Abstract
Reporting in Developmental Cell, Aramaki et al. (2013) identify T as a key mediator of primordial germ cell (PGC) specification in the embryo. Deconstruction of how Bmp and Wnt signals regulate T expression and targeting to regulatory elements of either mesodermal or PGC genes has implications for differentiation in vitro.
An enduring question in developmental biology is how cells in the early embryo use a limited set of cues to make fate decisions. As in real estate, the answer lies in timing and location. In this issue of Developmental Cell, Aramaki et al. (2013) demonstrate that the transcription factor T integrates signals from two major pathways, delivered in precise sequence, to designate the small cohort of cells that will carry the genome to the next generation.
Specification of these genomic heirs – primordial germ cells (PGCs) – occurs deterministically through inherited cytoplasmic factors in some organisms. However, a different mode of specification prevails in mammals. Pioneering transplantation experiments (Tam and Zhou, 1996) suggested that epiblast cells have equivalent germline potential if they land in the right place—the posterior corner of the proximal epiblast in the mouse. Bmp mutants pointed to signals that define the birthplace of germ cells, implicating Bmp4 in the extraembryonic ectoderm as an essential induction factor (Lawson et al., 1999). However, secreted ligands reach large swaths of cells, so it remained unclear how specific fates are established. Additionally, the limited number of PGCs in the embryo raised technical challenges to dissecting the molecular circuitry involved. Previous work by Mitinori Saitou and colleagues has broken ground in both aspects of this problem, establishing efficient PGC production methods from pluripotent cells and identifying the PGC specification transcriptional blueprint. cDNA libraries from single cells of the embryo proximal posterior region molecularly distinguished the earliest germ cells from mesodermal neighbors and revealed the first steps in PGC allocation (Saitou et al., 2002). Within that signature were the transcriptional regulators Blimp1 and Prdm14, the earliest markers of PGC commitment. Blimp and Prdm14 reporters pinpointed the critical period and cues for PGC competence: a pulse of Wnt3a is the priming factor for Bmp4 to coax the epiblast toward germline commitment (Ohinata et al., 2009).
How do two pedestrian signaling pathways like Wnt and Bmp ordain a small handful of epiblast cells as PGCs? Aramaki et al. (2013) sought the answer in downstream molecular machinery and timing of PGC fate decisions. A critical tool was the in vitro generation of PGCs, which routes embryonic stem cells (ESCs) through an epiblast-like cell (EpiLC) intermediate in a recapitulation of development (Hayashi et al., 2011). In both epiblasts and EpiLCs, the current study showed that Wnt3 and β-catenin are required for Blimp1 induction by Bmp4. Strikingly, they noticed that Blimp1 and Prdm14 transcripts increased more slowly than classical targets of Bmp or Wnt signaling, suggesting their indirect induction. Boolean logic applied to gene expression analysis of EpiLCs identified immediate response genes to combined Wnt3 and Bmp4. Among these, the mesoderm and notochord transcription factor T (Brachyury) stood out for its consistent expression in both mesoderm and nascent germ cells. Aramaki et al. (2013) went on to demonstrate that T is necessary and sufficient for induction of Blimp1, and chromatin immunoprecipitation (ChIP) revealed enrichment of T at loci near Blimp1 and Prdm14. Together these findings raise the tantalizing possibility that germline rather than mesoderm is the default T pathway in the epiblast when Bmp4 and Wnt3 are absent. Yet, T specificity to PGC genes was tightly regulated by timing of Wnt/β-catenin and Bmp4/Smad signals, with T-dependent expression of Blimp1 and Prdm14 precluded by early exposure to Wnt3. The authors propose a two-step model in which Wnt3 and Bmp4 synergize to induce T. The subsequent direction of T to either mesoderm genes or Blimp1 and Prdm14 is determined by the absence or presence, respectively, of Wnt3 and Bmp4 (Figure 1). It is reasonable to hypothesize that Wnt and Bmp-mediated transcription factors, Tcf1 and Smad, co-occupy PGC enhancers with T, but ChIP results did not concur. Alternatively, other Tcf family members may promote or inhibit transcription of PGC or mesoderm genes. Details of how Bmp4 antagonizes T targeting to mesodermal gene loci or promotes T occupancy of Blimp1 and Prdm1 remain to be clarified.
Figure 1. Model of PGC/mesoderm fate choice.
