Abstract
SARS-CoV from the coronaviridae family has been identified as the etiological agent of Severe Acute Respiratory Syndrome (SARS), a highly contagious upper respiratory disease that reached epidemic status in 2002. SARS-3CLpro, a cysteine protease indispensible to the viral life cycle, has been identified as one of the key therapeutic target against SARS. A combined ligand and structure based virtual screening was carried out against the Asinex Platinum collection. Multiple low micromolar inhibitors of the enzyme were identified through this search, one of which also showed activity against SARS-CoV in a whole cell CPE assay. Furthermore, multi nanosecond explicit solvent simulations were carried out using the docking poses of the identified hits to study the overall stability of the binding site interactions as well as identify important changes in the interaction profile that were not apparent from the docking study. Cumulative analysis of the evaluated compounds and the simulation studies led to the identification of certain protein-ligand interaction patterns which would be useful in further structure based design efforts.
INTRODUCTION
SARS-CoV (Severe acute respiratory syndrome-coronavirus) is a previously unidentified virus belonging to the coronaviridae family which has been recognized as the etiological agent for SARS, a highly infective upper respiratory tract disease. The disease was first diagnosed in the winter of 2002 amongst patients in the remote Guangdong province of southern China. This highly infectious disease with ~10% mortality rate, quickly reached pandemic status, spreading to over 37 countries worldwide and causing over ~9000 infections1–5. The initial symptoms of the disease are similar to those of flu and include cough, sore throat, respiratory distress, myalgia and fever. The symptoms of the disease may appear up to 10–13 days from the day of exposure and transmission may occur through sexual or non sexual contact with an infected person or by traveling to a region identified for local transmission of the disease. At the peak of the outbreak, a number of antiviral treatments designed against other viral diseases such as AIDS, Influenza and Hepatitis were evaluated for efficacy against the SARS-CoV infection. Reported treatments for patients infected with SARS involved isolation and administration of antipyretics, steroids and antivirals such as Ribavarin6–8. Although the initial outbreak of the virus was stymied about a year from its first discovery, recent findings report the isolation of SARS-CoV from animals including Chinese horse shoe bats9–11. They act as natural reservoirs for this virus and point to the possibility of an animal to human transmission of the wild type or mutated variant of SARS-CoV or other closely related corona viruses in the near future.
The imminent threat from this disease led to the discovery of a number of viral proteins12 which could be utilized as possible targets for the development of antiviral therapy. Historical evidence on antiviral drug discovery points towards a number of proteases13–15, critical to the viral replication cycle, that have been successfully targeted. Prominent examples include the viral protease from HIV (human immuno deficiency virus)16, 17, NS3/4A protease from Hepatitis C virus18, HSV (herpes simplex virus) protease19 and the rhinovirus protease20. Within the viral replication cycle, proteases are primarily utilized to either (a) process high molecular weight precursor proteins to produce functional proteins or (b) process structural proteins which are required for the morphogenesis and assembly of viral particles. Within the coronaviridae family, three proteases are usually utilized to fulfill these functional roles. The SARS-CoV differs from the rest of the members of the coronaviridae family by using only two proteases to accomplish these functions21–23. These include the PLP2pro 24, a Papain-like cysteine protease as well as the 3CLpro25–30, a Chymotrypsin-like cysteine protease also called as the main protease Mpro. In SARS-CoV, the 3CLpro enzyme is responsible for the processing of two large replicase polyproteins pp1a (~450 kDa) and pp1ab (~750 kDa) which function in the viral replication and transcription processes. Its integral role in the SARS-CoV lifecycle and the presence of a number of apo and ligand bound structures31–42 makes it an ideal target for structure-based drug discovery. Figure 1a shows the binding site of SARS-3CLpro with an irreversible peptidic inhibitor (PDB code – 2AMD) bound to it, forming a covalent Michael adduct with the catalytic cysteine (Cys145, shown in yellow). The S1’, S2 and S4 sites, all require hydrophobic groups (S1’ – ethyl, S2 – Leucine, S4 – Valine respectively in this case) of various flavors. In the natural substrate, the S1 site binds a Glutamine residue and is by far the most selective requirement. In this case, a cyclized glutamine derivative forms the key hydrogen bond through its side chain carbonyl with His163 (shown in blue).
Figure 1.
(a) Binding site of SARS-3CLpro (PDB code – 2AMD) showing a covalent peptidic inhibitor. The yellow and blue surface coloring refers to the position of Cys145 and His 163 respectively. (b) A representative set of small molecule SARS-3CLpro inhibitors reported in recent literature.
