Abstract
The identification of flavin-dependent thymidylate synthase (FDTS) as an essential enzyme and its occurrence in several pathogenic microbes opens opportunities for using FDTS enzyme as an excellent target for new antimicrobial drug discovery. In contrast to the human thymidylate synthase enzyme that utilizes methylene-tetrahydrofolate (CH2H4 folate) for the conversion of dUMP to dTMP, the microbial enzymes utilize an additional non-covalently bound FAD molecule for the hydride transfer from NAD(P)H. The structural and mechanistic differences between the human and microbial enzymes present an attractive opportunity for the design of antimicrobial compounds specific for the pathogens. We have determined the crystal structure of FDTS enzyme in complex with the methyl donor, CH2H4 folate. We describe here the structure of a FDTS mutant and compare it with other FDTS complex structures, including a FDTS-CH2H4 folate complex. We identified a conformational change essential for substrate binding and propose a strategy for the design of FDTS specific inhibitors.
Keywords: FDTS enzyme, Structure/function studies, Drug design, Crystallography
Introduction
The de novo biosynthesis of thymidylate (2′-deoxythymine-5′-monophosphate; dTMP), one of the four bases of DNA, requires the enzyme thymidylate synthase [1]. Two types of thymidylate synthases have been described and both of them use 2′-deoxyuridine-5′-monophosphate (dUMP) as the substrate [1,2]. The classical thymidylate synthases (TS) use N5,N10-methylene-5,6,7,8-tetrahydrofolate (CH2H4 folate) to reductively methylate dUMP producing dTMP, while the recently identified flavin-dependent thymidylate synthase (FDTS) uses a non-covalently bound flavin adenine nucleotide (FAD) for the reduction [2]. FDTS is found in ~30% of microbial genome. The two families of thymidylate synthases are mechanistically and structurally different [1-4]. Our recent studies have shown that, unlike the classical enzyme which uses a cysteine residue to form a covalent bond with dUMP, the flavin-dependent enzyme doesn’t use an enzymatic nucleophile for the reaction [3]. The uniqueness of the FDTS enzyme is also revealed by a novel fold of its structure [4]. The structures of FDTS from various organisms share similar fold, and the high level of sequence similarity of FDTS from other organisms indicates very similar structures for all of them [5-7].
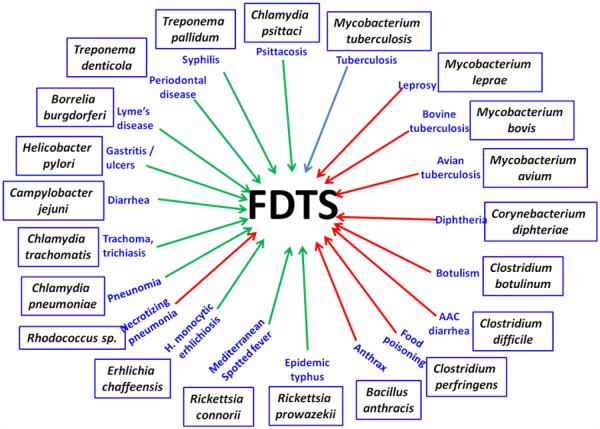
The rise in bacterial resistance has stimulated new interest in finding novel targets for the development of effective antimicrobial agents. The presence of FDTS in many pathogenic organisms (Figure 1) and its absence in human make FDTS as an attractive target for antimicrobials [2] and a number of studies are in progress to develop specific inhibitors for the FDTS enzymes [8,9]. The catalytic mechanism of classical enzyme is well understood and has facilitated the development of several inhibitors, some of which are in clinical use as anticancer drugs (e.g., 5-flouro-uracil, tomudex (Raltitrexed)) [1,10]. Several structures of the classical enzyme, including ternary complexes with various combinations of substrate and folate cofactor, along with their analogs are available [1,11]. Unfortunately, the inhibitors for the classical thymidylate synthase are not specific to the FDTS enzymes [12]. The complexity of the FDTS reaction mechanism and the conformational flexibility of the active site region make it difficult to perform rational drug design with the currently available information. There are opposing views regarding the most important methylene-transfer step, with some studies proposing an indirect methylene-transfer through an arginine residue [13] while other studies indicating a direct methylene transfer from CH2H4 folate to dUMP [3,6,12,14]. Therefore, it is important to understand the details of the FDTS mechanism and determine its structures in various complexes and intermediates.
