Abstract
A novel virus was detected in a sample collected from a Swedish moose (Alces alces). The virus was suggested as a member of the Hepeviridae family, although it was found to be highly divergent from the known four genotypes (gt1–4) of hepatitis E virus (HEV). Moose are regularly hunted for consumption in the whole of Scandinavia. Thus, the finding of this virus may be important from several aspects: (a) as a new diverged HEV in a new animal species, and (b) potential unexplored HEV transmission pathways for human infections. Considering these aspects, we have started the molecular characterization of this virus. A 5.1 kb amplicon was sequenced, and corresponded to the partial ORF1, followed by complete ORF2, ORF3 and poly(A) sequence. In comparison with existing HEVs, the moose HEV genome showed a general nucleotide sequence similarity of 37–63 % and an extensively divergent putative ORF3 sequence. The junction region between the ORFs was also highly divergent; however, two putative secondary stem–loop structures were retained when compared to gt1–4, but with altered structural appearance. In the phylogenetic analysis, the moose HEV deviated and formed its own branch between the gt1–4 and other divergent animal HEVs. The characterization of this highly divergent genome provides important information regarding the diversity of HEV infecting various mammalian species. However, further studies are needed to investigate its prevalence in the moose populations and possibly in other host species, including the risk for human infection.
Introduction
Hepatitis E virus (HEV) belongs to the Hepeviridae family and is a small non-enveloped 7.2 kb capped single stranded, positive sense RNA virus. The genome consists of three open reading frames 1–3 (ORF1–3) and terminates with a poly(A)-tail (Tam et al., 1991). The non-structural proteins involved in the viral replication are encoded by ORF1 in the two-thirds of the genome at the 5′ end (Koonin et al., 1992). ORF1 is followed by a short section called the junction region (JR), which is predicted to fold into two RNA stem–loop structures critical for RNA replication (Cao et al., 2010; Graff et al., 2005; Huang et al., 2007). The subsequent part of the genome is transcribed into a capped bi-cistronic subgenomic RNA (SgRNA) overlapping ORF2 and ORF3, with its 5′ terminal end positioned within the JR and terminating with poly(A) (Graff et al., 2006). ORF2 encodes the viral capsid protein with HEV RNA binding, assembly, virion packaging and host cell attachment properties (Mori & Matsuura, 2011). ORF3 encodes a small accessory protein with an unclear function, but with the ability to interact with ORF2 and cell related proteins (Korkaya et al., 2001; Zafrullah et al., 1997). It is estimated that two billion of the world’s population is at risk of being infected by HEV (Aggarwal & Jameel, 2008) and four genotypes 1–4 (gt1–4) infect humans. Gt1 and gt2 are restricted to humans, and have a mortality rate of 0.5 %, or up to 28 % in pregnant women; the reason for this is still unknown (Aggarwal, 2011; Navaneethan et al., 2008). Gt3 and gt4 have a wider host range with several animal species as hosts (Meng, 2011). HEV is transmitted mainly through the faecal–oral route and is an important causative agent of acute hepatitis in many developing countries, where it may cause large outbreaks (Okamoto, 2007), but is also present in developed countries with sporadic infections acquired locally within the country. These infections are caused by gt3 or gt4, often with unclear transmission routes (Purdy & Khudyakov, 2011). It seems that most HEV infections are asymptomatic and symptomatic cases alone only represent small part of the total (Kamar et al., 2012). This is probably reflected by the high seroprevalence against hepatitis E in the general population. In Sweden, the seroprevalence is estimated to be 9 % (Olsen et al., 2006), and between 5–30 % in other countries (Christensen et al., 2008; Dalton et al., 2007; Mansuy et al., 2009). However, there may be more clinical cases of hepatitis E than notified, since many nonA–nonD hepatitis cases are not examined due to unawareness of endemic hepatitis E. A growing number of animal HEV variants have been identified in a wide range of non-porcine animals and some of them have highly divergent genomes compared to gt1–4. Pigs appear to be the main HEV reservoir of gt3–4, with high HEV prevalence in many countries. In Sweden it is estimated that 30 % of the 2–4 months old pigs are gt3 HEV infected (Widén et al., 2011). Evidence for zoonotic transmission through ingestion of infected meat has been identified. Gt3–4 isolates with highly similar sequence identity were detected in patients and infected liver or meat from, for example, swine, wild boars and deer (Colson et al., 2010; Li et al., 2005; Meng, 2011; Takahashi et al., 2004; Tei et al., 2003).
Considering the limited understanding of the infection biology and host range of HEV, our aim was to investigate the existence of non-porcine HEV reservoirs for human infections and to better understand the relatedness of different HEV strains including the risk for humans that they may pose. The HEV prevalence in Swedish deer is unknown, but deer in other parts of Europe and Japan has been found to be HEV positive and been linked to human HEV infections (Pavio et al., 2010; Takahashi et al., 2004; Tei et al., 2003; Tomiyama et al., 2009). Here, we report for the first time the detection and genomic characterization of a new HEV in moose. The largest member of the deer family Cervidae, the moose (Alces alces), is regularly hunted and its meat consumed in Scandinavia. The knowledge that moose carry HEV opens additional unexplored transmission pathways of this virus to human populations. Based on the present investigation we propose that this newly detected moose virus is classified as a new member in the Hepeviridae family. Its infection biology should be studied in many aspects to clarify its potential as a zoonotic pathogen and shed more light on the increasingly complex Hepeviridae family.
Results
Detection and amplification of a HEV sequence from a moose sample
The knowledge that HEV can infect deer raised the possibility that moose might be infected with this virus. Indeed, when moose liver and kidney samples were screened by real-time PCR, one sample out of six was found to be HEV positive. This liver sample exhibited cycle threshold (Ct) values of 33.4 and 34.6 with the Jothikumar et al. (2006) and Gyarmati et al. (2007) assays. This HEV positive liver sample was collected from a diseased three-year-old pregnant female moose. The carcass was severely decomposed and therefore unsuitable for histopathological examination. However, the following could be detected: cachexia (emaciation), anaplasmosis, acute heart muscle degeneration and haemorrhage, cysticercosis and ear mites.
A 383 nt sequence covering part of the RNA dependent RNA polymerase was obtained using primer pair 1 (Table 1). This region is frequently used for genotyping (Zhai et al., 2006). According to nt blast, the closest match was gt3 with 88 % identity and covering only 41 % of the amplicon. To obtain more genome sequence, a series of PCRs were performed with three primer pairs 2–4 (Table 1). Primer pair 2 resulted in a 2.16 kb amplicon and primer pair 3 gave a 1.3 kb amplicon partially overlapped by the former amplicon. Primer pair 4 resulted in multiple products, and among these, only a 2.5 kb amplicon matched a HEV like sequence. By sequencing the amplicons, a 5.1 kb sequence was obtained corresponding to positions 2176–7227. All nt positions given for the sequences obtained in this study were referred to the Swedish swine gt3 HEV isolate SWX07-E1; accession number EU360977 (Xia et al., 2008).
Table 1. Primers used for moose HEV genome amplification and sequencing.
| Primer | Primer ID | Nucleotide sequence | Product size (kb) | Position*(nt) | Reference | |
| Primer sets | ||||||
| 1 | ESP | (Forward) | CATGGTAAAGTGGGTCAGGGTAT | 0.383 | 4248–4630 | Zhai et al. (2006) |
| EAP | (Reverse) | AGGGTGCCGGGCTCGCCGGA | ||||
| 2 | ESP | (Forward) | CATGGTAAAGTGGGTCAGGGTAT | 2.16 | 4248–6410 | Zhai et al. (2006) |
| HE041R | (Reverse) | GCCAATGGCGAGCCGACAGTGAA | Mizuo et al. (2002) | |||
| 3 | HEV5979F | (Forward) | CGAGGAGGAGGCTACGTCTGGTCTGGTA | 1.3 | 5979–7258 | This study |
| GenereRacer 3′Nest | (Reverse) | CGCTACGTAACGGCATGACAGTG | RACE kit (Invitrogen) | |||
| 4 | HEV108F | (Forward) | GCCTTGGCGAATGCTGTGGT | 2.5–4.4 | 108/2176–4585 | This study |
| HEV4585R | (Reverse) | GGACTCCTTCGGAGCCTGCAGCGTCCAA | ||||
| Sequencing primers | ||||||
| M13F | (Forward) | CAGGAAACAGCTATGAC | TOPO XL kit (Invitrogen) | |||
| HEV2491F | (Forward) | GGCTTGTCAATGCTGCAAACGCAGG | 2467–2491 | This study | ||
| HEV2683F | (Forward) | GGCTCCGGTTGGCCTATATCGAGGC | 2659–2683 | This study | ||
| HEV3499F | (Forward) | CCGTTCATGAGGCCCAGGGCGC | 3478–3499 | This study | ||
| HEV4147F | (Forward) | CCGTCTTGGCCCTTATCCAGC | 4127–4147 | This study | ||
| HEVF5 | (Forward) | CTTTGGAAYACTGTTTGGAATATGG | 4650–4674 | Xia et al. (2008) | ||
| HEV4835F | (Forward) | GCYTGTAYGCMGGCGWTGTC | 4816–4835 | This study | ||
| HEV5162F | (Forward) | GAGGGAATAACATTCAGGATGCGC | 5139–5162 | This study | ||
| HEV5597F | (Forward) | CGCCGACAGTACAATCTGTCAAC | 5575–5597 | This study | ||
| HEV5967F | (Forward) | GGYTGGCGCTCYGTYGAGAC | 5947–5967 | This study | ||
| HEV6345F | (Forward) | TGGCGGGCTCCCTACTGAGCTTGTGTCA | 6318–6345 | This study | ||
| HEV7013F | (Forward) | CCAGTTCCTGCTGACGTGCTTGAGGC | 6985–7013 | This study | ||
| HEV2491R | (Reverse) | CCTGCGTTTGCAGCATTGACAAGCC | 2467–2491 | This study | ||
| HEVR3 | (Reverse) | CGATATGCCGCCTCTAGCCTCTTGG | 2671–2695 | Xia et al. (2008) | ||
| HEV3499R | (Reverse) | CCGTTCATGAGGCCCAGGGCGC | 3478–3499 | This study | ||
| HEV3977R | (Reverse) | GACACTCTTACGATGGGCCGGTGCGG | 3952–3977 | This study | ||
| HEV4147R | (Reverse) | CCGTCTTGGCCCTTATCCAGC | 4127–4147 | This study | ||
| HEV4586R | (Reverse) | GGACTCCTTCGGAGCCTGCAGCGTCCAA | 4559–4586 | This study | ||
| HEV5304R | (Reverse) | CCAACCACCACCTCCGCCGCCGCCCG | 5279–5304 | This study | ||
| HEV5597R | (Reverse) | GTTGACAGATTGTACTGTCGGCG | 5575–5597 | This study | ||
| HEV6105R | (Reverse) | GCGAAACTCCACCTGTAGGGC | 6085–6105 | This study | ||
| HEV6841R | (Reverse) | CGGTCGTCGTGCCAGCCTGCCAATAG | 6816–6841 | This study | ||
| M13R | (Reverse) | CAGGAAACAGCTATGAC | TOPO XL kit (Invitrogen) |
Positions based on the Swedish reference gt3, SWX07-E1 (EU360977.1) genome (Xia et al., 2008).
