Abstract
Lamins A and C (products of the LMNA gene) are found in roughly equal amounts in peripheral tissues, but the brain produces mainly lamin C and little lamin A. In HeLa cells and fibroblasts, the expression of prelamin A (the precursor to lamin A) can be reduced by miR-9, but the relevance of those cell culture studies to lamin A regulation in the brain was unclear. To address this issue, we created two new Lmna knock-in alleles, one (LmnaPLAO-5NT) with a 5-bp mutation in a predicted miR-9 binding site in prelamin A's 3′ UTR, and a second (LmnaPLAO-UTR) in which prelamin A's 3′ UTR was replaced with lamin C's 3′ UTR. Neither allele had significant effects on lamin A levels in peripheral tissues; however, both substantially increased prelamin A transcript levels and lamin A protein levels in the cerebral cortex and the cerebellum. The increase in lamin A expression in the brain was more pronounced with the LmnaPLAO-UTR allele than with the LmnaPLAO-5NT allele. With both alleles, the increased expression of prelamin A transcripts and lamin A protein was greater in the cerebral cortex than in the cerebellum. Our studies demonstrate the in vivo importance of prelamin A's 3′ UTR and its miR-9 binding site in regulating lamin A expression in the brain. The reduced expression of prelamin A in the brain likely explains why children with Hutchinson–Gilford progeria syndrome (a progeroid syndrome caused by a mutant form of prelamin A) are spared from neurodegenerative disease.
INTRODUCTION
Lamins A and C (the A-type lamins) are alternatively spliced products of the LMNA gene (1). LMNA mutations have been linked to many diseases, including muscular dystrophy, cardiomyopathy, partial lipodystrophy, neuropathy and progeroid syndromes (2). Most of these mutations are located in sequences shared by prelamin A (the precursor to lamin A) and lamin C, but some, including the one for Hutchinson–Gilford progeria syndrome (HGPS; OMIM 176670), are located in sequences unique to prelamin A. The HGPS mutation has no effect on lamin C but leads to the synthesis of a mutant prelamin A that is toxic to cells and tissues (3,4).
Lamins A and C are expressed in roughly similar amounts in most tissues, but we recently encountered an exception: the brain produces mainly lamin C and little lamin A (5). The preferential synthesis of lamin C in the brain is intriguing because it suggested a new insight into the spectrum of disease phenotypes in HGPS. Children with HGPS have multiple disease phenotypes resembling premature aging, but they are spared from senile dementia. The absence of neurodegenerative disease could be due to the fact that the brain makes mainly lamin C and little of the toxic prelamin A.
We initially suspected that alternative splicing would explain the preferential synthesis of lamin C in the brain, but this was not the case. Lamin A expression in the brain was also low in ‘lamin A-only’ knock-in mice (6), where alternative splicing is absent and all of the output of the gene is channeled into prelamin A (5). Additional studies suggested that prelamin A expression might be regulated by miR-9, a microRNA that is expressed highly in the brain. When miR-9 was expressed in HeLa cells and mouse embryonic fibroblasts (MEFs), prelamin A transcripts were reduced but lamin C was unaffected (5).
The cell culture studies on miR-9 regulation of prelamin A left several questions unanswered. One was whether the experiments with cultured non-neuronal cells (i.e. HeLa cells and MEFs) were relevant to prelamin A regulation in the brain, particularly since microRNA-target interactions can be context-dependent (7–10). A second issue was whether other sequences in prelamin A's 3′ UTR, apart from the miR-9 binding site, might be important in regulating prelamin A expression. A third issue was whether the preferential expression of lamin C in the brain is crucial for brain homeostasis and whether lamin A expression in the brain would lead to adverse consequences. To address these issues, we created and characterized two new lines of Lmna knock-in mice with mutations in prelamin A's 3′ UTR—one with a 5-bp mutation in the predicted miR-9 binding site and a second in which prelamin A's 3′ UTR was replaced with lamin C's 3′ UTR.
RESULTS
We used sequence-replacement vectors to create two Lmna knock-in alleles, one (Lmnaplao-5nt) in which a predicted miR-9 seed-binding site in exon 12 (CCAAAG, within prelamin A's 3′ UTR) (5) was changed to ACCCTG, and a second (Lmnaplao-utr) in which prelamin A's 3′ UTR was replaced with lamin C's 3′ UTR (Fig. 1). Both vectors eliminated introns 10 and 11; consequently, the new alleles produced exclusively prelamin A and no lamin C (11). To be certain that the ‘ACCCT’ mutation in the LmnaPLAO-5NT allele would be effective in abolishing the effect of miR-9 on prelamin A transcripts, we performed reporter assays with a luciferase vector linked to prelamin A's 3′ UTR (Supplementary Material, Fig. S1). The ACCCT mutation in the 3′ UTR abolished the ability of miR-9 to reduce luciferase expression in HeLa cells and U-87 MG cells (a human glioblastoma cell line) (Supplementary Material, Fig. S1B and C).
