Abstract
Duchenne muscular dystrophy (DMD) is a lethal genetic disorder caused by loss of functional dystrophin protein. Accumulating evidence suggests that the deficiency of dystrophin leads to aberrant activation of many signaling pathways which contribute to disease progression. However, the proximal signaling events leading to the activation of various pathological cascades in dystrophic muscle remain less clear. TNF receptor-associated factor 6 (TRAF6) is an adaptor protein which acts as a signaling intermediate for several receptor-mediated signaling events leading to the context-dependent activation of a number of signaling pathways. TRAF6 is also an E3 ubiquitin ligase and an important regulator of autophagy. However, the role of TRAF6 in pathogenesis of DMD remains unknown. Here, we demonstrate that the levels and activity of TRAF6 are increased in skeletal muscle of mdx (a mouse model of DMD) mice. Targeted deletion of TRAF6 improves muscle strength and reduces fiber necrosis, infiltration of macrophages and the activation of proinflammatory transcription factor nuclear factor-kappa B (NF-κB) in 7-week-old mdx mice. Ablation of TRAF6 also increases satellite cells proliferation and myofiber regeneration in young mdx mice. Intriguingly, ablation of TRAF6 exacerbates muscle injury and increases fibrosis in 9-month-old mdx mice. TRAF6 inhibition reduces the markers of autophagy and Akt signaling in dystrophic muscle of mdx mice. Collectively, our study suggests that while the inhibition of TRAF6 improves muscle structure and function in young mdx mice, its continued inhibition causes more severe myopathy at later stages of disease progression potentially through repressing autophagy.
INTRODUCTION
Duchenne muscular dystrophy (DMD) is a devastating and ultimately fatal disease characterized by progressive muscle wasting and weakness. The absence of dystrophin is a key factor in developing DMD (1). Dystrophin is a critical component of dystrophin-glycoprotein complex (DGC), which links the cytoskeleton to the extracellular matrix thus maintaining muscle fiber membrane integrity (2). Although the primary genetic defect is known, the dystrophic process has not been clearly identified (3,4). Studies in animal models and humans have shown that the primary deficiency of dystrophin results in the activation of several pathological cascades such as extracellular matrix breakdown, oxidative stress, cycles of fiber degeneration and regeneration, inflammatory response and gradual replacement of muscle fibers with adipose and connective tissue (3,5–8).
Besides acting as a molecular scaffold serving mechanical function, accumulating evidence suggests that DGC also has an important signaling role in striated muscle. Loss of dystrophin in skeletal muscle leads to aberrant activation of a number of signaling pathways such as NF-κB, phosphatidylinositol 3-kinase (PI3K)/Akt and mitogen-activated protein kinases (9–14). Interestingly, many of these signaling pathways are activated even at pre-necrotic state and their modulation using molecular and pharmacological approaches considerably improves muscle pathology in models of DMD (9–11,13,14). However, given the progressive degenerative nature of DMD and the convoluted involvement of many secondary processes, developing a pan therapeutic strategy that proves beneficial during the course of the disease has been challenging. Despite the identification of many of the principal and auxiliary signaling pathways that contribute to myopathy, the proximal signaling events leading to the activation of such pathological cascades in dystrophic muscle remain unknown.
TNF receptor-associated factors (TRAFs) are a family of conserved adaptor proteins which act as signaling intermediates for several receptor-mediated signaling events leading to the context-dependent activation of a number of signaling pathways (15,16). TRAF6 functions as a signal transducer to activate IκB kinase (IKK) and subsequently NF-κB activation in response to proinflammatory cytokines, bacterial products, Toll/IL-1 family and from receptors such as receptor activator of NF-κB (RANK) and CD40 (16–19). TRAF6 is also an E3 ubiquitin ligase which undergoes autoubiquitination and catalyzes K63 polyubiquitination of TAK1 that is required for IKK activation (20,21). TRAF6 interacts with ubiquitin conjugating enzymes UBE2N/UBC13 and UBE2V1/UEV1A to stimulate the formation of polyubiquitin chains on IKK. This protein also causes the K63-linked polyubiquitination of Akt which leads to its translocation to cell membrane, phosphorylation and enzymatic activation (22). Other signaling proteins such as interleukin-1 receptor-associated kinase 1, Src family kinase and protein kinase C zeta have also been found to interact with TRAF6 further signifying a central role of TRAF6 in cross-talk between different signaling pathways (16,18,19). Moreover, TRAF6 interacts with scaffold protein p62/Sequestosome 1 which is involved in regulation of autophagy and trafficking of proteins to the proteasome (19,23–25). It has been also found that TRAF6 promotes the K63-linked ubiquitination of Beclin-1, which is critical for the induction of autophagy, in response to toll-like receptor 4 signaling (26). However, the role of TRAF6 signaling in muscular dystrophy remains unknown.
Accumulating evidence suggests that TRAF6 is a crucial regulator of skeletal muscle mass in catabolic states (27–29). Abundance and activation of TRAF6 are increased in skeletal muscle of mice in many atrophying conditions. Notably, inhibition of TRAF6 using genetic approaches attenuates muscle wasting in response to denervation, cancer cachexia and starvation (28,29). Furthermore, specific inhibition of TRAF6 also improves myofiber regeneration upon cardiotoxin-mediated injury (27). One of the mechanisms by which TRAF6 mediates muscle wasting is through stimulation of autophagy (28,29). Autophagy is an important homeostasis mechanism which is essential for clearing of dysfunctional organelles and preventing tissue damage (30). While basal level of autophagy is required for the maintenance of skeletal muscle mass, excessive autophagosome formation generally leads to muscle wasting (30–34). Interestingly, autophagy has been found to be impaired in muscle biopsies from patients with DMD and in skeletal muscle of mdx mice with concomitant accumulation of damaged organelles (35). However, the role of TRAF6 in regulation of autophagy in the ‘settings’ of muscular dystrophy remains unknown.
In this study, we have investigated the role and the mechanisms by which TRAF6 affects disease progression in mdx (a mouse model of DMD) mice. Our results show that depletion of TRAF6 attenuates injury and inflammation and improves muscle structure and regeneration in young mdx mice. In contrast, continued inhibition of TRAF6 causes fiber degeneration and fibrosis at later stages of mdx mouse development potentially through inhibition of autophagy.
RESULTS
TRAF6 levels are increased in skeletal muscle of mdx mice
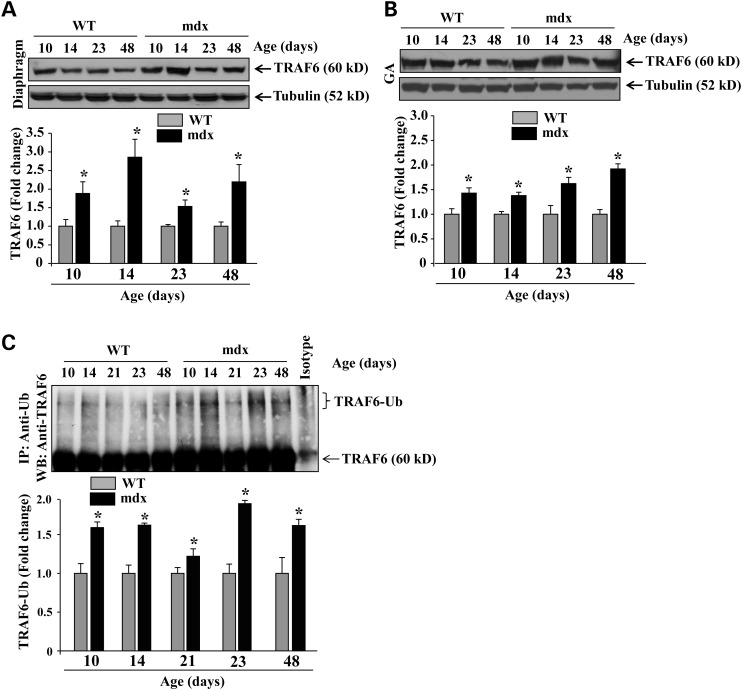
In mdx mice, muscle injury starts at around 2.5 weeks followed by peak necrotic phase in combination with inflammation between 3 and 4 weeks of age (36,37). Regeneration starts around the age of 6 weeks and continues while alternating with ongoing degeneration until 12 weeks of age (38,39). We first compared the levels of TRAF6 in skeletal muscle of wild-type (WT) and mdx mice at both pre-necrotic and necrotic stages. Diaphragm and gastrocnemius (GA) muscle from 10, 14, 23 and 48 days old WT (i.e. C57BL10) and mdx mice were isolated and processed to measure protein levels of TRAF6 by performing western blot. Consistent with our previously published results (29), the levels of TRAF6 were considerably higher in young mice and reduced at later stages of development. However, the levels of TRAF6 protein were markedly higher in both diaphragm and GA muscle of mdx mice compared with WT mice at all the ages (Fig. 1A and B). In a separate experiment, we measured protein levels of TRAF6 in quadriceps muscle of young (6-week) and old (9-month) WT and mdx mice. The levels of TRAF6 were significantly higher in quadriceps muscle of both 6-week- and 9-month-old mdx mice compared with their corresponding age-matched WT mice (Supplementary Material, Fig. S1). Increased levels of TRAF6 at 10 and 14 days in mdx mice also suggest that the levels of TRAF6 start increasing before the onset of fiber necrosis in mdx mice.
