Abstract
It has recently been shown that structural conditions on the reaction network, rather than a ‘fine-tuning’ of system parameters, often suffice to impart ‘absolute concentration robustness’ (ACR) on a wide class of biologically relevant, deterministically modelled mass-action systems. We show here that fundamentally different conclusions about the long-term behaviour of such systems are reached if the systems are instead modelled with stochastic dynamics and a discrete state space. Specifically, we characterize a large class of models that exhibit convergence to a positive robust equilibrium in the deterministic setting, whereas trajectories of the corresponding stochastic models are necessarily absorbed by a set of states that reside on the boundary of the state space, i.e. the system undergoes an extinction event. If the time to extinction is large relative to the relevant timescales of the system, the process will appear to settle down to a stationary distribution long before the inevitable extinction will occur. This quasi-stationary distribution is considered for two systems taken from the literature, and results consistent with ACR are recovered by showing that the quasi-stationary distribution of the robust species approaches a Poisson distribution.
Keywords: Markov chain, absolute concentration robustness, stability, quasi-stationary distributions, chemical reaction network theory
1. Introduction
The interaction networks of chemical reaction systems of cellular processes are notoriously complex. Despite this, hidden within the complexity there are often underlying structures that, if properly quantified, give great insight into the dynamical or stationary behaviour of the system. In this vein, Shinar & Feinberg [1] have presented conditions on the structure of biochemical reaction networks that are sufficient to guarantee absolute concentration robustness (ACR) on a particular species of the network. When the dynamics of the system are modelled using ordinary differential equations with mass-action kinetics, a species is said to possess ACR if its concentration has the same value at every positive equilibrium concentration permitted by the system of equations, regardless of total molar concentrations. Such a property, which allows cells to respond in a uniform, predictable way given varying environments, is fundamental to many biological processes, including signal transduction cascades and gene regulatory networks [2].
In the two-component EnvZ/OmpR osmoregulatory signalling system, for example, it is important that the amount of phosphorylated OmpR, OmpR-P, which regulates the transcription of the porins OmpF and OmpC, is kept within tight bounds. It has been observed that while OmpR-P is sensitive to the availability of ADP and ATP, it is relatively insensitive to changes in the overall concentrations of the signalling proteins EnvZ and OmpR [3]. Concentration robustness has also been experimentally observed and studied in the two-component KdpD/KdpE [4], PhoQ/PhoP [5] and CpxA/CpxR [6] signalling systems, and in the IDHKP-IDH glyoxylate bypass regulation system [7,8]. Structural sources of robustness have also been identified in the bacterial chemotaxis pathway [9–11].
Consistent with these empirical results, it has been shown that deterministic mathematical models (i.e. ODE models) of the EnvZ/OmpR system yield equilibrium concentrations of OmpR-P that are stable and do not depend on the overall concentrations of either EnvZ or OmpR [12]. This phenomenon of equilibrium concentrations being independent of total molar concentrations was the basis of Shinar & Feinberg [1] where they presented their conditions on the structure of biochemical reaction networks which are sufficient to guarantee ACR on a particular species of the network. Shinar and Feinberg relate the capacity of a network to exhibit ACR to a structural parameter called the deficiency, which is well studied in Chemical Reaction Network Theory [13–15]. They do not consider stability of such equilibria directly, but they do apply their results to several mass-action models of biochemical networks for which stability is known, including the EnvZ/OmpR signal transduction network and the IDHKP-IDH glyoxylate bypass regulatory system. An interesting recent addition to this framework is the work by Karp et al. [16], where the authors carry out a linear analysis of formal expressions in a reaction network to find ACR and more general steady-state invariants.
In this work, we consider stochastically modelled systems satisfying essentially the same network conditions used by Shinar and Feinberg, and we show that strikingly different conclusions are reached pertaining to the long-term dynamics of the systems. For a wide class of biochemical reaction networks with stable ACR equilibria, we show that trajectories are necessarily absorbed by a set of states that reside on the boundary of the positive orthant. Hence, there is necessarily an irreversible ‘extinction’ event in the system. One immediate corollary to this is that the models admit no stationary distributions with mass near the equilibrium of the deterministic model. Our results therefore demonstrate fundamentally different long-term dynamics than those observed in the corresponding deterministic models. Stochastic modelling of chemical reactions is particularly relevant in models of intracellular dynamics because critical proteins may have a low copy number per cell.
Depending upon the total molecular abundances of the constituent species, it may be that such an extinction is a rare event on the relevant timescales of the system. In this case, and under the assumptions used throughout this work, the process will very likely seem to settle down to an equilibrium distribution long before the resulting instability will appear. This distribution is called a quasi-stationary distribution, and ACR-like results may still be obtained by consideration of this distribution. In fact, in two examples provided here, we observe that the quasi-stationary distribution of the absolutely robust species limits in a natural way to a Poisson distribution with mean value given by the concentration predicted by the deterministic model.
Section 2 provides a motivating example for our main results. In §3, we formally introduce the concept of a species exhibiting ACR in the deterministic modelling context, and present both the main theoretical result from [1] together with the main result being introduced here, theorem 2. A detailed proof of theorem 2, and more general results, can be found in the electronic supplementary material. In §4, we consider the quasi-stationary distributions for the example models considered here. We close with a brief discussion.
