Abstract
The drivers of influenza seasonality remain heavily debated, especially in tropical/subtropical regions where influenza activity can peak in winter, during the rainy season, or remain constant throughout the year. We compared the epidemiological and evolutionary patterns of seasonal influenza epidemics in Hong Kong and Shenzhen, two adjacent cities in subtropical southern China. This comparison represents a unique natural experiment, as connectivity between these two cities has increased over the past decade. We found that, whilst summer influenza epidemics in Shenzhen used to peak 1–3 months later than those in Hong Kong, the difference decreased after 2005 (P<0.0001). Phylogenetic analysis revealed that influenza isolates from Shenzhen have become genetically closer to those circulating in Hong Kong over time (P = 0.045). Furthermore, although Shenzhen isolates used to be more distant from the global putative source of influenza viruses than isolates from Hong Kong (P<0.001), this difference has narrowed (P = 0.02). Overall, our study reveals that influenza activities show remarkably distinct epidemiological and evolutionary patterns in adjacent subtropical cities and suggests that human mobility patterns can play a major role in influenza dynamics in the subtropics.
Introduction
Influenza activity fluctuates throughout the year in temperate and tropical/subtropical regions, with temperate regions typically experiencing a single epidemic peak in winter, whilst tropical/subtropical areas are characterized by more complex dynamics with multiple epidemics throughout the year (Azziz Baumgartner et al., 2012; Bloom-Feshbach et al., 2013; Lee et al., 2009; Lipsitch & Viboud, 2009; Tamerius et al., 2011; Viboud et al., 2004, 2006a; Waicharoen et al., 2008). A number of factors could drive influenza seasonality including climate (Chan et al., 2009; Shaman et al., 2010; Tamerius et al., 2011, 2013), population movements (Alonso et al., 2007; Weinberger et al., 2012), social behaviour (Lipsitch & Viboud, 2009; Lowen & Palese, 2009) and the introduction of new viral variants (Bahl et al., 2011; Cheng et al., 2013; Finkelman et al., 2007; Rambaut et al., 2008). It is not clear, however, which of these potential factors influences the complex seasonality of influenza in tropical/subtropical regions.
The unique geographical and social/political environments of Hong Kong Special Administrative Region (SAR) and Shenzhen Special Economic Zone (SEZ), two neighbouring cities in mainland China, provide a unique opportunity to investigate hypotheses about the mechanisms of influenza seasonality and the role of human mobility. The two locales are directly adjacent and share the same subtropical climate but are separated by an immigration-control border. Over the past decade, changes in border policies have resulted in a more than sevenfold increase in population fluxes between the two cities (Hong Kong Tourism Board, 2013; The Government of the Hong Kong Special Administrative Region, 2013). In this study, we combined influenza epidemiological and genetic data from the two cities to assess seasonality differences and evolutionary distances between the two locales and to identify potential changes in epidemic dynamics over the past decade as cross-border traffic has increased.
Results
Recent trends in travel patterns from 2001 to 2009
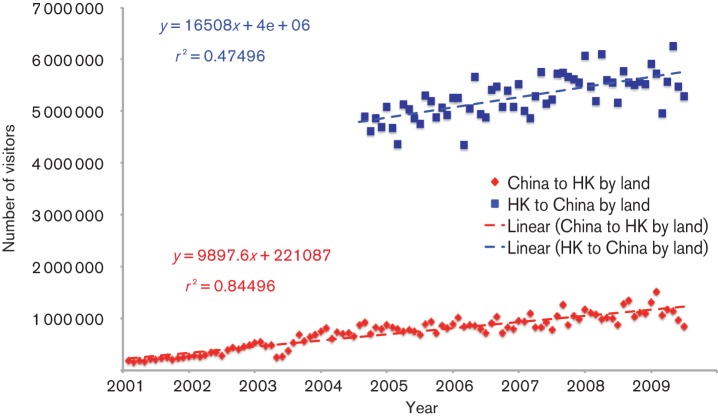
The number of visitors travelling between China and Hong Kong by land gradually increased during the study period from January 2001 to June 2009 (Fig. 1; test for trend: r2 = 0.845 for Chinese visitors to Hong Kong, P<0.001; r2 = 0.475 for Hong Kong residents to mainland China; P<0.001). We hypothesized that increased connectivity between Shenzhen and Hong Kong would promote epidemic synchrony and increase similarity in the viral populations between the two cities over time, which we could test based on the influenza epidemiological and genetic data.
Fig. 1.
Monthly Chinese visitor arrivals to Hong Kong by land and Hong Kong resident departures by land, from January 2001 to June 2009. Red diamonds and dashed line, Chinese visitor arrivals to Hong Kong by land and linear regression fit; blue squares and dashed line, Hong Kong resident departures to mainland China by land and linear regression fit.