Figure 1.
Activation of TRAF6 in skeletal muscle of mdx mice. Levels of TRAF6 protein in (A) diaphragm and (B) GA muscle of WT and mdx mice at different stages of development measured by western blotting. Representative immunoblots and densitometry quantification of data are presented here. (C) Diaphragm extracts from 10, 14, 21, 23 and 48 days old WT and mdx mice were immunoprecipitated with anti-ubiquitin followed by western blotting using anti-TRAF6. Representative immunoblot and quantification of fold change in the levels of ubiquitinated TRAF6 protein are depicted here. N = 3 or 4 at indicated age of mice. Error bars represent SD. *P < 0.05, values vary significantly from corresponding age-matched WT mice.
Since TRAF6 is an important E3 ubiquitin ligase which also undergoes autoubiquitination to induce cellular signaling (28,40,41), we next investigated the levels of ubiquitinated TRAF6 protein in skeletal muscle of WT and mdx mice. Diaphragm was isolated from 10, 14, 21, 23 and 48 days old mice and muscle extracts prepared were immunoprecipitated using anti-ubiquitin followed by western blot with anti-TRAF6. Results showed that the levels of ubiquitinated TRAF6 protein were significantly higher in mdx mice compared with WT mice at all the ages (Fig. 1C). Together, these data are suggestive that the levels and activation of TRAF6 are increased in skeletal muscle of mdx mice.
Targeted deletion of TRAF6 improves muscle strength in 7-week-old mdx mice
We have previously generated and characterized muscle-specific Traf6-knockout mice (Traf6mko) by crossing floxed TRAF6 (Traf6f/f) mice with muscle creatine kinase (MCK)-Cre mice (29). MCK deletes floxed Traf6 allele only in differentiated myofibers but not in other cell type such as satellite cells, endothelial cells and fibroblasts (27,29). For this study, we crossed TRAF6mko mice with mdx mice to obtain littermate mdx;Traf6f/f and mdx;Traf6mko mice. We first measured the levels of TRAF6 in skeletal muscle of mdx;Traf6f/f and mdx;Traf6mko mice at the age of 2 weeks (before the onset of fiber necrosis). Levels of TRAF6 were found to be significantly reduced (∼80%) in diaphragm and GA muscle of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 2A and B). The depletion of TRAF6 in mdx;Traf6mko mice was specific because levels TRAF2 and TRAF3 were comparable between mdx;Traf6f/f and mdx;Traf6mko mice (Fig. 2A). Furthermore, there was no significant change in the levels of TRAF6 in kidney, liver, spleen and cultured fibroblasts of mdx;Traf6f/f and mdx;Traf6mko mice, suggesting that MCK-Cre specifically depletes TRAF6 in skeletal muscle of mice (Supplementary Material, Fig. S2). We next investigated whether depletion of TRAF6 in skeletal muscle of mdx mice produces any developmental phenotype at prenecrotic state. Hematoxylin and Eosin (H&E) staining of GA muscle (Fig. 2C) and diaphragm (data not shown) showed that depletion of TRAF6 does not produce any overt phenotype in mdx mice at prenecrotic state.
Figure 2.
Targeted depletion of TRAF6 improves muscle strength in 7-week-old mdx mice. (A) Western blot analysis of TRAF2, TRAF3 and TRAF6 protein levels in diaphragm and GA muscle of 2-week-old mdx;Traf6f/f and mdx;Traf6mko mice. (B) Densitometry quantification of TRAF6 levels in diaphragm and GA muscle of 2-week-old mdx;TRAF6f/f and mdx;TRAF6mko mice. N = 4 in each group. (C) Representative photomicrographs of H&E-stained GA muscle sections of 2-week-old mdx;Traf6f/f and mdx;Traf6mko mice. Scale bar: 20 µm. Muscle strength was evaluated through a series of functional tests in 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice. Bar diagrams represent (D) forelimb grip strength normalized to body weight; (E) total four-limb grip strength; and (F) wire hanging time (in seconds). N = 8 in each group. Error bars represent SD. *P < 0.05, values vary significantly from mdx;Traf6f/f littermate.
We next performed systematic evaluation of muscle function in mdx;Traf6f/f and mdx;Traf6mko mice. We first sought to investigate whether depletion of TRAF6 affects muscle grip strength in mdx mice. Results showed that forelimb (Fig. 2D) and total four limb (Fig. 2E) grip strength were significantly higher in mdx;Traf6mko mice compared with mdx;Traf6f/f littermates at the age of 7 weeks. Furthermore, mdx;Traf6mko mice performed significantly better compared with mdx;Traf6f/f mice on a wire hanging test (Fig. 2F) providing initial evidence that inhibition of TRAF6 signaling improves muscle function in young mdx mice.
Depletion of TRAF6 improves muscle histopathology in young mdx mice
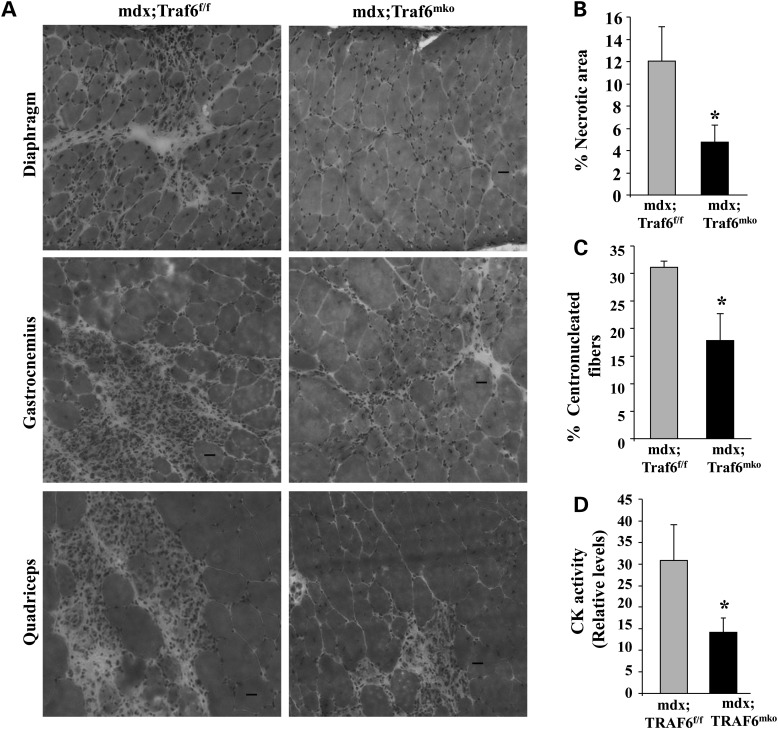
Diaphragm, GA, quadriceps muscles of 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice were isolated and processed for H&E staining. We observed typical features of dystrophic muscle including variability in fiber cross-section area, central nucleation, fiber necrosis and cellular infiltrates within muscle cross-sections in skeletal muscle of mdx;Traf6f/f mice. However, these dystrophic features were considerably reduced in skeletal muscles of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 3A and Supplementary Material, Fig. S3). Morphometric analysis of H&E-stained muscle section showed that the necrotic area (i.e. that contains only cellular infiltrate and no myofibers) was significantly reduced in skeletal muscle of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 3B). Furthermore, proportion of centronucleated myofibers was also significantly reduced in skeletal muscle of mdx;Traf6mko mice compared with mdx;Traf6f/f littermates (Fig. 3C). Since the serum creatine kinase (CK) level is an important marker of muscle injury, we also measured the levels of CK in plasma. Our results showed that serum CK levels were significantly lower in mdx;Traf6mko mice compared with littermate mdx;Traf6f/f mice (Fig. 3D). By performing immunostaining with Cy3-labeled anti-mouse IgG on muscle sections, we further evaluated sarcolemmal injury in mdx;Traf6f/f and mdx;Traf6mko mice. Consistent with serum CK activity, the number of IgG-filled fibers was significantly reduced in skeletal muscle of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Supplementary Material, Fig. S4). Taken together, these data are suggestive that the depletion of TRAF6 inhibits muscle injury in young mdx mice.