2. A motivating example
Consider the two-species activation/deactivation network
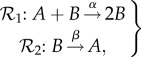 |
2.1 |
where A is the active form of a protein, B is the inactive form, and α and β are positive rate constants [1]. Note that the inactive form B regulates both the activation and deactivation steps of the mechanism. The usual differential equations governing the time evolution of the molar concentrations of A and B, denoted cA and cB here, are
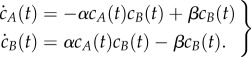 |
2.2 |
Setting the left-hand sides of the above equations to zero and solving yields the following values for the equilibrium concentrations:
| 2.3 |
where 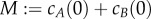 is the total conserved protein concentration. Equation (2.3) shows that the deterministically modelled system (2.1) has ACR for the protein A, as all positive equilibria, no matter the initial condition, must satisfy
is the total conserved protein concentration. Equation (2.3) shows that the deterministically modelled system (2.1) has ACR for the protein A, as all positive equilibria, no matter the initial condition, must satisfy 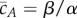 . It can also be easily checked that these equilibria are stable. The self-regulation of the mechanism (2.1), although quite simple, predicts the remarkable property that the concentration of the active protein is kept within tight bounds regardless of the value of the total protein concentration M.
. It can also be easily checked that these equilibria are stable. The self-regulation of the mechanism (2.1), although quite simple, predicts the remarkable property that the concentration of the active protein is kept within tight bounds regardless of the value of the total protein concentration M.
Now consider the usual stochastic model for the network (2.1), which treats the system as a continuous time Markov chain (CTMC) (see figure 1). Let XA(t) and XB(t) denote the individual counts of A and B, respectively, and model the reactions as discrete events which occur stochastically in time. Under usual assumptions on the rates, or propensities, of the reactions, the first reaction can only occur if XA(t) > 0 and XB(t) > 0, and the second only if XB(t) > 0, which implies no reaction may proceed if B is depleted completely. For a more thorough introduction to the stochastic models for biochemical systems, see the electronic supplemental material or [17].
Figure 1.
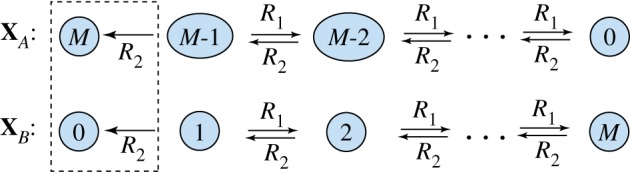
State space for the CTMC corresponding to (2.1). The chain X(t) = (XA(t), XB(t)) has a unique absorbing state at 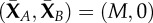 (boxed), where
(boxed), where 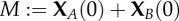 . (Online version in colour.)
. (Online version in colour.)
It is not hard to see that the long-term behaviour of this CTMC is different from that of the deterministic model (2.2). More specifically, it is possible for all of the inactive molecule to become active through the self-activation reaction B → A. This sends the chain to the state
| 2.4 |
where 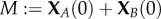 . After this time, neither reaction may occur and so no active molecules A may be deactivated. Therefore, rather than having trajectories of the system spend most of their time near the value (2.3), over a sufficiently long time frame the inevitable outcome of the system is convergence to (2.4).
. After this time, neither reaction may occur and so no active molecules A may be deactivated. Therefore, rather than having trajectories of the system spend most of their time near the value (2.3), over a sufficiently long time frame the inevitable outcome of the system is convergence to (2.4).
This disparity between the long-term predictions of the deterministic mass-action model and that of the stochastic model at first seems to be at odds with the established result that the stochastic chain 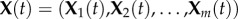 is well approximated by the corresponding deterministic model when molecular counts are high. However, such results are valid only on finite time intervals, and therefore stand silent on the long-term behaviour of the models [18,19]. Isolated examples of models exhibiting such a fundamental difference between the long-term behaviour of the corresponding deterministic and stochastic models are well known in the biochemical literature [20–23].
is well approximated by the corresponding deterministic model when molecular counts are high. However, such results are valid only on finite time intervals, and therefore stand silent on the long-term behaviour of the models [18,19]. Isolated examples of models exhibiting such a fundamental difference between the long-term behaviour of the corresponding deterministic and stochastic models are well known in the biochemical literature [20–23].
3. Results pertaining to absolute concentration robustness
The main problem we consider here is: what structural conditions on the reaction network yield an absorption event similar to that of (2.1) for the corresponding stochastic system? We are particularly interested in networks for which the deterministic model predicts ACR, and in this context we will use the original theorem due to Shinar & Feinberg [1]. We briefly introduce some terminology from chemical reaction network theory, including the network parameters n, ℓ and s.
We let n denote the number of vertices of the reaction network. These vertices are the linear combinations of the species at either end of a reaction arrow and are called complexes in the chemical reaction network literature. We will use this word, although the reference [1] instead uses ‘nodes’ to avoid confusion with the biological meaning of the word ‘complex’. Note that we may naturally associate a complex with a non-negative vector y in which the jth component of y is the multiplicity of species j in that complex. For example, the complex A + B in system (2.1) has associated vector (1, 1), whereas the complex 2B has associated vector (0, 2).
We let ℓ denote the number of connected components of the reaction network and associate with every reaction a reaction vector, which determines the counts of the molecules gained and lost in one instance of that reaction. For example, for the reaction y → y′, the reaction vector is y′ − y, where we have slightly abused notation by writing the vector associated with a complex in the place of the complex itself. We denote by s the dimension of the span of all of the reaction vectors. For the activation/deactivation network (2.1), there are four complexes, {A + B, 2B, B, A}, and two connected components, {A + B, 2B} and {B, A}. It follows that n = 4 and ℓ = 2. We also note that the reaction A + B → 2B has associated reaction vector (−1, 1), since the system loses one A molecule and gains a net of one B molecule owing to one instance of the reaction. The reaction vector for the reaction B → A is (1, −1) so that s = 1.
