Abstract
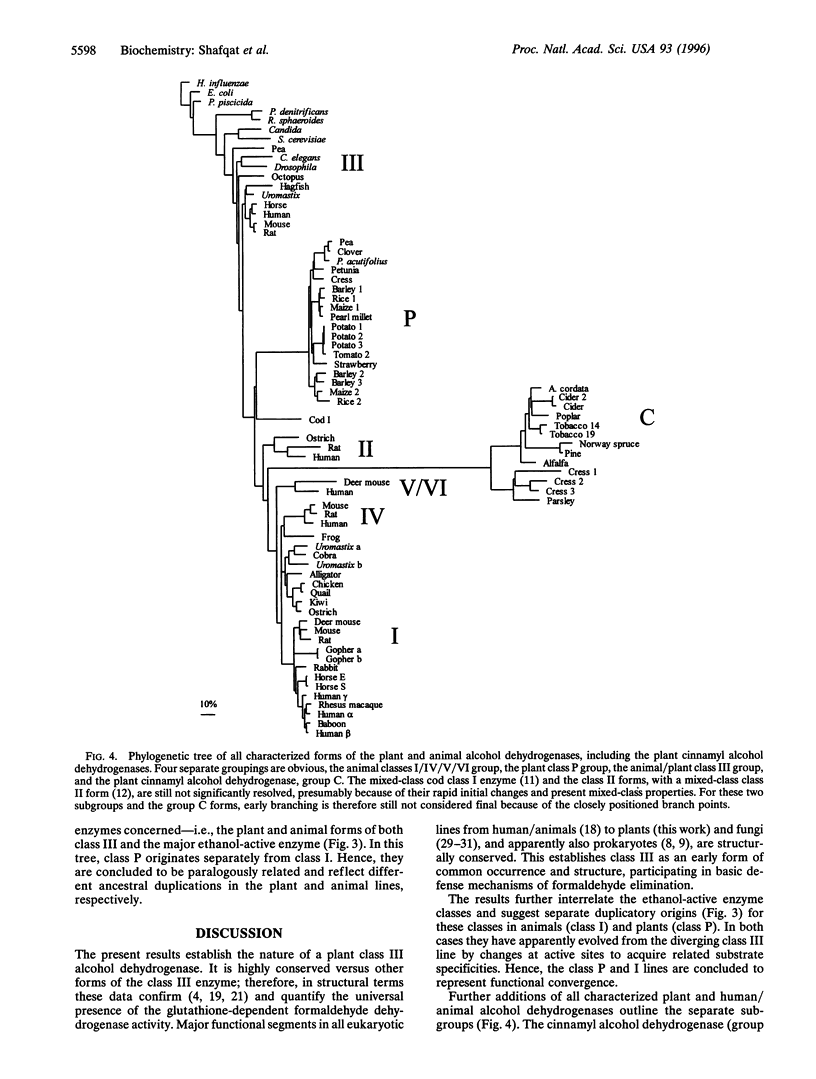
A plant class III alcohol dehydrogenase (or glutathione-dependent formaldehyde dehydrogenase) has been characterized. The enzyme is a typical class III member with enzymatic parameters and substrate specificity closely related to those of already established animal forms. Km values with the pea enzyme are 6.5 microM for NAD+, 2 microM for S-hydroxymethylglutathione, and 840 microM for octanol versus 9, 4, and 1200 microM, respectively, with the human enzyme. Structurally, the pea/human class III enzymes are closely related, exhibiting a residue identity of 69% and with only 3 of 23 residues differing among those often considered in substrate and coenzyme binding. In contrast, the corresponding ethanol-active enzymes, the long-known human liver and pea alcohol dehydrogenases, differ more (47% residue identities) and are also in functionally important active site segments, with 12 of the 23 positions exchanged, including no less than 7 at the usually much conserved coenzyme-binding segment. These differences affect functionally important residues that are often class-distinguishing, such as those at positions 48, 51, and 115, where the plant ethanol-active forms resemble class III (Thr, Tyr, and Arg, respectively) rather than the animal ethanol-active class I forms (typically Ser, His, and Asp, respectively). Calculations of phylogenetic trees support the conclusions from functional residues in subgrouping plant ethanol-active dehydrogenases and the animal ethanol-active enzymes (class I) as separate descendants from the class III line. It appears that the classical plant alcohol dehydrogenases (now called class P) have a duplicatory origin separate from that of the animal class I enzymes and therefore a paralogous relationship with functional convergence of their alcohol substrate specificity. Combined, the results establish the conserved nature of class III also in plants, and contribute to the molecular and functional understanding of alcohol dehydrogenases by defining two branches of plant enzymes into the system.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brändén C. I., Eklund H., Cambillau C., Pryor A. J. Correlation of exons with structural domains in alcohol dehydrogenase. EMBO J. 1984 Jun;3(6):1307–1310. doi: 10.1002/j.1460-2075.1984.tb01967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederlund E., Peralba J. M., Parés X., Jörnvall H. Amphibian alcohol dehydrogenase, the major frog liver enzyme. Relationships to other forms and assessment of an early gene duplication separating vertebrate class I and class III alcohol dehydrogenases. Biochemistry. 1991 Mar 19;30(11):2811–2816. doi: 10.1021/bi00225a011. [DOI] [PubMed] [Google Scholar]