Synchrony in Hong Kong and Shenzhen influenza epidemics from 1997 to 2009
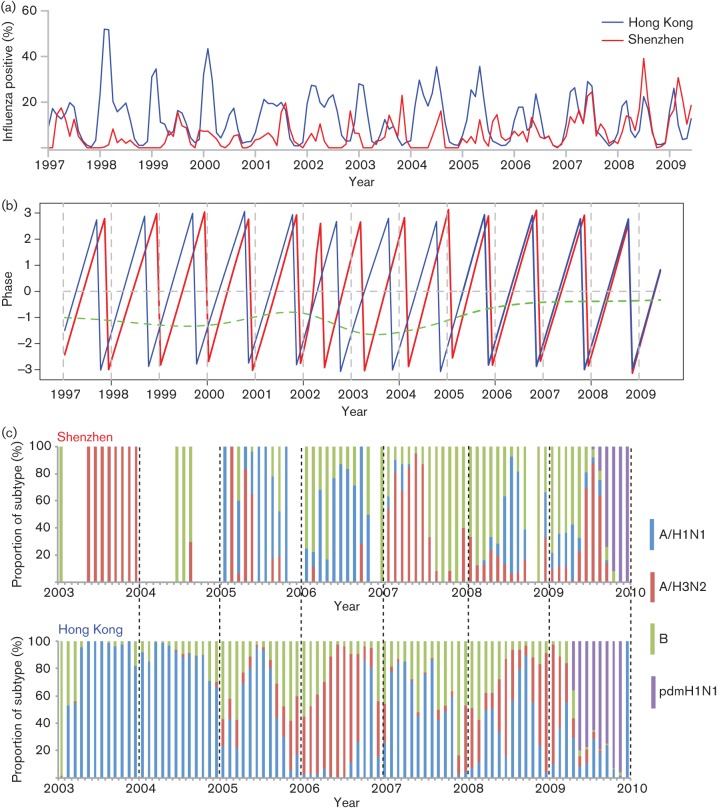
Epidemiological data were available for the pre-pandemic period January 1997 to June 2009, including 45 974 laboratory-confirmed influenza cases among 329 094 specimens tested in Hong Kong (14.0 % positive) and 1704 influenza positives among 21 768 specimens tested in Shenzhen (7.8 % positive). Influenza epidemics in Hong Kong exhibited both annual and semi-annual epidemic cycles, with a major peak occurring in winter/spring and a minor peak occurring in summer (Fig. 2a). In contrast, Shenzhen had a single dominant annual epidemic in summer without evidence of a semi-annual cycle (Fig. 2a). In some years, the epidemic peak in Shenzhen coincided with the summer peak in Hong Kong, but this pattern was not consistent (Fig. 2a).
Fig. 2.
Seasonality of influenza epidemics and monthly distribution of influenza virus subtypes in Shenzhen and Hong Kong. (a) Time series of influenza-positive specimens (percentage, calculated as the monthly number of influenza virus-positive specimens divided by the monthly number of all specimens tested) in Hong Kong (blue line) and Shenzhen (red line) from 1997 to June 2009 (before the spread of the pandemic 2009 H1N1). (b) Phases of time series of influenza-positive specimens in Shenzhen (red line) and Hong Kong (blue line) at a periodicity of 0.8–1.2 years, together with phase differences between Shenzhen and Hong Kong (green line). (c) Monthly distribution of influenza virus subtypes in Shenzhen (upper panel) and Hong Kong (lower panel) from 2003 to 2009. Blue bars, seasonal influenza A virus subtype H1N1 (A/H1N1); red bars, seasonal influenza A virus subtype H3N2 (A/H3N2); green bars, influenza B virus (B); purple bars, 2009 pandemic influenza A virus subtype H1N1 (pdmH1N1).
Wavelet phase synchrony analysis suggested that influenza epidemics in Shenzhen lagged behind those in Hong Kong by 2 months on average during 1997 to 2005 (mean 2.25 months, range 1.21–3.17 months). Since 2006, the timing of the annual epidemic cycles in Shenzhen and Hong Kong has become more similar, with a mean lag in Shenzhen of less than 1 month (mean 0.79 months, range 0.64–1.16 months; t-test for difference with earlier years, P<0.0001) (Fig. 2b).
Differences in subtype distribution from 2003 to 2009
We next determined whether the two adjacent cities had the same dominant influenza subtype during the same calendar year. Overall, there was little similarity in influenza genotype distribution between Shenzhen and Hong Kong (Fig. 2c). For instance, of the 7 years with available subtype information, A/H3N2 was dominant in 2003, early 2007 and early 2009 in Shenzhen, whilst A/H3N2 was dominant in 2006 and in winter 2008–2009 in Hong Kong. A/H1N1 was dominant in 2005, 2006 and late 2008 in Shenzhen, and in 2003–2005, 2007 and 2008 in Hong Kong. There were some similarities in A/H1N1 subtype dominance between the two cities, with an A/H1N1 epidemic occurring in both cities in 2005 and late 2008. Influenza B virus was more prevalent in the winter in both cities (Fig. 2c).