The deficiency of a network is defined to be 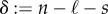 . The deficiency is known to only take non-negative values and has been used to show a variety of steady-state results for mass-action systems, both deterministic and stochastic [13,24–30]. For the network (2.1), we have
. The deficiency is known to only take non-negative values and has been used to show a variety of steady-state results for mass-action systems, both deterministic and stochastic [13,24–30]. For the network (2.1), we have
so the deficiency is one.
We say two complexes are strongly linked if there is a directed path of reactions from the first to the second, and also a directed path from the second back to the first. A strong linkage class of a reaction network is a maximal subset of complexes that are strongly linked to each other. A strong linkage class is furthermore called terminal if no complex in the class reacts to a complex in another strong linkage class. Complexes that do not belong to a terminal strong linkage class are called non-terminal. For example, in the network (2.1), the complexes A + B and B are non-terminal, whereas the complexes 2B and A are terminal. See also the network in figure 2, where the terminal and non-terminal complexes are labelled in different colours. Finally, we say that two complexes ‘differ only in species S’ if the difference between them is a non-zero multiple of a single species S. For example, in (2.1), the complexes A + B and B are both non-terminal and also differ only in species A.
Figure 2.
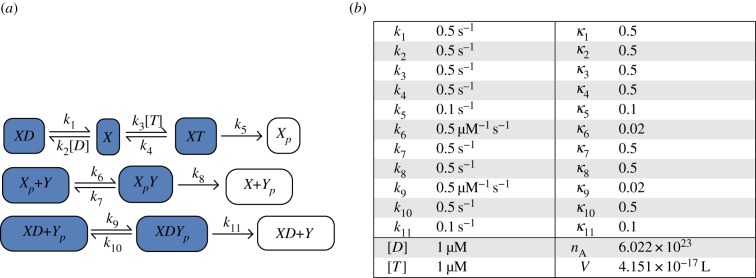
Hypothetical mechanism for the EnvZ/OmpR signal transduction system in Escherichia coli. The mechanism is represented in (a) where the terminal (white) and non-terminal (dark blue) complexes are labelled. The parameter values used for numerical simulations are given in (b). (Online version in colour.)
We can now state the main theorem of [1].
Theorem 3.1. —
Consider a deterministic mass-action system that admits a positive steady state and suppose that the deficiency of the underlying reaction network is one. If, in the network, there are two non-terminal complexes that differ only in species S, then the system has ACR in S.
We have already seen the system (2.1) has a positive steady state if M > β/α, that the underlying network has a deficiency of one, and that the non-terminal complexes A + B and B differ only in the species A. Thus, theorem 3.1 could be used to guarantee that the system exhibits ACR in A even if the equilibrium could not have been calculated explicitly.
Note that theorem 3.1 stands silent on whether or not the equilibria of the deterministically modelled systems are stable, either locally or globally. In fact, there do exist ACR models for which the set of equilibria is unstable. For example,
satisfies the requirements of theorem 3.1, but the equilibrium 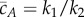 is unstable. The equilibria for the models considered in this article, however, are known to be globally stable.
is unstable. The equilibria for the models considered in this article, however, are known to be globally stable.
3.1. Stochastic differences in robustness
The network (2.1) demonstrates that biochemical reaction networks satisfying the hypotheses of theorem 3.1, and in which the ACR equilibria are known to be stable, may fail to exhibit similar stability when modelled stochastically. This disparity in the long-term dynamics is not restricted to only a few systems. In this section, we provide a theorem proved in the electronic supplemental material which demonstrates that a large class of biochemical reaction networks satisfying the assumptions of theorem 3.1, and thereby exhibit ACR when modelled deterministically, have absorbing boundary states. In that case, when the deterministic ACR equilibria are stable, the stochastic model exhibits fundamentally different long-term dynamics.
In order to state the result corresponding to theorem 3.1 for stochastically modelled systems, we need a few more basic definitions. A chemical reaction network is said to be conservative if there is a vector w with strictly positive components for which w · (y′ − y) = 0 for all reactions y → y′. Note that in this case, there is necessarily a conserved quantity M > 0 so that for the given species set {S1, S2, … , Sm} we have that
is invariant to the dynamics of the system. For example, we have already seen that 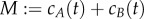 is a conserved quantity for the system (2.1).
is a conserved quantity for the system (2.1).
Enumerating the reactions arbitrarily, we denote by λk(x) the rate, or propensity, function of the reaction yk → y′k, and note that it is reasonable to assume that λk(x) > 0 if and only if xi ≥ yki, which says a reaction may only occur if there are a sufficient number of molecules in the system. We say any family of rate functions satisfying this simple condition is stoichiometrically admissible. For example, the rate function λk(x) = xB would be stoichiometrically admissible for the reaction B → A, but not for the reaction A + B → 2B. A common choice for the propensities λk(x) is stochastic mass-action
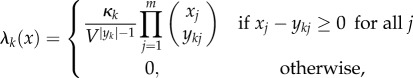 |
where 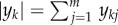 , κk is the rate constant and V is the volume of the reaction vessel. Note that under the assumption of mass-action kinetics, the propensity function is proportional to the number of ways in which one can choose the molecules necessary for the reaction to occur. Also, note that stochastic mass-action kinetics is stoichiometrically admissible.