- Danielsson O., Atrian S., Luque T., Hjelmqvist L., Gonzàlez-Duarte R., Jörnvall H. Fundamental molecular differences between alcohol dehydrogenase classes. Proc Natl Acad Sci U S A. 1994 May 24;91(11):4980–4984. doi: 10.1073/pnas.91.11.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsson O., Eklund H., Jörnvall H. The major piscine liver alcohol dehydrogenase has class-mixed properties in relation to mammalian alcohol dehydrogenases of classes I and III. Biochemistry. 1992 Apr 21;31(15):3751–3759. doi: 10.1021/bi00130a004. [DOI] [PubMed] [Google Scholar]
- Danielsson O., Jörnvall H. "Enzymogenesis": classical liver alcohol dehydrogenase origin from the glutathione-dependent formaldehyde dehydrogenase line. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9247–9251. doi: 10.1073/pnas.89.19.9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund H., Müller-Wille P., Horjales E., Futer O., Holmquist B., Vallee B. L., Hög J. O., Kaiser R., Jörnvall H. Comparison of three classes of human liver alcohol dehydrogenase. Emphasis on different substrate binding pockets. Eur J Biochem. 1990 Oct 24;193(2):303–310. doi: 10.1111/j.1432-1033.1990.tb19337.x. [DOI] [PubMed] [Google Scholar]
- Eklund H., Samama J. P., Jones T. A. Crystallographic investigations of nicotinamide adenine dinucleotide binding to horse liver alcohol dehydrogenase. Biochemistry. 1984 Dec 4;23(25):5982–5996. doi: 10.1021/bi00320a014. [DOI] [PubMed] [Google Scholar]
- Engeland K., Hög J. O., Holmquist B., Estonius M., Jörnvall H., Vallee B. L. Mutation of Arg-115 of human class III alcohol dehydrogenase: a binding site required for formaldehyde dehydrogenase activity and fatty acid activation. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2491–2494. doi: 10.1073/pnas.90.6.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estonius M., Hög J. O., Danielsson O., Jörnvall H. Residues specific for class III alcohol dehydrogenase. Site-directed mutagenesis of the human enzyme. Biochemistry. 1994 Dec 20;33(50):15080–15085. doi: 10.1021/bi00254a017. [DOI] [PubMed] [Google Scholar]
- Fernández M. R., Biosca J. A., Norin A., Jörnvall H., Parés X. Class III alcohol dehydrogenase from Saccharomyces cerevisiae: structural and enzymatic features differ toward the human/mammalian forms in a manner consistent with functional needs in formaldehyde detoxication. FEBS Lett. 1995 Aug 14;370(1-2):23–26. doi: 10.1016/0014-5793(95)00788-b. [DOI] [PubMed] [Google Scholar]
- Grima-Pettenati J., Feuillet C., Goffner D., Borderies G., Boudet A. M. Molecular cloning and expression of a Eucalyptus gunnii cDNA clone encoding cinnamyl alcohol dehydrogenase. Plant Mol Biol. 1993 Mar;21(6):1085–1095. doi: 10.1007/BF00023605. [DOI] [PubMed] [Google Scholar]
- Gutheil W. G., Holmquist B., Vallee B. L. Purification, characterization, and partial sequence of the glutathione-dependent formaldehyde dehydrogenase from Escherichia coli: a class III alcohol dehydrogenase. Biochemistry. 1992 Jan 21;31(2):475–481. doi: 10.1021/bi00117a025. [DOI] [PubMed] [Google Scholar]