Increase in similarity of viral populations in Shenzhen and Hong Kong over time, between 1995 and 2009
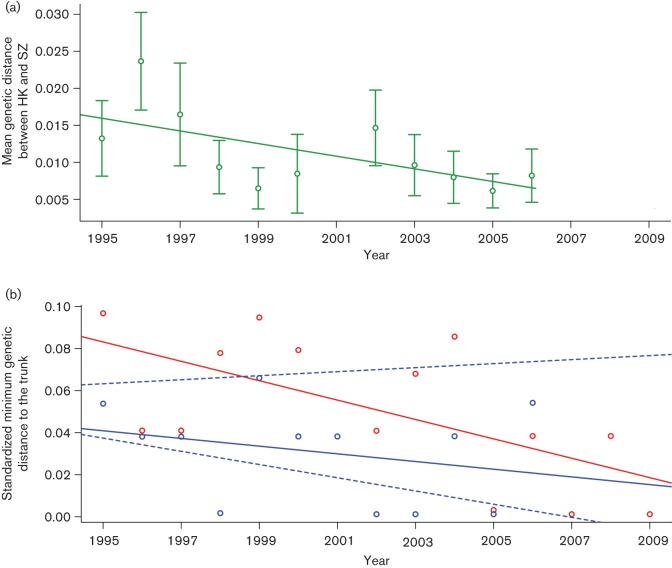
We measured the mean pairwise genetic distance of HA1 gene sequences between influenza A/H3N2 isolates in Shenzhen and Hong Kong during 1995–2006, a period with reasonable sequence availability. We found that the genetic distance between the two cities declined over time (Fig. 3a, time trend regression, r2 = 0.37, P = 0.046). Furthermore, we estimated the genetic distance from each HA sequence to the ‘trunk’ of a global phylogenetic tree (as defined by Cheng et al., 2013) and took the minimum distance by season and city. The minimum distance from the Hong Kong isolates to the trunk was small (Fig. 3b), indicating that the viruses were similar to the global pool, and this minimum distance did not change significantly over time (time trend regression, r2 = 0.08 P>0.05). In contrast, isolates from Shenzhen were generally more distant from the trunk (i.e. a larger minimum genetic distance, paired t-test, P<0.001) (Fig. 3b). Furthermore, the Shenzhen genetic distance declined significantly over time (time trend regression, r2 = 0.40, P = 0.016) and reached levels similar to those of Hong Kong. Taken together, this indicated that the isolates in Shenzhen became gradually more genetically similar to the putative source viruses, up to the level maintained by Hong Kong throughout the study period.
Fig. 3.
(a) Mean genetic distances between Shenzhen and Hong Kong between 1995 and 2009. Green dots and bar, mean and 95 % confidence interval of genetic distance between Shenzhen and Hong Kong; line, linear regression fit. (b) Standardized smallest distances from Shenzhen and Hong Kong isolates to the trunk of the global phylogeny between 1995 and 2009. Red dots and line, smallest distances from Shenzhen isolates to the trunk and linear regression fit; blue dots and solid line, smallest distances from Hong Kong isolates to the trunk and linear regression fit; blue dashed line, 95 % confidence interval of linear regression for 1000 subsampling mock datasets.
We performed several sensitivity analyses with the genetic data to test the impact of sampling intensity and design. As we did not know the month of isolation of the Shenzhen isolates, we considered whether differences in influenza seasonality between Shenzhen and Hong Kong might bias the genetic distance results. Using the Hong Kong data, there was no significant difference in minimum distance from the global trunk between winter and summer isolates (paired t-test matching estimates by year, P>0.05). Furthermore, similar to the main analysis, we did not find any time trend in minimum distance of summer or winter season isolates (Fig. S1, available in the online Supplementary Material, time trend regression, r2 = 0.001 for winter, P>0.05; r2 = 0.2 for summer, P>0.05).
