | 0.006 | |||||||
| CMP8 | 0.006 | |||||||
| CMP9 | 0.006 | |||||||
| CMP10 | 0.003 | |||||||
| CMP11 | 0.003 | |||||||
| CMP12 | 0.010 | |||||||
| CMP13 | 0.003 | |||||||
| CMP15 | 0.010 | |||||||
| CMP18 | 0.215 | |||||||
| CMP19 | 0.008 | |||||||
| CMP20 | 0.559 | 0.107 | 0.509 | 0.047 | 0.157 | |||
| CMP22 | 0.258 | 0.094 | ||||||
| CMP32 | 0.057 | 0.057 | ||||||
| CMP39 | 0.041 | |||||||
| CMP40 | 0.003 | 0.096 | ||||||
| CMP44 | 0.003 | 0.120 | ||||||
| CMP47 | 0.009 | 0.036 | 0.019 | 0.334 | ||||
| CMP49 | 0.003 | 0.250 | 0.399 | 0.125 | 0.116 | |||
| CMP50 | 0.099 | |||||||
| CMP54 | 0.314 | |||||||
| CMP57 | 0.006 | 0.343 | ||||||
| CMP60/CMP61 | 0.003 | 0.151 | ||||||
| CMP65 | 0.026 | 0.038 | ||||||
| CMP66 | 0.005 | |||||||
| CMP67 | 0.005 | |||||||
| CMP68 | 0.003 | |||||||
| CMP76 | 0.066 | |||||||
| CMP77 | 0.014 | 0.066 | 0.017 | |||||
| CMP80 | 0.003 | 0.055 | ||||||
| CMP81 | 0.012 | |||||||
| CMP82 | 0.025 | |||||||
| CMP83 | 0.524 | 0.172 | ||||||
| CMP84 | 0.005 | |||||||
| CMP85 | 0.003 | |||||||
| CMP86 | 0.012 | |||||||
| CMP87 | 0.096 | 0.003 | ||||||
| CMP88 | 0.003 | |||||||
| CMP89 | 0.052 | |||||||
| CMP90 | 0.048 | |||||||
| CMP91 | 0.012 | 0.035 | 0.073 | |||||
| CMP97 | 0.017 | |||||||
| CMP109 | 0.003 | |||||||
| CMP126 | 0.033 | |||||||
| CMP132 | 0.003 | |||||||
| CMP170 | 0.003 | |||||||
| CMP207 | 0.003 | |||||||
| Total | 349 | 84 | 198 | 106 | 383 | 121 | 229 | 311 |
| reference for haplotypes | this study | [32] | [32] | [32], Dutton et al. 2013,unpublished data | [32] | [31,34] | [30] | [29,30] |
| nesting females | 10 100 | 21 000 | 2985 | 23 925 | 518 | 574 | 3750 | |
| reference for nesting females | n.a. | [36,49] (Scott reef unquantified) | [36] | [44] | [36,49] (PNG unquantified) | [36,49] | [36] | [36] |
Table 2.
Mitochondrial control region diversity at Palmyra (in italic), as compared to other Indo-Pacific FGs from the published literature, with references. For standardization with other studies these measures were based on ∼384 bp long mtDNA segments, and recalculated for FGs described in the literature. One individual that was not measured was not included in this analysis.
| foraging ground | no. haplotypes | haplotype diversity (h) | nucleotide diversity (π) | CCL range (cm) | sample size | reference |
|---|---|---|---|---|---|---|
| Pacific | ||||||
| Hawaii, USA | 6 | 0.464 ± 0.018 | 0.003 ± 0.002 | not given | 788 | [12,30] |
| Yaeyama, Japan | 24 | 0.836 ± 0.022 | 0.033 ± 0.017 | 33.0–95.6a | 142 | [35] |
| Ginoza, Japan | 9 | 0.879 ± 0.043 | 0.035 ± 0.018 | 37.5–90.1a | 20 | [35] |
| Kanto, Japan | 8 | 0.696 ± 0.036 | 0.026 ± 0.013 | 33.0–105.2a | 145 | [35] |
| Gorgona, Colombia | 7 | 0.300 ± 0.080 | 0.011 ± 0.006 | 42.7–77.6 | 55 | [12] |
| Cocos Keeling Is. | 3 | 0.452 ± 0.070 | 0.001 ± 0.001 | 40–113.6 | 36 | [33] |
| Cobourg Peninsula | 15 | 0.785 ± 0.029 | 0.027 ± 0.014 | 66.6–103 | 91 | [33] |
| Aru Is. | 8 | 0.722 ± 0.059 | 0.037 ± 0.019 | 39.4–112.4 | 40 | [33] |
| Sir Edward Pellew Is. | 7 | 0.643 ± 0.035 | 0.008 ± 0.005 | 38.7–133.5 | 102 | [33] |
| AR/FB/FI | 22 | 0.713 ± 0.029 | 0.011 ± 0.006 | 33.7–78.2 | 194 | [33] |
| Palmyra juvenile (<65 cm) | 14 | 0.620 ± 0.037 | 0.011 ± 0.006 | 40.3–64.8 | 157 | this study |
| Palmyra subadult (65–84.9 cm) | 10 | 0.617 ± 0.041 | 0.011 ± 0.006 | 65.0–84.5 | 111 | this study |
| Palmyra adult (>84.9 cm) | 9 | 0.618 ± 0.037 | 0.005 ± 0.003 | 85.0–113.6 | 80 | this study |
| Palmyra male | 5 | 0.589 ± 0.060 | 0.003 ± 0.002 | 85.0–99.6 | 31 | this study |
| Palmyra female | 4 | 0.661 ± 0.070 | 0.008 ± 0.005 | 85.2–99.2 | 19 | this study |
| Palmyra all | 19 | 0.619 ± 0.023 | 0.009 ± 0.005 | 40.3–113.6 | 348 | this study |
aStraight carapace length only.
Nesting has been reported rarely at Palmyra but occurs regionally [44,45], and transient adults migrating through the area to breed might be confused with resident foraging turtles. Therefore, genetic differentiation among juveniles, subadults and adults was tested. Following Sterling et al. [45], individuals with CCL less than 65 cm were classified as juveniles, subadults were between 65 and 84.9 cm, and adults were greater than 85 cm. We also compared males with females. Individuals with CCL greater than or equal to 85 cm and tails greater than or equal to 30 cm long were classified as males, whereas those greater than or equal to 85 cm with tails less than or equal to 21 cm were considered females, with the caveat that laparoscopy was not carried out and visual assignment of gender must be interpreted with caution. Prior to carrying out MSA, it was necessary to determine whether the PANWR could be considered a mixed stock. To test the possibility of single origins, the pairwise tests described above were used to compare Palmyra to Indo-Pacific RMU rookeries [36] shown in table 1 and figure 1. Significance values were obtained from at least 10 000 permutations. All significant tests were corrected for multiple comparisons using the sequential Bonferroni procedure [54].