Figure 1.
Organisms carrying FDTS enzymes. The names of the organisms are shown in rectangular boxes and their diseases in blue letters. The green and blue arrows represent gram-negative and gram-positive organisms, respectively (M. tuberculosis shown with a grey arrow could belong to either of these categories). Sources: Centers for Disease Control and Prevention (www.cdc.gov). World Health Organization (www.who.int).
We have recently reported the first structures of the quaternary complexes of FDTS from Thermotoga maritima (TmFDTS) with FAD, dUMP and CH2H4 folate and CH2H4 folate mimics. Since several of the inhibitors of classical thymidylate synthase are based on the folate binding site and not selective for FDTS enzymes, it is expected that novel compounds utilizing the unique folate binding modes may provide new avenues for FDTS specific inhibitor design [15]. This emphasizes the importance of a proper understanding of the binding interactions near the folate binding site.
One of the residues implicated in the folate binding interaction in FDTS is histidine 53 (T. maritima numbering). This residue is fully conserved among the FDTS from various organisms and previous studies showed the essential role of this residue in NAD(P)H oxidation or methyl transfer [6]. The methylene transfer step is one of the least understood processes in the FDTS catalysis. The recent structures of the ternary complexes of TmFDTS with FAD, dUMP and CH2H4 folate and identified the folate binding site and proposed it as a binding site for NADPH [16]. One of the residues implicated in the folate binding interaction is histidine 53. We mutated this residue to aspartic acid (H53D) and present the structures of the H53D-FAD and H53D-FAD-dUMP complexes and a comparison with native enzyme structures. Earlier we reported the crystal structure of the H53A mutant and it complex with FAD, dUMP and CH2H4 folate [16]. We also reported that both the H53A and H53D mutants showed dTMP formation with dramatically reduced activity (Table S2 of reference 17).
Results and Discussion
We have crystallized and solved the structures of H53D mutant of the Thermotoga maritima FDTS with FAD and in complex with FAD and dUMP (Table 1). The structures of the H53D mutant complexes are very similar to the native enzyme, which forms a biologically active tetramer. An extensive array of hydrogen bonding and hydrophobic interactions stabilize the tetrameric structure with ~2000Å2 surface area buried per monomer. Previous crystallographic and activity studies have confirmed the presence of each active site at the interface of the three subunits [4,17]. The two interacting active sites in each side of the enzyme form a large active site grove spanning around 50Å. A tightly bound FAD molecule is observed in the all of the reported structures. However, a structure of the apoenzyme obtained by removing the FAD using high amounts of NaCl showed that FAD molecule is not essential for the stabilization of the tetramer [4]. The structures of the complexes presented here show that the substrate-binding loop can be stabilized in two conformations and this affects the binding of the molecules at the substrate binding site.
Table 1.
Crystallographic parameters, data collection and refinement statistics.