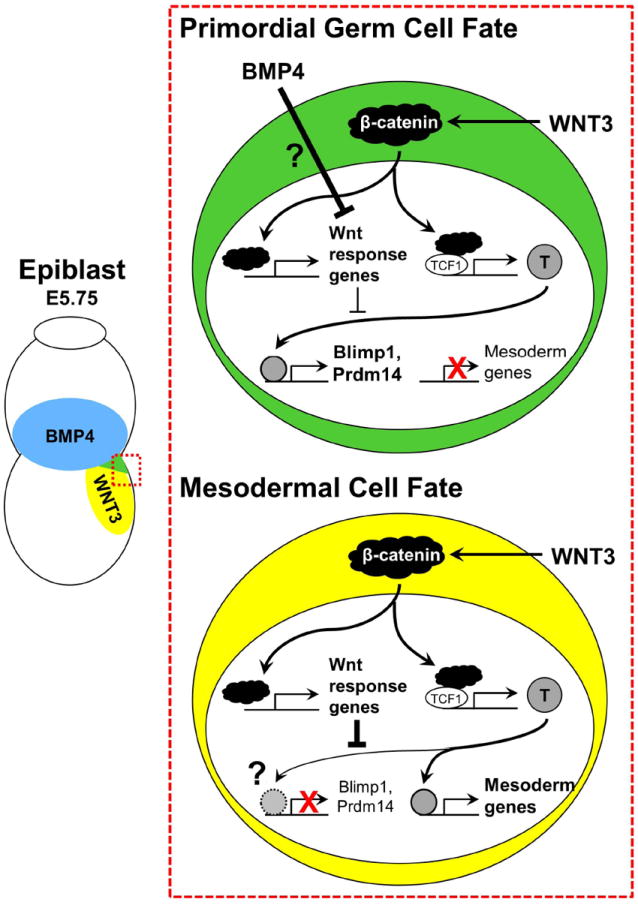
Mouse PGCs are specified in the posterior corner of the proximal epiblast where Bmp4 (blue) and Wnt3 (yellow) signals converge. Neighboring mesodermal cells do not receive high levels of Bmp signals, preventing their specification to the germ cell lineage.
Temporal and spatial coordination of major signaling pathways to lock down expression of lineage specific genes is an emerging theme in development. In the case of PGCs versus mesoderm, T hangs in the balance between Wnt and Bmp signaling. Elsewhere, Wnts and Bmps collaborate in different ways to dictate cell fate decisions. In zebrafish and mouse hematopoietic development, downstream transcription factors Tcfl2 and Smad colocalize with cell fate-specific transcription factors at genes critical for hematopoietic lineages (Trompouki et al., 2011). In human cells, precise timing of Wnt and Bmp signaling dictates hematopoietic versus mesenchymal cell fate specification from a common progenitor pool (Gertow et al., 2013). Aramaki et al. (2013) joins these studies in highlighting specific mechanisms employed by broad signaling networks in different cell contexts at distinct times in development. The temporal and geographic juxtaposition of blood islands in the extraembryonic mesoderm to the PGC birthplace in the proximal epiblast raises the question whether Bmp and Wnt signaling targets common transcription factors to lineage-specific genes via shared mechanisms. In the context of in vivo or in vitrostem cell biology, Bmp and Wnt synergies might suggest protracted lineage flexibility during the early commitment to mesoderm, PGC, or blood. Similarly, the requirement of T for PGC gene expression could explain the inefficiency of differentiation from mouse ESCs, and may guide strategies for improving human PGC derivation. As exemplified by Saitou and colleagues, interweaving approaches in the embryo and the dish toward understanding and recapitulating developmentally relevant intermediates is likely to be a successful strategy for in vitro differentiation in many tissues.
A broader implication from this work concerns the link between PGC specification and lineage determination in the embryo. Although modes of germ cell specification differ, both rely upon mechanisms of embryonic axis patterning. In PGC preformation, polarization of RNAs and proteins in the early embryo or oocyte ensures cytoplasmic inheritance of germ cell determinants in the right cells at the proper end of the embryo. Reliance of PGC induction upon a conserved primary axis determinant such as Wnt3 may arise as economical use of signals in the early embryo, or may represent a strategy for evolvability. Following the argument that germ cell formation by induction may be advantageous with changing body plans through evolution, a functional connection between early patterning and PGC formation allows portability of the germline. Invoking mesoderm transcription factors such as T in germ cell specification is hardly new – salamanders and crickets also induce PGCs from mesoderm (Ewen-Campen et al., 2013). Although highly conserved through evolution, shifting T expression and function could suggest a primary role in promoting cell motility. Indeed, in T mouse chimeras, a pileup of mutant cells in the primitive streak suggested T function in nascent mesoderm cell movement during gastrulation (Wilson et al., 1995). By extension, T targets in PGCs may include motility, adhesion, or cytoskeletal genes, thus eliciting a broader migratory gene program to equip newly minted germ cells for the next steps in their development: a multi-day migration to the gonads. Whether by borrowing T from the mesoderm or something more ancient, Bmp and Wnt create the perfect neighborhood for raising the cells of the next generation.
This is a commentary on article Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, Saitou M A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants.. Dev Cell. 2013;27(5):516-29.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yikihiro Y, Hiroko I, Yashuhiro M, Hamakubo T, Kato Y, Shirahige K, Saitou M. Dev Cell. 2013 doi: 10.1016/j.devcel.2013.11.001. this issue. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Donoughe S, Clarke DN, Extavour CG. Curr Biol. 2013;23:835–842. doi: 10.1016/j.cub.2013.03.063. [DOI] [PubMed] [Google Scholar]
- Gertow K, Hirst CE, Yu QC, Ng ES, Pereira LA, Davis RP, Stanley EG, Elefanty AG. Stem cell reports. 2013;1:53–65. doi: 10.1016/j.stemcr.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani MA. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Tam PP, Zhou SX. Developmental biology. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Manson L, Skarnes WC, Beddington RS. Development. 1995;121:877–886. doi: 10.1242/dev.121.3.877. [DOI] [PubMed] [Google Scholar]