, κk is the rate constant and V is the volume of the reaction vessel. Note that under the assumption of mass-action kinetics, the propensity function is proportional to the number of ways in which one can choose the molecules necessary for the reaction to occur. Also, note that stochastic mass-action kinetics is stoichiometrically admissible.
We say a complex yk is turned off at a particular state value x if xi < yki for at least one i; otherwise, we say the complex is turned on. Note that λk(x) = 0 for all x at which yk is turned off. Next, we say that a complex y is dominated by the complex y′ (denoted 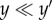 ) if
) if 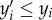 for all i = 1, … , m. In particular, whenever two complexes differ in only one species, one necessarily dominates the other. The intuition here is that if y′ is ever turned off, then so is y.
for all i = 1, … , m. In particular, whenever two complexes differ in only one species, one necessarily dominates the other. The intuition here is that if y′ is ever turned off, then so is y.
Finally, we remind the reader that a state X of a CTMC is recurrent if the chain satisfying X(0) = X returns to X with probability one, and that a recurrent state is positive recurrent if the expected value of the return time is finite. It is also a basic fact that stationary distributions only give mass to positive recurrent states, showing that the long-term dynamics are restricted to those states.
The following is the main theoretical result of the paper and should be compared with theorem 3.1. It, along with more general results allowing for higher deficiency, is proved in the electronic supplemental material.
Theorem 3.2. —
Consider a reaction network which is conservative, has a deficiency of one, and for which the deterministically modelled mass-action system admits a positive equilibrium for some choice of rate constants. Suppose that, in the network, there are two non-terminal complexes, y1 and y2 say, for which
. Then, for any choice of stoichiometrically admissible kinetics, all non-terminal complexes of the network are turned off at each positive recurrent state of the stochastically modelled system.
Hence, a trajectory of the stochastically modelled system will, with a probability of one, be absorbed by a set of states for which all of the non-terminal complexes are turned off. In particular, the propensity of reactions out of the complexes y1 and y2 will be zero.
For instance, in the simple system (2.1), we have 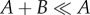 , and theorem 3.2 states that the complexes A + B and B are turned off at any positive recurrent state. This is easily verified since the only positive recurrent state is given by (2.4). In the electronic supplementary material, we also provide a theorem characterizing the limiting behaviour of the system after the absorption event. In particular, we show that the reduced network is weakly reversible and has a deficiency of zero, and so admits a stationary distribution which is a product of Poisson random variables [25]. We also provide in the electronic supplementary material an example in which the conclusions of the theorem do not hold if only the conservation requirement is dropped.
, and theorem 3.2 states that the complexes A + B and B are turned off at any positive recurrent state. This is easily verified since the only positive recurrent state is given by (2.4). In the electronic supplementary material, we also provide a theorem characterizing the limiting behaviour of the system after the absorption event. In particular, we show that the reduced network is weakly reversible and has a deficiency of zero, and so admits a stationary distribution which is a product of Poisson random variables [25]. We also provide in the electronic supplementary material an example in which the conclusions of the theorem do not hold if only the conservation requirement is dropped.
3.2. EnvZ/OmpR signalling system
In order to demonstrate theorem 3.2 on a more complicated example, we now consider a model of the two-component EnvZ/OmpR signalling system in Escherichia coli [3], which was also studied in [1]. The histidine kinase EnvZ is sensitive to extracellular osmolarity, the input of the system, and in its active phosphorylated form EnvZ-P is able to phosphorylate the response regulator OmpR into the active form OmpR-P. OmpR-P in turn signals the transcription of the porins OmpF and OmpC. The level of OmpR-P can therefore be thought of as the output of the system. EnvZ is also known to play a role in the regulation of the level of OmpR-P through dephosphorylation [31].
Multiple mechanisms have been proposed for the EnvZ/OmpR system [3,12,32]. We will consider the mechanism given in figure 2 which was proposed in [1,12]. Here, it is imagined that ADP (D) and ATP (T) interact with EnvZ (X) to produce bound complexes, but that only ATP can successfully transfer the phosphate group (P) to EnvZ to form EnvZ-P (Xp). EnvZ-P may then transfer the phosphate group to OmpR (Y) to form OmpR-P (Yp) while the modified EnvZ–ADP complex regulates the dephosphorylation of OmpR-P. Assuming [D], [T] and [P] are of sufficient quantity to be relatively unchanged by the course of the reaction, we may incorporate them into the rate constants, yielding the network contained in figure 2.
Note that the non-terminal complexes XD and XD + Yp differ only in the species Yp. As the network also has deficiency one (see the electronic supplemental material), by theorem 3.1 we may conclude that the deterministically modelled mass-action system exhibits ACR in Yp. It is shown in the electronic supplemental materials of [1] that the ACR value is
| 3.1 |
The corresponding equilibrium is stable and, consequently, the deterministic model predicts that the active form of the response regulator, OmpR-P, is robust to the overall level of the signalling proteins EnvZ and OmpR. That is to say, the prediction is that the system will exhibit a similar response regardless of differences in these internal characteristics.
We now consider the stochastic model for the network in figure 2. It can be seen that, in addition to satisfying the assumptions of theorem 3.1, the network has the conservation relations
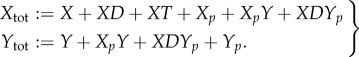 |
3.2 |
As the sum of these two relations has support on all species, it follows that the network is conservative. By theorem 3.2, the stochastic model converges in finite time to a state (or set of states) for which all non-terminal complexes are turned off.