| H53D+FAD | H53D+FAD+dUMP | |
|---|---|---|
|
| ||
| Crystallographic parameters | ||
| Space group | P212121 | P212121 |
| Unit-cell dimensions (Å) | 53.99, 116.64, 141.38Å 90.0, 90.0, 90.0° |
54.37, 116.67, 141.42Å 90.0, 90.0, 90.0° |
|
| ||
| Data collection statistics | ||
| Resolution limits (Å) | 38.2-2.03 | 38.3-1.85 |
| No. of observed reflections | 354413 | 278376 |
| No. of unique reflections | 58450 | 72637 |
| Completeness overall/outer shell |
99.8/99.2 | 93.4/62.9 |
| Rsyma (%) overall/outer shell |
7.1/97.1 | 3.3/56.8 |
| I/σ Overall/outer shell |
20.1/1.9 | 28.6/1.97 |
|
| ||
| Refinement statistics | ||
| Resolution limits | 38.2-2.05 | 38.3-1.85 |
| Number of reflections/% | 55525/99.8 | 69005/93.4 |
| Reflections used for Rfree | ||
| Rfactorb (%) | 19.6 | 17.0 |
| Rfree (%) | 24.9 | 21.1 |
| Model contents/average B(Å2) | ||
| Protein atoms | 7221/44.3 | 7417/31.5 |
| Ligand atoms | 140/77.3 | 181/52.0 |
| Ions/buffer atoms | 13/69.1 | 14/61.5 |
| Water molecules | 150/46.3 | 305/38.2 |
| RMS deviations | ||
| Bond length (Å) | 0.014 | 0.015 |
| Bond angle (°) | 1.571 | 1.756 |
Rsym=Σ | Iavg-Ii|/Σ Ii
Rfactor=Σ|Fp-Fpcalc |/ΣFp, where Fp and Fpcalc are observed and calculated structure factors; Rfree is calculated with 5% of the data.
FAD binding site
FAD acts as the reducing agent in the FDTS reaction. The ribityl and the AMP groups are strongly bound in the active site with the catalytically important flavin ring exposed to the solvent [4]. In the dUMP complexes, flavin ring of the FAD molecule stacks with the pyrimidine ring of the dUMP. It has been reported that the flavin ring of the FAD molecule is generally disordered in structures without the dUMP [4]. This is true for the current H53D complex with FAD. However, in the viral enzyme and the coryne enzyme flavin ring is stabilized by stacking interaction with the histidine 53 side chain [6,18]. Interestingly, the flavin ring uses the si-face and re-face for the stacking interaction in the viral and coryne enzymes, respectively. In the reported structure of the quaternary complex with FAD, dUMP and CH2H4 folate, the flavin ring uses the re-face to stack with the histidine side chain. It is also interesting to note that during the folate stacking histidine 53 side chain flips to the opposite side (torsion angle N-Cα-Cβ-Cγ=~−172° for viral and coryne enzymes and ~−56° for the folate bound complex). It is important to note that flavin ring uses the si-face to stack with dUMP [4] as well as the CH2H4 folate [16]. The folate/FAD-dependent tRNA T54 methyltransferase (TrmFO), which catalyzes the same net reaction as the FDTS enzyme, the re-face of the flavin is stacked with the folate [19]. Our earlier studies with two mutants of FDTS (E144R and R174K (ref 17) (R174K+FAD+dUMP work is not published)) with FAD and in complex with FAD and dUMP indicated that the flavin is able to rotate in the active site during the formation of the dUMP complex [16].
The details mentioned above show that isoalloxazine (flavin) ring of FAD binds in a big pocket that tolerates large movements of the isoalloxazine ring. Importantly, the isoalloxazine ring is able to rotate in the binding pocket and utilize same face of the ring to bind to substrate and cofactors. This is in contrast to the relatively rigid binding mode observed for the isoalloxazine ring in most of the enzymes that use FAD as the cofactor [20-23]. The presence of the large active site cavity in FDTS that tolerates major conformational movements of the ligands makes the design of specific inhibitors very challenging.
The FAD molecules in the H53D+FAD complex show very weak density for the whole FAD molecules and no density for the flavin ring (Table 2, Figure 2a). The FAD molecules in the H53D+FAD+dUMP complex also showed weak electron density indicating poor binding (Table 2, Figure 2b). This is in contrast to the flavin ring only disorder observed for the native enzyme with FAD complex and the very good electron density observed for FAD and dUMP in the FAD-dUMP complex (Table 2) [4].
Table 2.