Current drug design efforts43, 44 against this protease can be classified primarily into two categories: Peptidic molecules with reactive warheads45 and non-peptidic small molecule inhibitors. The peptidic ligands usually involves a sequence of amino acids mimicking the natural substrate of the enzyme attached at the terminus to a “warhead” group: Aldehyde39, 41, Michael acceptors35, 41, 46, epoxy ketone47, halo-methyl ketones48 etc. These inhibitors act through a two step procedure45. First, the ligand forms a non-covalent Michaelis complex, placing the warhead group in close proximity to the catalytic cysteine. The second step involves an attack by the nucleophilic thiolate anion of the catalytic cysteine onto the reactive atom of the warhead group, forming a covalent adduct that inactivates the enzyme. The non-peptidic small molecule inhibitors form a non-covalent complex by competitively displacing the enzyme’s substrate from the binding site and preventing enzyme processing. Small molecule inhibitors of SARS-3CLpro have been identified through in-silico approaches such as pharmacophore searching49, structure-based virtual screening36, 46, 50, 51, structure-based discovery11, 35, 48, 52–54 as well as high throughput screening applications55, 56. While readers may refer to more extensive reviews57–59 on the inhibitor discovery against SARS-CoV, Figure 1b summarizes some of the inhibitor classes56, 60–66 that have been reported within the last couple of years and points towards the high level of research interest around this prominent antiviral target.
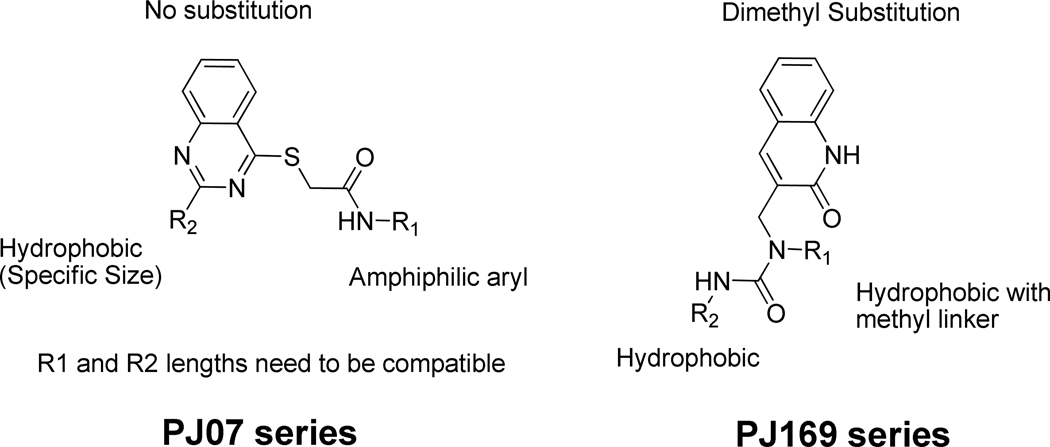
Previously, we have reported a structure-based virtual screening effort67 to identify small molecule inhibitors of the SARS-3CLpro enzyme. The effort resulted in the identification of two inhibitors PJ07 and PJ169 (Figure 1b) of micro molar potency (PJ07 IC50 18.2 µM, PJ169 IC50 17.2 µM), originating from novel chemical scaffolds. The current effort focuses on exploring the chemical space around these scaffolds from commercially available compound collections. The results provide insights into the protein-ligand interaction patterns within these scaffolds, establishing them for synthetic modification efforts and potential development towards leads against SARS-CoV.
We implemented a combined ligand and structure based protocol to carry out this screening exercise. Our aim was to evaluate the effects of diverse substitution patterns on the biological activity while preserving the core scaffolds of the identified hits. Therefore, substructure searching became the method of choice as the ligand based screening filter. Substructure searching allows the user to put specific constraints on the nature of the search results by querying for the presence of given substructure(s) within the identified hits and systematically exploring the activity landscape based on variations to a specific region of the chemical series in question while keeping the other regions defined by the substructure, constant. This methodology has seen a number of uses starting from screening of chemical databases68, 69, sequential filters as part of a more complex screening cascade70, 71, exploration of immediate chemical space around identified hits72 as well as lead optimization and synthesis73, 74.
The rationally curtailed dataset was put through a set of cascading filters involving (a) docking using a validated protocol and (b) ranking based on a combination of docking pose based descriptors and docking score to generate a final selection of molecules for biological evaluation. Since docking provides a static snapshot of the dynamic ligand binding process, several studies have suggested the use of molecular dynamics simulations (MD)75–78 in enhancing the understanding of the structural variations of such multi body complexes under physiological conditions. Explicit solvent MD simulations of the binding poses of the identified biological hits were therefore carried out. They helped in evaluating the stability of the binding site interactions as well as, identify perturbations in the interaction profiles that would not be possible through docking studies. Based on the analysis of substitution patterns and observations from the simulation studies, a number of critical inferences could be made that would be helpful in future structure based design efforts against this target.