Moose HEV sequence properties
The moose HEV sequence contained three putative ORFs, which is characteristic for HEV. The 5′-region of the amplified sequence was located at the putative proline hinge region in the hypervariable region of ORF1 and extended through the partially overlapping ORF2–3 region to the poly(A) at the 3′ terminal end. The sequence was highly divergent from other described HEV variants and had the highest similarity to gt1–4 and unclassified wild boars (gt1–4-Uwb) group (Table 2).
Table 2. Comparative genome analysis of moose HEV and other HEV strains.
| 5.1 kb moose HEV sequence | Partial ORF1 | ORF2 | ORF3 | |
| Genome position: 2176–7227*†‡ | 2176–5144*‡ | 5185–7152*‡ | 5168–5515*‡ | |
| Actual size: 5057 nt | 2966 nt | 1968 nt | 348 nt | |
| Sequence identity (%) | Sequence identity (%) | Sequence identity (%) | Sequence identity (%) | |
| HEV variant (number of compared strains*) | Nt | Nt / aa | Nt / aa | Nt / aa |
| Genotype 1 (3) | 61.9–62.9 | 62.8–63.4 / 66.6–67.1 | 65.7–66.1 / 72.4–73.0 | 53.3–53.9 / 30.3 |
| Genotype 2 (1) | 61.5 | 62.4 / 66.6 | 64.8 / 70.9 | 51.7 / 26.1 |
| Genotype 3 (5) | 62.9–63.1 | 61.8–62.8 / 65.5–66.5 | 66.3–67.6 / 72.9–74.0 | 52.8–55.0 / 29.4–31.1 |
| Genotype 4 (3) | 61.7–62.3 | 61.6–61.9 / 65.0–66.1 | 65.2–66.0 / 72.9–73.2 | 54.7–55.6 / 31.9–34.5 |
| Rabbit (1) | 60.9 | 59.8 / 63.5 | 65.7 / 72.0 | 54.7 / 34.5 |
| Unclassified wild boars (Uwb) (2) | 61.7–62.3 | 61.4–61.6 / 64.6–65.7 | 65.8–66.6 / 72.0–73.6 | 54.2–55.8 / 29.4–35.3 |
| Rat (1) | 52.1 | 52.9 / 53.4 | 55.7 / 56.0 | 32.8 / 17.0 |
| Ferret (1) | 52.6 | 54.0 / 54.5 | 55.3 / 54.9 | 35.8 / 16.8 |
| Bat (1) | 48.5 | 50.4 / 47.0 | 46.5 / 45.4 | 19.7 / 7.0 |
| Avian (1) | 46.1 | 50.2 / 45.4 | 44.7 / 41.6 | 23.3 / 11.0 |
| Trout (1) | 36.6 | 39.2 / 29.1 | 27.0 / 15.5 | 22.2 / 8.4 |
Strain information is summarized in Table S1.
Poly(A) sequence excluded.
Positions of the Swedish gt3, SWX07-E1 (EU360977.1) genome.
Characterization of the partial ORF1 sequence
The nt sequence 2176–5144 corresponding to the partial ORF1 sequence was analysed by blast and matched gt1 strains, but when translated to 987 aa, the blast search matched gt3 and gt4 strains. The moose HEV ORF1 stop codon (TAA) was shared with animal HEVs (rat, ferret, bat and trout) except for avian forms, which use TGA similar to the gt1–4-Uwb group.
Characterization of the proline hinge region.
When the recovered moose HEV sequence was translated into aa, the first 79 N-terminal residues (position 717–795) corresponded to the proline hinge region. Although high sequence diversity was observed between the moose HEV and SWX07-E1, the number of prolines was similar, 19 compared to 21 (Fig. S1, available in the online Supplementary Material) indicating the functional importance of conserved numbers of prolines. This region has an unknown function, but may play a role in host adaptation (Purdy et al., 2012).
Characterization of the X domain.
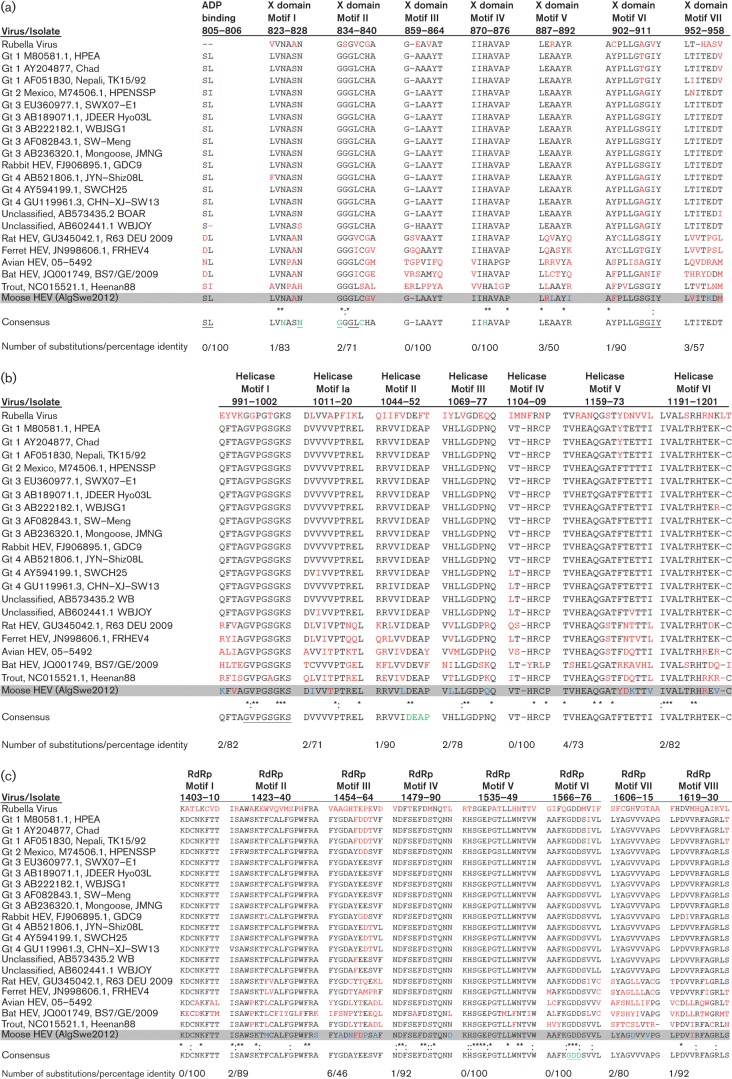
Alignment of the aa sequence of the moose HEV identified 164 residues corresponding to the characteristic putative X domain (macro domain), residues 795–958. When this region was blast searched, it matched most closely to gt4. Previous studies suggested that this domain has poly ADP-ribose-binding properties associated with replication and transcription (Egloff et al., 2006; Neuvonen & Ahola, 2009). The seven characteristic X domain motifs, here designated motifs I–VII (Koonin et al., 1992) were present in the moose HEV sequence (Fig. 1a). Potential ADP-binding and active sites were identified by the blastp. Upstream of motif I, two residues S805 and L806, present in moose HEV and in the gt1–4 strains, were according to blastp homologues to residues D and I, which have putative ribose binding properties. Motif I diverged with one residue from gt1–4 and was identical with rat, ferret and bat HEV. Motif II was unique to the moose HEV and diverged with only L837 compared to ferret HEV, while Motif III and IV remained conserved with the consensus sequence. Motif V contained three substitutions whereof L889 and I892 were unique making this motif more hydrophobic than in other HEVs, while motif VI diverged with F903 compared to the consensus sequence. Motif VI residues S-GIY908–911 appeared according to blastp as homologues to STGVY related to the putative ADP-binding site. Motif VII had three substitutions: two of them, V953 and M958, were partially shared with some animal HEVs, while K956 was unique to moose HEV.