2.1.3. Mixed stock analysis
Bayesian MSAs [27] were carried out to investigate PANWR natal origins at Indo-Pacific RMU rookeries [36] shown in table 1 and figure 1. RMUs with available genetic data were used as possible sources for the PANWR: (i) Pacific, Northwest; (ii) Pacific, Southwest; (iii) Pacific, West Central combined with neighbouring Pacific, South Central (to address issues with small rookery sample sizes); (iv) Pacific, West/Southeast Asia; (v) Indian, Southeast; (vi) Pacific, North Central; and (vii) Pacific, East (table 1 and figure 1). The Bayes program requires that sequences not found at any of the sources be removed; thus, five haplotypes unique to Palmyra (CMP97, 109, 132, 170, 207) comprising 3% of its green turtles were excluded. Seven chains, one per region, were run with 50 000 Markov chain Monte Carlo steps per chain. Each chain was initiated with a 95% contribution from one region per chain. The first half of the steps were discarded as burn-in, whereas the remaining 25 000 steps were used to calculate the posterior distribution of all chains combined. Four MSAs were carried out. The first (MSA1) had equal prior probabilities for each region, whereas, in the second (MSA2), priors were weighted to reflect annual numbers of nesting females [39,47] as shown in table 1. Following Proietti et al. [55], in MSA3, priors were weighted according to particle drift trajectories intersecting with each RMU (the percentage of particles arriving at the PANWR from each RMU), and in MSA4, priors were set considering both population size and particle modelling (the percentages used in MSA3 weighted by population size). In all analyses, Gelman and Rubin diagnostics confirmed chain convergence to the posterior density, with all shrink factors less than or equal to 1.0. Pearson's linear correlation tests were used to compare MSA estimates using Statplus v. 2009 (http://www.analystsoft.com/en/products/statplusmacle/).
2.2. Particle tracking
Hindcast output was extracted from the Global Hybrid Coordinate Ocean Model (Global HYCOM) [56] to examine how surface currents might influence the probability of green turtles from different RMUs reaching the PANWR. Global HYCOM output has a spatial resolution of 0.08° (approx. 6–9 km grid spacing), a snapshot of current velocity at 00 : 00 h each day, and is forced using wind stress, wind speed, heat flux and precipitation. HYCOM assimilates satellite altimetry data, sea surface temperature and in situ measurements from a global array of expendable bathythermographs, Argo floats, and moored buoys to produce hindcast model output. Thus, Global HYCOM accurately resolves mesoscale processes such as meandering currents, fronts, filaments and oceanic eddies as well as the transport of surface drifters.
Ichthyop v. 2.21 particle tracking software was used to identify possible migratory corridors post-hatchling turtles might use to reach the PANWR. Virtual particles were released within a 5.0° × 5.0° zone centred on Palmyra. Unlike previous simulations using Ichthyop [2] in which particles were released at a ‘start’ location and tracked forward through time, in this study, particles were released at their ‘final’ location (Palmyra) and tracked backwards through time. Particles were advected using a Runge–Kutta fourth-order, time-stepping method whereby particle position was calculated each half an hour [57]. This backtracking method has been used successfully to characterize population connectivity among green turtle rookeries and FGs throughout the Atlantic basin [7]. In total, 73 000 particles were released (40 particles per day) between 2012 and 2008. Latitude and longitude were recorded for each particle at 5 day intervals. HYCOM output is available from 2003 to present. Thus, to explore how annual variability in ocean circulation could influence recruitment to the PANWR, we plotted backtracking trajectories for particles released each year (2012, 2011, 2010, 2009, and 2008). To qualitatively examine how drift time might influence our estimates of connectivity, we tracked particles for the duration of available HYCOM output. Thus, particles released in 2012 were backtracked for 7 total years; those released in 2011 were backtracked for 6 total years, etc. However, to standardize drift times across years for weighting the genetic MSA, only the proportion of particles (calculated so that they would sum to 100%) that arrived at the PANWR from individual rookeries located in the seven RMUs within 3 years of drift was used. Three years of drift was chosen to maximize the annual variability depicted in ocean circulation while giving particles sufficient time to disperse; moreover, additional years of drift did not greatly alter the outcome of dispersal trajectories.
3. Results
3.1. Genetic analysis
3.1.1. Diversity and differentiation
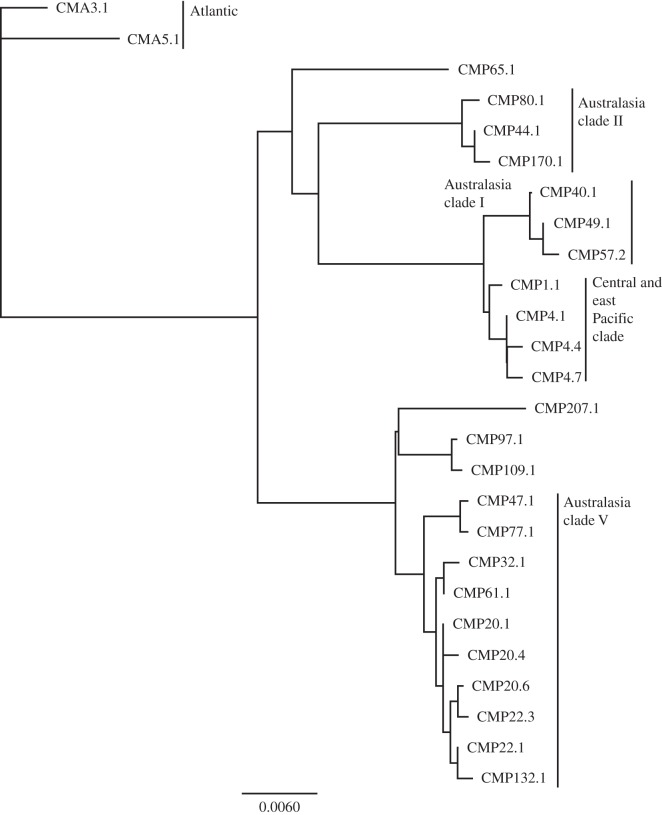
In total, 19 distinct haplotypes were identified, three of which were previously unknown (table 1 and figure 2). The three new haplotypes were assigned standardized names and accessioned on GenBank (CMP109.1, GU121961.1; CMP132.1, KF282705; CMP207.1, KF282706). The most common PANWR haplotype was CMP20 (found in 56% of PANWR turtles), reported from rookeries throughout the Pacific including at four RMUs (figure 1 and table 1). The second most common haplotype at Palmyra was CMP22 (25.8%). Among rookeries, CMP22 is known only from the Pacific West Central and Pacific South Central RMUs (table 1). The Pacific West Central RMU was the only one containing another PANWR haplotype, CMP32 (5.7% at Palmyra), whereas the Pacific South Central RMU uniquely contained the CMP65 haplotype (2.6% at Palmyra). The three most common Palmyra haplotypes as well as the newly discovered CMP132.1 fell within a closely related clade (figure 2). The phylogenetic relationships of CMP20 and CMP22 subhaplotypes were poorly resolved and possibly non-monophyletic. The remaining new haplotypes (CMP 109.1, CMP 207.1) joined CMP97.1 in another clade (figure 2). Seven turtles with the endemic eastern Pacific haplotype CMP4 were also found at the PANWR, comprising 2% of the sample (table 1 and figure 2). All had tapered carapaces characteristic of the eastern Pacific, but only three adults/subadults were darker greenish-black in carapace colour. Of the remaining four turtles carrying the haplotype, two juveniles were closer to the common golden-brown coloration, and a subadult and a juvenile were characterized by greyer coloration. The remaining PANWR haplotypes were rare (less than 5%) and belonged to Australasia clades I, II and V (figure 2), but none belonged to Australasia clades III and IV consisting of rare haplotypes from Malaysia, Australia and New Caledonia [32,33].