Comparison of the temperature factors of various protein chains, FAD molecules and dUMP molecules in the mutant and native enzymea
| H53D+FAD complex | H53D+FAD+dUMP complex | |||
|---|---|---|---|---|
| Protein B-factors |
FAD B-factors | Protein B-factors |
FAD B-factors |
dUMP B-factors |
| Chain A 44.6 | FAD-A 66.4 | Chain A 31.6 | FAD-A 65.0 | UMP-A 23.9 |
| Chain B 46.2 | FAD-B 74.7 | Chain B 34.7 | FAD-B 64.2 | UMP-B 26.9 |
| Chain C 42.6 | FAD-C 83.3 | Chain C 29.4 | Only one SO4 43.8 |
UMP-C 40.2 |
| Chain D 43.8 | FAD-D 85.8 | Chain D 30.4 | Only one SO4 48.2 |
Only PO4 57.6 |
|
Native+FAD complex (PDB
code:
1O2A) |
Native+FAD+dUMP complex
(PDB
code: 1O26) |
|||
|
Protein
B-factors |
FAD B-factors |
Protein
B-factors |
FAD
B-factors |
dUMP
B-factors |
| Chain A 28.9 | FAD-A 40.7 | Chain A 32.3 | FAD-A 22.2 | UMP-A 22.2 |
| Chain B 30.0 | FAD-B 55.1 | Chain B 33.6 | FAD-B 20.0 | UMP-B 20.2 |
| Chain C 27.9 | FAD-C 55.0 | Chain C 31.1 | FAD-C 22.6 | UMP-C 23.0 |
| Chain D 31.4 | FAD-D 53.1 | Chain D 32.3 | FAD-D 20.7 | UMP-D 21.7 |
The temperature factors generally show the positional movement of the atoms in the crystal structure. The molecules that are tightly bound in the protein show lower temperature factors compared to the molecules that interact weakly with the protein.
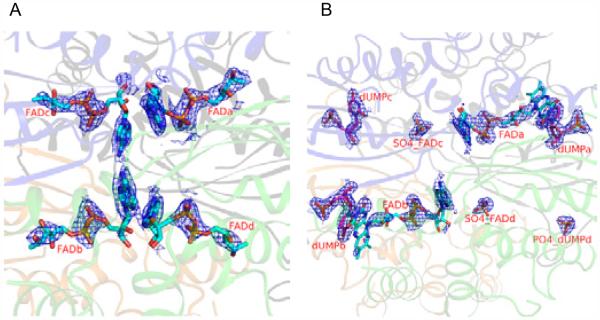
Figure 2.
A view of the electron density map at the four active sites of H53D+FAD and H53D+FAD+dUMP complexes.
A) Electron density for FAD molecules H53D+FAD complex. B) Electron density for FAD and dUMP in the H53D+FAD+dUMP complex. The 2Fo-Fc electron density around the ligand atoms are contoured at 1.0 sigma. Ribbon drawings for the protein chains and stick representation for FAD (SO4) and dUMP (PO4) molecules. The SO4 or PO4 densities observed in these binding sites are labeled (e.g., SO4_FADc indicates SO4 at the FAD binding site in chain C).
Substrate binding site
In general, dUMP and analogs are strongly bound in the enzyme with several direct and water mediated hydrogen bonds to the protein. Furthermore, the pyrimidine ring of dUMP is stacked to the flavin ring of FAD in complexes with FAD. It has also been reported that substrate induced conformational changes near the active site is important in the stabilization of the substrate binding site [4]. A main difference between the current and the reported structures is the very weak electron density observed for the dUMP (Table 2, Figure 2b). Only two of the active sites showed good electron density for dUMP, while the third active site showed weak density for dUMP, the fourth one showed very weak density only for the phosphate group. It is not clear whether differences in electron density between the four active sites indicate any allosteric interaction amongst the active sites.
Open and closed confirmations
There are several mechanisms proposed for the FDTS catalysis with various suggestions for the binding and release of the substrate and other cofactors [3]. Unfortunately, the large conformational flexibility of the FDTS active site makes it difficult to give a structural perspective to the biochemical results. It has been reported that the conformational changes during FAD and dUMP binding brings various conserved residues into close proximity to these molecules.