Fig. 1.
Comparison of aligned motifs within derived domains (Koonin et al., 1992) from different HEV isolates and rubella virus. The moose HEV is highlighted in grey. Red letters represent non-consensus residues, asterisks mark identical residues, : marks HEV-specific residues and blue letters represent unique moose HEV residues. (a) The X domain; underlined letters represent putative ADP ribose binding sites and green letters represent the active site. (b) The helicase domain; underlined letters represent putative ADP ribose binding sites and green letters represent Mg2+ binding sites. (c) The RNA dependent RNA polymerase domain; green letters represent the active site.
Characterization of the RNA helicase.
The HEV RNA helicase has been shown to have NTPase and RNA unwinding activities and has been classified into the helicase superfamily SF-1 (Karpe & Lole, 2010a, b). The putative RNA helicase in the moose HEV was 233 aa corresponding to residues 971–1203. Nt and protein blast with this region showed the closest match with gt1. All six signature motifs typical for the helicase superfamilies were identified (Gorbalenya et al., 1989) (Fig. 1b). Motif I of the moose HEV showed only a unique K991, followed by V993 substitution and eight conserved residues with nucleotide binding properties (Karpe & Lole, 2010b). Motif Ia contained one unique I1012 and a T1015 shared with avian and trout HEV. Motif II had one unique L1048, followed by the conserved residues DEAP1049–52, which have been shown to be involved in magnesium ion binding (Karpe & Lole, 2010b). Residues L1070 and Q1076 made motif III unique, while motif IV was identical to the consensus sequence. Two out of four residue substitutions, K1170 and V1173 were unique, making motif V most diverged among the six motifs. This motif was also highly divergent in the bat and trout HEV strains. In motif VI there were two residues divergent from the corresponding region of gt1–4-Uwb: R1198 that was shared with trout and avian HEV strains, and one unique V1200 resulting in an increased hydrophobicity of the motif.
Characterization of the RNA dependent RNA polymerase (RdRp).
The putative moose HEV RdRp was estimated to be formed of 488 aa, corresponding to aa positions 1218–1705 and a blastp search of this region resulted in a match with gt3 strains. The eight motifs in the HEV RdRp (Koonin et al., 1992), were all present in the moose HEV RdRp (Fig. 1c). Motif I was highly conserved among the HEVs including in the moose HEV, with substitutions only in the avian and bat HEV strains. Two unique aas, M1430 and S1440, made the moose HEV motif II differ from the other HEVs. Among the motifs, motif III was most divergent with six residue substitutions in the moose HEV of which four were unique, A1456, N1458, P1461 and A1463. In the highly conserved motif IV the moose HEV had one unique substitution, D1490. Motifs V and VI were also highly conserved among the HEVs, with the active site GDD1570–72 situated in motif VI. Motif VII in moose HEV had two unique substitutions, D1610 and V1613, and motif VIII had one I1623 substitution.
Identifying ORF2/3 start codons in the junction region (JR)
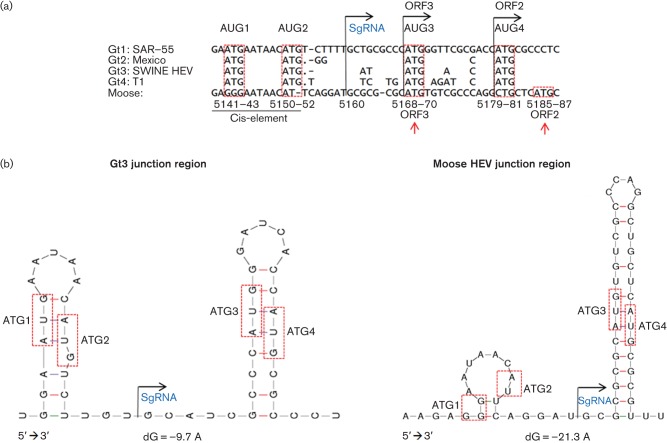
The junction region of the HEV genome contains a cis-reactive element which may be the promoter for the 2 kb bi-cistronic SgRNA for ORF2 and ORF3 (Graff et al., 2005). This region also contains loop structures important for virus replication (Cao et al., 2010). The start of this putative SgRNA could be located at nucleotide position 5160 (Fig. 2a). Two start codons, designated AUG3 and AUG4, representing the authentic start codons for ORF3 and ORF2 (Huang et al., 2007) were identified in the stem of the second stem–loop structure (Fig. 2a, b). Moose HEV had a different cis-reactive sequence element compared to other genotypes (Fig. 2a) and lacked the otherwise conserved AUG1 and AUG2. Mfold prediction of the cis-element revealed a first loop structure with truncated stem in the moose HEV (Fig. 2b). AUG3 was retained in the moose HEV, while AUG4 was three nucleotides downstream compared to AUG4 of gt1–4 strains, giving a longer stem length and a one nucleotide smaller loop (Fig. 2a, b). The SgRNA was estimated to be 2014 nt long, excluding the poly(A) sequence.
Fig. 2.
Identification of putative SgRNA with ORF2 and ORF3 start codons. (a) MSA of gt1–4 and moose HEV junction region. The SgRNA start position is indicated. The cis-element is underlined. Thr ORF2–3 start codons are marked, but with red arrows for moose HEV. (b) Mfold predicted two RNA secondary structures using gt3 or the moose HEV junction region. The ORF3 and ORF2 start codons were in the second stem structure of both HEVs. The ATG1–2 degeneration in moose HEV shown in (a) caused the truncated stem structure shown in (b). Position of ATG codons is shown by red dashed boxes.
Characterization of ORF2
ORF2, which encodes the viral capsid, was deduced to be at nt positions 5185–7152, and was in an alternative reading frame than ORF1. The 1968 nt sequence translated to a presumed 655 aa capsid protein, which was five aa shorter than ORF2 of gt1–4. ORF2 of the moose HEV had the highest nt (64.8–67.6 %) and aa (70.9–74 %) similarity to ORF 2 in gt1–4, rabbit HEV and unclassified wild boar HEV strains (Table 2). The moose HEV ORF2 had two aa deletions and four substitutions in the N-terminal signal sequence involved in translocating ORF2 to the ER (Ahmad et al., 2011) (Table 3a, Fig. S3). The subsequent region was followed by a region of 12 arginines compared to 14 in the consensus sequence (Fig. S3) and most likely binds to the HEV RNA genome (Mori & Matsuura, 2011). This region was absent in trout HEV for unknown reasons. The crystal structure of the capsid protein has been separated into three domains, designated the shell (S), middle (M) and protruding (P) domains (Xing et al., 2010). All three domains (Fig. S3) of the deduced moose HEV ORF2 were compared with the corresponding regions of the other HEV variants and showed several unique aa substitutions; four in the S domain, six substitutions, one G-deletion between residue position 404–405 in the M domain and 23 substitutions in the P domain (Table 3a). Three conserved asparagines representing potential glycosylation sites (Zafrullah et al., 1999) were also identified in the moose HEV ORF2 (Table 3a). The stop codon of the moose HEV ORF2 was TAG, which was also observed in ferret and gt1–2 while the stop codon in gt4, Uwb and RAT HEV was TGA, and in gt3, bat, avian and trout, it was TAA.
Table 3. Moose HEV sequences. (a) ORF2 region with consensus sequence derived from multiple sequence alignment (Fig. S3) with strains taken from Table S1. Each region was derived from Mori & Matsuura (2011). (b) ORF3 properties with consensus sequence derived from aligned HEV strain sequences taken from Table S1. Each region was derived from Ahmad et al. (2011). In both (a) and (b), underlined residues indicate moose HEV substitutions while grey letters indicate unique residues for moose HEV .