Figure 2.
Neighbour-joining tree of subhaplotypes (approx. 857 bp) found at the PANWR, with respect to rookery clades. Branch lengths are proportional to sequence divergence, and Atlantic haplotypes CMA3.1 and 5.1 were used as outgroups.
Analysis of the shorter control region segment showed that haplotype diversity (h = 0.619 ± 0.023) and nucleotide diversity (π = 0.009 ± 0.005) were average compared with other FGs and similar to Sir Edward Pellew Island, Australia, although the number of haplotypes and sample size were relatively high (table 2). Pairwise comparisons showed no significant differentiation among individuals captured in different years at the PANWR (see the electronic supplementary material, table S1). The tests also showed no difference between females and males, or between juveniles, subadults and adults (see the electronic supplementary material, table S1). The PANWR was highly differentiated from other Indo-Pacific FGs (table 3), and from the RMUs (table 1, p < 0.0001). The FG AMOVA showed the percentage of variation among populations was lower than within populations (38.47% and 61.53%, respectively; FST = 0.385, p < 0.0001).
Table 3.
Control region pairwise exact test p-values (below diagonal) and pairwise FST values (above diagonal) among Indo-Pacific green turtle FGs. The Palmyra study site is shown in bold. Asterisks indicate statistically significant comparisons prior to corrections. Values that were no longer significant after sequential Bonferroni corrections are shown in italics. Differences between tests are highlighted in grey. References as in table 2.
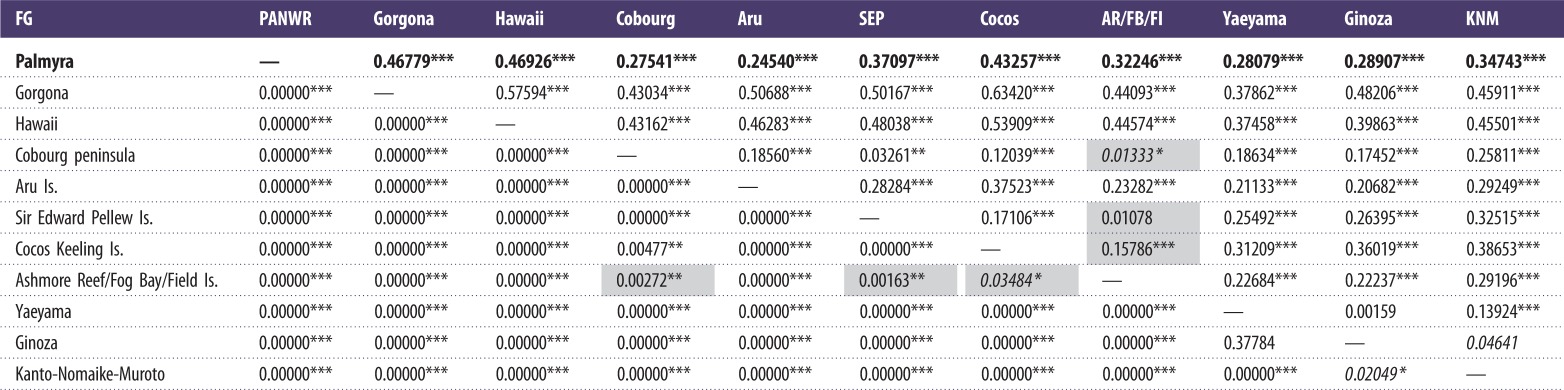 |
*p < 0.05, **p < 0.01, ***p < 0.001.
The longer control region fragment revealed 24 subhaplotypes. Haplotype diversity was 0.627 ± 0.023, and nucleotide diversity was 0.007 ± 0.004. Three of the variants identified for CMP4 (CMP4.1 (n = 5), 4.4 (n = 1) and 4.7 (n = 1; Dutton et al. 2013, unpublished data) were found in our sample. While 193 of the CMP20 sequences were CMP20.1, one sequence was CMP20.4 and another was CMP20.6. Similarly, one of the 90 CMP22 sequences had a new subhaplotype (CMP22.3). The other haplotypes had single subhaplotypes designated with their original CMP nomenclature followed by ‘.1’ (e.g. CMP20.1), except for CMP57.2 (Dutton et al. 2013, unpublished data). When the longer sequences were examined, there continued to be no significant differentiation among: (i) years, (ii) juveniles, subadults and adults or (iii) females and males (see the electronic supplementary material, table S1).
3.1.2. Mixed stock analysis
MSA estimates for even and weighted priors were highly correlated (R > 0.9999, p < 0.0001), and CIs were comparatively narrow. PANWR natal origins were constrained almost exclusively (approx. 97%) to the Pacific, West Central and Pacific, South Central combined RMUs (table 4).
Table 4.
Mixed stock analysis of Palmyra green sea turtle control region haplotypes using Bayesian methods with equal priors (MSA1), priors weighted to reflect population size (MSA2), priors weighted considering particle modelling (MSA3), and priors weighted by particle modelling and population size (MSA4). Mean values are shown with standard deviation (s.d.). The 2.5% and 97.5% values indicate the upper and lower bounds of the 95% CI.