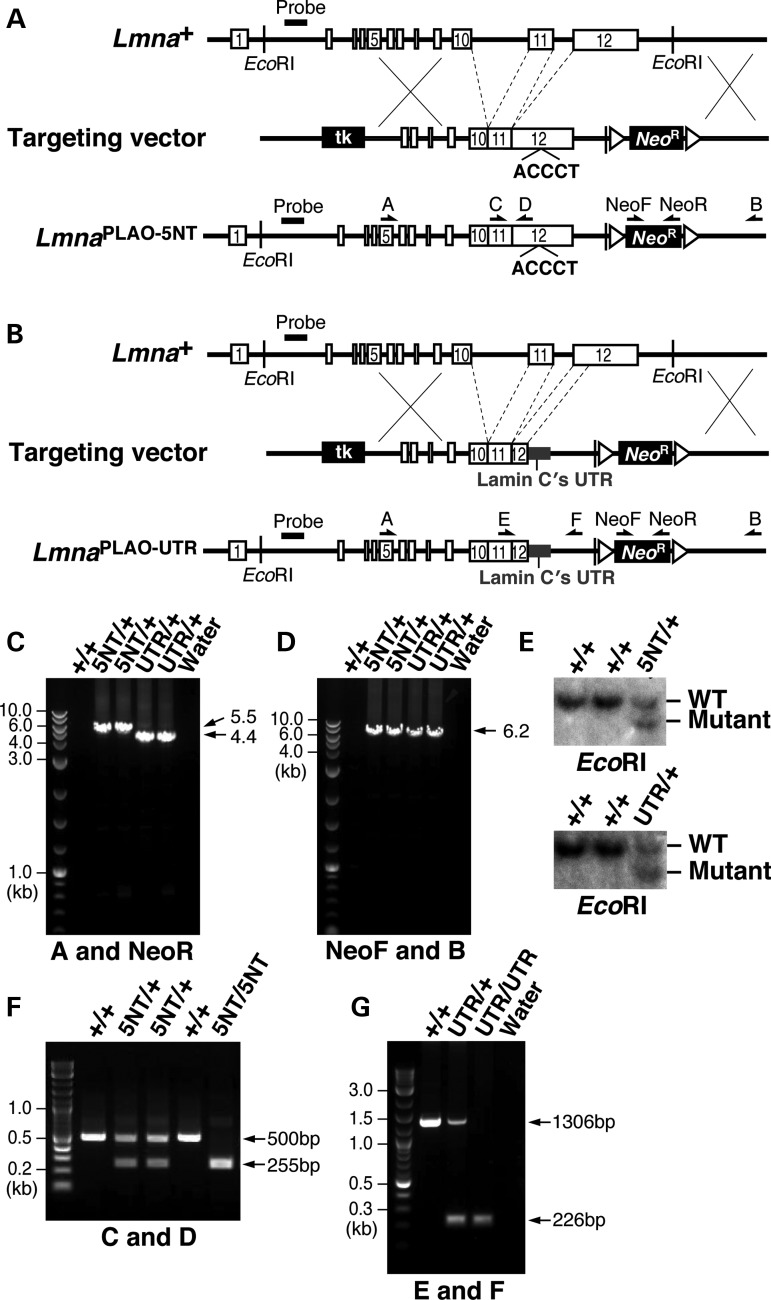
Figure 1.
Knock-in mice expressing prelamin A transcripts with mutations in the 3′ UTR. (A) Strategy to create the LmnaPLAO-5NT allele, in which 5-nt of the seed-binding sequence in a predicted miR-9 binding site in prelamin A's 3′ UTR (CCAAAG) (5) were replaced with ACCCTG. Introns 10 and 11 were also deleted; consequently, the targeted allele yields exclusively prelamin A (and no lamin C) (11). Exons are depicted as white boxes; loxP sites are represented by arrowheads; the position of the 5′ flanking probe and EcoRI sites for Southern blotting are also identified. Primer locations for the 5′ (primers A and NeoR) and 3′ (primers NeoF and B) long-range PCRs and genotyping PCRs (primers C and D) are indicated with arrows. (B) Strategy to create the Lmnaplao-utr allele, in which prelamin A's 3′ UTR was replaced with lamin C's 3′ UTR (grey). Introns 10 and 11 were deleted. Primer locations for the 5′ (primers A and NeoR) and 3′ (primers NeoF and B) long-range PCRs and genotyping PCRs (primers E and F) are indicated with arrows. (C) The 5′ long-range PCRs with the LmnaPLAO-5NT and Lmnaplao-utr alleles yield 5.5 kb (lanes 3 and 4) and 4.4 kb (lanes 5 and 6) fragments, respectively. No PCR product was amplified from wild-type DNA (lane 2). (D) The 3′ long-range PCRs with the LmnaPLAO-5NT and Lmnaplao-utr alleles yield a 6.2 kb fragment (lanes 3–6); no product was amplified from wild-type DNA (lane 2). (E) Southern blot analysis to identify targeted mouse ES cell clones for the LmnaPLAO-5NT and Lmnaplao-utr alleles. Genomic DNA was digested with EcoRI, and the blot was hybridized with a 5′ flanking probe (A and B). (F) Genotyping PCR for the LmnaPLAO-5NT allele yields a 255 bp fragment, whereas wild-type DNA yields a 500 bp fragment. (G) Genotyping PCR with the Lmnaplao-utr allele yields a 226 bp fragment, whereas wild-type DNA yields a 1.3 kb fragment.
We used western blots to assess relative amounts of lamins A and C in the cerebral cortex, cerebellum, heart, liver and kidney of wild-type, Lmnaplao-5nt/plao-5nt, Lmnaplao-utr/plao-utr and Lmnaplao/plao mice. [Lmnaplao/plao mice lack introns 10 and 11 and therefore produce exclusively prelamin A, but they have no mutations in prelamin A's 3′ UTR (11).] Large amounts of both lamins A and C were present in the peripheral tissues of wild-type mice; in the brain, lamin C was abundant, but lamin A expression was low (Fig. 2A). Lamin A levels were higher in Lmnaplao/plao mice than in wild-type mice (including in the brain), reflecting the fact that all of the output of the LmnaPLAO allele is channeled into prelamin A (rather than into both lamin C and prelamin A). Like Lmnaplao/plao mice, Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice expressed exclusively lamin A. In all three lines, the amount of lamin A in peripheral tissues, relative to actin, was similar, but there were obvious differences in the brain. In the cerebral cortex of Lmnaplao-5nt/plao-5nt mice, the level of lamin A was 2.28 ± 0.33-fold higher, relative to actin, than in Lmnaplao/plao mice (P < 0.005; Fig. 2B); in the cerebellum, the level of lamin A was 1.66 ± 0.27-fold higher (P < 0.05). The differences were more striking in Lmnaplao-utr/plao-utr mice. In those mice, the lamin A level in the cerebral cortex was 4.30 ± 0.84-fold higher than in Lmnaplao/plao mice (P < 0.0005); in the cerebellum, the lamin A level was 1.99 ± 0.24-fold higher (P < 0.005).