Figure 3.
Depletion of TRAF6 improves muscle histopathology in 7-week-old mdx mice. (A) Skeletal muscle of 7-week-old mdx;Traf6mko and littermate mdx; TRAF6f/f mice were isolated and processed for H&E staining. Representative photomicrographs of H&E-stained diaphragm (top), GA (middle) and quadriceps (bottom) muscles displaying manifestations of pathology. Scale bar: 20 µm. (B) Percentage necrotic area in quadriceps muscle of 7-week-old mdx;Traf6f/f and mdx;Traf6mko littermates. (C) Percentage of fibers with one or more centrally localized nuclei in quadriceps muscle of 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice. (D) Relative fold change in CK activity in serum of 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice. N = 7 in each group. Error bars represent SD. *P < 0.05, values vary significantly from mdx;Traf6f/f mice.
Inhibition of TRAF6 reduces macrophage accumulation, NF-κB activation and expression of inflammatory cytokines in dystrophic muscle of young mdx mice
Inflammation is a major pathological feature that contributes significantly to disease progression in DMD (3,5,6,42). To understand whether TRAF6 plays a role in exacerbating inflammatory response in skeletal muscle of mdx mice, by performing immunohistochemistry with F4/80 (a marker for macrophages) antibody, we studied the accumulation of macrophages in diaphragm of 7–8 weeks old mdx;Traf6f/f and mdx;Traf6mko mice. Interestingly, concentration of F4/80+ macrophages was found to be significantly reduced in diaphragm of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 4A and B, and Supplementary Material, Fig. S5). NF-κB is a major proinflammatory transcription factor that leads to the expression of a wide variety of inflammatory cytokines, chemokines and matrix degrading enzymes (43,44). To understand the mechanisms by which deletion of TRAF6 improves muscle inflammation in mdx mice, we studied the activation of NF-κB by performing electrophoretic mobility shift assay (EMSA). DNA-binding activity of NF-κB was considerably reduced in both diaphragm and quadriceps muscle of mdx;Traf6mko mice compared with mdx;Traf6f/f littermates (Fig. 4C and D). Furthermore, transcript levels of proinflammatory cytokines TNF-α, IL-1β and IL-6, and a matrix degrading enzyme, matrix metalloproteinase-9 (MMP-9) were significantly reduced in diaphragm of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 4E). These data demonstrate that TRAF6 mediates NF-κB activation and inflammatory response in myofibers of mdx mice.
Figure 4.
Muscle-specific depletion of TRAF6 attenuates inflammatory processes in dystrophic muscle of 7-week-old mdx mice. (A) Representative photomicrographs demonstrating F4/80+ macrophages in diaphragm section of 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice. Scale bar: 20 µm. (B) Quantification of F4/80+ cells in diaphragm of 7-week-old mdx;Traf6mko and mdx; TRAF6f/f mice. N = 7 or 8 in each group. (C) Representative EMSA gels presented here demonstrate that the DNA-binding activity of NF-κB is reduced in diaphragm (upper panel) and quadriceps (lower panel) muscle in 7-week-old mdx;Traf6mko mice compared with littermate mdx; TRAF6f/f mice. (D) Quantification of fold change in DNA-binding activity of NF-κB in diaphragm and quadriceps muscle of 7-week-old mdx;Traf6mko mice and mdx; Traf6f/f littermates. N = 3 in each group. (E) Relative mRNA levels of IL-1β, IL-6, TNFα and MMP-9 measured by QRT-PCR in diaphragm of 7-week-old mdx; TRAF6f/f and mdx;Traf6mko mice. N = 4 in each group. Error bars represent SD. *P < 0.05, values significantly different from littermate mdx;Traf6f/f mice.
Depletion of TRAF6 improves myofiber regeneration in 7-week-old mdx mice
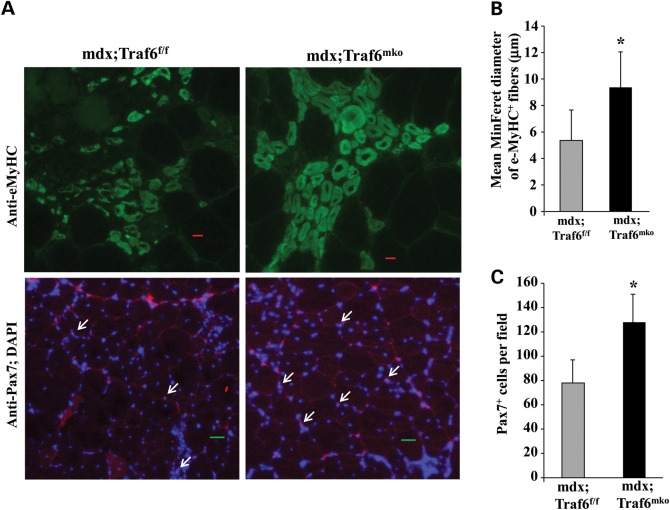
Accumulating evidence suggests that NF-κB and inflammatory cytokines inhibit skeletal muscle regeneration in mdx mice (9,45,46). We have also recently reported that inhibition of TRAF6 improves myofiber regeneration upon cardiotoxin-mediated injury (27). We next sought to investigate whether the inhibition of TRAF6 improves muscle regeneration in mdx mice. We performed immunostaining on quadriceps muscle sections with an antibody that recognizes embryonic (developmental) myosin heavy chain (eMyHC). Our analysis showed that eMyHC-positive fibers were generally scattered and smaller in size in skeletal muscle mdx;Traf6f/f mice. In contrast, the eMyHC-positive fibers were more tightly packed in mdx;Traf6mko compared with mdx;Traf6f/f mice (Fig. 5A, top). Furthermore, the mean minimum Feret (MinFeret) diameter of eMyHC-positive fibers was significantly higher in mdx;Traf6mko mice compared with TRAF6f/f mice (Fig. 5B). Since satellite cells are mainly responsible for repair of injured myofibers in adults, we also investigated whether depletion of TRAF6 in skeletal muscle of mdx mice affects the number of satellite cells. Quadriceps muscle sections from 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice were stained for Pax7, a marker for quiescent and proliferating satellite cells (47–49), and nuclei were counterstained with DAPI (Fig. 5A, bottom). Results showed that the number of satellite cells was significantly increased in quadriceps of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 5C). These results suggest that the inhibition of TRAF6 improves satellite cells proliferation and myofiber regeneration in mdx mice.
Figure 5.
Depletion of TRAF6 improves myofiber regeneration in young mdx mice. (A) Quadriceps muscle from 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice were isolated and processed for e-MyHC (top) and Pax7 (bottom) staining. Nuclei were identified by co-staining with DAPI. Representative photomicrographs are presented here. Arrows point to Pax7+ cells. (B) Quantification of mean minimum Feret (MinFeret) diameter of eMyHC+ fibers in quadriceps muscle of mdx;Traf6f/f and mdx;Traf6mko mice. (C) Average number of Pax7+ cells per field (∼0.15 mm2) in quadriceps muscle of 7-week-old mdx;Traf6f/f and mdx;Traf6mko mice. N = 8 in each group. Scale bar: 20 μm. Error bars represent SD. *P < 0.05, values significantly different from mdx;Traf6f/f mice.