| RMU | MSA | mean | s.d. | 2.5% | median | 97.5% |
|---|---|---|---|---|---|---|
| Indian, Southeast | MSA1 | 0.001 | 0.002 | 0.000 | 0.000 | 0.005 |
| MSA2 | 0.001 | 0.002 | 0.000 | 0.000 | 0.006 | |
| MSA3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| MSA4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Pacific, West Pacific, SE Asia | MSA1 | 0.008 | 0.006 | 0.000 | 0.008 | 0.023 |
| MSA2 | 0.010 | 0.006 | 0.000 | 0.009 | 0.025 | |
| MSA3 | 0.001 | 0.003 | 0.000 | 0.000 | 0.013 | |
| MSA4 | 0.003 | 0.005 | 0.000 | 0.000 | 0.018 | |
| Pacific, South and West Central | MSA1 | 0.975 | 0.013 | 0.945 | 0.977 | 0.994 |
| MSA2 | 0.978 | 0.011 | 0.952 | 0.979 | 0.994 | |
| MSA3 | 0.984 | 0.012 | 0.958 | 0.986 | 1.000 | |
| MSA4 | 0.978 | 0.013 | 0.949 | 0.979 | 0.999 | |
| Pacific, Southwest | MSA1 | 0.008 | 0.007 | 0.000 | 0.007 | 0.024 |
| MSA2 | 0.010 | 0.007 | 0.000 | 0.008 | 0.026 | |
| MSA3 | 0.003 | 0.006 | 0.000 | 0.000 | 0.019 | |
| MSA4 | 0.007 | 0.007 | 0.000 | 0.005 | 0.024 | |
| Pacific, Northwest | MSA1 | 0.001 | 0.002 | 0.000 | 0.000 | 0.005 |
| MSA2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| MSA3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| MSA4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Pacific, North Central | MSA1 | 0.003 | 0.005 | 0.000 | 0.000 | 0.016 |
| MSA2 | 0.000 | 0.002 | 0.000 | 0.000 | 0.003 | |
| MSA3 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| MSA4 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Pacific, East | MSA1 | 0.005 | 0.008 | 0.000 | 0.000 | 0.028 |
| MSA2 | 0.002 | 0.006 | 0.000 | 0.000 | 0.022 | |
| MSA3 | 0.012 | 0.010 | 0.000 | 0.010 | 0.035 | |
| MSA4 | 0.012 | 0.010 | 0.000 | 0.011 | 0.035 |
3.2. Particle tracking
The paths drifting objects take to reach Palmyra within 3 years are shown in figure 3. In principle, connectivity between the PANWR and turtle rookeries associated with all RMUs spanning the equatorial Pacific is possible via surface currents. The only RMU in which no connectivity was predicted was the North Central Pacific (Hawaii). Annual variability in ocean circulation indicates that particles are reaching Palmyra from rookeries (see the electronic supplementary material, figure S1, white squares) along a near-Equatorial swathe traversing the Pacific. Particles are primarily arriving at the PANWR from the east, but some years transport also comes from the west. Although transport of the particles from the west to the PANWR was sporadic, when it occurred it was quite rapid; some particles travelled more than 8000 km in a year or less (see the electronic supplementary material, figure S1). The proportion of particles that reached the PANWR within 3 years from rookeries within each RMU (figure 3, white squares) was 49.74% arriving from the South Central and West Central Pacific, 49.52% from the East Pacific, 0.57% from the Southwest Pacific, 0.15% from the West Pacific/Southeast Asia, 0.02% from the Northwest Pacific, and 0.01% from the Southeast Indian Ocean.
Figure 3.
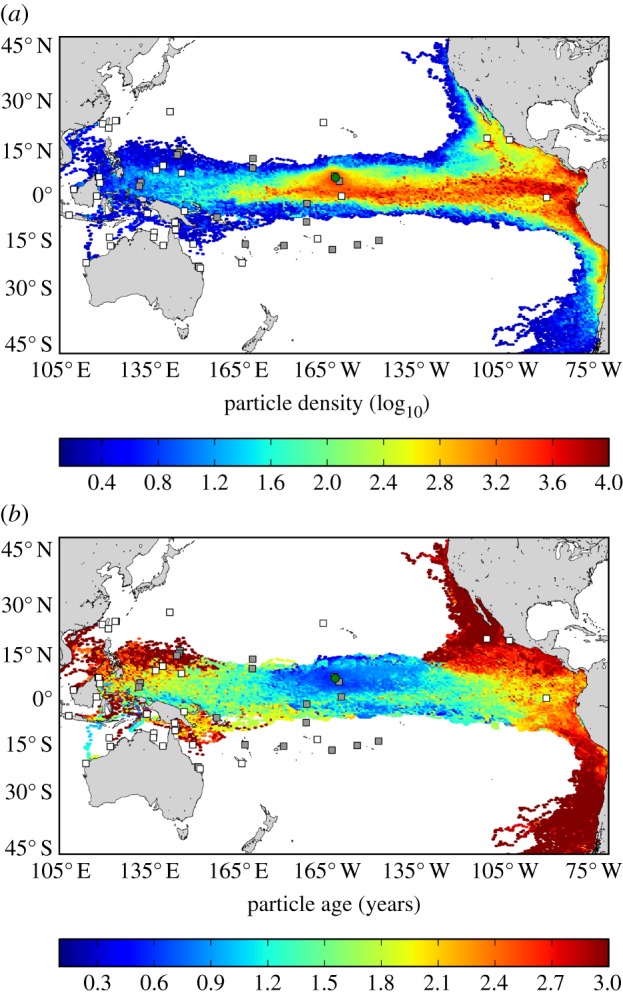
Distribution of 73 000 particles tracked in reverse for 3 years from Palmyra Atoll (green circle) relative to green turtle nesting sites, including those analysed genetically (white squares) as well as those not yet subject to genetic analysis (grey squares). (a) Shading indicates the number of particles at a particular location throughout the 3 year simulations (counted at 5 day intervals). Thus, this map identifies connectivity ‘hot spots’ between oceanic locations and Palmyra Atoll. Note the logarithmic scale. (b) Shading indicates the average number of years a particle would have to drift before reaching Palmyra Atoll from a particular location. (Online version in colour.)
4. Discussion
4.1. Palmyra and regional rookeries
Combining genetic analysis and dispersal simulations suggests that green turtle population distribution in the Indo-Pacific arises from a complex suite of factors. Surprisingly, source population size and ocean circulation processes, two factors that are considered to be very important in predicting marine connectivity [1], do not fully account for the proportion of haplotypes observed among green turtles at Palmyra Atoll. For instance, rookery sizes in the West Pacific/Southeast Asia and Pacific Southwest RMUs dwarf others by an order of magnitude (table 1) and yet at most 5% of the turtles at Palmyra originate from these regions (table 4). Although in the Atlantic population size strongly influenced the composition of FGs such as the Bahamas [58], as, at Palmyra, it was not a determining factor in Australasia [33], Colombia [12] or Japan [35]. Further, our genetic and particle modelling estimates disagree primarily in that the latter indicate nearly half of the particles arrive at Palmyra from the East Pacific RMU. By contrast, the upper genetic estimate of turtles from this RMU is 3.5% (table 4). The finding of a few eastern Pacific haplotypes at the PANWR is consistent with other studies that revealed only their occasional presence across large areas of the Pacific ([35,43] and Dutton et al. 2013, unpublished data). This supports the MSA results, whose validity is also highlighted by narrow CIs and robustness independent of weighting scheme (table 4). We therefore suggest that this discrepancy between genetic estimates and transport predictions is the result of biological processes absent from the particle model.
Our findings support the growing consensus that additional, possibly combined factors, such as mortality [4,5], sporadic meteorological and oceanographic events [6], and swimming and foraging behaviours [2,3] may play key roles in driving distributions and connectivity of marine populations. Studies increasingly indicate that marine animals do not randomly search out food patches, but rather follow somewhat-fixed migratory routes that coincide with typically productive oceanic regions [2,59,60]. For example, young loggerhead turtles in the north Atlantic possess a navigation strategy in which, when they encounter magnetic fields characteristic of specific oceanic regions, they adopt swimming directions that bring them into ocean currents leading to suitable nursery habitat [2]. Additionally, these behaviours generally appear to keep turtles from drifting outside of their normal oceanic range. Recent experiments showing similar responses to magnetic fields in Pacific salmon further suggest that this might be a widespread behaviour among marine animals [61]. Such behaviour would also tend to reduce temporal variability in distributions introduced by ocean circulation variation [62], as found at the PANWR (see the electronic supplementary material, figure S1), because animals would follow ‘average’ paths. This behaviour could potentially minimize effects of climate variation including extreme events such as El Niño, known to affect ocean currents bathing Pacific FGs [12].