We also considered whether sampling intensity might bias estimates of minimum distance to the trunk. The number of sequenced isolates increased in Shenzhen over time, and there was a significant association between the available sequences each year and the minimum distance to the trunk (i.e. fewer isolates, greater minimum distance from the trunk). However, in a multivariate regression of distance against time and the number of sequences, the number of sequences dropped out of the model. Furthermore, we subsampled the Hong Kong viral sequence data to create 1000 mock datasets that were equivalent in size to the Shenzhen dataset. In these 1000 sparser Hong Kong datasets, there was no significant trend in minimum distance from the trunk, as in the original data (Fig. 3b, time trend regression, r2 ≤0.18, 95 % confidence intervals of slope −0.003 to 0.001, P>0.05). Because the observed value of the time trend for the Shenzhen dataset (r2 = 0.37) falls outside the simulated range (95 % confidence intervals 7.2×10−5 to 0.18), this would indicate that the trend is robust and is unlikely to be the result of sparse sampling. Overall, these sensitivity analyses suggested that the decreasing influenza genetic distance to the trunk in Shenzhen, and the comparatively low distance maintained by Hong Kong, are not artefacts of sampling intensity.
Discussion
In this study, we relied on datasets obtained from different sources. To maximize information from individual datasets and analyses, we use slightly different study periods throughout the paper, as indicated. Overall, we concentrated on influenza dynamics and travel patterns from the mid-1990s to the emergence of the A/H1N1 pandemic virus in China in June 2009. We showed that, despite the geographical proximity and shared climate of Shenzhen and Hong Kong, the two cities experienced distinct influenza seasonality and epidemic timing, and viral genetic populations differed in most years (at both the subtype and genetic levels). Furthermore, epidemic synchrony between the two cities has increased in the more recent years, in parallel to increasing similarity in genetic composition of the viral populations, which we suspect was driven by a substantial increase in population mixing as border control measures were relaxed between mainland China and Hong Kong SAR.
The ‘handover’ of Hong Kong from the UK to China took place on 1 July 1997 (Roger, 1997); residents from mainland China were allowed to visit Hong Kong on business visas or in group tours. Following the severe acute respiratory syndrome outbreak in Hong Kong from March to June 2003, and to promote the economy of Hong Kong and Macau, an ‘Individual Visit Scheme’ was introduced in four cities of Guangdong province on 28 July 2003 and in Shenzhen 1 month later, allowing individual travellers to visit Hong Kong and Macau (The Government of the Hong Kong Special Administrative Region, 2013). Since then, the scheme has been expanded to include 49 mainland China cities, including all cities in Guangdong province, the capital cities of each of the 31 Chinese provinces and other major cities. Because visitors travelling by land between mainland China and Hong Kong have to go through Shenzhen due to the geography of this region, more flexible visiting schemes have increased population mixing between residents of Hong Kong and Shenzhen.
The characteristics of seasonal influenza epidemics are thought to be influenced by environmental factors, population mixing and host immunity. As Shenzhen and Hong Kong are immediately adjacent, they essentially share the same subtropical climate with synchronous peaks and troughs in temperature and humidity (Fig. S2; Diebel & Norda, 2013a, b). We cannot rule out, however, minor difference in microclimates, especially as Hong Kong represents a rather large and geographically diverse area. If local climate was a major driver of influenza seasonality in the region, we would expect similar influenza seasonal patterns in the two cities. Our epidemiological data showed that it is not the case, and hence we conclude that factors other than climate play a major role in influenza dynamics in subtropical locales, as suspected previously (Tamerius et al., 2013).
Influenza dominant subtypes often differed between the two cities in the same year, although, in general, A/H3N2 epidemics tended to be earlier and more intense in both cities, whilst A/H1N1 and B epidemics tended to occur later and be milder. Lack of agreement in subtype dominance could result from the relative separation of the two populations in the earlier years of the study, or from demographic and cultural differences between residents of the two cities. It is accepted that influenza A/H1N1 and B viruses tend to be more dominant in younger individuals, whilst A/H3N2 viruses affect all age groups (Turbelin et al., 2013). Shenzhen has a large population of migrant Chinese workers who come to Shenzhen temporarily for work purposes, and, as a result, the population is skewed towards younger adults (Cheng et al., 2013; Garske et al., 2011). Temporary workers return home in large numbers for the Lunar New Year holiday, potentially leading to different contact patterns and social networks between the two cities, which could in turn affect influenza seasonal patterns. More detailed demographic analyses of influenza infections in the two cities would be required to understand such demographic drivers. An age-stratified analysis of influenza seasonality might also help to determine the importance of the population structure in these two cities.
Hong Kong and Shenzhen are both located in subtropical South-east Asia, which has been proposed as a possible source of new influenza virus variants for temperate regions (Rambaut et al., 2008; Russell et al., 2008), although the global circulation of influenza is still debated (Bahl et al., 2011; Bedford et al., 2010; Cheng et al., 2013). Recent studies show that epidemics in Hong Kong and Shenzhen probably resulted from multiple introductions from tropical/subtropical and temperate regions throughout the year, casting doubts on the ‘source–sink’ model from South-east Asia (Bahl et al., 2011; Cheng et al., 2013). Our phylogenetic analysis shows that the viruses from Hong Kong were closer to the global putative source virus pool than the viruses from Shenzhen, and that the genetic distance did not change significantly over time. These findings suggest that, in general, Hong Kong is better connected to the rest of the world than Shenzhen and might therefore be more important as a hub for the global dynamics of influenza virus. However, with increasing population mixing between these two cities and, to a lesser extent, between Shenzhen and the rest of the world, Shenzhen has become more closely linked to the global dynamics of influenza virus.