Inhibition of TRAF6 exaggerates myopathy in 9-month-old mdx mice
Upon confirming that the inhibition of TRAF6 improves muscle histopathology and regeneration in 7-week-old mdx mice, we next sought to determine whether the inhibition of TRAF6 will also be effective in alleviating muscle pathology in older mdx mice. We first performed histological analysis of skeletal muscle at the age of 5 months. Intriguingly, we did not find any noticeable difference in muscle histopathology or level of fibrosis in skeletal muscle of 5-month-old mdx;Traf6f/f and mdx;Traf6mko mice (data not shown). We next analyzed skeletal muscle of these mice at the age of 9 months. We first performed western blot and confirmed that the levels of TRAF6 are repressed in skeletal muscle of 9-month-old mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Supplementary Material, Fig. S6, Supplemental data file). Surprisingly, 9-month-old mdx;Traf6mko mice showed increased muscle histopathology compared with mdx;Traf6f/f mice (Fig. 6A). Skeletal muscle of TRAF6mko mice showed increased necrotic area, cellular infiltrate and variability in fiber size (Fig. 6A–C). To detected injured/permeable fibers in mdx mice, we also performed immunostaining with Cy3-labeled anti-mouse IgG on GA muscle sections (Fig. 6D). The number of IgG-filled fibers was significantly higher in mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 6E).
Figure 6.
Inhibition of TRAF6 exacerbates myopathy in 9-month-old mdx mice. Diaphragm and GA muscles were isolated from 9-month-old mdx; Traf6f/f and mdx;Traf6mko mice and processed for H&E staining. (A) Representative photomicrographs of H&E-stained transverse sections of diaphragm (top) and GA (bottom) muscle. Scale bar: 20 µm. (B) Percentage necrotic area in H&E-stained sections of GA muscle of 9-month-old mdx;Traf6f/f and mdx;Traf6mko littermates. (C) Quantification of variability in fiber cross-sectional area (CSA) in GA muscle of 9-month-old mdx;Traf6f/f and mdx;Traf6mko mice. (D) GA muscle sections from 9-month-old mdx;Traf6f/f and mdx;Traf6mko mice were immunostained with Cy3-labeled goat anti-mouse IgG to detect permeable/damaged fibers. Representative photomicrographs are presented here. (E) Quantification of IgG-positive fibers in GA muscle sections from 9-month-old mdx;Traf6f/f and mdx;Traf6mko mice. N = 7 or 8 in each group. Scale bar: 20 μm. Error bars represent SD. *P < 0.05, values vary significantly from mdx;Traf6f/f mice.
Muscle-specific depletion of TRAF6 increases fibrosis in 9-month-old mdx mice
Fibrosis is a pathogenic factor characterized by chronic inflammation with persistent production of profibrotic cytokines and excessive deposition of ECM proteins, including collagens and fibronectin which can impair tissue function (50). Fibrosis is also a major pathological feature in muscular dystrophy which progressively deteriorates locomotor capacity, posture maintenance and the vital function of respiratory muscles (51). Progressive fibrosis is observed in diaphragm of the mdx mice which recapitulates clinical signs of DMD patients (37). We measured the level of fibrosis by performing Trichrome staining on diaphragm and GA muscle sections. Accumulation of collagens was significantly increased in both diaphragm and GA muscle of 9-month-old mdx;Traf6mko mice compared with age-matched mdx;Traf6f/f littermates (Fig. 7A and B). We next sought to investigate whether the deterioration in muscle histopathology observed in 9-month-old mdx;Traf6mko mice is reflected in muscle function. Indeed, forelimb grip strength and total limb grip strength of mdx;Traf6mko mice were found to be significantly reduced compared with littermate mdx;Traf6f/f mice (Fig. 7C and D). Together, these results demonstrate that continued inhibition of TRAF6 exacerbates pathology and caused loss of muscle strength in old mdx mice.
Figure 7.
Muscle-specific depletion of TRAF6 increases fibrosis in 9-month-old mdx mice. (A) Diaphragm and GA muscle were isolated from 9-month-old mdx; Traf6f/f and mdx;Traf6mko mice and transverse muscle sections made were processed for Mason's Trichrome staining for detection of collagen accumulation. Representative photomicrographs are presented here. Scale bar: 20 µm. (B) Quantification of fibrotic area in diaphragm and GA muscle of 9-month-old mdx;Traf6f/f mice and mdx;Traf6f/f littermates. Muscle strength of 9-month-old mdx;Traf6f/f and mdx;Traf6mko mice was evaluated. Bar diagrams representing (C) forelimb grip strength and (D) total four-limb grip strength normalized to body weight. N = 8 in each group. Scale bar: 20 μm. Error bars represent SD. *P < 0.05, values significantly different from mdx; Traf6f/f mice.
Inhibition of TRAF6 reduces the markers of autophagy and Akt signaling in dystrophic muscle of mdx mice
Recent studies have shown that inhibition of autophagy is one of the major causes for myopathy in various models of muscular dystrophy, including mdx mice (35). Since TRAF6 is an important positive regulator of autophagy, we next sought to determine whether the inhibition of TRAF6 further represses autophagy in mdx mice. Elevation in the level of LC3B-II protein is a hallmark of autophagy (30). We measured the levels of LC3B-I and LC3B-II protein in skeletal muscle of mdx;Traf6f/f and mdx;Traf6mko mice. The levels of LC3B-II were significantly reduced in skeletal muscle of 6-week-old mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 8A and B). Moreover, the levels of another autophagy-related protein Beclin1 were significantly reduced in myofibers of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 8A and B). p62 protein undergoes degradation through autophagy-lysosomal system (29,30). To further confirm the role of TRAF6 in induction of autophagy in myofibers of mdx mice, we also measured the levels of p62 protein. Results showed that the levels of p62 protein were significantly higher in skeletal muscle of mdx;Traf6mko mice compared with mdx;Traf6f/f littermates (Fig. 8A and B). Moreover, these markers of autophagy were also similarly affected in dystrophic muscle of 5-month-old mdx;Traf6mko compared with TRAF6f/f littermates (data not shown). TRAF6 has also been shown to induce the expression of ubiquitin-proteasome system (UPS) and autophagy-related molecules in skeletal muscle in many catabolic conditions (28,29). To understand whether TRAF6 regulates the expression of the components of UPS and autophagy in dystrophic muscle, we performed QRT-PCR assay. Our results showed that the mRNA levels of LC3B, Beclin1 and Atrogin-1 are significantly reduced in dystrophic muscle of 9-month-old mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Supplementary Material, Fig. S7). Together, these results suggest that the inhibition of TRAF6 represses autophagy and UPS which may be responsible for accumulation of damaged proteins and organelles leading to increased myopathy at later stages of disease progression in mdx mice.
Figure 8.
Role of TRAF6 in activation of autophagy and Akt signaling pathway. GA muscle of 6-week-old mdx;Traf6f/f and mdx;Traf6mko mice were processed for western blotting. (A) Representative immunoblots present here demonstrate the levels of LC3B I and LC3B II and Beclin1, p62 and an unrelated protein GAPDH (glyceraldehyde 3-phosphate dehydrogenase). (B) Densitometry quantification of LC3B I & II, Beclin1 and p62 protein levels in GA muscle of 6-week-old mdx;Traf6f/f and mdx;Traf6mko mice. (C) Representative immunoblots presented here demonstrate the levels of phosphorylated (p) and total Akt, mTOR, p70 S6 kinase (p70 S6K) and GAPDH protein. (D) Quantification of phosphorylated versus total Akt, mTOR, p70 S6K ratio in GA muscle of 6-week-old mdx;Traf6f/f and mdx;Traf6mko mice. N = 6 in each group. Error bars represent SD. *P < 0.05, values significantly different from mdx;Traf6f/f mice.
Previous studies have shown that the Akt-mTOR signaling pathway inhibits autophagy in skeletal muscle (30). There is also evidence that TRAF6 activates Akt in some cell types (22). Moreover, it has been also reported that the activity of Akt-mTOR pathway is increased in skeletal muscle of mdx mice compared with WT controls (11,52). We investigated whether TRAF6 affects the activation of Akt in skeletal muscle of mdx mice. Protein extracts prepared from GA muscle of mdx;Traf6f/f and mdx;Traf6mko mice were subjected to immunoblotting to detect phosphorylated and total protein levels of Akt, mTOR and p70S6 kinase. Results showed that the phosphorylation of Akt, mTOR and p70S6 kinase was significantly reduced in myofibers of mdx;Traf6mko compared with mdx;Traf6f/f mice (Fig. 8C and 8D). Collectively, these results indicate that TRAF6 is involved in the activation of Akt; however, TRAF6 regulates autophagy independent of Akt signaling.