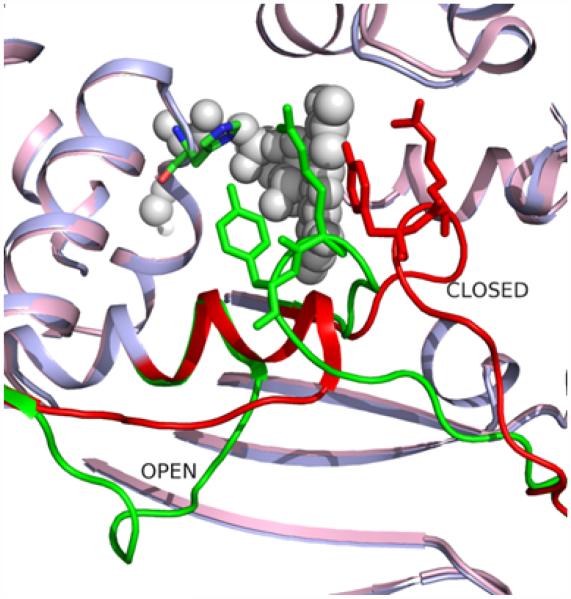
We compared the native enzyme structure with the FAD complex, with FAD and dUMP complex, and FAD, dUMP and CH2H4 folate complex and identified two major conformational changes during various binding processes (Figure 3). Various combinations of these conformational changes take place during the binding of the substrate and/or cofactors. The close to open conformational change of the 90-loop/substrate-binding loop is very important because this conformational change brings important residues to the substrate binding site [4]. In the open conformation of the substrate-binding loop, residues from Ser88 to Arg90 make hydrogen-bonding interactions with the substrate. While the Ser88 Oγ and Gly89 N atoms H-bonds to the phosphate group of the substrate, the Arg90 side chain H-bonds to one of the oxygen atoms of the pyrimidine base. The Ser88 and Arg90 are highly conserved residues [16].
Figure 3.
The open and closed conformations of the 30-loop and 90-loop/substrate binding loop. Ribbon drawings for the protein chains (PDB code: 1O26 (light blue) and PDB code: 1O24 (light pink). The open and closed conformations are shown in green and red. The His53, Arg90, Tyr91 residues are shown by stick representation and the FAD molecules in spheres.
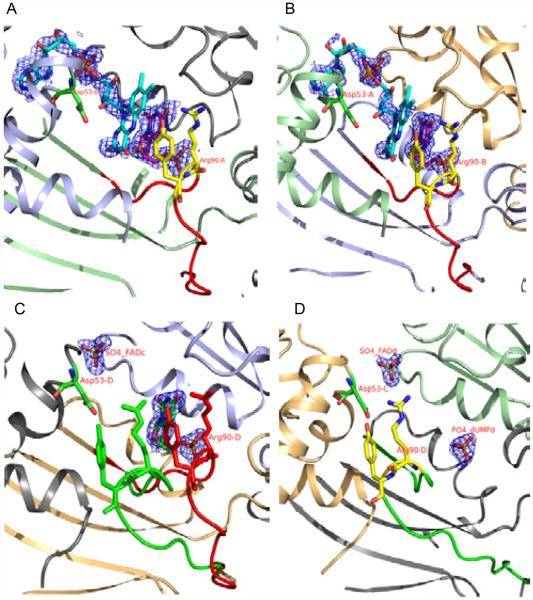
A comparison of the active sites of the H53D+dUMP complex shows that the substrate-binding loop conformational change plays an important role in the stabilization of the dUMP binding (Table 2, Figure 4). The active sites that show good electron density for dUMP (chains A and B) showed closed conformation for the substrate-binding loop. The dUMP molecule in chain C showed weaker density and the substrate-binding loop showed double conformation. The open confirmation observed in chain D showed very weak density for dUMP with density for the phosphate group only. This shows that the open conformation of the substrate-binding loop doesn’t favor the substrate binding. These conformational changes may also be important for the binding and release of the substrate and product.
Figure 4.
The various states of the 90-loop/substrate-binding loop captured in the four actives sites of the H53D+FAD+dUMP complex.
a. Active site of chain A. b. Active site of chain B. c. Active site of chain C. d. Active site of chain D. The 2Fo-Fc electron density around the ligand atoms are contoured at 1.0 sigma. Ribbon drawings for the protein chains and stick representation for FAD (SO4) and dUMP (PO4) molecules. The SO4 or PO4 density observed in the FAD and dUMP binding sites are labeled (naming convention similar to Figure 3). The Asp53 and Arg90 are labeled with their chain numbers. The open and closed conformations of the substrate-binding loop are colored in green and red. The views from a to d show movement of the 90-loop/substrate-binding loop from the closed state (chain A) to the open state (chain D). Please refer to table 2 for additional information.