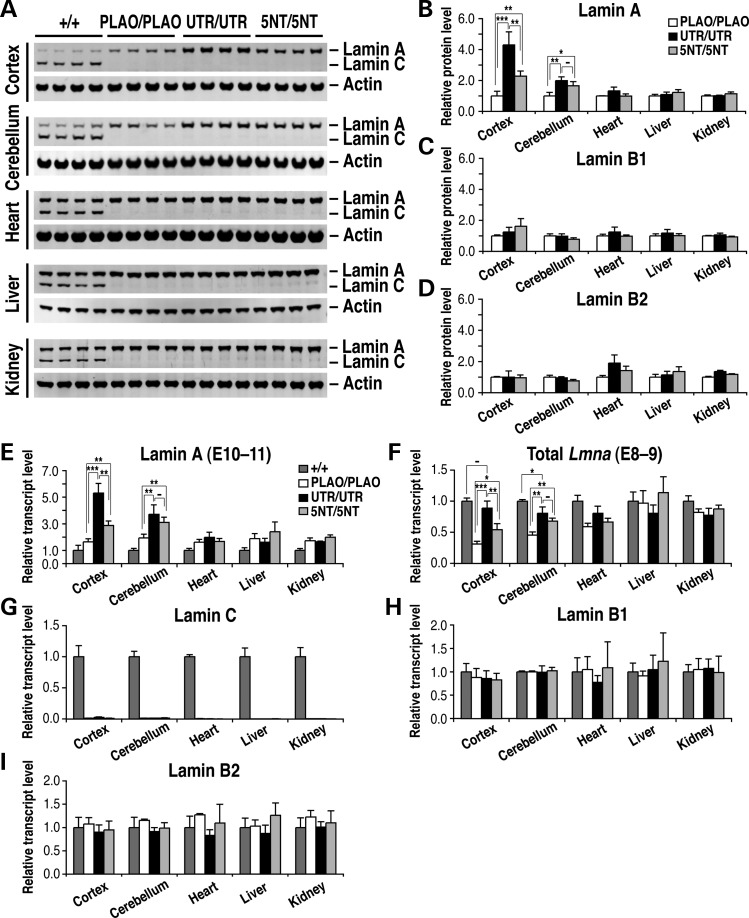
Figure 2.
Higher lamin A expression in the brain of Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice. (A) Western blot analysis of lamins A and C expression in the cerebral cortex, cerebellum, heart, liver and kidney from Lmna+/+ (+/+), Lmnaplao/plao (PLAO/PLAO) (11), Lmnaplao-utr/plao-utr (UTR/UTR) and Lmnaplao-5nt/plao-5nt (5NT/5NT) mice with antibodies against lamin A/C and actin. All samples are from 1-month-old mice. (B–D) Quantification of lamin protein levels relative to actin (A; Supplementary Material, Fig. S2). Values represents mean ± SD. *P < 0.05; **P < 0.005; ***P < 0.0005; (−) P > 0.05. (E–I) Lamin transcript levels, as judged by quantitative reverse transcription polymerase chain reaction in the same tissues analyzed in (A)–(D). Transcript levels were normalized to cyclophilin A and compared with levels in wild-type mice (set at 1.0). Values represent the mean ± SD.
Consistent changes were observed at the RNA level (Fig. 2E). Prelamin A transcripts in the cerebral cortex of Lmnaplao-5nt/plao-5nt mice were 1.75 ± 0.21-fold higher than in Lmnaplao/plao mice (P < 0.005). In Lmnaplao-utr/plao-utr mice, prelamin A transcripts in the cerebral cortex were 3.24 ± 0.44-fold higher than in Lmnaplao/plao mice (P < 0.0005). Prelamin A transcripts in the cerebellum were 1.60 ± 0.19-fold higher in Lmnaplao-5nt/plao-5nt mice than in Lmnaplao/plao mice (P < 0.005) and 1.92 ± 0.36-fold higher in Lmnaplao-utr/plao-utr mice than in Lmnaplao/plao mice (P < 0.005). There were no substantial differences in prelamin A transcript levels in the heart, liver, and kidney of Lmnaplao-5nt/plao-5nt, Lmnaplao-utr/plao-utr and Lmnaplao/plao mice.
The changes in lamin A expression in Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice were not accompanied by substantial changes in the expression of lamins B1 or B2. Quantitative analyses of western blots uncovered minor fluctuations in lamins B1 and B2 levels in the tissues of Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice (Fig. 2C and D; Supplementary Material, Fig. S2), but the significance of these changes is questionable because consistent changes were not observed at the RNA level (Fig. 2H and I).
To determine if the amounts of lamin A expression in the central nervous system of Lmnaplao-utr/plao-utr and Lmnaplao-5nt/plao-5nt mice were similar to the total amount of A-type lamins (i.e. lamins A and C) in the brain of wild-type mice, we quantified total Lmna transcripts with PCR primers common to lamins A and C (Fig. 2F). Total Lmna transcript levels in the cerebral cortex of Lmnaplao-5nt/plao-5nt mice were 54.3 ± 9.6% of the levels in wild-type mice (P < 0.0005). In Lmnaplao-utr/plao-utr mice, total Lmna transcript levels in the cerebral cortex were 88.8 ± 11.3% of the levels in wild-type mice (P > 0.05). In the cerebellum, total Lmna transcripts in Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice were somewhat lower than in wild-type mice (68.0 ± 4.7 and 80.7 ± 10.0%, respectively; P < 0.05).
Mice that were heterozygous for the knock-in mutations exhibited consistent changes in lamin A expression (Supplementary Material, Fig. S3). Lamin A expression in the cerebral cortex and cerebellum was significantly higher in Lmnaplao-5nt/+ and Lmnaplao-utr/+ mice than in Lmnaplao/+ mice. In peripheral tissues, the amounts of lamin A, relative to actin, were similar in Lmnaplao-5nt/+, Lmnaplao-utr/+ mice and Lmnaplao/+ mice.
A recent study by Senyuk et al. (12) reported that miR-9 is expressed in myeloid cells and promotes myeloid differentiation. To assess the impact of the LmnaPLAO-5NT knock-in mutation on lamin A expression in myeloid cells, we used western blots to compare the relative amounts of lamins A and C in bone marrow myeloid cells and peritoneal macrophages (Supplementary Material, Fig. S4A). Lamin A expression was lower than lamin C in both bone marrow myeloid cells and peritoneal macrophages in wild-type mice. Lamin A expression was higher in Lmnaplao/+ mice than in wild-type mice, as expected. However, unlike the situation in the brain, the levels of lamin A expression were not further increased in Lmnaplao-5nt/+ mice (Supplementary Material, Fig. S4A), demonstrating that miR-9 does not play a significant role in regulating prelamin A expression in bone marrow myeloid cells or peritoneal macrophages. Of note, the levels of miR-9 expression in myeloid cells were far lower than those in the cerebral cortex and rather similar to the miR-9 levels in other peripheral tissues Supplementary Material, Fig. S4B.