DISCUSSION
Recent studies using genetic mouse models and pharmacological approaches have provided strong evidence that the modulation of activity of specific signaling pathways has enormous potential to reduce the severity of disease progression in muscular dystrophy (9,10,53). While the role of anomalous myogenic signaling has become increasingly clear, the proximal signaling events which lead to the activation of different pathways in dystrophic muscle remain less understood. Furthermore, the effect of long-term inhibition of different signaling pathway in disease progression in models of DMD has not been yet investigated. Since TRAF6 is an important upstream regulator of many proinflammatory signaling pathways, we investigated the role of TRAF6 in pathogenesis of DMD. Our study demonstrates that the expression and activity of TRAF6 are increased in dystrophin-deficient skeletal muscle (Fig. 1). While muscle-specific depletion of TRAF6 signaling reduces injury and improves muscle histopathology (Fig. 3) and regeneration (Fig. 5) in young mice (7–8 weeks), long-term inhibition of TRAF6 results in deterioration of myopathy and loss of function in mdx mice (Figs 6 and 7).
One of the mechanisms by which muscle-derived TRAF6 signaling causes dystrophy in young mdx mice is by promoting chronic inflammatory response. Initial fiber necrosis due to sarcolemmal instability causes severe inflammatory response in skeletal muscle which includes infiltration of macrophages and neutrophils in dystrophic muscle and increased levels of proinflammatory cytokines leading to hastening of fiber necrosis and disease progression in DMD (10,53). Muscle-derived factors appear to be some of the important contributors of chronic inflammation in mdx myofibers supported by the findings that the activation of proinflammatory transcription factors such as NF-κB and activator protein-1 (AP-1) and levels of various inflammatory cytokines (e.g. TNF-α and IL-1β) start increasing before the onset of fiber necrosis (9,11,13,14). Our results demonstrate that muscle-specific inhibition of TRAF6 leads to diminished activation of NF-κB in dystrophic muscle of young mdx mice (Fig. 4C and 4D). It is notable that in addition to proinflammatory cytokines, NF-κB also induces the expression of a number of chemokines which causes extravasation and migration of leukocytes (43,44). Consistently, we have found that the accumulation of macrophages and transcript levels of inflammatory cytokines were also significantly reduced in skeletal muscle of mdx;Traf6mko mice compared with mdx;Traf6f/f mice (Fig. 4). These results are consistent with previous reports demonstrating that muscle-specific depletion of TRAF6 inhibits activation of NF-κB in the conditions of denervation and cancer cachexia and cardiotoxin-mediated muscle injury (27,29). Although depletion of TRAF6 in skeletal muscle reduces inflammation, in the present study, we have not investigated the role of TRAF6 in immune cells. It is possible that TRAF6 signaling emanating from immune compartments might also contribute to the inflammatory processes in dystrophic muscles and concurrent inhibition of TRAF6 in skeletal muscle and immune cells may lead to further improvement in dystrophinopathy. Future research should clarify the role of the TRAF6 signaling in immune cells in pathogenesis of DMD.
Another mechanism by which TRAF6 signaling causes dystrophy is through the repression of myofiber regeneration. Inhibition of TRAF6 increases the formation of new myofibers in mdx mice (Fig. 5). These results are in agreement with our previous findings demonstrating that the depletion of TRAF6 improves whereas its overexpression represses the regeneration of myofibers in response to acute injury in WT mice (27). Muscle regeneration after injury is a complex process which involves the activation of quiescent satellite cells, their differentiation into myoblasts and finally fusion of myoblasts to damaged myofibers (54,55). Although the exact mechanisms remain enigmatic, the results of the present study suggest that TRAF6 signaling from injured myofibers limit the activation of satellite cells and may interfere with their differentiation and fusion into myotubes. It has been consistently observed that inflammatory cytokines such as TNF-α, IL-1β and IL-6 inhibit the differentiation of myoblasts by repressing the levels of MyoD (56–60). Furthermore, there is a positive feed-back loop between inflammatory cytokines and NF-κB transcription factor. Elevated levels of inflammatory cytokines activate NF-κB, which in turn induces the expression of inflammatory cytokines leading to sustained activation of NF-κB (43,44). Acharyya et al. (9) have previously reported that NF-κB inhibits myofiber regeneration in mdx mice. Our results demonstrate that TRAF6 is an upstream activator of NF-κB and expression of inflammatory cytokines in dystrophic muscle (Fig. 4). Therefore, it is reasonable to speculate that the inhibition of TRAF6 improves muscle regeneration through blocking NF-κB and reducing repertoire of inflammatory cytokines in dystrophic muscle.
A striking observation of the present study is that the long-term inhibition of TRAF6 signaling deteriorates muscle pathology in mdx mice. Analysis of myofibers of 9-month-old mdx;Traf6mko mice revealed the presence of significantly increased number of necrotic fibers and replacement of muscle fibers by collagens (Figs 6 and 7). Although the exact mechanisms remain enigmatic, inhibition of autophagy appears to be one of the important reasons for increased myopathy in mdx;Traf6mko mice. Autophagy is an important homeostasis mechanism which is critical for clearing dysfunctional organelles and to preventing tissue damage (30,61). Recently, the role of autophagy in pathogenesis of muscular dystrophy has been investigated using multiple approaches. Autophagy has been found to be impaired in muscle biopsies from patients with DMD and in mdx mice with concomitant accumulation of damaged organelles (35). Notably, reactivation of autophagy by feeding low-protein diet improved muscle strength and various pathological features including fiber necrosis, pathological hypertrophy and fibrosis in mdx mice (35). Similarly, forced activation of autophagy using AMPK agonist, AICAR (5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside), led to improvements in mdx diaphragm histopathology and in force-generating capacity (62).
TRAF6 is an essential component of autophagy in mammalian cells. TRAF6 interacts with LC3B through p62. It has been recently demonstrated that TRAF6 causes the Lys-63-linked ubiquitination of Beclin1 (the mammalian homologue of yeast Atg6), which is essential for autophagosome formation in response to Toll-like receptor 4 signaling (26). We have previously reported that muscle-specific depletion of TRAF6 inhibits expression of autophagy-related molecules and autophagosome formation in atrophying skeletal muscle (28,29). Our results demonstrate that the levels of autophagy markers are considerably reduced in dystrophic muscle of mdx;Traf6mko compared with mdx;Traf6f/f mice, suggesting that TRAF6 is required for the activation of autophagy in mdx mice (Fig. 8A and B). While autophagy is also inhibited in young mdx;Traf6mko mice, there is still significant improvement in muscle pathology compared with mdx;Traf6f/f littermates. We envision that inflammation plays a predominant role in initial muscle injury and regeneration and that there may not be sufficient load of damaged organelles to clear through autophagy pathway in young mdx mice. It is notable that excessive autophagy causes muscle wasting in many catabolic states (30). Thus, the initial inhibition of autophagy in young mdx;TRAF6mko mice may be a protective mechanism to preserve skeletal muscle mass. However, due to the progressive nature of myopathy in mdx mice, autophagy becomes essential for clearance of defunct cellular organelles at later stages and hence continued inhibition of autophagy exaggerates dystrophic phenotype in mdx mice. Moreover, our results demonstrate that depletion of TRAF6 inhibits both NF-κB and Akt signaling in skeletal muscle of mdx mice (Figs 4C and D and 8C and D). While specific inhibition of NF-κB has been shown to ameliorate dystrophy in young mdx mice (9), the effects of long-term inhibition of NF-κB have not been yet investigated. Both NF-κB and Akt are known to play critical roles in cell survival mechanisms (43,63). Therefore, it is possible that prolonged inhibition of NF-κB and Akt in mdx;Traf6mko mice also directly induces muscle cell death. Indeed, it has been previously reported that forced activation of Akt prevents myofiber degeneration and improves regeneration in mdx mice (64).
Akt signaling is also an important regulator of autophagy in skeletal muscle. Activation of Akt inhibits autophagy through repressing the activity of FOXO family transcription factors (30). Recently, De Palma et al. (35) have suggested that the activation of Akt signaling is responsible for inhibition of autophagy in mdx mice. However, our results demonstrate that TRAF6 regulates autophagy independent of Akt signaling. Muscle-specific inhibition of TRAF6 also inhibited Akt pathway in myofibers of mdx mice (Fig. 8C and D). It is also notable that the inhibition of TRAF6 signaling did not affect the activation of Akt kinase in regenerating myofibers in response to cardiotoxin-mediated injury (27). However, in some other cell types, TRAF6 has been found to be an important upstream activator of Akt kinase (22). Together, these findings further highlight the role of TRAF6 in context-dependent activation of various signaling pathways.