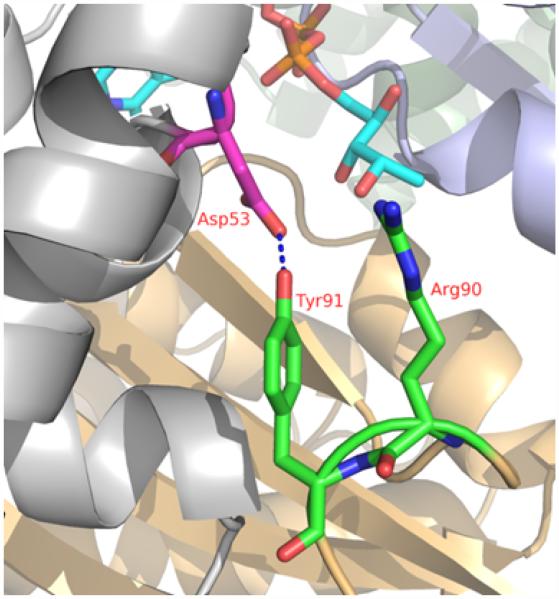
A closer examination of the open and closed conformation of the substrate-binding loop shows that the open conformation is stabilized by hydrogen bonding interaction of the tyrosine 91 hydroxyl group to the mutated aspartic acid (Figure 5). Similar hydrogen bonding interaction of the tyrosine 91 from the open loop with histidine 53 is observed in the native enzyme FAD complex (PDB code: 1O2A). This hydrogen bonding interaction is absent in the closed conformation and the distance between the corresponding atoms in the closed conformation is around 8 Å. The structural changes accompanying the open conformation also brings the conserved arginine 90 to the vicinity of tyrosine 47. In the closed conformation of the substrate-binding loop, arginine 90 side chain is involved in hydrogen bonding interactions with the substrate and protein atoms from the neighboring protein chain. These interactions stabilize the substrate binding site. The tyrosine 47 and 91 residues generally show good conservation among the FDTS enzymes [16].
Figure 5.
The vicinity of the H53D site with the open conformation of the substrate-binding loop. The Asp53, Arg90, and Tyr91 residues are shown by stick representation and labeled. The hydrogen bond between Asp53 and Tyr91 is shown by blue dashed line. The FAD molecule is shown in stick (cyan) for reference. Ribbon drawing is for protein chains.
The observed stabilization of the closed conformation substrate-binding loop in the mutated protein suggests the possibility of using chemical compounds to lock the open conformation of the substrate-binding loop. Since closed conformation of the substrate-binding loop is very important for substrate binding, design of chemicals to lock the open conformation may be a good strategy to develop inhibitors specific for the FDTS enzymes. The recently discovered 150-cavity in group-1 influenza A neuraminidase provided a target for rational structure-based drug development and novel techniques have been developed to lock open the 150-loop as a strategy for the inhibition [24,25]. An analysis of the reported structures of various FDTS enzymes shows that FDTS tolerates large movements of the ligands in the binding pocket, thus making the design of specific inhibitors very challenging.
Conclusions
FDTS is an essential enzyme found in several pathogenic microbes. Because of the structural and mechanistic differences between FDTS and the human enzyme and the crucial role of FDTS enzyme in bacterial cells, the FDTS enzymes have been proposed as a priority target for developing new anti-microbial compounds [2,26]. Unfortunately, because of the complex nature of the FDTS reaction catalysis and the non-specificity of the known TS inhibitors for FDTS enzyme, it has been difficult to develop FDTS specific inhibitors. We have shown that conformational changes of active site are important for the binding of the substrate and various cofactors. Our data shows that the closed conformation of the substrate-binding loop is essential for substrate binding. We propose the development of compounds that can lock the open conformation of the substrate-binding loop as a strategy for FDTS specific inhibitor design.