Note that, as the species X, XD, XT, XpY and XDYp are also non-terminal complexes, theorem 3.2 guarantees that each will be zero after the inevitable absorption event. As Xtot is conserved, we may also conclude that Xp = Xtot at this time. Finally, as Xp + Y is also non-terminal, we may also conclude that Y = 0. In this way, the unique sink of the stochastic chain is seen to be
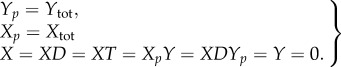 |
3.3 |
4. Time until absorption and quasi-stationary distributions
A disparity between the long-term behaviour of deterministic and stochastic models of some reaction networks is well known in the literature [20–23]. The activation/deactivation network (2.1) is a canonical example that mimics the well-studied stochastic susceptible–infected–susceptible (SIS) epidemic model [33–36], where A and B correspond to the number of healthy and infected individuals, respectively. In that setting, the state (2.4) corresponds to a true ‘extinction state’. A similar extinction effect is also achieved in many population biology models where stochastic effects may irreversibly drive a population to zero [37].
Although the chain associated with (2.1) will inevitably converge to the extinction state (2.4), such an event may be exceedingly rare on biologically reasonable timescales. In such situations, the absorbing state is not of practical concern, and, in fact, the process may seem to settle to a stationary distribution. This distribution is called the quasi-stationary probability distribution, and it is useful in analysing the transient behaviour of a model before the absorption event. We suspect that stable ACR-like behaviour may still be attained in the present context by consideration of this distribution.
Suppose we denote the absorbing states of a process X(t) by ∂A. If we define 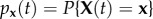 , where x is an arbitrary state in the state space, and
, where x is an arbitrary state in the state space, and 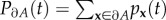 , then the transition probabilities conditioned upon non-extinction, qx(t), are given for t ≥ 0 by
, then the transition probabilities conditioned upon non-extinction, qx(t), are given for t ≥ 0 by
| 4.1 |
The limiting vector 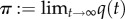 , if it exists, is the quasi-limiting distribution of the process, which equals the quasi-stationary distribution in the present context. See [38] for proofs of the facts that such a distribution exists, and is unique, in the setting of our theorem 3.2. See [37] for a recent survey article on quasi-stationary distributions in the context of population processes.
, if it exists, is the quasi-limiting distribution of the process, which equals the quasi-stationary distribution in the present context. See [38] for proofs of the facts that such a distribution exists, and is unique, in the setting of our theorem 3.2. See [37] for a recent survey article on quasi-stationary distributions in the context of population processes.
Reconsider the activation/deactivation system (2.1). We begin by reimagining the chain 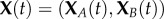 as a birth–death process following XB(t) with the extinction state XB = 0 corresponding to (2.4). The first reaction corresponds to a ‘birth’ since B is increased by one while the second reaction corresponds to a ‘death’ since B is decreased by one. The chain for XA can then be determined by the conservation relation
as a birth–death process following XB(t) with the extinction state XB = 0 corresponding to (2.4). The first reaction corresponds to a ‘birth’ since B is increased by one while the second reaction corresponds to a ‘death’ since B is decreased by one. The chain for XA can then be determined by the conservation relation 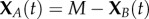 . Under mass-action kinetics, the corresponding birth and death propensities, λ(i) and μ(i), respectively, are given by
. Under mass-action kinetics, the corresponding birth and death propensities, λ(i) and μ(i), respectively, are given by
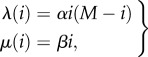 |
4.2 |
where i = 0, … , M corresponds to the state with i molecules of B. Note that λ(0) = λ(M) = μ(0) = 0 so that XB = 0 is an absorbing state and XB = M is a reflecting state. Up to rescaling of the rate constants, this chain is identical to the stochastic SIS epidemic model considered in [33–35].
We are interested in whether the quasi-stationary distribution displays a form of robustness in XA with respect to changes in overall molecularity M. Furthermore, we are interested in the case of large M, as that is when the time to extinction is large. In the electronic supplemental material, we prove that as M → ∞, the quasi-stationary distribution for XA approaches the Poisson distribution
| 4.3 |
which has previously been shown to be the limit of one commonly used approximation to the quasi-stationary distribution [39]. In figure 3, we graphically demonstrate the convergence by providing realizations of the quasi-stationary distribution for different values of β and M.
Figure 3.
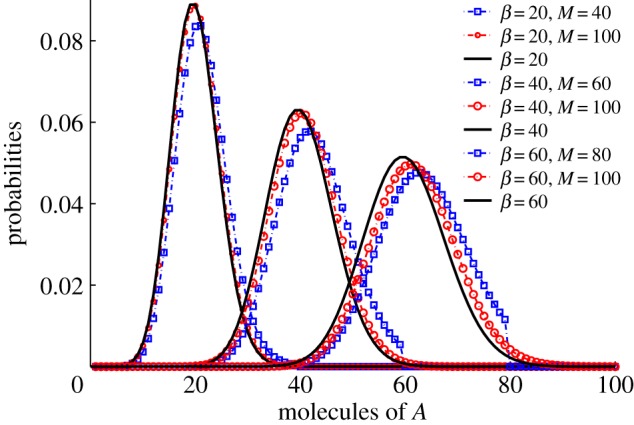
Quasi-stationary distributions of XA with α = 1 and various values of β and M. As M → ∞, the quasi-stationary distributions approach the overlain Poisson distributions (4.3) (solid line). The iterative procedure of [33,34,40] was used to construct the plots. (Online version in colour.)