- Hjelmqvist L., Hackett M., Shafqat J., Danielsson O., Iida J., Hendrickson R. C., Michel H., Shabanowitz J., Hunt D. F., Jörnvall H. Multiplicity of N-terminal structures of medium-chain alcohol dehydrogenases. Mass-spectrometric analysis of plant, lower vertebrate and higher vertebrate class I, II, and III forms of the enzyme. FEBS Lett. 1995 Jul 3;367(3):237–240. doi: 10.1016/0014-5793(95)00572-q. [DOI] [PubMed] [Google Scholar]
- Holmquist B., Moulis J. M., Engeland K., Vallee B. L. Role of arginine 115 in fatty acid activation and formaldehyde dehydrogenase activity of human class III alcohol dehydrogenase. Biochemistry. 1993 May 18;32(19):5139–5144. doi: 10.1021/bi00070a024. [DOI] [PubMed] [Google Scholar]
- Hurley T. D., Bosron W. F., Hamilton J. A., Amzel L. M. Structure of human beta 1 beta 1 alcohol dehydrogenase: catalytic effects of non-active-site substitutions. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8149–8153. doi: 10.1073/pnas.88.18.8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H., Hög J. O. Nomenclature of alcohol dehydrogenases. Alcohol Alcohol. 1995 Mar;30(2):153–161. [PubMed] [Google Scholar]
- Jörnvall H., Persson B., Jeffery J. Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenases. Eur J Biochem. 1987 Sep 1;167(2):195–201. doi: 10.1111/j.1432-1033.1987.tb13323.x. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., Ghosh D. Short-chain dehydrogenases/reductases (SDR). Biochemistry. 1995 May 9;34(18):6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- Kaiser R., Fernández M. R., Parés X., Jörnvall H. Origin of the human alcohol dehydrogenase system: implications from the structure and properties of the octopus protein. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11222–11226. doi: 10.1073/pnas.90.23.11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat V., Kennedy K., Feuillet C., McKie J. H., Gorrichon L., Baltas M., Boudet A. M., Grima-Pettenati J., Douglas K. T. Site-directed mutagenesis of a serine residue in cinnamyl alcohol dehydrogenase, a plant NADPH-dependent dehydrogenase, affects the specificity for the coenzyme. Biochemistry. 1995 Sep 26;34(38):12426–12434. doi: 10.1021/bi00038a041. [DOI] [PubMed] [Google Scholar]
- Lutz R. A., Bull C., Rodbard D. Computer analysis of enzyme-substrate-inhibitor kinetic data with automatic model selection using IBM-PC compatible microcomputers. Enzyme. 1986;36(3):197–206. doi: 10.1159/000469292. [DOI] [PubMed] [Google Scholar]
- McKie J. H., Jaouhari R., Douglas K. T., Goffner D., Feuillet C., Grima-Pettenati J., Boudet A. M., Baltas M., Gorrichon L. A molecular model for cinnamyl alcohol dehydrogenase, a plant aromatic alcohol dehydrogenase involved in lignification. Biochim Biophys Acta. 1993 Sep 3;1202(1):61–69. doi: 10.1016/0167-4838(93)90063-w. [DOI] [PubMed] [Google Scholar]
- Moulis J. M., Holmquist B., Vallee B. L. Hydrophobic anion activation of human liver chi chi alcohol dehydrogenase. Biochemistry. 1991 Jun 11;30(23):5743–5749. doi: 10.1021/bi00237a016. [DOI] [PubMed] [Google Scholar]