Our study had several limitations. There were more influenza diagnostic tests performed in Hong Kong during the study period and hence a larger number of viral sequences were available from this city. It is possible that sampling bias in Shenzhen could have influenced our results if subpopulations that were not representative of the general population were tested preferentially, such as specific age or risk groups. We did not have demographic information available to evaluate this possibility, although to date there has been no indication that the genetic makeup of influenza viruses varies by age or severity status (Lavenu et al., 2006). Similarly, the number of diagnostic tests and sampling in Shenzhen increased dramatically during the study period, perhaps increasing the chances of detecting virus strains that are closer to the trunk of the global phylogenetic tree. As a sensitivity analysis, we repeatedly ‘downsampled’ the data from Hong Kong to determine the probability that the observed minimum distance from the tree trunk observed in Shenzhen could arise by chance as a result of the relatively small sample size. This was not the case. Furthermore, the isolation month was unavailable for Shenzhen sequences, preventing us from controlling for potential genetic variations in the virus within a calendar year. To test whether precise isolation date was important, we performed a sensitivity analysis of distance from the trunk using Hong Kong data stratified by season. We did not find any significant relationship between the time of year and distance from the trunk. Taken together, these well-established downsampling methods and sensitivity analyses confirm that our findings are not artefacts of uneven sampling but instead truly reflect differences in disease dynamics between cities and over time. An additional limitation of our study relates to the comparison of a variety of datasets from different sources, which were not consistently available throughout the entire study period, so that we had to consider slightly different time periods in different analyses. It is particularly unfortunate that publicly available influenza sequence data were very sparse for Hong Kong after 2006, precluding detailed analyses of most recent years. A final limitation is the lack of information on travel statistics by air and boat, and on population mixing between Shenzhen and other regions of the world, which could also affect influenza dynamics in Shenzhen. We strongly suspect, however, that population travel has increased in Shenzhen across the board in recent years.
In conclusion, we demonstrated that the seasonal patterns, subtype distributions and genetic distances of influenza isolates in the adjacent cities of Shenzhen and Hong Kong show important differences but have become more similar in recent years. This pattern echoes historical studies of influenza epidemics in Iceland, indicating increasing synchrony with epidemics in the USA and Denmark as air travel to and from the island increased over the 20th century (Weinberger et al., 2012). In contrast, there was no trend in influenza epidemic synchrony within the USA as domestic travel increased by 300 % between 1972 and 2002 (Viboud et al., 2006b). Overall, the relationship between population mixing and influenza dynamics has not been fully elucidated, especially with regard to the impact on influenza seasonality in the subtropics. More studies from less connected regions of South America, Africa and Asia are needed before the global drivers of influenza dynamics can be fully understood. Finally, our study carries important public health implications. Our results suggest that, with increasing global population mixing, new influenza variants could disseminate more quickly in subtropical regions where environmental fluctuations are weaker, potentially diminishing the short window of time available for preparation of routine influenza vaccines.
Methods
Data sources.
For the sake of simplicity, we have used the term ‘city’ to qualify Hong Kong SAR (Hong Kong) and Shenzhen SEZ (Shenzhen) in the remainder of the text. Both Hong Kong and Shenzhen have conducted influenza surveillance and sequencing since the 1990s, and the travel data between these two cities is available from January 2001. In this study, we collected epidemiological data, molecular data and travel data from both cities for as many years as possible. Our study period ended in June 2009, before the major peak of 2009 pandemic A/H1N1 activity in southern China, in order to concentrate on inter-pandemic influenza dynamics. Because we analysed a variety of datasets from different sources, data availability dictated that we use slightly different time periods throughout the paper. The epidemiological and molecular datasets have been described in detail previously (Cheng et al., 2013; Yang et al., 2008) and are summarized briefly here.
Influenza epidemiological data.
Between January 1997 and June 2009, influenza laboratory surveillance data from Hong Kong were obtained from the Department of Health and accessed through the Centre for Health Protection website (http://www.chp.gov.hk). Nasopharyngeal swabs were collected from influenza-like illness patients visiting general practitioners or outpatient clinics, and from patients hospitalized with acute respiratory diseases (Yang et al., 2008). Respiratory specimens were cultured for haemagglutination and PCR tests.
Monthly influenza laboratory surveillance data were obtained from the Shenzhen Center for Disease Control between January 1997 and June 2009. In Shenzhen, nasopharyngeal swabs were collected from outpatients and patients hospitalized with upper respiratory infections (before 2003) or influenza-like illness (after 2003) and cultured for haemagglutination and PCR tests (Cheng et al., 2013).