Whether young east Pacific green turtles use geomagnetic cues to assess their location and orient their swimming to avoid being swept into the west central Pacific, where foraging areas might not be recognized, is not known. However, even a simple behaviour, such as westward drift eliciting northwards swimming by turtles from the Galapagos Islands, could bring them into contact with the North Equatorial Countercurrent, which would transport them eastward to South America. This transport possibility is well supported by genetic analysis of a green turtle FG near Colombia showing that most of the turtles arrive from the Galapagos [12].
Inclusion of mortality in dispersal models can also greatly alter perceived patterns of distribution [4], and it is possible that transport from east Pacific rookeries is greatly reduced by high mortality rates characteristic of young turtles. However, survival is likely increased with transport away from near-shore environments (where predator abundance is high), and there is no reason to believe that pelagic stage mortality differs substantively between RMUs. Thus, dispersal via ocean currents and active swimming is likely to be favourable for hatchlings of most populations [63,64]. Rather, drifting turtles might adopt swimming behaviour that promotes retention within the first area possessing adequate food availability; a possibility that could favour many turtles staying within the same broad region as their natal site.
Consistent with a hypothesis of transport via ocean currents combined with swimming, a large proportion of turtles was found by genetic MSA to come to Palmyra from the West and South Central combined RMUs. The atoll is bathed by the Equatorial Countercurrent flowing from the west, which could carry pelagic turtles from the West Central Pacific RMU to the PANWR. There was only a small genetic sample available from the South Central RMU (n = 13, Dutton et al. 2013, unpublished data), and further genetic characterization is required to substantiate its connectivity to the PANWR. Indeed, there are some rookeries between the West Central and South Central RMUs that have not been characterized genetically, and are therefore not yet included in any RMU [36] (figure 3). Although relatively small in terms of nesting females, they are still possible sources for the PANWR, where orphan/new haplotypes, an indication that regional rookeries are insufficiently characterized [33,35], were found. However, at 3% frequency, these orphan/new haplotypes (CMP97, 109, 132, 170, 207) are not a large part of the sample. Further, our MSA estimates had narrow CIs as discussed above, and were robust to different weighting schemes, highlighting their reliability. This is likely due to the informative nature of the data and the presence of rookery-specific haplotypes [65], as reported throughout the region [12,33,35]. Large immature green turtles are known to actively swim among different FGs in the Atlantic [66], and swimming by young loggerhead turtles is also reported from Japan [67]. Linkages between Palmyra and the South Central RMU could highlight the importance of directed swimming by turtles of various stage-classes [2,66,68] to reach the small and isolated atoll.
4.2. Palmyra and regional feeding grounds
Genetic analysis provided useful insights concerning the lack of genetic structure at the PANWR and links to other FGs. Palmyra was distinct from all other characterized FGs, as was Gorgona in Colombia [12], highlighting their uniqueness. Australasian and Japanese FGs were also distinct from most other FGs, although some aggregations were made up of more than one FG [33,35]. The lack of distinctiveness among stage-classes at the PANWR may reflect a lack of juvenile migrations [33]. The study also provided data about cryptic males in that we detected no significant differentiation between males and females at this FG. At Palmyra, the lack of genetic variation among years was consistent with other areas ([35,39,47,69], but see [40]). Because temporal constancy is an MSA assumption, this validated the method's use. The results of pairwise comparisons were not substantively affected by using the longer mitochondrial sequences, however, it is possible that the subhaplotypes will be more useful once longer sequences are available from other regional sites [53].
Finally, our study provided information regarding the correspondence between genotype and phenotype for eastern Pacific turtles sometimes referred to as ‘black turtles’. Early morphological studies noted that eastern Pacific turtles were not always characterized by the darker coloration [70, 71]. Genetic studies revealed that, although turtles with the characteristic phenotype generally carry the CMP4 and other haplotypes endemic to eastern Pacific rookeries, the relationship is not absolute. At the Gorgona FG, all CMP4 and other endemic eastern Pacific haplotypes were carried by black–green colour morphotypes, whereas turtles with western or central Pacific sequences had more variable and different colorations [12]. However, turtles nesting at Revillagigedos in Mexico and foraging in San Diego Bay, USA may carry endemic eastern Pacific haplotypes but have different phenotypes (Dutton et al. 2013, unpublished data). Phenotypic characteristics (colour and shape) are highly variable and not reliable diagnostics for identifying these individuals (Dutton et al. 2013, unpublished data). At the PANWR, this was also the case; all turtles carrying CMP4 had a tapered carapace characteristic of eastern Pacific populations, but different colorations were present. Furthermore, some individuals identified in the field as possible eastern Pacific turtles based on phenotype had western Pacific haplotypes.
4.3. Conservation applications
Turtles leaving the protected waters of the isolated and mostly uninhabited PANWR may face significant threats and dangers, underscoring the need to understand their population distribution for comprehensive conservation. Our study provides a clearer understanding of where green turtle migratory pathways and dispersal routes are concentrated in the Pacific, and the utility of environmental parameters such as ocean current dispersal modelling for predicting origins and possible occurrence. Space-based conservation management in open ocean systems is challenging, and our results may help pinpoint the geographical regions needing additional monitoring. Dispersal modelling showed that ‘lost years’ pathways might be of high conservation concern, because many are located outside of protected areas and exclusive economic zones (EEZs). Such ‘commons’ may be subject to overexploitation [72] and may be more difficult to manage than hotspots nearer to the coast and within the EEZ of sovereign nations [25,26]. In terms of conservation priority, the Palmyra FG is highly distinct from all others, indicating an argument for its protection. Further, the study is helpful in defining the southern boundary of the North Central Pacific RMU, as it supports Dutton et al.'s [30] conclusions by showing no connectivity between these sites. Special attention should be paid to fibropapillomatosis, a disease with high incidence in areas such as Hawaii, which has not yet been observed on the Atoll [45]. Perhaps the lack of connectivity has slowed spread of the disease from highly infected areas to Palmyra. Habitat loss, climate change, harvest and fishery bycatch affect rookeries in the West Central and South Central RMUs [44], and thus Palmyra by extension. A possible next step will be to incorporate FGs such as the PANWR into RMUs [36], and our research furnishes information needed for these and other regional management initiatives. In conclusion, this study provides data necessary for conservation and management in protected and threatened areas, and assists with regional management of these highly migratory, transboundary and threatened marine turtles.