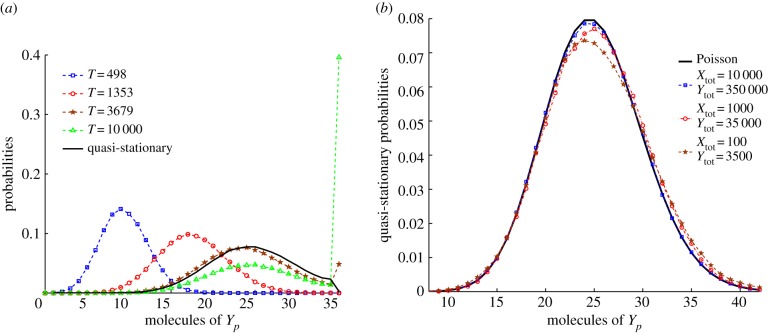
Now consider the EnvZ/OmpR signalling system from figure 2. As in the simple activation/deactivation network, the expected time before entering the state (3.3) may be very large. We therefore consider the quasi-stationary distribution of the process. This distribution cannot be computed using a simple analytic formula, so we approximate it numerically and point the reader to the electronic supplementary material for the details of our computational methods. We use the parameter values given in the table in figure 2b, which have been chosen to be in close agreement with the values in [32]. It was found in [41] that a typical E. coli cell has roughly 100 total molecules of EnvZ and 3500 molecules of OmpR. Therefore, for our simulations we chose a ratio of Ytot : Xtot of 35 : 1. Based on the deterministic model, the anticipated mean of the ACR species Yp in the quasi-stationary distribution is 25.
The results of the simulations are shown in figure 4. In figure 4a we see that, for low molecularity of EnvZ and OmpR (1 and 35, respectively), the chains converge to the boundary state (3.3), even while the quasi-stationary distribution becomes apparent. In figure 4b, we see that the quasi-stationary distribution of Yp appears to converge to a Poisson distribution centred around the deterministic steady state as the total molecularity grows.
Figure 4.
Results of numerical simulation for the EnvZ/OmpR mechanism of figure 2. In (a), we display the approximate probability distribution of the chain X(t) at four time points, computed by averaging 105 independent realizations of the process with Xtot = 1 and Ytot = 35, and initial conditions X(0) = Xtot and Y(0) = Ytot. The corresponding quasi-stationary distribution is overlain (solid). In (b), we display approximations of the quasi-stationary distributions for various values of Ytot and Xtot in the ratio 35 : 1. Details of the simulations are provided in the electronic supplementary material. For comparison, a Poisson distribution with mean 25 is overlain (black). Convergence to the Poisson distribution as the total molecularity increases is apparent. (Online version in colour.)
5. Discussion
Robustness and stability in the face of varying environments are of fundamental importance to the proper functioning of many biological processes, and understanding this behaviour is one arena where mathematics can play a role in elucidating biological phenomena. In this paper, together with its electronic supplemental material, we have outlined a class of structural conditions that are sufficient to guarantee convergence of trajectories of stochastically modelled systems to an absorbing boundary set. Notably, these conditions overlap significantly with a set of structural conditions that are known to confer ACR on the corresponding deterministic models. For such ACR models with stable equilibria, our results present a significant disparity in the predictions for the limiting behaviour of the systems.
This work highlights several points. First, it is surprising that the long-term behaviour of such a large class of systems considered in [1] is fundamentally different for the stochastic and deterministic models. Second, deterministic models are typically unable to capture trapping phenomena such as those described here unless they are artificially modified, for instance by adding degradation terms. We have shown here that for a wide range of models no such ad hoc modifications are necessary for stochastically modelled systems. Third, in regions where the time to absorption for the stochastic model is large relative to the timescale of the system, the proper object of study when considering the stochastic analogue of ACR behaviour is the quasi-stationary distribution, as opposed to the stationary distribution.
This work suggests a number of promising avenues for future research. First, finding more general conditions for which the conclusions of theorem 3.2 hold will be a focus. In particular, weakening the requirement that the system possesses a conservation relation will allow the results to be applicable to more models arising in ecology and population processes, which typically do not satisfy such an assumption, though do often possess low numbers of the constituent species [42]. Second, the question of when equilibria exhibiting ACR in the deterministic modelling context are stable or unstable is, to the best of the authors' knowledge, currently open and has to date received surprisingly little consideration in the literature. Third, we have observed in the two examples considered here the recurrence of the Poisson distribution when analysing a certain limiting behaviour of their quasi-stationary distributions. It is a suspicion of the authors that this phenomenon applies more generally to other systems satisfying ACR, and this will be investigated.
Results of the type presented here are not only of theoretical interest. In particular, they are exactly the types of results required in order to automate the multi-scale reduction methods for stochastic models of biochemical processes currently being explored in the probability literature [43]. For example, in order to determine the behaviour of species operating on a timescale that is slower than other species, it is necessary to first understand the long-term dynamics of the species on the fast timescale in order to perform either the necessary stochastic averaging, or to recognize that some species will have gone extinct.
This work is part of a growing research field in which mathematical methods are developed in order to rise above the bewildering complexity of biochemical processes. Algebraic methods, for example, have been successfully employed in a number of areas related to the equilibria of mass-action systems, where the steady states of such systems form a real algebraic variety [26,44,45]. We believe there is significant progress to be made towards the understanding of stochastically modelled systems through analysis of the underlying network, and we hope to uncover the important substructures hidden in the complexity of biochemical networks that inform system behaviour, both on short and long time frames.