The higher levels of lamin A in the cerebral cortex of Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice were easily detectable by immunohistochemistry (Fig. 3). Lamin A-positive cells were abundant throughout the cerebral cortex in both lines of mice, but were particularly striking in Lmnaplao-utr/plao-utr mice (Fig. 3). In wild-type and Lmnaplao/plao mice, lamin A staining in the cerebral cortex was minimal; only a few cells were positive for lamin A, reflecting the fact that capillary endothelial cells express lamin A (5).
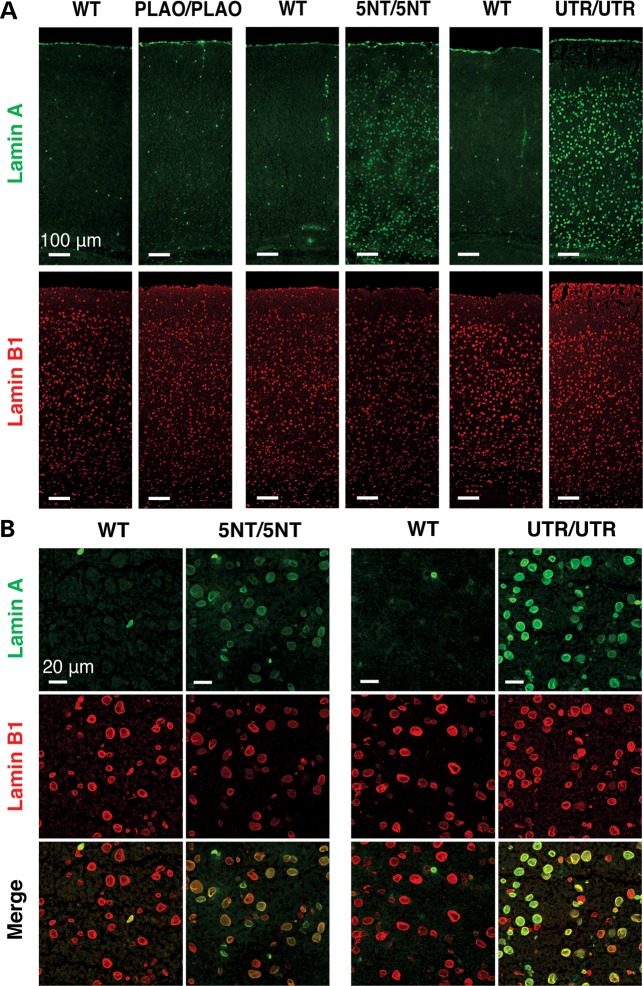
Figure 3.
Immunofluorescence microscopy of the cerebral cortex from Lmnaplao/plao (PLAO/PLAO), Lmnaplao-5nt/plao-5nt (5NT/5NT), Lmnaplao-utr/plao-utr (UTR/UTR) and wild-type (WT) littermate mice, revealing increased lamin A expression in the mice harboring mutations in prelamin A's 3′ UTR. (A and B) Low- and high-magnification images of the cerebral cortex stained with antibodies against lamin A (green) and lamin B1 (red). The scattered lamin A-positive cells in wild-type and Lmnaplao/plao mice are endothelial cells (5). Scale bar in (A), 100 µm; (B), 20 µm.
We used immunohistochemistry to test the possibility that the impact of the LmnaPLAO-5NT and Lmnaplao-utr mutations might vary in different regions of the brain. In the cortex of Lmnaplao-utr/+ mice, most lamin C-positive cells expressed lamin A; the same was the case in Lmnaplao-5nt/+ mice, although the intensity of lamin A expression in those mice, relative to lamin C, was lower (Supplementary Material, Fig. S5A). In the corpus callosum and in the arbor vitae of the cerebellum, the expression of lamin A was similar in Lmnaplao-5nt/+ and Lmnaplao-utr/+ mice (Supplementary Material, Fig. S5A and B). In some regions of the brain, e.g. the striatum, lamin A expression was low in both Lmnaplao-5nt/+ and LmnaPLAO-UTR/+ mice (Supplementary Material, Fig. S5 and Table S1). In the striatum, miR-9 expression was robust, as judged by in situ hybridization (Supplementary Material, Fig. S6A). Consistent with that finding, the magnitude of the increase in prelamin A transcripts (as measured by qRT-PCR) in the striatum of Lmnaplao-utr/plao-utr mice was similar to that in the cortex (Supplementary Material, Fig. S6B and C). We suspect that our inability to observe a significant increase in lamin A expression in the striatum of Lmnaplao-utr/+ mice by immunohistochemistry probably relates to the low levels of Lmna expression in that site, given that lamin C expression was also low in the same tissue sections (Supplementary Material, Fig. S5).
To further define lamin A expression in the brain, we stained brain sections from Lmnaplao-5nt/+, Lmnaplao-5nt/plao-5nt, Lmnaplao-utr/+ and Lmnaplao-utr/plao-utr mice with an antibody against lamin A—along with an antibody against NeuN (neuronal nuclei), Olig2 (oligodendrocyte lineage transcription factor 2) or GFAP (glial fibrillary acidic protein) (Fig. 4; Supplementary Material, Fig. S7). In the cerebral cortex and hippocampus of Lmnaplao-5nt/+ and Lmnaplao-utr/+ mice, all NeuN-positive neurons were stained positively for lamins A and C, but lamin A staining was more intense in Lmnaplao-utr/+ mice (Fig. 4A; Supplementary Material, Fig. S7A). In both Lmnaplao-5nt/+ and Lmnaplao-utr/+ mice, lamin A expression was observed in most Olig2-positive oligodendrocytes (Fig. 4B, yellow arrows). The expression of lamin A in GFAP/lamin C-positive astrocytes in Lmnaplao-5nt/+ and Lmnaplao-utr/+ mice was less evident, likely due to low expression of A-type lamins in astrocytes (Fig. 4C; Supplementary Material, Fig. S7B, yellow arrows).