Acknowledgements
We thank Eric Dougherty and Ellen Trimarco for laboratory support, and are grateful to Pete Ersts for preparing figure 1 and for other valuable assistance. We thank Katherine Holmes for help in the field. We also thank the Palmyra Atoll Research Consortium, The Nature Conservancy and USFWS for facilitating this research. Oregon State University provided computational support for running ocean dispersal simulations. We thank the following people for research support: G. Balazs, A. Clarry, A. Farkas, P. Farkas, K. Frey, A. Gomez, K. Maison, D. McCauley, M. Rice, J. Vander Veur, T. Work and the Telljohann family. We are grateful to Graeme Hays and two anonymous reviewers for comments that greatly improved the manuscript.
Funding statement
This research was approved by the American Museum of Natural History's Institutional Animal Care and Use Committee (IACUC), under permits authorized by the National Oceanic and Atmospheric Administration (NOAA/NMFS permit no. 10027) as well as the PANWR, USFWS (USFWS special use permit nos. 12533-08013, 12533-09018, 12533-10008, 12533-11008, 12533-12008). Samples were collected by the AMNH under awards nos. NA07NMF4540185 and NA10NMF4540299 from NOAA. The statements, findings, conclusions and recommendations are those of the author(s) and do not necessarily reflect the views of the NOAA, or the US Department of Commerce. The use of trade names or products does not constitute endorsement by the US Government. The study was also supported by grants from the Royal Caribbean Ocean Fund and the Regina Bauer Frankenberg Foundation for Animal Welfare. This is Palmyra Atoll Research Consortium publication number PARC-0099.
References
- 1.Cowen RK, Gawarkiewicz G, Pineda J, Thorrold SR, Werner FE. 2007. Population connectivity in marine systems: an overview. Oceanography 20, 14–21. ( 10.5670/oceanog.2007.26) [DOI] [Google Scholar]
- 2.Putman NF, Verley P, Shay TJ, Lohmann KJ. 2012. Simulating transoceanic migrations of young loggerhead sea turtles: merging magnetic navigation behavior with an ocean circulation model. J. Exp. Biol. 215, 1863–1870. ( 10.1242/jeb.067587) [DOI] [PubMed] [Google Scholar]
- 3.Staaterman E, Paris CB. 2013. Modelling larval fish navigation: the way forward. ICES J. Mar. Sci. ( 10.1093/icesjms/fst103) [DOI] [Google Scholar]
- 4.Cowen RK, Lwiza KMM, Sponaugle S, Paris CB, Olson DB. 2000. Connectivity of marine populations: open or closed? Science 287, 857–859. ( 10.1126/science.287.5454.857) [DOI] [PubMed] [Google Scholar]
- 5.Putman NF, Mansfield KL, He R, Shaver DJ, Verley P. 2013. Predicting the distribution of oceanic-stage Kemp's ridley sea turtles. Biol. Lett. 9, 20130345 ( 10.1098/rsbl.2013.0345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monzón-Argüello C, Dell'Amico F, Morinière P, Marco A, López-Jurado LF, Hays GC, Scott R, Marsh R, Lee PLM. 2012. Lost at sea: genetic, oceanographic and meteorological evidence for storm-forced dispersal. J. R. Soc. Interface 9, 1725–1732. ( 10.1098/rsif.2011.0788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putman NF, Naro-Maciel E. 2013. Finding the ‘lost years’ in green turtles: insights from ocean circulation models and genetic analysis. Proc. R. Soc. B 280, 20131468 ( 10.1098/rspb.2013.1468) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen BW, Karl SA. 2007. Population genetics and phylogeography of sea turtles. Mol. Ecol. 16, 4886–4907. ( 10.1111/j.1365-294X.2007.03542.x) [DOI] [PubMed] [Google Scholar]
- 9.Carr AF. 1967. So excellent a fishe: a natural history of sea turtles. Garden City, NY: Natural History Press. [Google Scholar]
- 10.Reich KJ, Bjorndal KA, Bolten AB. 2007. The ‘lost years’ of green turtles: using stable isotopes to study cryptic lifestages. Biol. Lett. 3, 712–714. ( 10.1098/rsbl.2007.0394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Limpus CJ, Reed PC. 1985. The green turtle, Chelonia mydas in Queensland: a preliminary description of the population structure in a coral reef feeding ground. In Biology of Australasian frogs and reptiles (eds Grigg G, Shine R, Ehmann H.), pp. 47–52. Chipping Norton, UK: Surrey Beatty and Sons and The Royal Zoological Society Of New South Wales. [Google Scholar]
- 12.Amorocho DF, Abreu-Grobois FA, Dutton PH, Reina RD. 2012. Multiple distant origins for green sea turtles aggregating off Gorgona Island in the Colombian eastern Pacific. PLoS ONE 7, e31486 ( 10.1371/journal.pone.0031486) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whiting S, Miller JD. 1998. Short term foraging ranges of adult green turtles (Chelonia mydas). J. Herpetol. 32, 330–337. ( 10.2307/1565446) [DOI] [Google Scholar]
- 14.Kennett R, Munungurritj N, Yunupingu D. 2004. Migration patterns of marine turtles in the Gulf of Carpentaria, northern Australia: implications for Aboriginal management. Wildl. Res. 31, 241–248. ( 10.1071/WR03002) [DOI] [Google Scholar]
- 15.Hirth HF. 1997. Synopsis of the biological data on the green turtle Chelonia mydas (Linnaeus 1758). Washington, DC: Fish and Wildlife Service, U.S. Department of the Interior. [Google Scholar]
- 16.Limpus CJ, Miller JD, Parmenter CJ, Reimer D, McLachlan N, Webb R. 1992. Migration of green (Chelonia mydas) and loggerhead (Caretta caretta) turtles to and from eastern Australian rookeries. Wildl. Res. 19, 347–358. ( 10.1071/WR9920347) [DOI] [Google Scholar]
- 17.FitzSimmons NN, Limpus CJ, Norman JA, Goldizen AR, Miller JD, Moritz C. 1997. Philopatry of male marine turtles inferred from mitochondrial DNA markers. Proc. Natl Acad. Sci. USA 94, 8912–8917. ( 10.1073/pnas.94.16.8912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FitzSimmons NN, Moritz C, Limpus CJ, Pope L, Prince R. 1997. Geographic structure of mitochondrial and nuclear gene polymorphisms in Australian green turtle populations and male-biased gene flow. Genetics 147, 1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James MC, Eckert SA, Myers RA. 2005. Migratory and reproductive movements of male leatherback turtles (Dermochelys coriacea). Mar. Biol. 147, 845–853. ( 10.1007/s00227-005-1581-1) [DOI] [Google Scholar]