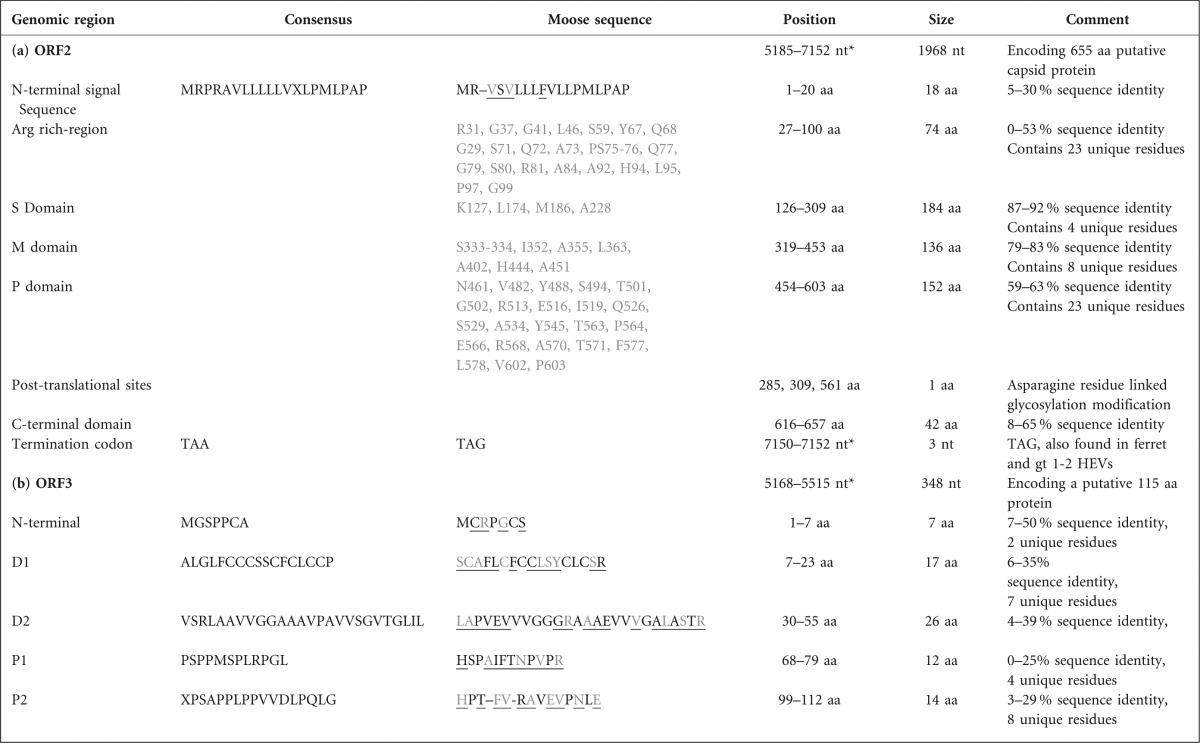
Positions based on the reference gt3 SWX07-E1 (EU360977.1) genome.
Characterization of ORF3
The putative ORF3 protein was identified by MSA and Mfold analysis to be at nt positions 5168–5515 and this 348 nt sequence corresponded to an 115 aa protein (Fig. 2, Table 3b). It showed nt (22–56 %) and aa (8–35 %) sequence similarity with other HEVs (Table 2). Four domains characterize ORF3 (Ahmad et al., 2011) and all were present in the moose HEV (Table 3b). The highly diverged ORF3 N-terminal region contained two unique residues. The following hydrophobic domain 1 (D1) with a region of cystines has been associated with binding to the cytoskeleton, microtubules and mitogen-activated protein kinase (Kannan et al., 2009; Kar-Roy et al., 2004; Zafrullah et al., 1997). The subsequent hydrophobic domain 2 (D2) has been associated with haemopexin binding (Ratra et al., 2008), and both domains contained seven and eight unique residues. The two overlapping motifs PMSP and SPLR, as part of the P1 domain, acting as potential kinase substrates (Zafrullah et al., 1997) were only observed in gt1 and some gt3s, while moose HEV displayed many unique residue substitutions making the target serine absent for possible kinase phosphorylation. The two overlapping PXXP motifs in the P2 domain, which are associated with SH3 protein domain binding and ORF3 interaction (Korkaya et al., 2001), were detected in the gt1–4, but mutated in the moose HEV and in other animal HEVs.
3′UTR properties
The 3′UTR was 82 nt in length, spanned positions 7153–7227 and terminated with 26 nt poly(A). This region was highly diverged with 54 % sequence identity to gt3 SWX07-E1 and is thought to fold into stem–loop and hairpin structures and play a role in HEV replication (Ahmad et al., 2011).
The phylogenetic relationship of moose HEV with other HEVs
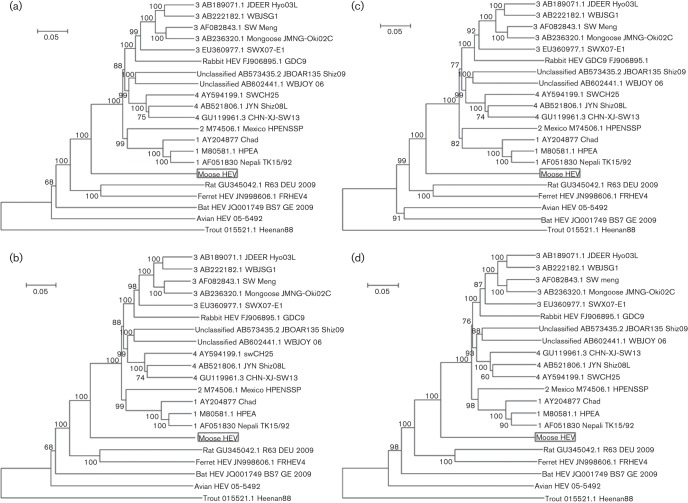
The moose HEV deviated in its own phylogenetic tree branch, sharing the same ancestor with the gt1–4-Uwb group. This occurred regardless if the generated phylogenetic trees were based on different regions in ORF1 or ORF2 or both ORFs combined (Fig. 3a–d). The 2.16 kb based phylogenetic tree (Fig. 3b) was similar to the 5.1 kb and ORF2 based trees (Fig. 3a, d), indicating that this region alone was sufficient for phylogenetic analysis. Only one phylogenetic tree (partial ORF1) was different, with a shared ancestor between avian and bat HEVs (Fig. 3c). An alternative method for investigating genetic relationships with other HEVs through aa p-distances (Smith et al., 2013) was also evaluated. It resulted with the highest p-distance value of 0.83 when moose HEV was compared to trout HEV, while the values were between 0.22–0.52 for other HEV variants (Table 4). Higher p-distance values in animal HEVs corresponded well with the phylogenetic trees (Fig. 3a–d).
Fig. 3.
Phylogenetic nt relationship between HEVs based on a neighbour-joining approach using a Tamura-Nei model with estimated γ-parameter and 1000 bootstraps. (a) Concatenated ORF1 in-frame with ORF2, γ = 0.38. (b) Partial 2.16 kb ORF1 fused in-frame with ORF2, γ = 0.38. (c) Partial ORF1, γ = 0.42. (d) ORF2, γ = 0.3. Node numbers 1–4 represent gt1–4. Bar, genetic distance.
Table 4. Amino acid sequence percentage identity (lower left) and amino acid p-distance (upper right) relationship matrix of HEVs based on partial ORF1 in-frame fused with ORF2 with removed non-residue-coding junction region.
Moose HEV values are underlined separating the animal HEVs from Gt1–4-Unclassified wild boar 1–2 group. The p-distance is the proportion of amino acid sites at which the two sequences to be compared are different. It is obtained by dividing the number of amino acid differences by the total number of sites compared. The p-distance separating the genotypes is with a P-distance value of at least 0.06.