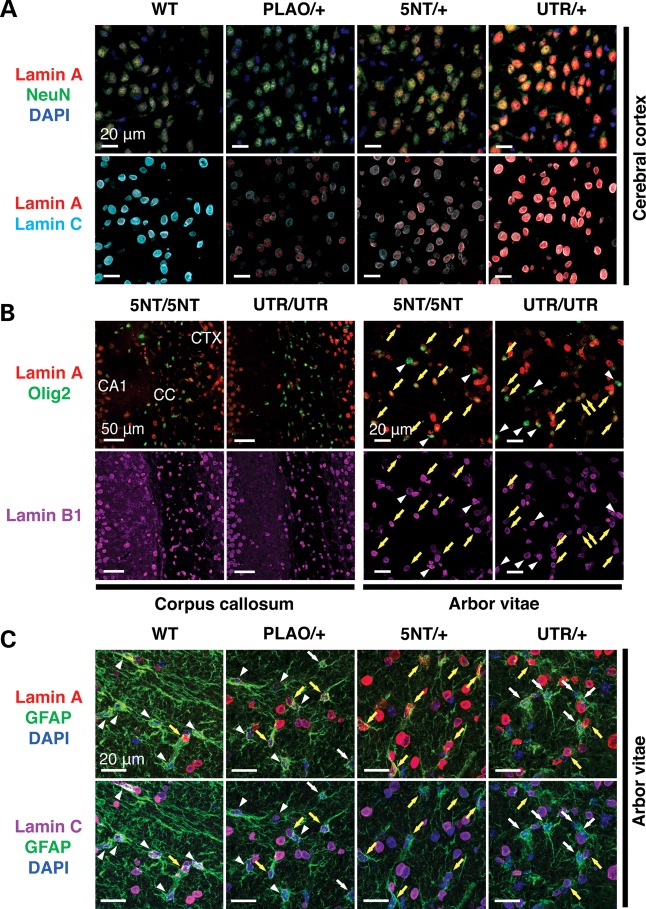
Figure 4.
Lamin A expression in different cell types in the brain of Lmnaplao-5nt/+, Lmnaplao-5nt/plao-5nt, Lmnaplao-utr/+ and Lmnaplao-utr/plao-utr mice. (A) Immunofluorescence microscopy of the cerebral cortex from Lmnaplao-5nt/+ (5NT/+) and Lmnaplao-utr/+ (UTR/+) mice stained with antibodies against lamin A (red), lamin C (cyan) and NeuN (green). (B) Immunofluorescence microscopy of the corpus callosum or the arbor vitae from Lmnaplao-5nt/plao-5nt (5NT/5NT) and Lmnaplao-utr/plao-utr (UTR/UTR) mice stained with antibodies against lamin A (red), Olig2 (green) and lamin B1 (magenta). (C) Immunofluorescence microscopy of the arbor vitae from Lmnaplao-5nt/+ (5NT/+) and Lmnaplao-utr/+ (UTR/+) mice stained with antibodies against lamin A (red), lamin C (magenta) and GFAP (green). White arrowheads indicate cells in which lamin A expression was undetectable, even though these cells expressed lamin C (C) or lamin B1 (B); white arrows indicate cells in which neither lamin A nor lamin C was detectable; yellow arrows indicate lamin C-expressing cells in which lamin A expression was robust. Scale bars: as indicated in the figures. CTX, cerebral cortex; CC, corpus callosum.
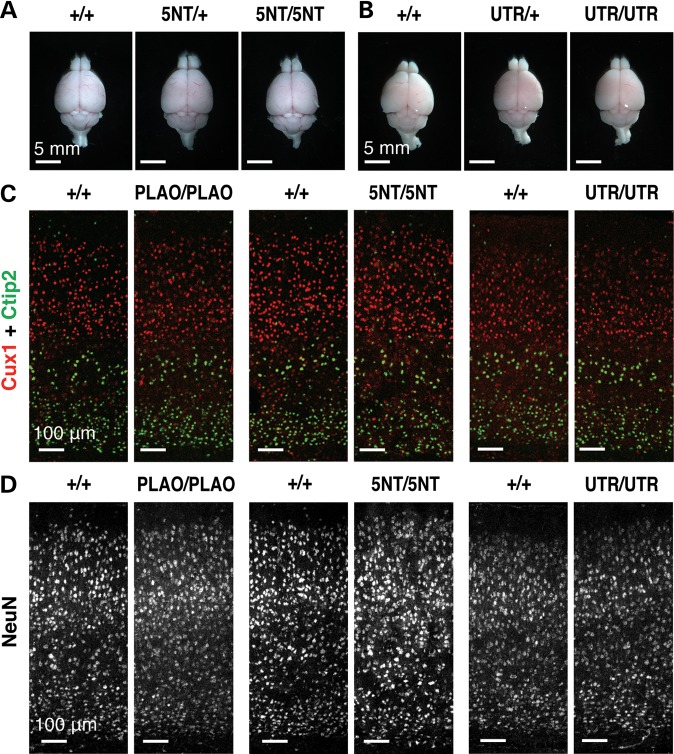
The health and vitality of Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice were indistinguishable from wild-type littermates (Supplementary Material, Fig. S8A and B). At 1 month of age, the size and morphology of the brain in Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr, and wild-type mice were similar (Fig. 5A and B). We found no evidence for abnormal layering of neurons or lower numbers of neurons in brains of knock-in mice (Fig. 5C and D; Supplementary Material, Fig. S8C–E). In addition, we found no evidence that the expression of lamin A (rather than lamin C) in the brain resulted in misshapen cell nuclei or mislocalization of other nuclear membrane proteins (Fig. 3B; Supplementary Material, Fig. S9). Older Lmnaplao-5nt/plao-5nt (at 6.5 months of age) and Lmnaplao-utr/plao-utr mice (at 9.5 months of age) remained healthy (Supplementary Material, Fig. S10); neither loss of neurons nor increased cell death was observed (Supplementary Material, Fig. S10D and E). Lmnaplao-utr/plao-utr mice were also indistinguishable from wild-type mice by SHIRPA (SmithKline Beecham, Harwell, Imperial College and Royal London Hospital phenotype assessment) behavioral screens and by rotarod tests (13,14) (Supplementary Material, Fig. S11 and Table S2).