- 20.Schofield G, et al. 2013. Satellite tracking large numbers of individuals to infer population level dispersal and core areas for the protection of an endangered species. Divers. Distrib. 19, 834–844. ( 10.1111/ddi.12077) [DOI] [Google Scholar]
- 21.Hays GC, Fossette S, Katselidis KA, Schofield G, Gravenor MB. 2010. Breeding periodicity for male sea turtles, operational sex ratios, and implications in the face of climate change. Conserv. Biol. 24, 1636–1643. ( 10.1111/j.1523-1739.2010.01531.x) [DOI] [PubMed] [Google Scholar]
- 22.NMFS, USFWS. 1998. Recovery plan for US Pacific populations of the green turtle (Chelonia mydas). Silver Spring, MD: National Marine Fisheries Service, U.S. Dept. of Commerce. [Google Scholar]
- 23.Wallace BP, et al. 2011. Global conservation priorities for marine turtles. PLoS ONE 6, e24510 ( 10.1371/journal.pone.0024510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godley BJ, Barbosa C, Bruford M, Broderick AC, Catry P, Coyne MS, Formia A, Hays GC, Witt MJ. 2010. Unravelling migratory connectivity in marine turtles using multiple methods. J. Appl. Ecol. 47, 769–778. ( 10.1111/j.1365-2664.2010.01817.x) [DOI] [Google Scholar]
- 25.Dutton PH, Squires D. 2008. Reconciling biodiversity with fishing: a holistic strategy for Pacific sea turtle recovery. Ocean Dev. Int. Law 39, 1–23. ( 10.1080/00908320701831849) [DOI] [Google Scholar]
- 26.Dutton PH, Squires D. 2011. A holistic strategy for Pacific sea turtle conservation. In Conservation and sustainable management of sea turtles in the Pacific Ocean (eds Dutton P, Squires D, Mahfuzuddin A.), pp. 37–59. Honolulu, HI: University of Hawaii Press. [Google Scholar]
- 27.Pella J, Masuda M. 2001. Bayesian methods for analysis of stock mixtures from genetic characters. Fish. Bull. 99, 151–167. [Google Scholar]
- 28.Bolker BM, Okuyama T, Bjorndal KA, Bolten AB. 2007. Incorporating multiple mixed stocks in mixed stock analysis: ‘many-to-many’ analyses. Mol. Ecol. 16, 685–695. ( 10.1111/j.1365-294X.2006.03161.x) [DOI] [PubMed] [Google Scholar]
- 29.Chassin-Noria O, Abreu-Grobois A, Dutton PH, Oyama K. 2004. Conservation genetics of the east Pacific green turtle (Chelonia mydas) in Michoacan, Mexico. Genetica 121, 195–206. ( 10.1023/B:GENE.0000040394.47843.e4) [DOI] [PubMed] [Google Scholar]
- 30.Dutton P, Balazs G, LeRoux R, Murakawa S, Zarate P, Martines L. 2008. Composition of Hawaiian green turtle foraging aggregations: mtDNA evidence for a distinct regional population. Endang. Species Res. 5, 37–44. ( 10.3354/esr00101) [DOI] [Google Scholar]
- 31.Cheng I-J, Dutton PH, Chen C-L, Chen H-C, Chen Y-H, Shea J-W. 2008. Comparison of the genetics and nesting ecology of two green turtle rookeries. J. Zool. 276, 375–384. ( 10.1111/j.1469-7998.2008.00501.x) [DOI] [Google Scholar]
- 32.Dethmers KEM, et al. 2006. The genetic structure of Australasian green turtles (Chelonia mydas): exploring the geographical scale of genetic exchange. Mol. Ecol. 15, 3931–3946. ( 10.1111/j.1365-294X.2006.03070.x) [DOI] [PubMed] [Google Scholar]
- 33.Dethmers KEM, Jensen MP, FitzSimmons NN, Broderick D, Limpus CJ, Moritz C. 2010. Migration of green turtles (Chelonia mydas) from Australasian feeding grounds inferred from genetic analyses. Mar. Freshw. Res. 61, 1376 ( 10.1071/MF10084) [DOI] [Google Scholar]
- 34.Nishizawa H, Abe O, Okuyama J, Kobayashi M, Arai N. 2011. Population genetic structure and implications for natal philopatry of nesting green turtles Chelonia mydas in the Yaeyama Islands, Japan. Endang. Species Res. 14, 141–148. ( 10.3354/esr00355) [DOI] [Google Scholar]
- 35.Nishizawa H, et al. 2013. Composition of green turtle feeding aggregations along the Japanese archipelago: implications for changes in composition with current flow. Mar. Biol. 160, 2671–2685. ( 10.1007/s00227-013-2261-1) [DOI] [Google Scholar]
- 36.Wallace BP, et al. 2010. Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PLoS ONE 5, e15465 ( 10.1371/journal.pone.0015465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowen BW, et al. 2004. Natal homing in juvenile loggerhead turtles (Caretta caretta). Mol. Ecol. 13, 3797–3808. ( 10.1111/j.1365-294X.2004.02356.x) [DOI] [PubMed] [Google Scholar]
- 38.Jensen MP, FitzSimmons NN, Dutton PH. 2013. Molecular genetics of sea turtles. In The biology of sea turtles, vol. 3 (eds Wyneken J, Lohmann KJ, Musick JA.), pp. 135–154. Boca Raton, FL: CRC Press. [Google Scholar]
- 39.Naro-Maciel E, Becker JH, Lima EHSM, Marcovaldi MA, DeSalle R. 2007. Testing dispersal hypotheses in foraging green sea turtles (Chelonia mydas) of Brazil. J. Hered. 98, 29–39. ( 10.1093/jhered/esl050) [DOI] [PubMed] [Google Scholar]
- 40.Bjorndal KA, Bolten AB. 2008. Annual variation in source contributions to a mixed stock: implications for quantifying connectivity. Mol. Ecol. 17, 2185–2193. ( 10.1111/j.1365-294X.2008.03752.x) [DOI] [PubMed] [Google Scholar]
- 41.Blumenthal JM, et al. 2009. Turtle groups or turtle soup: dispersal patterns of hawksbill turtles in the Caribbean. Mol. Ecol. 18, 4841–4853. ( 10.1111/j.1365-294X.2009.04403.x) [DOI] [PubMed] [Google Scholar]
- 42.Pritchard P. 1999. Status of the black turtle. Conserv. Biol. 13, 1000–1003. ( 10.1046/j.1523-1739.1999.98432.x) [DOI] [Google Scholar]
- 43.Parker D, Dutton PH, Balazs GH. 2011. Oceanic diet and distribution of haplotypes for the green turtle, Chelonia mydas, in the Central North Pacific. Pac. Sci. 65, 419–431. ( 10.2984/65.4.419) [DOI] [Google Scholar]
- 44.Maison KA, Kelly IK, Frutchey KP. 2010. Green turtle nesting sites and sea turtle legislation throughout Oceania. Honolulu, HI: National Marine Fisheries Service, U.S. Department of Commerce. [Google Scholar]