- Parés X., Cederlund E., Moreno A., Hjelmqvist L., Farrés J., Jörnvall H. Mammalian class IV alcohol dehydrogenase (stomach alcohol dehydrogenase): structure, origin, and correlation with enzymology. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1893–1897. doi: 10.1073/pnas.91.5.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson B., Zigler J. S., Jr, Jörnvall H. A super-family of medium-chain dehydrogenases/reductases (MDR). Sub-lines including zeta-crystallin, alcohol and polyol dehydrogenases, quinone oxidoreductase enoyl reductases, VAT-1 and other proteins. Eur J Biochem. 1994 Nov 15;226(1):15–22. doi: 10.1111/j.1432-1033.1994.tb20021.x. [DOI] [PubMed] [Google Scholar]
- Plapp B. V., Ganzhorn A. J., Gould R. M., Green D. W., Jacobi T., Warth E., Kratzer D. A. Catalysis by yeast alcohol dehydrogenase. Adv Exp Med Biol. 1991;284:241–251. doi: 10.1007/978-1-4684-5901-2_26. [DOI] [PubMed] [Google Scholar]
- Ras J., Van Ophem P. W., Reijnders W. N., Van Spanning R. J., Duine J. A., Stouthamer A. H., Harms N. Isolation, sequencing, and mutagenesis of the gene encoding NAD- and glutathione-dependent formaldehyde dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in which GD-FALDH is essential for methylotrophic growth. J Bacteriol. 1995 Jan;177(1):247–251. doi: 10.1128/jb.177.1.247-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasnauskas K., Jomantiene R., Januska A., Lebediene E., Lebedys J., Janulaitis A. Cloning and analysis of a Candida maltosa gene which confers resistance to formaldehyde in Saccharomyces cerevisiae. Gene. 1992 Dec 1;122(1):207–211. doi: 10.1016/0378-1119(92)90052-q. [DOI] [PubMed] [Google Scholar]
- Shafqat J., Hjelmqvist L., Jörnvall H. Liver class-I alcohol dehydrogenase isozyme relationships and constant patterns in a variable basic structure. Distinctions from characterization of an ethanol dehydrogenase in cobra, Naja naja. Eur J Biochem. 1996 Mar 1;236(2):571–578. doi: 10.1111/j.1432-1033.1996.00571.x. [DOI] [PubMed] [Google Scholar]
- Sun H. W., Plapp B. V. Progressive sequence alignment and molecular evolution of the Zn-containing alcohol dehydrogenase family. J Mol Evol. 1992 Jun;34(6):522–535. doi: 10.1007/BF00160465. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uotila L., Koivusalo M. Purification of formaldehyde and formate dehydrogenases from pea seeds by affinity chromatography and S-formylglutathione as the intermediate of formaldehyde metabolism. Arch Biochem Biophys. 1979 Aug;196(1):33–45. doi: 10.1016/0003-9861(79)90548-4. [DOI] [PubMed] [Google Scholar]
- Wehner E. P., Rao E., Brendel M. Molecular structure and genetic regulation of SFA, a gene responsible for resistance to formaldehyde in Saccharomyces cerevisiae, and characterization of its protein product. Mol Gen Genet. 1993 Mar;237(3):351–358. doi: 10.1007/BF00279438. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Harry D. E. Molecular phylogeny and evolutionary rates of alcohol dehydrogenases in vertebrates and plants. Mol Biol Evol. 1993 Nov;10(6):1215–1226. doi: 10.1093/oxfordjournals.molbev.a040073. [DOI] [PubMed] [Google Scholar]