Figure 5.
Lmnaplao-5nt/plao-5nt and Lmnaplao-utr/plao-utr mice have no detectable neuropathology. (A and B) Brains from 1-month-old Lmna+/+ (+/+), Lmnaplao-5nt/+ (5NT/+), Lmnaplao-5nt/plao-5nt (5NT/5NT), Lmnaplao-utr/+ (UTR/+) and Lmnaplao-utr/plao-utr (UTR/UTR) mice, viewed from the top. Scale bar, 5 mm. (C and D) Immunofluorescence microscopy of the cerebral cortex from 1-month-old Lmnaplao/plao (PLAO/PLAO), Lmnaplao-5nt/plao-5nt (5NT/5NT), Lmnaplao-utr/plao-utr (UTR/UTR) and wild-type (+/+) littermate mice stained with antibodies against Cux1 (red) and Ctip2 (green) (C) or NeuN (white) (D). Scale bar, 100 µm.
DISCUSSION
Earlier studies by Jung et al. (5) with transfected cells suggested that the low levels of lamin A expression in the brain might be due to regulation of prelamin A transcripts by miR-9. A subsequent study suggested the same possibility (15). While intriguing, these reports dealt solely with cultured cells, and whether the experiments were actually relevant to lamin A expression in the brain was unclear. In the current study, we addressed that issue by creating two new lines of Lmna knock-in mice—one in which a 5-nucleotide (nt) mutation was introduced into the predicted miR-9 binding site in prelamin A's 3′ UTR (Lmnaplao-5nt/plao-5nt) and a second in which prelamin A's 3′ UTR was replaced with lamin C's 3′ UTR (Lmnaplao-utr/plao-utr). Both mouse models produced increased amounts of lamin A in the brain, providing strong support for the role of miR-9 in regulating prelamin A expression in the central nervous system. The discovery that miR-9 is crucial in determining prelamin A production is clinically important: the limited expression of prelamin A in the brain likely explains why children with HGPS are spared from neurodegenerative disease.
The LmnaPLAO-5NT and Lmnaplao-utr alleles lacked introns 10 and 11, eliminating any possibility of the alternative splicing event that normally leads to lamin C synthesis. The fact that both knock-in mice were ‘prelamin A-only’ models was a boon for our studies, making it possible for us to quantify the impact of the 3′ UTR mutations on prelamin A/lamin A expression without having to worry about effects on prelamin A/lamin C splicing. As a control, we simultaneously evaluated prelamin A/lamin A expression levels in prelamin A-only mice lacking any 3′ UTR mutations (Lmnaplao/plao mice) (11). Lamin A expression in the brain of Lmnaplao/plao mice was low.
The expression of prelamin A transcripts and lamin A protein in the cerebral cortex and cerebellum of Lmnaplao-5nt/plao-5nt mice was significantly higher than in Lmnaplao/plao mice. The sole difference between these models was the 5-nt mutation in prelamin A's 3′ UTR. We are confident that this mutation was effective in eliminating miR-9-mediated regulation because it abolished the ability of miR-9 to inhibit the expression of a luciferase reporter (linked to prelamin A's 3′ UTR). Also, this mutation resulted in significantly increased expression of prelamin A transcripts in the brain (where miR-9 expression is robust) but had no effect on prelamin A transcripts in peripheral tissues (where miR-9 is absent).
The increase in prelamin A/lamin A expression in the cerebral cortex was more substantial in Lmnaplao-utr/plao-utr mice than in Lmnaplao-5nt/plao-5nt mice. The mutation in the Lmnaplao-utr allele was obviously more extensive, eliminating both the miR-9 binding site and ∼1 kb of additional sequences in prelamin A's 3′ UTR. The higher levels of lamin A expression in the brain of Lmnaplao-utr/plao-utr mice, compared with Lmnaplao-5nt/plao-5nt mice, suggest that other sequences in prelamin A's 3′ UTR play a role in regulating prelamin A. The possibility is certainly plausible because large stretches of prelamin A's 3′ UTR (and not simply the region surrounding the miR-9 binding site) have been conserved in mammalian evolution. Interestingly, the effects of the Lmnaplao-utr allele on prelamin A/lamin A expression varied in different regions of the brain. The increase in prelamin A expression in the cerebral cortex of Lmnaplao-utr/plao-utr mice was greater than in Lmnaplao-5nt/plao-5nt mice, but in the cerebellum, the increase in prelamin A expression was fairly similar in the two lines of mice. These findings suggest that the regulation of lamin A in the cerebellum could depend primarily on the miR-9 binding site, whereas lamin A regulation in the cortex could involve both the miR-9 site and other, as-yet-undefined, 3′ UTR sequences. It is also possible that the differences in lamin A regulation in different parts of the brain relate, at least in part, to distinct impacts of the two knock-in mutations on different cell types and to different populations of cells in different regions of the brain. By immunohistochemistry, lamin A expression in NeuN-positive cortical neurons was increased in both Lmna knock-in lines, and the effects were clearly greater in Lmnaplao-utr/plao-utr mice. However, the increase in lamin A expression in Olig2-positive oligodendrocytes was less impressive in both knock-in lines, and the increase in lamin A in GFAP-positive astrocytes was nearly undetectable. As a result, we could not discern significant differences between the two knock-in lines with respect to glial cell expression of lamin A.
Our studies provide new insights regarding the functional importance of the A-type lamins in the brain. First, we observed no neuropathology in Lmnaplao/plao mice, which cannot make lamin C and produce little lamin A in brain parenchymal cells. Thus, it would appear that very low levels of A-type lamins in the brain are not associated with adverse consequences. This is very different from the situation with striated muscle, where an absence of lamins A and C causes myopathic changes and early death (16). Second, we observed no pathology or behavioral abnormalities in Lmnaplao-utr/plao-utr mice, which produce prelamin A (rather than lamin C) in brain parenchymal cells, demonstrating that lamin A production is not overtly toxic in the brain.