Acknowledgements
We gratefully acknowledge the American Institute of Mathematics (AIM) for hosting a workshop at which this research was initiated.
Funding statement
D.F.A. was supported by NSF grant nos. DMS-1009275 and DMS-1318832. M.D.J. was supported by NSF grant no. DMS-1009275 and NIH grant no. R01-GM086881. G.A.E. was supported by NSF grant nos. DMS-1122478 and DMS-1129008.
References
- 1.Shinar G, Feinberg M. 2010. Structural sources of robustness in biochemical reaction networks. Science 327, 1389–1391. ( 10.1126/science.1183372) [DOI] [PubMed] [Google Scholar]
- 2.Blanchini F, Franco E. 2011. Structurally robust biological networks. BMC Syst. Biol. 5, 74 ( 10.1186/1752-0509-5-74) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batchelor E, Goulian M. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl Acad. Sci. USA 100, 691–696. ( 10.1073/pnas.0234782100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kremling A, Heermann R, Centler F, Jung K, Gilles ED. 2004. Analysis of two-component signal transduction by mathematical modeling using the KdpD/KdpE system of Escherichia coli. Biosystems 78, 23–37. ( 10.1016/j.biosystems.2004.06.003) [DOI] [PubMed] [Google Scholar]
- 5.Miyashiro T, Goulian M. 2008. High stimulus unmasks positive feedback in an autoregulated bacterial signaling circuit. Proc. Natl Acad. Sci. USA 105, 17 457–17 462. ( 10.1073/pnas.0807278105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siryaporn A, Goulian M. 2008. Cross-talk suppression between the CpxA–CpxA and EnvZ–OmpR two-component systems in E. coli. Mol. Microbiol. 70, 494–506. ( 10.1111/j.1365-2958.2008.06426.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaPorte DC, Thorsness PE, Koshland DE. 1985. Compensatory phosphorylation of isocitrate dehydrogenase. J. Biol. Chem. 260, 10 563–10 568. [PubMed] [Google Scholar]
- 8.Shinar G, Rabinowitz JD, Alon U. 2009. Robustness in glyoxylate bypass regulation. PLoS Comput. Biol. 5, e1000297 ( 10.1371/journal.pcbi.1000297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alon U, Surette MG, Barkai N, Leibler S. 1999. Robustness in bacterial chemotaxis. Nature 397, 168–171. ( 10.1038/16483) [DOI] [PubMed] [Google Scholar]
- 10.Barkai N, Leibler S. 1997. Robustness in simple biochemical networks. Nature 387, 913–917. ( 10.1038/43199) [DOI] [PubMed] [Google Scholar]
- 11.Steuer R, Waldherr S, Sourjik V, Kollmann M. 2011. Robust signal processing in living cells. PLoS Comput. Biol. 7, 1–10. ( 10.1371/journal.pcbi.1002218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinar G, Milo R, Martinez MR, Alon U. 2007. Input–output robustness in simple bacterial signaling systems. Proc. Natl Acad. Sci. USA 104, 19 931–19 935. ( 10.1073/pnas.0706792104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg M. 1972. Complex balancing in general kinetic systems. Arch. Ration. Mech. Anal. 49, 187–194. ( 10.1007/BF00255665) [DOI] [Google Scholar]
- 14.Horn F. 1972. Necessary and sufficient conditions for complex balancing in chemical kinetics. Arch. Ration. Mech. Anal. 49, 172–186. ( 10.1007/BF00255664) [DOI] [Google Scholar]
- 15.Horn F, Jackson R. 1972. General mass action kinetics. Arch. Ration. Mech. Anal. 47, 187–194. ( 10.1007/BF00251225) [DOI] [Google Scholar]
- 16.Karp R, Millán MP, Dasgupta T, Dickenstein A, Gunawardena J. 2012. Complex-linear invariants of biochemical networks. J. Theor. Biol. 311, 130–138. ( 10.1016/j.jtbi.2012.07.004) [DOI] [PubMed] [Google Scholar]
- 17.Anderson DF, Kurtz TG. 2011. Continuous time Markov chain models for chemical reaction networks. In Design and analysis of biomolecular circuits: engineering approaches to systems and synthetic biology (eds Koeppl H, Densmore D, Setti G, di Bernardo M.), pp. 3–42. New York, NY: Springer. [Google Scholar]
- 18.Kurtz TG. 1972. The relationship between stochastic and deterministic models for chemical reactions. J. Chem. Phys. 57, 2976–2978. ( 10.1063/1.1678692) [DOI] [Google Scholar]
- 19.Kurtz TG. 1978. Strong approximation theorems for density dependent Markov chains. Stoch. Proc. Appl. 6, 223–240. ( 10.1016/0304-4149(78)90020-0) [DOI] [Google Scholar]
- 20.Allen L. 2003. An introduction to stochastic processes with applications to biology. London, UK: Pearson. [Google Scholar]
- 21.Keizer J. 1987. Statistical thermodynamics of nonequilibrium processes. Berlin, Germany: Springer. [Google Scholar]
- 22.Oppenheim I, Shuler KE, Weiss G-H. 1977. Stochastic theory of nonlinear rate processes with multiple stationary states. Physica A 88, 191–214. ( 10.1016/0378-4371(77)90001-2) [DOI] [Google Scholar]