Materials and Methods
Chemicals
All chemicals were reagent grade and used as purchased without further purification, unless specified.
Protein expression and purification
The H53D mutant of FDTS from T. maritima (TM0449, GenBank accession number NP228259) was expressed and purified as previously described [27].
Crystallization and structure determination
The crystals of the H53D mutant with FAD and with FAD and dUMP were crystallized at 22°C in 50-60% (w/v) PEG 200 and 100 mM Tris buffer, pH 8.0. The FAD molecule stays bound during purification and no further FAD was included in the crystallization trials. The dUMP complex was prepared by treating the FAD complex with 10 mM dUMP. The crystals were flash cooled directly from the drop. Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9-2 using Q315 detector. The wavelengths used for the data collection of the H53D with FAD and the dUMP complexes were 0.9795 Å and 1.0 Å, respectively. All data were integrated using the XDS package [28]. These crystals belonged to the P212121 space group.
Structures of the complexes were solved by molecular replacement (MOLREP [29]) or rigid body refinement using the T. maritima tetramer (PDB code: 1O26) as the search template. Model building and refinement were performed by Coot [30] and REFMAC [31]. The Ramachandran statistics for the final structures showed no outliers (Table 1). The figures were generated using PyMOL graphic program [32].
Coordinates
Coordinates for the complexes have been deposited in the Protein Data Bank (accession codes: 4KAR (H53D+FAD complex) and 4KAS (H53D+FAD+dUMP complex).
Acknowledgements
I thank S. A. Lesley, H. Klock, and E. Ambing (The Genomics Institute of the Novartis Research Foundation) for the protein samples and Q. Xu and A. Kumar for critical reading of the manuscript. I thank members of the SMB group at SSRL for helpful discussions and support. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences.
References
- 1.Finer-Moore JS, Santi DV, Stroud RM. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme: thymidylate synthase mutagenesis, function, and structure. Biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 2.Myllykallio H, Lipowski G, Leduc D, Filee J, Forterre P, et al. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 3.Koehn EM, Fleischmann T, Conrad JA, Palfey BA, Lesley SA, et al. An unusual mechanism of thymidylate biosynthesis in organisms containing the thyX gene. Nature. 2009;458:919–923. doi: 10.1038/nature07973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathews II, Deacon AM, Canaves JM, McMullan D, Lesley SA, et al. Functional analysis of substrate and cofactor complex structures of a thymidylate synthase-complementing protein. Structure. 2003;11:677–690. doi: 10.1016/s0969-2126(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 5.Wang K, Wang Q, Chen J, Chen L, Jiang H, et al. Crystal structure and enzymatic characterization of thymidylate synthase X from Helicobacter pylori strain SS1. Protein Sci. 2011;20:1398–1410. doi: 10.1002/pro.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graziani S, Xia Y, Gurnon JR, Van Etten JL, Leduc D, et al. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria Chlorella virus-1. J Biol Chem. 2004;279:54340–54347. doi: 10.1074/jbc.M409121200. [DOI] [PubMed] [Google Scholar]
- 7.Sampathkumar P, Turley S, Ulmer JE, Rhie HG, Sibley CH, et al. Structure of the Mycobacterium tuberculosis flavin dependent thymidylate synthase (MtbThyX) at 2.0A resolution. J Mol Biol. 2005;352:1091–1104. doi: 10.1016/j.jmb.2005.07.071. [DOI] [PubMed] [Google Scholar]