| HEV variant | Gt 1 | 1 | 1 | 2 | 3 | 3 | 3 | 3 | 3 | Rabbit | 4 | 4 | 4 | Uwb1 | Uwb2 | Moose | Ferret | Rat | Bat | Av | Trout |
| Gt 1 M80581.1 HPEA | . | 0.02 | 0.01 | 0.09 | 0.09 | 0.10 | 0.10 | 0.09 | 0.09 | 0.11 | 0.09 | 0.10 | 0.10 | 0.11 | 0.12 | 0.28 | 0.41 | 0.43 | 0.48 | 0.51 | 0.82 |
| Gt 1 AY204877 Chad | 98.6 | . | 0.02 | 0.09 | 0.09 | 0.09 | 0.09 | 0.09 | 0.09 | 0.11 | 0.09 | 0.10 | 0.10 | 0.11 | 0.12 | 0.28 | 0.41 | 0.43 | 0.48 | 0.51 | 0.82 |
| Gt 1 AF051830 Nepali TK15/92 | 99.0 | 98.0 | . | 0.09 | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.11 | 0.10 | 0.10 | 0.11 | 0.12 | 0.12 | 0.28 | 0.41 | 0.43 | 0.49 | 0.51 | 0.82 |
| Gt 2 M74506.1 Mexico HPENSSP | 90.4 | 90.0 | 90.0 | . | 0.10 | 0.11 | 0.11 | 0.11 | 0.10 | 0.12 | 0.11 | 0.11 | 0.11 | 0.12 | 0.13 | 0.29 | 0.41 | 0.44 | 0.49 | 0.52 | 0.83 |
| Gt 3 EU360977.1 SW07-E1 | 89.9 | 90.0 | 89.0 | 88.0 | . | 0.03 | 0.03 | 0.03 | 0.03 | 0.07 | 0.08 | 0.08 | 0.09 | 0.08 | 0.10 | 0.28 | 0.40 | 0.43 | 0.49 | 0.52 | 0.83 |
| Gt 3 AB189071.1 JDEER-Hyo03L | 89.2 | 89.0 | 89.0 | 88.0 | 96.0 | . | 0.01 | 0.01 | 0.02 | 0.07 | 0.08 | 0.08 | 0.08 | 0.09 | 0.10 | 0.29 | 0.40 | 0.43 | 0.49 | 0.51 | 0.83 |
| Gt 3 AB222182.1 WbJSG1 | 89.2 | 89.0 | 89.0 | 88.0 | 96.0 | 98.0 | . | 0.01 | 0.02 | 0.06 | 0.08 | 0.08 | 0.09 | 0.09 | 0.11 | 0.29 | 0.40 | 0.43 | 0.49 | 0.52 | 0.83 |
| Gt 3 AF082843.1 SW Meng | 89.3 | 90.0 | 89.0 | 88.0 | 96.0 | 99.0 | 98.2 | . | 0.01 | 0.07 | 0.08 | 0.08 | 0.08 | 0.09 | 0.11 | 0.28 | 0.40 | 0.43 | 0.49 | 0.51 | 0.82 |
| Gt 3 AB236320.1 Mongoose JMNG-Ok | 89.7 | 90.0 | 89.0 | 88.0 | 96.0 | 98.0 | 98.2 | 98.6 | . | 0.06 | 0.08 | 0.08 | 0.08 | 0.09 | 0.11 | 0.29 | 0.40 | 0.43 | 0.49 | 0.51 | 0.83 |
| Rabbit HEV FJ906895.1 GDC9 | 87.9 | 88.0 | 87.0 | 86.0 | 93.0 | 92.0 | 92.7 | 92.3 | 93.0 | . | 0.10 | 0.10 | 0.11 | 0.11 | 0.13 | 0.29 | 0.40 | 0.43 | 0.49 | 0.52 | 0.83 |
| Gt 4 AB521806.1 JYN Shiz08L | 89.2 | 89.0 | 89.0 | 87.0 | 91.0 | 91.0 | 90.9 | 91.0 | 91.0 | 88.3 | . | 0.02 | 0.03 | 0.08 | 0.10 | 0.28 | 0.40 | 0.42 | 0.49 | 0.51 | 0.83 |
| Gt 4 AY594199.1 SWCH25 | 89.2 | 89.0 | 89.0 | 87.0 | 91.0 | 91.0 | 91.1 | 91.2 | 91.0 | 88.5 | 97.5 | . | 0.02 | 0.08 | 0.09 | 0.29 | 0.39 | 0.42 | 0.49 | 0.51 | 0.83 |
| Gt 4 GU119961.3 CHN-XJ-SW13 | 88.8 | 89.0 | 88.0 | 87.0 | 91.0 | 91.0 | 90.5 | 90.9 | 91.0 | 88.2 | 97.1 | 97.0 | . | 0.09 | 0.10 | 0.29 | 0.40 | 0.43 | 0.49 | 0.52 | 0.83 |
| Unclassified AB573435.2 JBOAR1 (UWB1) | 87.4 | 88.0 | 87.0 | 86.0 | 90.0 | 90.0 | 89.6 | 90.0 | 90.0 | 87.8 | 90.7 | 91.0 | 90.4 | . | 0.09 | 0.28 | 0.40 | 0.43 | 0.48 | 0.51 | 0.82 |
| Unclassified AB602441.1 WBJOY (UWB2) | 86.6 | 86.0 | 86.0 | 86.0 | 88.0 | 88.0 | 88.1 | 88.1 | 88.0 | 85.5 | 89.3 | 90.0 | 89.4 | 89.0 | . | 0.29 | 0.39 | 0.42 | 0.49 | 0.52 | 0.83 |
| Moose HEV | 69.7 | 70.0 | 69.0 | 69.0 | 69.0 | 69.0 | 69.0 | 69.4 | 69.0 | 68.4 | 69.2 | 69.0 | 68.4 | 69.0 | 67.8 | . | 0.41 | 0.43 | 0.49 | 0.52 | 0.81 |
| Ferret HEV JN998606.1 FRHEV4 | 56.7 | 56.0 | 56.0 | 56.0 | 57.0 | 57.0 | 57.1 | 57.4 | 57.0 | 56.8 | 56.9 | 57.0 | 57.0 | 57.0 | 57.2 | 55.8 | . | 0.24 | 0.48 | 0.52 | 0.83 |
| Rat GU345042.1 R63 DEU 2009 | 54.6 | 54.0 | 54.0 | 54.0 | 54.0 | 55.0 | 54.7 | 54.9 | 55.0 | 54.4 | 55.1 | 55.0 | 54.9 | 55.0 | 55.0 | 54.8 | 73.9 | . | 0.51 | 0.54 | 0.82 |
| Bat HEV JQ001749 BS7 GE 2009 | 48.2 | 48.0 | 48.0 | 47.0 | 47.0 | 48.0 | 47.6 | 47.7 | 48.0 | 47.1 | 47.9 | 48.0 | 47.6 | 48.0 | 47.4 | 47.5 | 48.8 | 46.0 | . | 0.50 | 0.83 |
| Avian HEV 05-5492 (Av) | 45.7 | 46.0 | 45.0 | 45.0 | 45.0 | 45.0 | 45.1 | 45.3 | 45.0 | 44.7 | 45.5 | 45.0 | 45.1 | 45.0 | 44.8 | 44.6 | 44.9 | 44.0 | 48.0 | . | 0.82 |
| Trout HEV 015521.1 Heenan88 | 15.3 | 15.0 | 15.0 | 15.0 | 15.0 | 15.0 | 15.3 | 15.3 | 15.0 | 14.7 | 14.9 | 15.0 | 15.1 | 15.0 | 14.8 | 16.1 | 14.7 | 15.0 | 15.0 | 16.5 | . |
Discussion
A new member in the Hepeviridae family
In this study we describe, to the best of our knowledge, the first detection of a hepatitis E like virus in moose. Even though the sequences were highly divergent from known gt1–4, three HEV specific ORFs could be detected in the genome of this novel virus. The ORFs encode the non-structural proteins in ORF1, capsid protein in ORF2 and the multifunctional phospho-protein in ORF3. This observation together with ORF2/3 start codon and phylogenetic analysis (Figs 2 and 3), supported the classification of this new virus as a member of the Hepeviridae family. Even though there were several nucleotide substitutions in primer binding sites, real-time PCR assays were still able to detect the virus (Fig. S2). Future screening should benefit from redesigning the primers and probes for optimal detection. Primer pair 4 (Table 1) should in theory result in a larger HEV amplicon covering a larger sequence from the putative 5′ terminal methyltransferase to RdRp, but only a smaller amplicon was obtained starting from the putative proline hinge region. Although several primer pair combinations were tested, we were unsuccessful in amplifying the remaining moose HEV sequence at the 5′ end. This suggested an incomplete virus genome and may be due to degradation, or by high sequence divergence in combination with low virus concentration. The 2.16 kb amplicon (Table 1) was useful for phylogenetic characterization (Fig. 3b), since it covered regions in RdRp and ORF2 commonly used for HEV genotyping (Mizuo et al., 2002; Zhai et al., 2006). The secondary structure analysis of the JR (Fig. 2) should be valuable for identifying the ORF2/3 start codons in other uncharacterized novel divergent HEVs.
Four genotypes are recognized according to the ICTV Hepeviridae study group and there are still no consistent criteria for classification for novel HEVs. A recently introduced HEV species concept suggested that aa p-distances would provide an approach for evaluating taxonomic relationship (Smith et al., 2013). The authors presented aa p-distances of 0.25–0.50 between HEV variants and as a comparison they used Picornaviridae classification that did not differ by more than 0.58 between genera (Knowles et al., 2012). The p-distances of moose HEV did not surpass 0.58 and corresponded to an aa identity of 45–70 % to other HEVs (Table 4). The trout HEV was the only exception that surpassed 0.58 with values 0.81–0.83 corresponding to 15–17 % aa identity to other HEVs (Table 4). This was in line with the values of 0.60–0.90 observed by Smith et al. (2013). Therefore, the HEV genus cut off value may be in the 0.50–0.60 interval and adjusted by additional HEVs discovered in the future. We propose moose HEV as a new species, together with four other proposed species (HEV variants infecting humans and pigs; variants infecting rat and ferrets; variants from bat and those from poultry; Smith et al., 2013) as members of the Hepevirus genus when using the cut off value of 0.58. The trout HEV with values >0.58, would then be classified as a member of a separate genus. More sequences from other moose HEV strains will confirm the separate phylogenetic branch deviation (Fig. 3a–d) of the moose HEV, and will show if moose HEV forms its own group consisting of separate genotypes, as has been shown for avian HEV (Bányai et al., 2012; Bilic et al., 2009; Sprygin et al., 2012; Zhao et al., 2010) and rat HEV (Li et al., 2013).
Host specificity, virulence and zoonotic aspects
The diverged moose HEV genome most likely reflects host adaptation, and sequenced regions that attract the most attention are ORF1; motifs I, II, V and VII of the X domain; motifs I, Ia, II, III, V, VI and VII of the helicase domain; and motifs II, III, IV and VII of the RdRp (Fig. 1a–c). These regions contain mutations compared to the consensus sequence and some are unique to the moose HEV. As a comparison, almost the entire of motif III in the RdRp of rubella virus was degenerate and even within the HEVs there was great aa diversity, implicating that this motif is suited to a more flexible adaptation (Fig. 1c). The D1461 in this motif was conserved in gt1–2, gt4 and in rabbit HEV. Gt1–2 and 4 are considered to be more pathogenic than gt3 (Abdelwahab et al., 2012) and might share common potential virulence factors. Previous study of chimeric HEVs containing JR, ORF2 and 3′UTR from gt3 or 4 in the backbone of gt1 failed to establish infection in pigs, suggesting that the 5′UTR and ORF1 may also be involved in cross-species infection (Feagins et al., 2011). The gt1 blast match of the moose HEV helicase domain may explain it as one factor by which ORF1 contributes to host restriction. Reinfecting pigs with the chimaeric gt1 backbone replaced with either the gt3 or gt4 helicase might confirm this hypothesis.