- 23.Vellela M, Qian H. 2007. A quasistationary analysis of a stochastic chemical reaction: Keizer's paradox. Bull. Math. Bio. 69, 1727–1746. ( 10.1007/s11538-006-9188-3) [DOI] [PubMed] [Google Scholar]
- 24.Anderson DF. 2011. A proof of the global attractor conjecture in the single linkage class case. SIAM J. Appl. Math. 71, 1487–1508. ( 10.1137/11082631X) [DOI] [Google Scholar]
- 25.Anderson DF, Craciun G, Kurtz TG. 2011. Product-form stationary distributions for deficiency zero chemical reaction networks. Bull. Math. Biol. 72, 1947–1970. ( 10.1007/s11538-010-9517-4) [DOI] [PubMed] [Google Scholar]
- 26.Craciun G, Dickenstein A, Shiu A, Sturmfels B. 2009. Toric dynamical systems. J. Symb. Comput. 44, 1551–1565. ( 10.1016/j.jsc.2008.08.006) [DOI] [Google Scholar]
- 27.Feinberg M. 1987. Chemical reaction network structure and the stability of complex isothermal reactors: I. The deficiency zero and deficiency one theorems. Chem. Eng. Sci. 42, 2229–2268. ( 10.1016/0009-2509(87)80099-4) [DOI] [Google Scholar]
- 28.Feinberg M. 1988. Chemical reaction network structure and the stability of complex isothermal reactors: II. Multiple steady states for networks of deficiency one. Chem. Eng. Sci. 43, 1–25. ( 10.1016/0009-2509(88)87122-7) [DOI] [Google Scholar]
- 29.Feinberg M. 1995. The existence and uniqueness of steady states for a class of chemical reaction networks. Arch. Ration. Mech. Anal. 132, 311–370. ( 10.1007/BF00375614) [DOI] [Google Scholar]
- 30.Feinberg M. 1995. Multiple steady states for chemical reaction networks of deficiency one. Arch. Ration. Mech. Anal. 132, 371–406. ( 10.1007/BF00375615) [DOI] [Google Scholar]
- 31.Zhu Y, Qin L, Yoshida T, Inouye M. 2000. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc. Natl Acad. Sci. USA 97, 7808–7813. ( 10.1073/pnas.97.14.7808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igoshin OA, Alves R, Savageau MA. 2008. Hysteretic and graded responses in bacterial two-component signal transduction. Mol. Microbiol. 68, 1196–1215. ( 10.1111/j.1365-2958.2008.06221.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kryscio RJ, Lefévre C. 1989. On the extinction of the S-I-S stochastic logistic epidemic. J. Appl. Probab. 27, 685–694. ( 10.2307/3214374) [DOI] [Google Scholar]
- 34.Nasell I. 1996. The quasi-stationary distribution of the closed endemic SIS model. Adv. Appl. Probab. 28, 895–932. ( 10.2307/1428186) [DOI] [Google Scholar]
- 35.Nasell I. 1999. On the quasi-stationary distribution of the stochastic logistic epidemic. Math. Biosci. 156, 21–40. ( 10.1016/S0025-5564(98)10059-7) [DOI] [PubMed] [Google Scholar]
- 36.Weiss GH, Dishon M. 1971. On the asymptotic behavior of the stochastic and deterministic models of an epidemic. Math. Biosci. 11, 261–265. ( 10.1016/0025-5564(71)90087-3) [DOI] [Google Scholar]
- 37.Méléard S, Villemonais D. 2012. Quasi-stationary distributions and population processes. Probab. Surv. 9, 340–410. ( 10.1214/11-PS191) [DOI] [Google Scholar]
- 38.Darroch JN, Seneta E. 1967. On quasi-stationary distributions in absorbing continuous-time finite Markov chains. J. Appl. Probab. 4, 192–196. ( 10.2307/3212311) [DOI] [Google Scholar]
- 39.Hernández-Suárez CM, Castillo-Chavez C, López OM, Hernández-Cuevas K. 2010. An application of queuing theory to SIS and SEIS epidemic models. Math. Biosci. Eng. 7, 809–823. ( 10.3934/mbe.2010.7.809) [DOI] [PubMed] [Google Scholar]
- 40.Cavender JA. 1978. Quasi-stationary distributions of birth-and-death processes. Adv. in Appl. Probab. 10, 570–586. ( 10.2307/1426635) [DOI] [Google Scholar]
- 41.Cai SJ, Inouye M. 2002. EnvZ–OmpR interaction and osmoregulation in Escherichia coli. J. Biol. Chem. 277, 24 155–24 161. ( 10.1074/jbc.M110715200) [DOI] [PubMed] [Google Scholar]
- 42.Smith HL, Thieme HR. 2011. Dynamical systems and population persistence. Graduate Studies in Mathematics. Providence, RI: American Mathematical Society. [Google Scholar]
- 43.Kang HW, Kurtz TG. 2013. Separation of time-scales and model reduction for stochastic reaction network. Ann. Appl. Probab. 23, 529–583. ( 10.1214/12-AAP841) [DOI] [Google Scholar]
- 44.Manrai A, Gunawardena J. 2009. The geometry of multisite phosphorylation. Biophys. J. 95, 5533–5543. ( 10.1529/biophysj.108.140632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millán MP, Dickenstein A, Shiu A, Conradi C. 2012. Chemical reaction systems with toric steady states. Bull. Math. Biol. 74, 1027–1065. ( 10.1007/s11538-011-9685-x) [DOI] [PubMed] [Google Scholar]