In total, there were 329 094 respiratory specimens tested in Hong Kong and 21 768 specimens tested in Shenzhen during the study period. Influenza genotyping has been performed in both Hong Kong and Shenzhen since 2003. The influenza virus samples used in this study were collected as part of an ongoing and nationally approved influenza surveillance programme. As no patient data were used in this study, written consent was not required.
Influenza virus sequence data.
Publicly available influenza haemagglutinin (HA) nucleotide sequences were obtained from the NCBI influenza website, following a previous phylogenetic study of influenza in Shenzhen, southern China (Cheng et al., 2013). We extracted all influenza HA sequences from Hong Kong sampled between 1995 and 2009. The great majority of Hong Kong sequences, however, were from 2006 and there were too few sequences for analysis of evolutionary patterns in 2007–2009 (n<5 annually). All Hong Kong sequences were provided with the month and year of isolation (Cheng et al., 2013). For the Shenzhen isolates, collection dates were between 1995 and 2009 with only the year of isolation available (Cheng et al., 2013).
Travel data.
Publicly accessible information about the monthly number of mainland Chinese visitor arrivals to Hong Kong by land during January 2001 and June 2009 and the monthly number of Hong Kong residents travelling to mainland China by land during July 2004 and June 2009 were downloaded from the open-access website of the Hong Kong Tourism Board (Hong Kong Tourism Board, 2013). Because Shenzhen is situated immediately north of Hong Kong and going through Shenzhen is the only way to travel by land between Hong Kong and mainland China, these data reflect the actual level of population mixing between the two cities.
Statistical approaches
Epidemic timing.
We compiled monthly proxies of influenza virus incidence in Shenzhen and Hong Kong, calculated as the monthly number of influenza virus-positive specimens divided by the monthly number of respiratory specimens tested. Time series were square-root transformed prior to analysis to stabilize the variance and account for different surveillance intensities in these two cities. We used wavelet analysis to estimate epidemic timing, an approach that has been used previously to measure synchrony in influenza activity between different locations or between different incidence proxies (Grenfell et al., 2001; Viboud et al., 2006c; Weinberger et al., 2012; Yang et al., 2008). As a measure of epidemic synchrony between Shenzhen and Hong Kong, we estimated the monthly phase angle difference in wavelet-reconstructed time series, after extraction of the main annual cycle (0.8–1.2 year periods; Morlet continuous wavelet) (Grenfell et al., 2001). We also used the global wavelet spectrum to estimate the amplitude of annual and semi-annual epidemic cycles in each city (r package, developed by Johansson et al., 2009).
Phylogenetic analysis.
All genetic analyses of influenza virus in Shenzhen and Hong Kong described below focused on the A/H3N2 subtype, which was the most prevalent during the study period in both cities, and for which background global sequences are more widely available. All available HA sequences of human influenza A/H3N2 viruses were subject to phylogeny reconstruction by a maximum-likelihood (ML) method implemented in RAxML (version 7.2.8), using the general-time-reversal model of nucleotide substitution with a gamma distribution (Γ) of among-site rate variation (Stamatakis, 2006). A total of 200 independent searches for ML phylogenies were performed with different random maximum-parsimony starting trees and the phylogeny with the highest likelihood score was selected for further analysis (Cheng et al., 2013).
Measures of genetic distance.
Genetic distance between the influenza A/H3N2 viruses in Shenzhen and Hong Kong was determined from the mean pairwise nucleotide distances between HA genes sampled in the two cities each year from 1995 to 2006. Linear regression analysis was performed to estimate the trend in genetic distance over time.
The trunk of the phylogeny of influenza A/H3N2 viruses theoretically represents the putative source for global epidemics (Cheng et al., 2013). Hence, the shortest genetic distance to the trunk was calculated separately for the Shenzhen and Hong Kong viruses in each year in order to determine their respective proximity to the putative source. We then evaluated time trends in shortest genetic distance estimates.
Because there were sampling differences between the two cities, most notably more sequences were available from Hong Kong and the temporal resolution of sequence data were more refined in this city, we evaluated the sensitivity of our results to sampling intensity. First, we stratified Hong Kong estimates of shortest genetic distance to the trunk by semester, considering extended winter (October–March) and summer (April–September) seasons. We then tested: (i) whether the summer estimates were different from the winter estimates by paired t-test, and (ii) whether there was a significant change over time in summer and winter estimates by linear regression. Secondly, the estimates for the shortest distance to the trunk were evaluated to determine whether they were influenced by the number of isolates tested in each city and each year. To do this, we generated 1000 random subsets of the Hong Kong data equivalent in size to the smaller Shenzhen dataset, separately for each year. Next, we tested changes over time in distance estimates derived from these downsampled Hong Kong data. Statistical analyses were performed in r version 2.15.3 (R Development Core Team, 2013).