In summary, our study has provided novel mechanistic insights in dystrophic progression by identifying the role of TRAF6 in mdx mice. It is now increasingly clear that muscular dystrophy is a complex disorder and that a single therapeutic intervention may not be sufficient to treat patients with muscular dystrophy. Recent studies further support this notion by providing experimental evidence that combinatorial approaches correcting more than one parameter of pathological cascade can produce more robust improvement in myopathy in mouse models of muscular dystrophy (65). Our results demonstrate that the inhibition of TRAF6 is effective in reducing inflammatory response and initial fiber necrosis in mdx mice. However, it is also evident that the inhibition of TRAF6 can also produce deleterious effects due to further repression of autophagy in dystrophic muscle. Nevertheless, it is reasonable to speculate that a combinatorial approach involving pharmacological inhibitors of TRAF6 and activators of autophagy can provide an effective treatment strategy for patients with DMD.
MATERIALS AND METHODS
Mice
WT (strain: C57BL/10 ScSn) and mdx (strain: C57BL/10 ScSn DMDmdx) mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Floxed Traf6 (Traf6f/f) and muscle-specific Traf6-knockout (Traf6mko) mice have been previously described (28,29). Traf6mko mice were crossed with mdx mice for five to six generations to generate littermate mdx;Traf6f/f and mdx;Traf6mko mice. All genotypes were determined by PCR analysis from tail DNA. Mice were housed in the animal facility of the University of Louisville under conventional conditions with constant temperature and humidity and fed a standard diet. All experiments with animals were approved by the Institutional Animal Care and Use Committee of the University of Louisville.
Creatine kinase (CK) assay
The serum level of CK was determined using a commercially available kit (Stanbio Laboratory, TX, USA).
Histology and morphometric analysis
Skeletal muscle tissues were isolated, frozen in isopentane cooled in liquid nitrogen and sectioned in a microtome cryostat. For the assessment of tissue morphology, 10 µm thick transverse sections of each muscle were stained with H&E, and the staining was visualized on a microscope (Eclipse TE 2000-U), a digital camera (Digital Sight DS-Fi1) and NIS Elements BR 3.00 software (all from Nikon). The images were stored as JPEG files, and image levels were equally adjusted using Photoshop CS2 software (Adobe). Pictures of the whole muscle sections were captured and the percentage of centrally nucleated fibers was counted in the entire muscle section. To quantify the variation in fiber size, fiber cross-sectional area was measured for every fiber in each section using Nikon NIS Elements BR 3.00 software (Nikon). Variability in cross-sectional areas between samples was expressed as the mean of the standard deviations for each population. Necrotic area in H&E-stained sections was determined by measuring percentage area filled with cellular infiltrate in whole muscle section. Mean minimum feret diameter of eMyHC+ fibers was determined after measuring cross-sectional area for each fiber. The extent of fibrosis in muscle cryosections was determined using a Trichrome staining kit following a protocol suggested by manufacturer (American Master Tech).
For immunohistochemical detection of macrophages, rat anti-mouse F4/80 (Serotec) was used at 1:500 dilution and horseradish peroxidase-labeled streptavidin biotin technique (DAKO K5001 with E468) was used as a detection system. The concentrations of F4/80+ cells were expressed as the number of cells per volume of each section.
Indirect immunofluorescence
For immunohistochemistry study, muscle sections were blocked in 1% bovine serum albumin in phosphate-buffered saline (PBS) for 1 h, and incubated with anti-Pax7 (1:20, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) or anti-E-MyHC (1:150, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA) in blocking solution at 4°C overnight under humidified conditions. The sections were washed briefly with PBS before incubation with Alexa Fluor® 488 or 594-conjugated secondary antibody (1:3000, Invitrogen) for 1 h at room temperature and then washed three times for 5 min with PBS. The slides were mounted using fluorescence medium (Vector Laboratories) and visualized at room temperature on Nikon Eclipse TE 2000-U microscope (Nikon), a digital camera (Nikon Digital Sight DS-Fi1) and Nikon NIS Elements BR 3.00 software (Nikon). Image levels were equally adjusted using Abode Photoshop CS2 software (Adobe). Damaged/permeabilized fibers in muscle cryosections were identified by immunostaining with Cy3-labelled goat anti-mouse IgG (1:3000, Invitrogen).
Immunoprecipitation and western blotting
Quantitative estimation of specific protein was done by western blot using a method as previously described (66). Briefly, individual tissues were washed with PBS and homogenized in lysis buffer A [50 mm Tris-Cl (pH 8.0), 200 mm NaCl, 50 mm NaF, 1 mm dithiotheritol (DTT), 1 mm sodium orthovanadate, 0.3% IGEPAL and protease inhibitors]. Approximately, 100 µg protein was resolved on each lane on 10% SDS–PAGE, electrotransferred onto nitrocellulose membrane and probed using anti-TRAF6 (1:500 MBL International), anti-TRAF2 (1:500; Santa Cruz Biotechnology, Inc.), anti-TRAF3 (1:500; Santa Cruz Biotechnology, Inc), anti-LC3B (1:500; Cell signaling Technology), anti-Beclin (1:500; Cell signaling Technology), anti-p62 (1:500; MBL International), anti-phospho-Akt (1:500; Cell Signaling Technology), anti-Akt (1:500; Cell Signaling Technology), anti-phospho-mTOR (1:500; Cell Signaling Technology), anti-mTOR (1:500; Cell Signaling Technology), anti-phospho-p70-S6K (1:500; Cell Signaling Technology), anti-p70-S6K (1:500; Cell Signaling Technology), anti-GADPH (1:2000; Cell Signaling Technology) and anti-tubulin (1:2000, Cell Signaling Inc) and detected by chemiluminescence.
To study the ubiquitination of TRAF6, muscle extract (400 μg protein) was incubated overnight with 1 μg of anti-Ubiquitin antibody (Santa Cruz Biotechnology, Inc.) in 600 μl of lysis buffer, protein A-Sepharose beads were added and the mixture was incubated at 4°C for an additional 2 h. The beads were washed four times with lysis buffer and finally suspended in 2× Laemmli sample buffer. Proteins were resolved on 10% SDS–PAGE gel and immunoblotted using TRAF6 antibody (1:500; MBL International).
Electrophoretic mobility shift assay (EMSA)
The DNA binding activity of NF-κB transcription factor was measured using EMSA as detailed (14). Briefly, 20 µg of nuclear extract prepared from skeletal muscle was incubated with 16 fmol 32P-γ ATP-end-labeled NF-κB consensus double-stranded oligonucleotide (Promega, MA, USA) for 20 min at 37°C. The incubation mixture included 2–3 µg of poly dI.dC in a binding buffer [25 mm HEPES (pH 7.9), 0.5 mm EDTA, 0.5 mm dithiothreitol, 1% IGEPAL, 5% glycerol, 50 mm NaCl]. The DNA–protein complex thus formed was separated from free oligonucleotides on a 7.5% native polyacrylamide gel. The gel was dried, and radioactive bands were visualized and quantitated by PhosphorImager using ImageQuant TL software (GE Healthcare, Piscataway, NJ, USA).
Quantitative real-time PCR (QRT-PCR)
Real-time PCR for individual genes was performed using an ABI Prism 7300 Sequence Detection System (Applied Biosystems) using a method as previously described (60,66). Briefly, the first-strand cDNA reaction (0.5 μl) from GA muscle of individual control or mdx mice (n = 4 in each group) was subjected to real-time PCR amplification using gene-specific primers. The primers were designed according to ABI primer express instructions using Vector NTI software and were purchased from Sigma-Genosys (Spring, TX, USA). The sequences of the primers used are as follows: TNF-α, 5′-GCA TGA TCC GCG ACG TGGAA-3′ (forward) and 5′-AGATCCATGCCGTTGGCC AG-3′(reverse); IL-1β, 5′-CTCCATGAGCTTTGTACAAGG-3′ (forward) and 5′-TGCTGATGTACCAGTTGGGG-3′ (reverse); IL-6, 5′-CCTTCTTGGGACTGATGCTGG-3′ (forward) and 5′-GCCTCCGACTTGTGAAGTGGT-3′ (reverse); MMP-9, 5′-GCGTGTCTGGAGATTCGACTT G-3′ (forward) and 5′-CATGGTCCACCTTGTTCACCTC-3′ (reverse); LC3B, 5′-CTGGTGAATGGGCACAGCATG-3′ (forward) and 5′-CGTCCGCTGGTAACATCCCTT-3′ (reverse); Beclin1, 5′-TGAAATCAATGCTGCCTGGG-3′ (forward) and 5′-CCAGAACAGTATAACGGCAACTCC-3′ (reverse); p62, 5′-AGCACAGGCACAGAAGACAAGAGT-3′ (forward) and 5′-AATGTGTCCAGTCATCGTCTCCTC-3′ (reverse); Atrogin-1, 5′-GTCGCAGCCAAGAAGAGAAAGA-3′ (forward) and 5′-TGCTATCAGCTCCAACAGCCTT-3′ (reverse); MuRF1, 5′-TAACTGCATCTCCATGCTGGTG-3′ (forward) and 5′-TGGCGTAGAGGGTGTCAAACTT-3′ (reverse); and beta-actin, 5′-CAGGCATTGCTGACAGGATG-3′ (forward) and 5′-TGCTGATCCACATCTGCTGG-3′ (reverse).