We previously reported that lamins B1 and B2 are expressed highly in the developing brain and that both proteins are crucial for neuronal migration (17–19). Neither lamin A nor lamin C is expressed in migrating neurons, but large amounts of lamin C are expressed after birth. From the current studies, we now understand why lamin A synthesis is low in brain parenchymal cells, but the physiologic ‘rationale’ for limiting lamin A expression in the brain is still elusive. The nuclear lamins are known to interact with a variety of inner nuclear membrane proteins as well as proteins in the chromatin (20,21). One possibility is that lamin C is better suited for interacting with the spectrum of nuclear envelope and chromatin proteins that are expressed in the brain. Another possibility is that lamin A expression in the brain, while well tolerated in laboratory mice, would be suboptimal in extenuating circumstances, e.g. in response to injury or metabolic stress.
Mice lacking miR-9 exhibit defective neuronal migration and impaired neuronal survival (22). Neither of these phenotypes were encountered in mice lacking the miR-9 binding site in prelamin A's 3′ UTR. Of course, this discrepancy is not surprising, given that miR-9 almost certainly has many dozens of targets, the majority of which await characterization. Once all of the miR-9 targets are defined and understood, it is possible that the rationale for miR-9 regulation of prelamin A expression will come into sharper focus.
MATERIALS AND METHODS
Generation of knock-in mice with mutations in prelamin A's 3′ UTR
The LmnaPLAO-5NT allele was created with a sequence-replacement vector identical to the one used to create the LmnaPLAO allele (11), except that the miR-9 seed-binding sequence in prelamin A's 3′ UTR (CCAAAG) was changed to ACCCTG. The mutation was introduced into the 5′ arm of the LmnaPLAO targeting vector by site-directed mutagenesis (QuikChange kit, Stratagene) with primer 5′-AGCAGGCCTGAAGACCCTGAAAAATTTATC-3′ (and a complementary reverse primer). The vector used to create the Lmnaplao-utr vector was also similar to the LmnaPLAO vector, except that the sequences encoding prelamin A's 3′ UTR in the 5′ arm of the vector were replaced with lamin C's 3′ UTR (using the In-Fusion Advantage PCR cloning kit from Clontech). Lamin C's 3′ UTR was amplified with primers 5′-TGCAGCATCATGTAAGGCCCAGCCCACAAGGGTA-3′ and 5′-GACACCACAGCATCTGGCATTCCAAAACAT-3′ and then inserted into the 5′ arm of the LmnaPLAO vector with primers 5′-AGATGCTGTGGTGTCTCTTTGTTG-3′ and 5′-TTACATGATGCTGCAGTTCTGGGAGCTCT-3′. The integrity of the vectors was verified by restriction endonuclease digestion and DNA sequencing.
After electroporating the vectors into 129/OlaHsd embryonic stem (ES) cells, targeted clones were identified by long-range PCR (TaKaRa LA Taq polymerase, Clontech) with primers A and NeoR on the 5′ end and primers NeoF and B on the 3′ end (see Supplementary Material, Table S3A for the primer sequences; Fig. 1A and B). The targeting events were confirmed by Southern blotting with EcoRI-cleaved genomic DNA and a 5′ flanking probe (11). The probe detected a 10.4-kb fragment in a wild-type Lmna allele; a 9.4-kb fragment in the LmnaPLAO-5NT allele and an 8.6-kb fragment in the Lmnaplao-utr allele.
Targeted ES cell clones were injected into C57BL/6 blastocysts, and the resulting chimeras were bred with C57BL/6 females to generate heterozygous Lmna knock-in mice, which were then intercrossed to generate homozygotes. Genotyping was performed by PCR with primers C and D for the LmnaPLAO-5NT allele and primers E and F for the Lmnaplao-utr allele (Fig. 1A and B).
All mice were fed a chow diet and housed in a virus-free barrier facility with a 12-h light/dark cycle. Animal protocols were approved by UCLA's Animal Research Committee.
Western blots
Protein extracts from mouse tissues were prepared as described (5,23). Briefly, snap-frozen mouse tissues were powdered with a chilled metal mortar and pestle, resuspended in ice-cold PBS containing 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 mm NaF and protease inhibitors (Roche) and homogenized with a glass tissue grinder (Kontes). The cell pellets were resuspended in urea buffer [9 m urea, 10 mm Tris–HCl (pH 8.0), 1 mm NaF, 1 mm PMSF, 10 μm ethylenediaminetetraacetic acid 0.2% β-mercaptoethanol and a Roche Protease Inhibitors Cocktail Tablet], sonicated and centrifuged at 18,000g for 5 min to isolate the urea-soluble protein fraction. Protein extracts were size-fractionated on 4–12% gradient polyacrylamide Bis-Tris gels (Invitrogen) and transferred to a nitrocellulose membrane. Membranes were incubated with primary antibodies as indicated in Supplementary Material, Table S3B. Antibody binding was detected with infrared (IR)-dye-conjugated secondary antibodies (Rockland) and an Odyssey IR scanner (LI-COR).
Quantitative RT–PCR
Levels of lamin transcripts were assessed as described previously (19). Snap-frozen mouse tissues were homogenized in TRI reagent (Molecular Research Center), and total RNA was extracted according to the manufacturer's protocol. RNA was treated with DNase I (Ambion) and then reverse transcribed with oligo(dT), random primers and SuperScript III (Invitrogen). qPCRs were prepared with SYBR Green PCR Master Mix (Bioline) and performed on a 7900 Fast Real-Time PCR system (Applied Biosystems). Transcript levels were determined by the comparative cycle threshold method and normalized to levels of cyclophilin A.
Histology and immunofluorescence microscopy
Freshly harvested mouse tissues were fixed in 10% formalin (Evergreen) overnight at 4°C; tissues were then embedded in paraffin, sectioned (5 µm) and stained with hematoxylin and eosin. For immunohistochemical studies, mouse tissues were embedded in Optimum Cutting Temperature compound (Sakura Finetek), cryosectioned (10 µm) and fixed in ice-cold methanol. After rinsing with acetone, the sections were washed with 0.1% Tween-20 in tris-buffered saline and incubated with M.O.M. Mouse Ig Blocking Reagent (Vector Laboratories). Primary antibodies are listed in Supplementary Material, Table S3B. Binding of primary antibodies was detected with Alexa Fluor-labeled donkey antibodies against goat, rabbit or rat IgG (Invitrogen); the binding of mouse monoclonal antibodies was detected with M.O.M. Biotinylated Anti-Mouse IgG Reagent (Vector Laboratories) and Alexa Fluor-conjugated streptavidin (Invitrogen). DNA was stained with 4′,6-diamidino-2-phenylindole.