Acknowledgements
This research was conducted in the context of the MISMS Study, an ongoing international collaborative effort to understand influenza epidemiological and evolutionary patterns, led by the Fogarty International Center, National Institutes of Health (http://www.origem.info/misms/index.php). The MISMS study is funded by the International Influenza Unit, Office of Global Affairs, US Department of Health and Human Services. The Department of Health, Hong Kong, is acknowledged for access to the laboratory surveillance data from Hong Kong. The authors declare no conflicts of interest.
Footnotes
Two supplementary figures are available with the online version of this paper.
References
- Alonso W. J., Viboud C., Simonsen L., Hirano E. W., Daufenbach L. Z., Miller M. A. (2007). Seasonality of influenza in Brazil: a traveling wave from the Amazon to the subtropics. Am J Epidemiol 165, 1434–1442 10.1093/aje/kwm012 [DOI] [PubMed] [Google Scholar]
- Azziz Baumgartner E., Dao C. N., Nasreen S., Bhuiyan M. U., Mah-E-Muneer S., Al Mamun A., Sharker M. A., Uz Zaman R., Cheng P. Y. & other authors (2012). Seasonality, timing, and climate drivers of influenza activity worldwide. J Infect Dis 206, 838–846 10.1093/infdis/jis467 [DOI] [PubMed] [Google Scholar]
- Bahl J., Nelson M. I., Chan K. H., Chen R., Vijaykrishna D., Halpin R. A., Stockwell T. B., Lin X., Wentworth D. E. & other authors (2011). Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc Natl Acad Sci U S A 108, 19359–19364 10.1073/pnas.1109314108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Cobey S., Beerli P., Pascual M. (2010). Global migration dynamics underlie evolution and persistence of human influenza A (H3N2). PLoS Pathog 6, e1000918 10.1371/journal.ppat.1000918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom-Feshbach K., Alonso W. J., Charu V., Tamerius J., Simonsen L., Miller M. A., Viboud C. (2013). Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): a global comparative review. PLoS ONE 8, e54445 10.1371/journal.pone.0054445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. K., Mok H. Y., Lee T. C., Chu I. M., Lam W. Y., Sung J. J. (2009). Seasonal influenza activity in Hong Kong and its association with meteorological variations. J Med Virol 81, 1797–1806 10.1002/jmv.21551 [DOI] [PubMed] [Google Scholar]
- Cheng X., Tan Y., He M., Lam T. T., Lu X., Viboud C., He J., Zhang S., Lu J. & other authors (2013). Epidemiological dynamics and phylogeography of influenza virus in southern China. J Infect Dis 207, 106–114 10.1093/infdis/jis526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebel J., Norda J. (2013a). Average weather for Chek Lap Kok, Hong Kong. [Accessed 31 October 2013] http://weatherspark.com/averages/33929/Chek-Lap-Kok-Kowloon-Hong-Kong [Google Scholar]
- Diebel J., Norda J. (2013b). Average weather for Shenzhen, China. [Accessed 31 October 2013] http://weatherspark.com/averages/34108/Shenzhen-Guangdong-China [Google Scholar]
- Finkelman B. S., Viboud C., Koelle K., Ferrari M. J., Bharti N., Grenfell B. T. (2007). Global patterns in seasonal activity of influenza A/H3N2, A/H1N1, and B from 1997 to 2005: viral coexistence and latitudinal gradients. PLoS ONE 2, e1296 10.1371/journal.pone.0001296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garske T., Yu H., Peng Z., Ye M., Zhou H., Cheng X., Wu J., Ferguson N. (2011). Travel patterns in China. PLoS ONE 6, e16364 10.1371/journal.pone.0016364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell B. T., Bjørnstad O. N., Kappey J. (2001). Travelling waves and spatial hierarchies in measles epidemics. Nature 414, 716–723 10.1038/414716a [DOI] [PubMed] [Google Scholar]
- Hong Kong Tourism Board (2013). Visitor arrival statistics. January 2002–December 2012. [Accessed 22 April 2013] https://securepartnernet.hktb.com/en/my_partnernet/index.html?reqlogin=1&themeClassName=research_statistic [Google Scholar]