Approximately 25 μl of reaction volume was used for the real-time PCR assays which consisted of 2× (12.5 μl) Brilliant SYBR Green QPCR Master Mix (Stratagene), 400 nm of primers (0.5 μl each from the stock), 11 μl of water and 0.5 μl of template. The thermal conditions consisted of an initial denaturation at 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing and extension at 60°C for 1 min and, for a final step, a melting curve of 95°C for 15 s, 60°C for 15 s and 95°C for 15 s. All reactions were carried out in triplicate to reduce variation. The data were analyzed using SDS software version 2.0, and the results were exported to Microsoft Excel for further analysis. Data normalization was accomplished using two endogenous control (β-actin) and the normalized values were subjected to a 2−ΔΔCt formula to calculate the fold change between the control and experimental groups. The formula and its derivations were obtained from the ABI Prism 7900 Sequence Detection System user guide.
Grip strength and wire hanging measurements
A digital grip-strength meter (Columbus Instruments, Columbus, OH, USA) was used to measure forelimb or total four-limb grip strength in mice. Mice were acclimatized for 5 min before starting test. The mouse was allowed to grab the metal pull bar with the forepaws and in a separate experiment with or all four paws. The mouse tail was then gently pulled backward in the horizontal plane until it could no long grasp the bar. The force at the time of release was recorded as the peak tension. Each mouse was tested five times with a 20–40 s break between tests. The average peak tension from three best attempts normalized against total body weight was defined as forelimb grip strength. For wire hanging time evaluation, mice were placed on a grid in a starting upright position. The grid was then gradually inverted above a cage filled with bedding. Hanging time was determined as the longest time sustained hanging against gravity from three repetitions.
Statistical analysis
Results are expressed as mean ± standard deviation (SD). Statistical analysis used Student's t-test (two tailed) to compare quantitative data populations with normal distribution and equal variance. A value of P < 0.05 was considered statistically significant unless otherwise specified.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by funding from Muscular Dystrophy Association and National Institute of Health grants AG029623 and AR059810 to A.K.
Supplementary Material
REFERENCES
- 1.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E.P., Brown R.H., Jr, Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Rando T.A. The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies. Muscle Nerve. 2001;24:1575–1594. doi: 10.1002/mus.1192. [DOI] [PubMed] [Google Scholar]
- 4.Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl Acad. Sci. USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engvall E., Wewer U.M. The new frontier in muscular dystrophy research: booster genes. FASEB J. 2003;17:1579–1584. doi: 10.1096/fj.02-1215rev. [DOI] [PubMed] [Google Scholar]
- 6.Khurana T.S., Davies K.E. Pharmacological strategies for muscular dystrophy. Nat. Rev. Drug. Discov. 2003;2:379–390. doi: 10.1038/nrd1085. [DOI] [PubMed] [Google Scholar]
- 7.Chakkalakal J.V., Thompson J., Parks R.J., Jasmin B.J. Molecular, cellular, and pharmacological therapies for Duchenne/Becker muscular dystrophies. FASEB J. 2005;19:880–891. doi: 10.1096/fj.04-1956rev. [DOI] [PubMed] [Google Scholar]
- 8.Spencer M.J., Tidball J.G. Do immune cells promote the pathology of dystrophin-deficient myopathies? Neuromuscul. Disord. 2001;11:556–564. doi: 10.1016/s0960-8966(01)00198-5. [DOI] [PubMed] [Google Scholar]
- 9.Acharyya S., Villalta S.A., Bakkar N., Bupha-Intr T., Janssen P.M., Carathers M., Li Z.W., Beg A.A., Ghosh S., Sahenk Z., et al. Interplay of IKK/NF-kappaB signaling in macrophages and myofibers promotes muscle degeneration in Duchenne muscular dystrophy. J. Clin. Invest. 2007;117:889–901. doi: 10.1172/JCI30556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatnagar S., Kumar A. Therapeutic targeting of signaling pathways in muscular dystrophy. J. Mol. Med. 2010;88:155–166. doi: 10.1007/s00109-009-0550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dogra C., Changotra H., Wergedal J.E., Kumar A. Regulation of phosphatidylinositol 3-kinase (PI3K)/Akt and nuclear factor-kappa B signaling pathways in dystrophin-deficient skeletal muscle in response to mechanical stretch. J. Cell Physiol. 2006;208:575–585. doi: 10.1002/jcp.20696. [DOI] [PubMed] [Google Scholar]
- 12.Dogra C., Srivastava D.S., Kumar A. Protein-DNA array-based identification of transcription factor activities differentially regulated in skeletal muscle of normal and dystrophin-deficient mdx mice. Mol. Cell Biochem. 2008;312:17–24. doi: 10.1007/s11010-008-9716-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar A., Boriek A.M. Mechanical stress activates the nuclear factor-kappaB pathway in skeletal muscle fibers: a possible role in Duchenne muscular dystrophy. FASEB J. 2003;17:386–396. doi: 10.1096/fj.02-0542com. [DOI] [PubMed] [Google Scholar]
- 14.Kumar A., Khandelwal N., Malya R., Reid M.B., Boriek A.M. Loss of dystrophin causes aberrant mechanotransduction in skeletal muscle fibers. FASEB J. 2004;18:102–113. doi: 10.1096/fj.03-0453com. [DOI] [PubMed] [Google Scholar]
- 15.Chung J.Y., Park Y.C., Ye H., Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 16.Zapata J.M., Lefebvre S., Reed J.C. Targeting TRAfs for therapeutic intervention. Adv. Exp. Med. Biol. 2007;597:188–201. doi: 10.1007/978-0-387-70630-6_15. [DOI] [PubMed] [Google Scholar]
- 17.Bradley J.R., Pober J.S. Tumor necrosis factor receptor-associated factors (TRAFs) Oncogene. 2001;20:6482–6491. doi: 10.1038/sj.onc.1204788. [DOI] [PubMed] [Google Scholar]
- 18.Chung J.Y., Lu M., Yin Q., Lin S.C., Wu H. Molecular basis for the unique specificity of TRAF6. Adv. Exp. Med. Biol. 2007;597:122–130. doi: 10.1007/978-0-387-70630-6_10. [DOI] [PubMed] [Google Scholar]
- 19.Moscat J., Diaz-Meco M.T., Wooten M.W. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skaug B., Jiang X., Chen Z.J. The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 22.Yang W.L., Wang J., Chan C.H., Lee S.W., Campos A.D., Lamothe B., Hur L., Grabiner B.C., Lin X., Darnay B.G., et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura K., Kimple A.J., Siderovski D.P., Johnson G.L. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J. Biol. Chem. 2010;285:2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seibenhener M.L., Babu J.R., Geetha T., Wong H.C., Krishna N.R., Wooten M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wooten M.W., Geetha T., Seibenhener M.L., Babu J.R., Diaz-Meco M.T., Moscat J. The p62 scaffold regulates nerve growth factor-induced NF-kappaB activation by influencing TRAF6 polyubiquitination. J. Biol. Chem. 2005;280:35625–35629. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- 26.Shi C.S., Kehrl J.H. TRAF6 And A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hindi S.M., Paul P.K., Dahiya S., Mishra V., Bhatnagar S., Kuang S., Choi Y., Kumar A. Reciprocal interaction between TRAF6 and notch signaling regulates adult myofiber regeneration upon injury. Mol. Cell Biol. 2012;32:4833–4845. doi: 10.1128/MCB.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul P.K., Bhatnagar S., Mishra V., Srivastava S., Darnay B.G., Choi Y., Kumar A. The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol. Cell Biol. 2012;32:1248–1259. doi: 10.1128/MCB.06351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul P.K., Gupta S.K., Bhatnagar S., Panguluri S.K., Darnay B.G., Choi Y., Kumar A. Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J. Cell Biol. 2010;191:1395–1411. doi: 10.1083/jcb.201006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandri M. Autophagy in skeletal muscle. FEBS Lett. 2010;584:1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 31.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S.J., Di Lisi R., Sandri C., Zhao J., et al. Foxo3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 33.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S.H., Goldberg A.L. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. Foxo3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 35.De Palma C., Morisi F., Cheli S., Pambianco S., Cappello V., Vezzoli M., Rovere-Querini P., Moggio M., Ripolone M., Francolini M., et al. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis. 2012;3:e418. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulfield G., Siller W.G., Wight P.A., Moore K.J. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl Acad. Sci. USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stedman H.H., Sweeney H.L., Shrager J.B., Maguire H.C., Panettieri R.A., Petrof B., Narusawa M., Leferovich J.M., Sladky J.T., Kelly A.M. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352:536–539. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 38.Dangain J., Vrbova G. Muscle development in mdx mutant mice. Muscle Nerve. 1984;7:700–704. doi: 10.1002/mus.880070903. [DOI] [PubMed] [Google Scholar]
- 39.DiMario J.X., Uzman A., Strohman R.C. Fiber regeneration is not persistent in dystrophic (MDX) mouse skeletal muscle. Dev. Biol. 1991;148:314–321. doi: 10.1016/0012-1606(91)90340-9. [DOI] [PubMed] [Google Scholar]
- 40.Lamothe B., Besse A., Campos A.D., Webster W.K., Wu H., Darnay B.G. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. J. Biol. Chem. 2007;282:4102–4112. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamothe B., Webster W.K., Gopinathan A., Besse A., Campos A.D., Darnay B.G. TRAF6 ubiquitin ligase is essential for RANKL signaling and osteoclast differentiation. Biochem. Biophys. Res. Commun. 2007;359:1044–1049. doi: 10.1016/j.bbrc.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tidball J.G., Wehling-Henricks M. Damage and inflammation in muscular dystrophy: potential implications and relationships with autoimmune myositis. Curr. Opin. Rheumatol. 2005;17:707–713. doi: 10.1097/01.bor.0000179948.65895.1a. [DOI] [PubMed] [Google Scholar]
- 43.Kumar A., Takada Y., Boriek A.M., Aggarwal B.B. Nuclear factor-kappaB: its role in health and disease. J. Mol. Med. 2004;82:434–448. doi: 10.1007/s00109-004-0555-y. [DOI] [PubMed] [Google Scholar]
- 44.Li H., Malhotra S., Kumar A. Nuclear factor-kappa B signaling in skeletal muscle atrophy. J. Mol. Med. 2008;86:1113–1126. doi: 10.1007/s00109-008-0373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson J.M., Bakkar N., Guttridge D.C. NF-kappaB signaling in skeletal muscle health and disease. Curr. Top. Dev. Biol. 2011;96:85–119. doi: 10.1016/B978-0-12-385940-2.00004-8. [DOI] [PubMed] [Google Scholar]
- 46.Lu A., Proto J.D., Guo L., Tang Y., Lavasani M., Tilstra J.S., Niedernhofer L.J., Wang B., Guttridge D.C., Robbins P.D., et al. NF-kappaB negatively impacts the myogenic potential of muscle-derived stem cells. Mol. Ther. 2012;20:661–668. doi: 10.1038/mt.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuang S., Charge S.B., Seale P., Huh M., Rudnicki M.A. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuang S., Gillespie M.A., Rudnicki M.A. Niche regulation of muscle satellite cell self-renewal and differentiation. Cell Stem Cell. 2008;2:22–31. doi: 10.1016/j.stem.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 49.Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mann C.J., Perdiguero E., Kharraz Y., Aguilar S., Pessina P., Serrano A.L., Munoz-Canoves P. Aberrant repair and fibrosis development in skeletal muscle. Skelet. Muscle. 2011;1:21. doi: 10.1186/2044-5040-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernasconi P., Torchiana E., Confalonieri P., Brugnoni R., Barresi R., Mora M., Cornelio F., Morandi L., Mantegazza R. Expression of transforming growth factor-beta 1 in dystrophic patient muscles correlates with fibrosis. Pathogenetic role of a fibrogenic cytokine. J. Clin. Invest. 1995;96:1137–1144. doi: 10.1172/JCI118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peter A.K., Crosbie R.H. Hypertrophic response of Duchenne and limb-girdle muscular dystrophies is associated with activation of Akt pathway. Exp. Cell Res. 2006;312:2580–2591. doi: 10.1016/j.yexcr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 53.Shin J., Tajrishi M.M., Ogura Y., Kumar A. Wasting mechanisms in muscular dystrophy. Int. J. Biochem. Cell Biol. 2013;45:2266–2279. doi: 10.1016/j.biocel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charge S.B., Rudnicki M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 55.Kuang S., Rudnicki M.A. The emerging biology of satellite cells and their therapeutic potential. Trends Mol. Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 56.Langen R.C., Schols A.M., Kelders M.C., Wouters E.F., Janssen-Heininger Y.M. Inflammatory cytokines inhibit myogenic differentiation through activation of nuclear factor-kappaB. FASEB J. 2001;15:1169–1180. doi: 10.1096/fj.00-0463. [DOI] [PubMed] [Google Scholar]
- 57.Langen R.C., Van Der Velden J.L., Schols A.M., Kelders M.C., Wouters E.F., Janssen-Heininger Y.M. Tumor necrosis factor-alpha inhibits myogenic differentiation through MyoD protein destabilization. FASEB J. 2004;18:227–237. doi: 10.1096/fj.03-0251com. [DOI] [PubMed] [Google Scholar]
- 58.Guttridge D.C., Mayo M.W., Madrid L.V., Wang C.Y., Baldwin A.S., Jr NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- 59.Peterson J.M., Guttridge D.C. Skeletal muscle diseases, inflammation, and NF-kappaB signaling: insights and opportunities for therapeutic intervention. Int. Rev. Immunol. 2008;27:375–387. doi: 10.1080/08830180802302389. [DOI] [PubMed] [Google Scholar]
- 60.Dogra C., Changotra H., Mohan S., Kumar A. Tumor necrosis factor-like weak inducer of apoptosis inhibits skeletal myogenesis through sustained activation of nuclear factor-kappaB and degradation of MyoD protein. J. Biol. Chem. 2006;281:10327–10336. doi: 10.1074/jbc.M511131200. [DOI] [PubMed] [Google Scholar]
- 61.Bonaldo P., Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model Mech. 2013;6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pauly M., Daussin F., Burelle Y., Li T., Godin R., Fauconnier J., Koechlin-Ramonatxo C., Hugon G., Lacampagne A., Coisy-Quivy M., et al. AMPK Activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am. J. Pathol. 2012;181:583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 63.Manning B.D., Cantley L.C. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim M.H., Kay D.I., Rudra R.T., Chen B.M., Hsu N., Izumiya Y., Martinez L., Spencer M.J., Walsh K., Grinnell A.D., et al. Myogenic Akt signaling attenuates muscular degeneration, promotes myofiber regeneration and improves muscle function in dystrophin-deficient mdx mice. Hum. Mol. Genet. 2011;20:1324–1338. doi: 10.1093/hmg/ddr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamauchi J., Kumar A., Duarte L., Mehuron T., Girgenrath M. Triggering regeneration and tackling apoptosis: a combinatorial approach to treating congenital muscular dystrophy type 1 A. Hum. Mol. Genet. 2013;22:4306–4317. doi: 10.1093/hmg/ddt280. [DOI] [PubMed] [Google Scholar]
- 66.Srivastava A.K., Qin X., Wedhas N., Arnush M., Linkhart T.A., Chadwick R.B., Kumar A. Tumor necrosis factor-alpha augments matrix metalloproteinase-9 production in skeletal muscle cells through the activation of transforming growth factor-beta-activated kinase 1 (TAK1)-dependent signaling pathway. J. Biol. Chem. 2007;282:35113–35124. doi: 10.1074/jbc.M705329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.