Light microscopy images were captured with a Leica MZ6 dissecting microscope [a Plan 0.5× objective (air)] with a DFC290 digital camera (Leica) and a Nikon Eclipse E600 microscope [Plan Fluor 4×/0.13 NA or 10×/0.30 NA objectives (air)] with a DS-Fi2 camera (Nikon). The images were recorded with Leica Application Suite imaging software and NIS-Elements F (Nikon), respectively. Confocal microscopy was performed with a Zeiss LSM700 laser-scanning microscope [Plan Apochromat 10×/0.45 NA or 20×/0.80 NA objectives (air)]. Images along the z-axis were captured and merged images were generated with Zen 2010 software (Zeiss).
Behavioral analysis
The SHIRPA primary screen (13) was performed with adult Lmnaplao-utr/plao-utr mice in the UCLA Behavioral Testing Core facility. The tests included observation of mice in a viewing jar and measurements of transfer arousal, muscle tone, autonomic behaviors and righting reflex. Rotarod testing (14) was also performed. All tests were performed in a blinded fashion.
Statistical analysis
Statistical analyses were performed with GraphPad QuickCalcs (http://www.graphpad.com/ last accessed on 29 October, 2013). Differences in levels of lamin transcripts, proteins and luciferase activities were analyzed by a two-tailed Student's t-test.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the National Institutes of Health Grants (HL86683 to L.G.F., HL089781 to L.G.F. and AG035626 to S.G.Y.).
Supplementary Material
REFERENCES
- 1.Lin F., Worman H.J. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 2.Worman H.J., Fong L.G., Muchir A., Young S.G. Laminopathies and the long strange trip from basic cell biology to therapy. J. Clin. Invest. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M., et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 4.Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung H.J., Coffinier C., Choe Y., Beigneux A.P., Davies B.S., Yang S.H., Barnes R.H., 2nd, Hong J., Sun T., Pleasure S.J., et al. Regulation of prelamin A but not lamin C by miR-9, a brain-specific microRNA. Proc. Natl Acad. Sci. USA. 2012;109:E423–E431. doi: 10.1073/pnas.1111780109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffinier C., Jung H.J., Li Z., Nobumori C., Yun U.J., Farber E.A., Davies B.S., Weinstein M.M., Yang S.H., Lammerding J., et al. Direct synthesis of lamin A, bypassing prelamin a processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J. Biol. Chem. 2010;285:20818–20826. doi: 10.1074/jbc.M110.128835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 9.Gao F.B. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev. 2010;5:25. doi: 10.1186/1749-8104-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coolen M., Bally-Cuif L. MicroRNAs in brain development and physiology. Curr. Opin. Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Davies B.S., Barnes R.H., 2nd, Tu Y., Ren S., Andres D.A., Spielmann H.P., Lammerding J., Wang Y., Young S.G., Fong L.G. An accumulation of non-farnesylated prelamin A causes cardiomyopathy but not progeria. Hum. Mol. Genet. 2010;19:2682–2694. doi: 10.1093/hmg/ddq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senyuk V., Zhang Y., Liu Y., Ming M., Premanand K., Zhou L., Chen P., Chen J., Rowley J.D., Nucifora G., et al. Critical role of miR-9 in myelopoiesis and EVI1-induced leukemogenesis. Proc. Natl Acad. Sci. USA. 2013;110:5594–5599. doi: 10.1073/pnas.1302645110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers D.C., Fisher E.M., Brown S.D., Peters J., Hunter A.J., Martin J.E. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome. 1997;8:711–713. doi: 10.1007/s003359900551. [DOI] [PubMed] [Google Scholar]
- 14.Carter R.J., Morton J., Dunnett S.B. Motor coordination and balance in rodents. Curr. Protoc. Neurosci. 2001;Chapter 8:Unit 8.12. doi: 10.1002/0471142301.ns0812s15. [DOI] [PubMed] [Google Scholar]
- 15.Nissan X., Biondel S., Navarro C., Maury Y., Denis C., Girard M., Martinat C., De Sandre-Giovannoli A., Levy N., Peschanski M. Unique preservation of neural cells in Hutchinson-Gilford progeria syndrome Is due to the expression of the neural-specific miR-9 microRNA. Cell Rep. 2012;1:1–9. doi: 10.1016/j.celrep.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 16.van Engelen B.G., Muchir A., Hutchison C.J., van der Kooi A.J., Bonne G., Lammens M. The lethal phenotype of a homozygous nonsense mutation in the lamin A/C gene. Neurology. 2005;64:374–376. doi: 10.1212/01.WNL.0000149763.15180.00. [DOI] [PubMed] [Google Scholar]
- 17.Coffinier C., Chang S.Y., Nobumori C., Tu Y., Farber E.A., Toth J.I., Fong L.G., Young S.G. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl Acad. Sci. USA. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffinier C., Jung H.J., Nobumori C., Chang S., Tu Y., Barnes R.H., 2nd, Yoshinaga Y., de Jong P.J., Vergnes L., Reue K., et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell. 2011;22:4683–4693. doi: 10.1091/mbc.E11-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung H.J., Nobumori C., Goulbourne C.N., Tu Y., Lee J.M., Tatar A., Wu D., Yoshinaga Y., de Jong P.J., Coffinier C., et al. Farnesylation of lamin B1 is important for retention of nuclear chromatin during neuronal migration. Proc. Natl Acad. Sci. USA. 2013;110:E1923–E1932. doi: 10.1073/pnas.1303916110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schirmer E.C., Foisner R. Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 2007;313:2167–2179. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Dauer W.T., Worman H.J. The nuclear envelope as a signaling node in development and disease. Dev. Cell. 2009;17:626–638. doi: 10.1016/j.devcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 22.Shibata M., Nakao H., Kiyonari H., Abe T., Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fong L.G., Ng J.K., Meta M., Cote N., Yang S.H., Stewart C.L., Sullivan T., Burghardt A., Majumdar S., Reue K., et al. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc. Natl Acad. Sci. USA. 2004;101:18111–18116. doi: 10.1073/pnas.0408558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.