- Johansson M. A., Cummings D. A., Glass G. E. (2009). Multiyear climate variability and dengue–El Niño southern oscillation, weather, and dengue incidence in Puerto Rico, Mexico, and Thailand: a longitudinal data analysis. PLoS Med 6, e1000168 10.1371/journal.pmed.1000168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenu A., Leruez-Ville M., Chaix M. L., Boelle P. Y., Rogez S., Freymuth F., Hay A., Rouzioux C., Carrat F. (2006). Detailed analysis of the genetic evolution of influenza virus during the course of an epidemic. Epidemiol Infect 134, 514–520 10.1017/S0950268805005686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee V. J., Yap J., Ong J. B., Chan K. P., Lin R. T., Chan S. P., Goh K. T., Leo Y. S., Chen M. I. (2009). Influenza excess mortality from 1950–2000 in tropical Singapore. PLoS ONE 4, e8096 10.1371/journal.pone.0008096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Viboud C. (2009). Influenza seasonality: lifting the fog. Proc Natl Acad Sci U S A 106, 3645–3646 10.1073/pnas.0900933106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen A., Palese P. (2009). Transmission of influenza virus in temperate zones is predominantly by aerosol, in the tropics by contact: a hypothesis. PLoS Curr 1, RRN1002 10.1371/currents.RRN1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2013). r: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2010; [Accessed 30 April 2013] http://www.R-project.org/ [Google Scholar]
- Rambaut A., Pybus O. G., Nelson M. I., Viboud C., Taubenberger J. K., Holmes E. C. (2008). The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 10.1038/nature06945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger B. (1997). Hong Kong: the Road to 1997. Cambridge: Cambridge University Press [Google Scholar]
- Russell C. A., Jones T. C., Barr I. G., Cox N. J., Garten R. J., Gregory V., Gust I. D., Hampson A. W., Hay A. J. & other authors (2008). The global circulation of seasonal influenza A (H3N2) viruses. Science 320, 340–346 10.1126/science.1154137 [DOI] [PubMed] [Google Scholar]
- Shaman J., Pitzer V. E., Viboud C., Grenfell B. T., Lipsitch M. (2010). Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol 8, e1000316 10.1371/journal.pbio.1000316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Tamerius J., Nelson M. I., Zhou S. Z., Viboud C., Miller M. A., Alonso W. J. (2011). Global influenza seasonality: reconciling patterns across temperate and tropical regions. Environ Health Perspect 119, 439–445 10.1289/ehp.1002383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamerius J. D., Shaman J., Alonso W. J., Bloom-Feshbach K., Uejio C. K., Comrie A., Viboud C. (2013). Environmental predictors of seasonal influenza epidemics across temperate and tropical climates. PLoS Pathog 9, e1003194 10.1371/journal.ppat.1003194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Government of the Hong Kong Special Administrative Region (2013). Individual visit scheme. [Accessed 22 April 2013] http://www.tourism.gov.hk/english/visitors/visitors_ind.html [Google Scholar]
- Turbelin C., Souty C., Pelat C., Hanslik T., Sarazin M., Blanchon T., Falchi A. (2013). Age distribution of influenza like illness cases during post-pandemic A(H3N2): comparison with the twelve previous seasons, in France. PLoS ONE 8, e65919 10.1371/journal.pone.0065919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C., Boëlle P. Y., Pakdaman K., Carrat F., Valleron A. J., Flahault A. (2004). Influenza epidemics in the United States, France, and Australia, 1972–1997. Emerg Infect Dis 10, 32–39 10.3201/eid1001.020705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C., Alonso W. J., Simonsen L. (2006a). Influenza in tropical regions. PLoS Med 3, e89 10.1371/journal.pmed.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C., Miller M. A., Grenfell B. T., Bjørnstad O. N., Simonsen L. (2006b). Air travel and the spread of influenza: important caveats. PLoS Med 3, e503, author reply e502. [Erratum PLoS Med 2007 4, e32.] 10.1371/journal.pmed.0030503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud C., Bjørnstad O. N., Smith D. L., Simonsen L., Miller M. A., Grenfell B. T. (2006c). Synchrony, waves, and spatial hierarchies in the spread of influenza. Science 312, 447–451 10.1126/science.1125237 [DOI] [PubMed] [Google Scholar]
- Waicharoen S., Thawatsupha P., Chittaganpitch M., Maneewong P., Thanadachakul T., Sawanpanyalert P. (2008). Influenza viruses circulating in Thailand in 2004 and 2005. Jpn J Infect Dis 61, 321–323 [PubMed] [Google Scholar]
- Weinberger D. M., Krause T. G., Mølbak K., Cliff A., Briem H., Viboud C., Gottfredsson M. (2012). Influenza epidemics in Iceland over 9 decades: changes in timing and synchrony with the United States and Europe. Am J Epidemiol 176, 649–655 10.1093/aje/kws140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Wong C. M., Lau E. H., Chan K. P., Ou C. Q., Peiris J. S. (2008). Synchrony of clinical and laboratory surveillance for influenza in Hong Kong. PLoS ONE 3, e1399 10.1371/journal.pone.0001399 [DOI] [PMC free article] [PubMed] [Google Scholar]