Previous aa substitution studies on certain aa pairs in the ORF2 P domain resulted in lack of virion attachment activity in HEV susceptible Huh7 and A549 cells (Yamashita et al., 2009). A single hydrophobic aa substitution (T562V) in one of these aa pairs (aa pair T562 and N560) was found in moose HEV and bat HEV. Substitutions like this may disturb virion binding, leading to altered host specificity. The study of bat HEV observed that ORF3 in many animal HEVs was highly divergent (Drexler et al., 2012), including the moose HEV (Tables 2 and 3b), and may reflect changes in host range and potential virulence properties. The 5′ terminal end region in moose HEV could not be analysed in this study and it would be interesting to detect other putative ORFs, like in ferret and rat (Johne et al., 2010; Raj et al., 2012).
In general, the detection of new viruses in animals usually raises the question regarding their zoonotic potential. Currently, the zoonotic transmission of HEV gt3–4 has been frequently reported, in association with the consumption of pig, wild boar and deer products in industrialized countries (Li et al., 2005; Takahashi et al., 2004; Tei et al., 2003). Moose meat is consumed on a regular basis and therefore the zoonotic potential of HEV in moose is important to study. Experimental cross-species studies have been attempted with divergent HEVs like rat and avian HEV, but with negative results (Cossaboom et al., 2012; Huang et al., 2004), indicating limited host range. It is tempting to speculate that the moose HEV also has a narrow host range and features like a highly divergent genome indicates that this may be so. The application of cell culture and animal models could be useful for testing its cross-species transmission properties.
More divergent animal HEVs and gt1–4 that are more closely related to human HEV do exist in parallel leading to competition, but this also opens possibilities for recombination to occur within the host (Shukla et al., 2011; Wang et al., 2010). An example is the human gt1–2 vs gt 3–4 infections, and another example is the divergent rat HEV (Johne et al., 2010) and the recently discovered rat gt3-like HEV (Lack et al., 2012). Could similar evolution also occur in the Cervidae family, including in moose? The moose HEV may be the more divergent type with narrow host specificity, while the deer gt3 may represent the flexible variant with a wider host range. It would be interesting to see if this moose HEV could infect other species of Cervidae and if the deer gt3 could establish infection in moose. Previous studies revealed that gt3 HEVs are present in both domestic swine and wild boars in Scandinavia with high sequence similarity. HEV isolates taken from a Swedish patient revealed high sequence similarity with porcine HEV isolates. The strains appeared to be phylogenetically clustered into specific geographical clades; country- and even county-specific, thus opening the possibility to derive the geographical origin of HEV strains (Norder et al., 2009; Widén et al., 2011). A geographical clustering for rat HEV was observed in Germany, which when combined with serological analysis demonstrated antigenic differences between rat HEV and HEV gt3 antigen, indicating aa divergence in the immunogenic region corresponding to the P domain of ORF2 (Johne et al., 2012). The moose HEV P domain showed 23 unique residues compared to other HEVs (Table 3a), indicating that antigenic differences might also exist. It would be interesting to investigate these observations in more detail for the moose HEV as well.
This HEV positive moose was emaciated and had an Anaplasma phagocytophilum infection in combination with other infections and infestations. What significance the infections had for the condition and myocardial injury of the moose is not possible to determine. Anaplasma infections are common tick-borne infections in animals and exhibit immunosuppressive properties (Rikihisa, 2011; Stuen, 2007).
In summary, more investigation is needed to better understand the infection biology, epidemiology and clinical manifestation of moose HEV in moose and humans. It is important that the public should be aware of that handling and consumption of HEV contaminated moose meat/organs may pose a risk for HEV infection.
Methods
Homogenization of liver samples.
Six Swedish moose samples taken from liver and/or kidney were homogenized in 2 ml grinding tubes (Eppendorf) containing 2 mm zirconia beads (BioSpec Products) and 600 µl buffer RLT from an RNeasy Mini kit (Qiagen).
RNA extraction and cDNA synthesis.
Total RNA was extracted from homogenized liver/kidney samples with an RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. The concentration and quality of RNA was determined by NanoDrop (NanoDrop Technologies). The 20 µl cDNA synthesis mix consisted of 1 µl Oligo dT(20) (Invitrogen) or 1 µl GeneRACER Oligo dT(24) (Invitrogen), used for priming cDNA synthesis with 3 µl RNA, 1 µl (40 U) of RNaseOUT (Invitrogen) and 1 µl (200 U) of Superscript III, RNase H− reverse transcriptase (Invitrogen). One microlitre (end concentration 5 %) polymerase GC melt from an Advantage GC 2 polymerase mix kit (Clontech) was also added to facilitate amplification of high GC-content regions and reduce secondary structure formation in the HEV genome (Xia et al., 2008). The cDNA reaction was kept at 50 °C for 60 min, followed by 15 min incubation at 70 °C. The reaction was finalized with 2 U of Escherichia coli RNase H (Invitrogen) for 20 min at 37 °C.
HEV real-time PCR and PCR assay.
Three microlitres of extracted and purified RNA from moose liver and kidney were analysed by real-time (RT)-PCR using an Ag-Path-ID one-step RT-PCR kit (Applied Biosystems), with a total volume of 12.5 µl containing 250 nM JVHEV forward respective reverse primers and 100 nM Cy5 based probe targeting the overlapping ORF2/3 region (Jothikumar et al., 2006) and 0.4 × enzyme mix, or with primers targeting a sequence downstream of the ORF2 region with 500 nM forward and 250 nM reverse primers, 250 nM, 260 nM FAM based probe (Gyarmati et al., 2007) and 1× enzyme mix. Both methods have been shown to be more sensitive in comparison with other HEV detection methods (Vasickova et al., 2012) and combining both methods increases the chance of detecting clinical HEV positive samples. The samples were loaded into a Rotor-Gene 3000 (Corbett Research) with the following thermal steps: 45 °C for 10 min, followed by 95 °C for 15 min, cycled 55 times between 95 °C 15 s, 60 °C 60 s. Fluorescence was monitored during the annealing step of each cycle. The diluted full HEV genome of gt3 SWX07-E1 isolate cloned into a plasmid (Xia et al., 2008) with a known concentration was used as a control and for the generation of standard curves.
To acquire more sequence information, we used PCR primer pair 1 (Table 1) targeting part of the RdRp giving a 383 nt fragment commonly used for genotyping (Zhai et al., 2006) with 30 µl PCR mix: 6 µl of synthesized HEV positive cDNA template, 1.2U Platinum Taq polymerase (Invitrogen), 1× PCR RXN buffer, 1 mM MgCl2, 0.2 µM of each ESP and EAP primer, 5 % DMSO and 0.2 mM dNTP. The cycling parameters were 95 °C 3 min, cycled 40 times 94 °C 1 min, 55 °C 1 min, 72 °C 1 min and finishing with 72 °C for 10 min. To obtain more sequence information the forward primer ESP (Zhai et al., 2006) and ORF2, HE041 reverse primer (Mizuo et al., 2002) were used in conventional PCR. The PCR mix contained 1 µl cDNA as template with 0.15 µl Phusion Hot Start High-Fidelity DNA polymerase (Thermo Scientific, Finnzyme) with provided 1× GC buffer, 0.3 µl 0.2 mM dNTP, 0.5 µM ESP forward primer, 0.5 µM HE041 reverse primer and 0.45 µl DMSO (final 3 %) were also added in a total PCR volume of 15 µl. The PCR program had the following profile: 98 °C for 2 min, then cycled 40 times 98 °C 20 s, 65 °C 30 s, 72 °C 2 min and finished with 72 °C for 10 min. An overlap PCR primer walking strategy was used to further extend the 5′ side of the moose HEV genome by primer pair 4 (Table 1). The PCR profile was similar to that mentioned above, but with combined Tm and extension temperature of 72 °C for 3 min.
All amplified PCR products were verified by agarose gel (LE Agarose, Semkem) electrophoresis in gels of different percentages depending on the fragment size.
Purification, cloning and sequencing of amplicons and RACE amplification.
The amplified putative HEV amplicons were purified according to the manufacturer’s instructions with a Wizard SV gel and PCR clean-up system (Promega) for larger amplicons and a PureLink Quick gel extraction kit (Invitrogen) for smaller amplicons. Size and single bands of purified amplicons were confirmed in agarose gels. The purified amplicons were sequenced by using same amplicon PCR primers in forward or reverse for confirming the HEV positive sequence. blast searches at both the nt and aa level was used to identify the sequenced amplicons. The phusion PCR generated products lacking 3′terminal overhangs required for TOPO XL cloning (Invitrogen). Therefore, poly(A) overhangs were synthesized before the cloning procedure in a 10 µl reaction mix with final concentration of 0.2 µM dATP, 1× PCR RXN buffer, 2.4 mM MgCl2, 0.5 U platinium taq polymerase (Invitrogen) and 8.22 µl of purified PCR product. The reaction was incubated in 72 °C for 15 min and put on ice according to the manufacturer’s instructions. The 3′UTR terminal end was amplified with primer pair 3 (Table 1) using a RACE kit (Invitrogen) resulted in a 1.3 kb overlapping PCR product according to PCR program profile 98 °C 2 min, 98 °C 10 s, 65 °C 30 s, 72 °C 2 min and 72 °C 10 min.