- 8.Basta T, Boum Y, Briffotaux J, Becker HF, Lamarre-Jouenne I, et al. Mechanistic and structural basis for inhibition of thymidylate synthase ThyX. Open Biol. 2012;2:120120. doi: 10.1098/rsob.120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esra Onen F, Boum Y, Jacquement C, Spanedda MV, Jaber N, et al. Design, synthesis and evaluation of potent thymidylate synthase X inhibitors. Bioorg Med Chem Lett. 2008;18:3628–3631. doi: 10.1016/j.bmcl.2008.04.080. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham D, Zalcberg J, Smith I, Gore M, Pazdur R, et al. ‘Tomudex’ (ZD1694): a novel thymidylate synthase inhibitor with clinical antitumour activity in a range of solid tumours. ‘Tomudex’ International Study Group. Ann Oncol. 1996;7:179–182. doi: 10.1093/oxfordjournals.annonc.a010546. [DOI] [PubMed] [Google Scholar]
- 11.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 12.Mishanina TV, Koehn EM, Conrad JA, Palfey BA, Lesley SA, et al. Trapping of an intermediate in the reaction catalyzed by flavin-dependent thymidylate synthase. J Am Chem Soc. 2012;134:4442–4448. doi: 10.1021/ja2120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin J, Roshick C, Iliffe-Lee E, McClarty G. Catalytic mechanism of Chlamydia trachomatis flavin-dependent thymidylate synthase. J Biol Chem. 2005;280:5456–5467. doi: 10.1074/jbc.M412415200. [DOI] [PubMed] [Google Scholar]
- 14.Hunter JH, Gujjar R, Pang CK, Rathod PK. Kinetics and ligand-binding preferences of Mycobacterium tuberculosis thymidylate synthases, ThyA and ThyX. PLoS One. 2008;3:e2237. doi: 10.1371/journal.pone.0002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chernyshev A, Fleischmann T, Kohen A. Thymidyl biosynthesis enzymes as antibiotic targets. Appl Microbiol Biotechnol. 2007;74:282–289. doi: 10.1007/s00253-006-0763-1. [DOI] [PubMed] [Google Scholar]
- 16.Koehn EM, Perissinotti LL, Moghram S, Prabhakar A, Lesley SA, et al. Folate binding site of flavin-dependent thymidylate synthase. Proc Natl Acad Sci U S A. 2012;109:15722–15727. doi: 10.1073/pnas.1206077109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leduc D, Graziani S, Lipowski G, Marchand C, Le Maréchal P, et al. Functional evidence for active site location of tetrameric thymidylate synthase X at the interphase of three monomers. Proc Natl Acad Sci U S A. 2004;101:7252–7257. doi: 10.1073/pnas.0401365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. FDTS from Corynebacterium glutamicum-PDB code: 3FNN.
- 19.Nishimasu H, Ishitani R, Yamashita K, Iwashita C, Hirata A, et al. Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase. Proc Natl Acad Sci U S A. 2009;106:8180–8185. doi: 10.1073/pnas.0901330106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park HW, Kim ST, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 21.Sridhar Prasad G, Kresge N, Muhlberg AB, Shaw A, Jung YS, et al. The crystal structure of NADPH:ferredoxin reductase from Azotobacter vinelandii. Protein Sci. 1998;7:2541–2549. doi: 10.1002/pro.5560071207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraaije MW, van den Heuvel RH, van Berkel WJ, Mattevi A. Covalent flavinylation is essential for efficient redox catalysis in vanillyl-alcohol oxidase. J Biol Chem. 1999;274:35514–35520. doi: 10.1074/jbc.274.50.35514. [DOI] [PubMed] [Google Scholar]
- 23.Cunane LM, Chen ZW, Shamala N, Mathews FS, Cronin CN, et al. Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism. J Mol Biol. 2000;295:357–374. doi: 10.1006/jmbi.1999.3290. [DOI] [PubMed] [Google Scholar]
- 24.Amaro RE, Swift RV, Votapka L, Li WW, Walker RC, et al. Mechanism of 150-cavity formation in influenza neuraminidase. Nat Commun. 2011;2:388. doi: 10.1038/ncomms1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudrawar S, Dyason JC, Rameix-Welti MA, Rose FJ, Kerry PS, et al. Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase. Nat Commun. 2010;1:113. doi: 10.1038/ncomms1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chernyshev A, Fleischmann T, Kohen A. Thymidyl biosynthesis enzymes as antibiotic targets. Appl Microbiol Biotechnol. 2007;74:282–289. doi: 10.1007/s00253-006-0763-1. [DOI] [PubMed] [Google Scholar]
- 27.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, et al. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci U S A. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32. The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.