- 45.Sterling EJ, Mcfadden KW, Holmes KE, Vintinner EC, Arengo F, Naro-Maciel E. 2013. Ecology and conservation of marine turtles in a central Pacific foraging ground. Chel. Conserv. Biol. 12, 2–16. ( 10.2744/CCB-1014.1) [DOI] [Google Scholar]
- 46.Herbst LH, Jacobson ER. 2003. Practical approaches for studying sea turtle disease and health. In The biology of sea turtles, vol. 2 (eds Lutz PL, Musick JA, Wyneken J.), pp. 385–410. Boca Raton, FL: CRC Press. [Google Scholar]
- 47.Naro-Maciel E, Bondioli ACV, Martin M, de Pádua Almeida A, Baptistotte C, Bellini C, Marcovaldi MÂ, Santos AJB, Amato G. 2012. The interplay of homing and dispersal in green turtles: a focus on the southwestern Atlantic. J. Hered. 103, 792–805. ( 10.1093/jhered/ess068) [DOI] [PubMed] [Google Scholar]
- 48.Leroux RA, et al. 2012. Re-examination of population structure and phylogeography of hawksbill turtles in the wider Caribbean using longer mtDNA sequences. J. Hered. 103, 806–820. ( 10.1093/jhered/ess055) [DOI] [PubMed] [Google Scholar]
- 49.SWOT. 2011. The state of the world's sea turtles (SWOT) report, vol. VI Arlington, TX: SWOT. [Google Scholar]
- 50.Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. ( 10.1111/j.1755-0998.2010.02847.x) [DOI] [PubMed] [Google Scholar]
- 51.Nei M. 1987. Molecular evolutionary genetics. New York NY: Columbia University Press. [Google Scholar]
- 52.Raymond M, Rousset F. 1995. An exact test for population differentiation. Evolution 49, 1280–1283. ( 10.2307/2410454) [DOI] [PubMed] [Google Scholar]
- 53.Shamblin BM, Bjorndal KA, Bolten AB, Hillis-Starr ZM, Lundgren I, Naro-Maciel E, Nairn CJ. 2012. Mitogenomic sequences better resolve stock structure of southern Greater Caribbean green turtle rookeries. Mol. Ecol. 21, 2330–2340. ( 10.1111/j.1365-294X.2012.05530.x) [DOI] [PubMed] [Google Scholar]
- 54.Rice WR. 1989. Analyzing tables of statistical tests. Evolution 43, 223–225. ( 10.2307/2409177) [DOI] [PubMed] [Google Scholar]
- 55.Proietti M, Reisser J, Kinas P, Kerr R, Monteiro D, Marins L, Secchi E. 2012. Green turtle Chelonia mydas mixed stocks in the western South Atlantic, as revealed by mtDNA haplotypes and drifter trajectories. Mar. Ecol. Prog. Ser. 447, 195–209. ( 10.3354/meps09477) [DOI] [Google Scholar]
- 56.Chassignet EP, Hurlburt HE, Smedstad OM, Halliwell GR, Hogan PJ, Wallcraft AJ, Baraille R, Bleck R. 2007. The HYCOM (hybrid coordinate ocean model) data assimilative system. J. Mar. Syst. 65, 60–83. ( 10.1016/j.jmarsys.2005.09.016) [DOI] [Google Scholar]
- 57.Lett C, Verley P, Mullon P, Parada C, Brochier T, Penven P, Blank B. 2008. A Lagrangian tool for modelling ichthyoplankton dynamics. Environ. Model. Softw. 23, 1210–1214. ( 10.1016/j.envsoft.2008.02.005) [DOI] [Google Scholar]
- 58.Lahanas PN, Bjorndal KA, Bolten AB, Encalada SE, Miyamoto MM, Valverde RA, Bowen BW. 1998. Genetic composition of a green turtle (Chelonia mydas) feeding ground population: evidence for multiple origins. Mar. Biol. 130, 345–352. ( 10.1007/s002270050254) [DOI] [Google Scholar]
- 59.Wilson RP, Griffiths IW, Legg PA, Friswell M, Bidder OR, Halsey LG, Lambertucci SA, Shepard ELC. 2013. Turn costs change the value of animal search paths. Ecol. Lett. 16, 1145–1150. ( 10.1111/ele.12149) [DOI] [PubMed] [Google Scholar]
- 60.Costa DP, Breed GA, Robinson PW. 2012. New insights into pelagic migrations: implications for ecology and conservation. Annu. Rev. Ecol. Evol. Syst. 43, 73–96. ( 10.1146/annurev-ecolsys-102710-145045) [DOI] [Google Scholar]
- 61.Putman NF, Scanlan MM, Billman EJ, O' Neil JP, Couture RB, Quinn TP, Lohmann KJ, Noakes DLG. In press Inherited magnetic map guides ocean navigation in juvenile Pacific salmon. Curr. Biol. [DOI] [PubMed]
- 62.Hays GC, Fossette S, Katselidis KA, Mariani P, Schofield G. 2010. Ontogenetic development of migration: Lagrangian drift trajectories suggest a new paradigm for sea turtles. J. R. Soc. Interface 7, 1319–1327. ( 10.1098/rsif.2010.0009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Putman NF, Bane JM, Lohmann KJ. 2010. Sea turtle nesting distributions and oceanographic constraints on hatchling migration. Proc. R. Soc. B 277, 3631–3637. ( 10.1098/rspb.2010.1088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Putman NF, Scott R, Verley P, Marsh R, Hays GC. 2012. Natal site and offshore swimming influence fitness and long-distance ocean transport in young sea turtles. Mar. Biol. 159, 2117–2126. ( 10.1007/s00227-012-1995-5) [DOI] [Google Scholar]
- 65.Karl SA, Toonen RJ, Grant WS, Bowen BW. 2012. Common misconceptions in molecular ecology: echoes of the modern synthesis. Mol. Ecol. 21, 4171–4189. ( 10.1111/j.1365-294X.2012.05576.x) [DOI] [PubMed] [Google Scholar]
- 66.Godley B, Lima E, Åkesson S, Broderick A, Glen F, Godfrey M, Luschi P, Hays G. 2003. Movement patterns of green turtles in Brazilian coastal waters described by satellite tracking and flipper tagging. Mar. Ecol. Prog. Ser. 253, 279–288. ( 10.3354/meps253279) [DOI] [Google Scholar]
- 67.Okuyama J, Kitagawa T, Zenimoto K, Kimura S, Arai N, Sasai Y, Sasaki H. 2011. Trans-Pacific dispersal of loggerhead turtle hatchlings inferred from numerical simulation modeling. Mar. Biol. 158, 2055–2063. ( 10.1007/s00227-011-1712-9) [DOI] [Google Scholar]
- 68.Gaspar P, Benson SR, Dutton PH, Réveillère A, Al E. 2012. Oceanic dispersal of juvenile leatherback turtles: going beyond passive drift modeling. Mar. Ecol. Prog. Ser. 457, 265–284. [Google Scholar]
- 69.Bass AL, Epperly SP, Braun-McNeill J. 2004. Multi-year analysis of stock composition of a loggerhead turtle (Caretta caretta) foraging habitat using maximum likelihood and Bayesian methods. Conserv. Genet. 5, 783–796. ( 10.1007/s10592-004-1979-1) [DOI] [Google Scholar]
- 70.Carr AF. 1961. Pacific turtle problem. Nat. Hist. 70, 64–71. [Google Scholar]
- 71.Pritchard PCH. 1971. Galapagos sea turtles: preliminary findings. J. Herpetol. 5, 1–9. ( 10.2307/1562836) [DOI] [Google Scholar]
- 72.Hardin G. 1968. The tragedy of the commons. Science 162, 1243–1248. ( 10.1126/science.162.3859.1243) [DOI] [PubMed] [Google Scholar]