All sequencing reactions were carried out with a Big Dye Terminator Cycle Sequencing Ready reaction kit version 3.1 (Applied Biosystems) with program profile 95 °C 15 s, 50 °C 10 s, 60 °C 4 min cycling 25 times. Sequences were analysed in Lasergene 8 (dnastar). All primers for amplicon amplification and sequencing are referred to Table 1 and the papers of Koonin et al. (1992), Xing et al. (2010) and Ahmad et al. (2011) were used as guidance for identifying ORF1, ORF2 and ORF3 domain/motif regions.
Junction region analysis.
A multiple sequence alignment of JR containing the 5′-end, putative cis-reactive element and putative start codons for ORF2 and 3 were represented by AUG1–4 (Fig. 2). A secondary structure analysis with Mfold for the JR containing ORF2–3 start codons were investigated for a gt3 (AB481229.1) in parallel with the moose HEV with the similar approach as in Huang et al. (2007).
Phylogenetic analysis.
All phylogenetic trees (Fig. 3) and aa p-distance calculation (Table 4) was performed with mega 5.0 (Tamura et al., 2011). Multiple HEV sequences (Table S1), were aa aligned and reconverted back to nucleotides. The Tamura-Nei evolutionary distance model was used for generating neighbour-joining phylogenetic trees with 0.30–0.42 γ-parameter values and 1000 bootstraps.
Acknowledgements
We would like to thank the Swedish hunters, the Departments of POV, ESS, DOA, the Parasitology section of VIP (SVA), the Wild Tech project (EU 7th Framework Program for Research and Technological Development, grant agreement no. 222633), Roland Mattsson for providing samples and Ann-Sophie Olofson for excellent technical assistance. This study was supported by the Swedish Research Council Formas (grant 2009-747). Part of this study was supported under the project ‘PREDEMICS’ (7th Framework Program, grant agreement no. 278433) and the Wild Tech project.
Footnotes
Three supplementary figures and one supplementary table are available with the online version of this paper.
References
- Abdelwahab S., Rewisha E., Hashem M., Sobhy M., Galal I., Allam W. R., Mikhail N., Galal G., El-Tabbakh M. & other authors (2012). Risk factors for hepatitis C virus infection among Egyptian healthcare workers in a national liver diseases referral centre. Trans R Soc Trop Med Hyg 106, 98–103 10.1016/j.trstmh.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Aggarwal R. (2011). Clinical presentation of hepatitis E. Virus Res 161, 15–22 10.1016/j.virusres.2011.03.017 [DOI] [PubMed] [Google Scholar]
- Aggarwal R., Jameel S. (2008). Hepatitis E vaccine. Hepatol Int 2, 308–315 10.1007/s12072-008-9071-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I., Holla R. P., Jameel S. (2011). Molecular virology of hepatitis E virus. Virus Res 161, 47–58 10.1016/j.virusres.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai K., Tóth A. G., Ivanics E., Glávits R., Szentpáli-Gavallér K., Dán A. (2012). Putative novel genotype of avian hepatitis E virus, Hungary, 2010. Emerg Infect Dis 18, 1365–1368 10.3201/eid1808.111669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilic I., Jaskulska B., Basic A., Morrow C. J., Hess M. (2009). Sequence analysis and comparison of avian hepatitis E viruses from Australia and Europe indicate the existence of different genotypes. J Gen Virol 90, 863–873 10.1099/vir.0.007179-0 [DOI] [PubMed] [Google Scholar]
- Cao D., Huang Y. W., Meng X. J. (2010). The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J Virol 84, 13040–13044 10.1128/JVI.01475-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P. B., Engle R. E., Hjort C., Homburg K. M., Vach W., Georgsen J., Purcell R. H. (2008). Time trend of the prevalence of hepatitis E antibodies among farmers and blood donors: a potential zoonosis in Denmark. Clin Infect Dis 47, 1026–1031 10.1086/591970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson P., Borentain P., Queyriaux B., Kaba M., Moal V., Gallian P., Heyries L., Raoult D., Gerolami R. (2010). Pig liver sausage as a source of hepatitis E virus transmission to humans. J Infect Dis 202, 825–834 10.1086/655898 [DOI] [PubMed] [Google Scholar]
- Cossaboom C. M., Córdoba L., Sanford B. J., Piñeyro P., Kenney S. P., Dryman B. A., Wang Y., Meng X. J. (2012). Cross-species infection of pigs with a novel rabbit, but not rat, strain of hepatitis E virus isolated in the United States. J Gen Virol 93, 1687–1695 10.1099/vir.0.041509-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton H. R., Thurairajah P. H., Fellows H. J., Hussaini H. S., Mitchell J., Bendall R., Banks M., Ijaz S., Teo C. G., Levine D. F. (2007). Autochthonous hepatitis E in southwest England. J Viral Hepat 14, 304–309 10.1111/j.1365-2893.2006.00800.x [DOI] [PubMed] [Google Scholar]
- Drexler J. F., Seelen A., Corman V. M., Fumie Tateno A., Cottontail V., Melim Zerbinati R., Gloza-Rausch F., Klose S. M. & other authors (2012). Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol 86, 9134–9147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff M. P., Malet H., Putics A., Heinonen M., Dutartre H., Frangeul A., Gruez A., Campanacci V., Cambillau C. & other authors (2006). Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80, 8493–8502 10.1128/JVI.00713-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins A. R., Córdoba L., Sanford B. J., Dryman B. A., Huang Y. W., LeRoith T., Emerson S. U., Meng X. J. (2011). Intergenotypic chimeric hepatitis E viruses (HEVs) with the genotype 4 human HEV capsid gene in the backbone of genotype 3 swine HEV are infectious in pigs. Virus Res 156, 141–146 10.1016/j.virusres.2010.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E., Blinov V. M., Donchenko A. P., Koonin E. V. (1989). An NTP-binding motif is the most conserved sequence in a highly diverged monophyletic group of proteins involved in positive strand RNA viral replication. J Mol Evol 28, 256–268 10.1007/BF02102483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Nguyen H., Yu C., Elkins W. R., St Claire M., Purcell R. H., Emerson S. U. (2005). The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J Virol 79, 6680–6689 10.1128/JVI.79.11.6680-6689.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff J., Torian U., Nguyen H., Emerson S. U. (2006). A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J Virol 80, 5919–5926 10.1128/JVI.00046-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyarmati P., Mohammed N., Norder H., Blomberg J., Belák S., Widén F. (2007). Universal detection of hepatitis E virus by two real-time PCR assays: TaqMan and Primer-Probe Energy Transfer. J Virol Methods 146, 226–235 10.1016/j.jviromet.2007.07.014 [DOI] [PubMed] [Google Scholar]
- Huang F. F., Sun Z. F., Emerson S. U., Purcell R. H., Shivaprasad H. L., Pierson F. W., Toth T. E., Meng X. J. (2004). Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J Gen Virol 85, 1609–1618 10.1099/vir.0.79841-0 [DOI] [PubMed] [Google Scholar]
- Huang Y. W., Opriessnig T., Halbur P. G., Meng X. J. (2007). Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J Virol 81, 3018–3026 10.1128/JVI.02259-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R., Heckel G., Plenge-Bönig A., Kindler E., Maresch C., Reetz J., Schielke A., Ulrich R. G. (2010). Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16, 1452–1455 10.3201/eid1609.100444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R., Dremsek P., Kindler E., Schielke A., Plenge-Bönig A., Gregersen H., Wessels U., Schmidt K., Rietschel W. & other authors (2012). Rat hepatitis E virus: geographical clustering within Germany and serological detection in wild Norway rats (Rattus norvegicus). Infect Genet Evol 12, 947–956 10.1016/j.meegid.2012.02.021 [DOI] [PubMed] [Google Scholar]
- Jothikumar N., Cromeans T. L., Robertson B. H., Meng X. J., Hill V. R. (2006). A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J Virol Methods 131, 65–71 10.1016/j.jviromet.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Kamar N., Bendall R., Legrand-Abravanel F., Xia N. S., Ijaz S., Izopet J., Dalton H. R. (2012). Hepatitis E. Lancet 379, 2477–2488 10.1016/S0140-6736(11)61849-7 [DOI] [PubMed] [Google Scholar]
- Kannan H., Fan S., Patel D., Bossis I., Zhang Y. J. (2009). The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol 83, 6375–6382 10.1128/JVI.02571-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar-Roy A., Korkaya H., Oberoi R., Lal S. K., Jameel S. (2004). The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J Biol Chem 279, 28345–28357 10.1074/jbc.M400457200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y. A., Lole K. S. (2010a). RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J Virol 84, 9637–9641 10.1128/JVI.00492-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y. A., Lole K. S. (2010b). NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J Virol 84, 3595–3602 10.1128/JVI.02130-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles N. J., Hovi T., Hyypiä T., King A. M. Q., Lindberg A. M., Pallansch M. A., Palmenberg A. C., Simmonds P., Skern T. & other authors (2012). Picornaviridae. In Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses, pp 855–880 Edited by King A. M. Q., et al. San Diego: Elsevier [Google Scholar]
- Koonin E. V., Gorbalenya A. E., Purdy M. A., Rozanov M. N., Reyes G. R., Bradley D. W. (1992). Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci U S A 89, 8259–8263 10.1073/pnas.89.17.8259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H., Jameel S., Gupta D., Tyagi S., Kumar R., Zafrullah M., Mazumdar M., Lal S. K., Xiaofang L. & other authors (2001). The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J Biol Chem 276, 42389–42400 10.1074/jbc.M101546200 [DOI] [PubMed] [Google Scholar]
- Lack J. B., Volk K., Van Den Bussche R. A. (2012). Hepatitis E virus genotype 3 in wild rats, United States. Emerg Infect Dis 18, 1268–1273 10.3201/eid1808.120070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. C., Chijiwa K., Sera N., Ishibashi T., Etoh Y., Shinohara Y., Kurata Y., Ishida M., Sakamoto S. & other authors (2005). Hepatitis E virus transmission from wild boar meat. Emerg Infect Dis 11, 1958–1960 10.3201/eid1112.051041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T. C., Ami Y., Suzaki Y., Yasuda S. P., Yoshimatsu K., Arikawa J., Takeda N., Takaji W. (2013). Characterization of full genome of rat hepatitis E virus strain from Vietnam. Emerg Infect Dis 19, 115–118 10.3201/eid1901.121007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansuy J. M., Abravanel F., Miedouge M., Mengelle C., Merviel C., Dubois M., Kamar N., Rostaing L., Alric L. & other authors (2009). Acute hepatitis E in south-west France over a 5-year period. J Clin Virol 44, 74–77 10.1016/j.jcv.2008.09.010 [DOI] [PubMed] [Google Scholar]
- Meng X. J. (2011). From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res 161, 23–30 10.1016/j.virusres.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuo H., Suzuki K., Takikawa Y., Sugai Y., Tokita H., Akahane Y., Itoh K., Gotanda Y., Takahashi M. & other authors (2002). Polyphyletic strains of hepatitis E virus are responsible for sporadic cases of acute hepatitis in Japan. J Clin Microbiol 40, 3209–3218 10.1128/JCM.40.9.3209-3218.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y., Matsuura Y. (2011). Structure of hepatitis E viral particle. Virus Res 161, 59–64 10.1016/j.virusres.2011.03.015 [DOI] [PubMed] [Google Scholar]
- Navaneethan U., Al Mohajer M., Shata M. T. (2008). Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int 28, 1190–1199 10.1111/j.1478-3231.2008.01840.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvonen M., Ahola T. (2009). Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol 385, 212–225 10.1016/j.jmb.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norder H., Sundqvist L., Magnusson L., Østergaard Breum S., Löfdahl M., Larsen L. E., Hjulsager C. K., Magnius L., Böttiger B. E., Widén F. (2009). Endemic hepatitis E in two Nordic countries. Euro Surveill 14, 19211. [DOI] [PubMed] [Google Scholar]
- Okamoto H. (2007). Genetic variability and evolution of hepatitis E virus. Virus Res 127, 216–228 10.1016/j.virusres.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Olsen B., Axelsson-Olsson D., Thelin A., Weiland O. (2006). Unexpected high prevalence of IgG-antibodies to hepatitis E virus in Swedish pig farmers and controls. Scand J Infect Dis 38, 55–58 10.1080/00365540500321470 [DOI] [PubMed] [Google Scholar]
- Pavio N., Meng X. J., Renou C. (2010). Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet Res 41, 46 10.1051/vetres/2010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy M. A., Khudyakov Y. E. (2011). The molecular epidemiology of hepatitis E virus infection. Virus Res 161, 31–39 10.1016/j.virusres.2011.04.030 [DOI] [PubMed] [Google Scholar]
- Purdy M. A., Lara J., Khudyakov Y. E. (2012). The hepatitis E virus polyproline region is involved in viral adaptation. PLoS ONE 7, e35974 10.1371/journal.pone.0035974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V. S., Smits S. L., Pas S. D., Provacia L. B., Moorman-Roest H., Osterhaus A. D., Haagmans B. L. (2012). Novel hepatitis E virus in ferrets, the Netherlands. Emerg Infect Dis 18, 1369–1370 10.3201/eid1808.111659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratra R., Kar-Roy A., Lal S. K. (2008). The ORF3 protein of hepatitis E virus interacts with hemopexin by means of its 26 amino acid N-terminal hydrophobic domain II. Biochemistry 47, 1957–1969 10.1021/bi7016552 [DOI] [PubMed] [Google Scholar]
- Rikihisa Y. (2011). Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev 24, 469–489 10.1128/CMR.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P., Nguyen H. T., Torian U., Engle R. E., Faulk K., Dalton H. R., Bendall R. P., Keane F. E., Purcell R. H., Emerson S. U. (2011). Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108, 2438–2443 10.1073/pnas.1018878108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Purdy M. A., Simmonds P. (2013). Genetic variability and the classification of hepatitis E virus. J Virol 87, 4161–4169 10.1128/JVI.02762-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprygin A. V., Nikonova Z. B., Zinyakov N. G. (2012). Avian hepatitis E virus identified in Russian chicken flocks exhibits high genetic divergence based on the ORF2 capsid gene. Avian Pathol 41, 459–463 10.1080/03079457.2012.711464 [DOI] [PubMed] [Google Scholar]
- Stuen S. (2007). Anaplasma phagocytophilum - the most widespread tick-borne infection in animals in Europe. Vet Res Commun 31 (Suppl 1), 79–84 10.1007/s11259-007-0071-y [DOI] [PubMed] [Google Scholar]
- Takahashi K., Kitajima N., Abe N., Mishiro S. (2004). Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330, 501–505 10.1016/j.virol.2004.10.006 [DOI] [PubMed] [Google Scholar]
- Tam A. W., Smith M. M., Guerra M. E., Huang C. C., Bradley D. W., Fry K. E., Reyes G. R. (1991). Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185, 120–131 10.1016/0042-6822(91)90760-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei S., Kitajima N., Takahashi K., Mishiro S. (2003). Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362, 371–373 10.1016/S0140-6736(03)14025-1 [DOI] [PubMed] [Google Scholar]
- Tomiyama D., Inoue E., Osawa Y., Okazaki K. (2009). Serological evidence of infection with hepatitis E virus among wild Yezo-deer, Cervus nippon yesoensis, in Hokkaido, Japan. J Viral Hepat 16, 524–528 10.1111/j.1365-2893.2009.01107.x [DOI] [PubMed] [Google Scholar]
- Vasickova P., Kralik P., Slana I., Pavlik I. (2012). Optimisation of a triplex real time RT-PCR for detection of hepatitis E virus RNA and validation on biological samples. J Virol Methods 180, 38–42 10.1016/j.jviromet.2011.12.007 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang W., Ni B., Shen H., Song Y., Wang X., Shao S., Hua X., Cui L. (2010). Recombination analysis reveals a double recombination event in hepatitis E virus. Virol J 7, 129 10.1186/1743-422X-7-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widén F., Sundqvist L., Matyi-Toth A., Metreveli G., Belák S., Hallgren G., Norder H. (2011). Molecular epidemiology of hepatitis E virus in humans, pigs and wild boars in Sweden. Epidemiol Infect 139, 361–371 10.1017/S0950268810001342 [DOI] [PubMed] [Google Scholar]
- Xia H., Liu L., Linde A. M., Belák S., Norder H., Widén F. (2008). Molecular characterization and phylogenetic analysis of the complete genome of a hepatitis E virus from European swine. Virus Genes 37, 39–48 10.1007/s11262-008-0246-9 [DOI] [PubMed] [Google Scholar]
- Xing L., Li T. C., Mayazaki N., Simon M. N., Wall J. S., Moore M., Wang C. Y., Takeda N., Wakita T. & other authors (2010). Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J Biol Chem 285, 33175–33183 10.1074/jbc.M110.106336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T., Mori Y., Miyazaki N., Cheng R. H., Yoshimura M., Unno H., Shima R., Moriishi K., Tsukihara T. & other authors (2009). Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci U S A 106, 12986–12991 10.1073/pnas.0903699106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M., Ozdener M. H., Panda S. K., Jameel S. (1997). The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol 71, 9045–9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrullah M., Ozdener M. H., Kumar R., Panda S. K., Jameel S. (1999). Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J Virol 73, 4074–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L., Dai X., Meng J. (2006). Hepatitis E virus genotyping based on full-length genome and partial genomic regions. Virus Res 120, 57–69 10.1016/j.virusres.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Zhou E. M., Dong S. W., Qiu H. K., Zhang L., Hu S. B., Zhao F. F., Jiang S. J., Sun Y. N. (2010). Analysis of avian hepatitis E virus from chickens, China. Emerg Infect Dis 16, 1469–1472 10.3201/eid1609.100626 [DOI] [PMC free article] [PubMed] [Google Scholar]