METHODS
Database preparation and ligand based searching
The Asinex platinum collection (Feb 2007 release) comprising of ~130,000 molecules was used to generate a Unity database within Sybyl6.9 (Tripos Inc., St. Louis, MO) for similarity searching. The 2D coordinates file in the .sdf format was downloaded and submitted for database creation. Similarity searching was carried out using the dbsimilar script from the Selector module of Sybyl6.9. The similarity scores (ranging from 100 to 0) to the given query molecules were computed using the Tanimoto Coefficient (Tc). The Tc for a target molecule defined by a bits set, of which c bits set are common to a query molecule with b bits set is defined as
| (1) |
The substructure searching was carried out using the dbsearch command from the Unity search module using queries generated within Sybyl. 3D coordinate generation for docking of selected molecules was carried out using the Concord (Tripos Inc., St Louis, MO) standalone utility. The structures were refined using 2000 steps of conjugate gradient to a RMS convergence of 0.01 Kcal/mol-Å. The Gasteiger-Huckel partial charge method and the Tripos force field were used for minimization to produce structures for docking.
Docking protocol
The docking parameters utilized in this study have been validated previously67. The dimeric structure of SARS-3CLpro (PDB code 2AMD) co-crystallized with a covalently bound ligand was utilized for all docking runs. To prepare the protein for docking, the ligand-protein covalent bond was deleted and protonation and tautomer assignment for the protein was carried out using the Pprep (Schrodinger, LLC, Portland, OR) utility. The resulting structure was then submitted to a restrained molecular mechanics refinement using the OPLS2001 force field incorporated in the Impref (Schrodinger, LLC, Portland, OR) protein structure refinement utility and the final refined structure was used for docking. Docking was carried out in Gold 3.0.1 (CCDC, Cambridge, UK) using (a) Automatic GA (genetic algorithm) settings at the rigorous 200% accuracy level, (b) Gold score function for pose ranking and (c) Docking constraints (Supplementary Figure 1): Three hydrophobic constraints, defined as spheres of 2Å radius defined around the centroid of the heavy atoms of the hydrophobic groups and a protein hydrogen bond donor constraint (defined using the N(ε)–H of His163). The poses were rescored using a consensus score
| (2) |
Where Sn, Smax and Smin refers to the docking scores for a given molecule n and the maximum and minimum scores attained within the docked set of molecules for the given scoring function (Gscore and Dscore generated from the Cscore module of Sybyl) respectively. Docking pose based descriptors were generated in Silver 1.1 (CCDC, Cambridge, UK) using the co-crystallized ligand pose from the crystal structure (PDB code 2AMD). It consisted of (a) Three hydrophobic occupancy descriptors (defined as spheres of 2 Å radius defined around the heavy atom centroid of relevant hydrophobic groups with ≥ 75% occupancy of the sphere volume defined as the criteria for fulfillment) and (b) One hydrogen bond donor descriptor (defined using the N(ɛ)–H of His 163 and had the geometric criteria of D-H-A angle ≥ 120° and D-A distance ≤ 3.5 Å for fulfillment).
Molecular dynamics simulation
The docked ligand/SARS-3CLpro complexes of the biological hits were used to perform explicit solvent MD simulations. The parallelized Desmond Molecular Dynamics System v2.2 (D. E. Shaw Research, New York, NY) and associated analysis tools, available within the Schrodinger suite (Schrodinger, LLC, Portland, OR) were used for this purpose.
The model system for simulation was prepared using the ‘system builder’ utility. The OPLS-AA/2005 force field parameters were assigned for all the simulation systems. Each inhibitor-enzyme complex was placed in the center of a cubic box with a dimension of 94.4 Å ensuring 10 Å solvent buffers (~23,400 water molecules) between the solute and the boundary of simulation box in each direction. The SPC model was used to describe the water molecules. Na+ and Cl− ions were added at physiological concentration of 0.15 M to ensure the overall neutrality of the systems. Any solvent molecules that overlapped with the solute structures were removed. The final size of the solvated system was close to ~33,000 atoms. The crystallographic waters were retained as part of the simulation system.
The model systems were relaxed using a six step default protocol implemented in Desmond, utilized to prepare systems for production-quality simulation. The relaxation protocol implemented in Desmond involved a series of minimizations followed by a series of short MD runs to relax the model systems. The solvated systems were first minimized for 10 steps of steepest-descent followed by the limited-memory Broyden-Fletcher-Goldfarb-Shanno (LBFGS) minimization over a maximum of 2000 steps, keeping all the protein and ligand atoms fixed. The convergence criterion and force constant on the solute was set to 50.0 kcal mol−1 Å−1. In the second step, all positional restraints were removed and the system was minimized to a convergence criterion of 5 kcal mol−1 Å−1 using the same sequence as the last step. In the third step, a 12 ps simulation was performed using the NVT ensemble using a Berendsen thermostat79, a temperature of 10 K, a fast temperature relaxation constant of 0.1 ps, velocity rescaling at every 1 ps and restraints on all non-hydrogen solute atoms with a force constant of 50.0 kcal mol−1 Å−1. Next, two short simulations of 12 and 24 ps were performed using the NPT ensemble using a Berendsen barostat79, maintaining pressure at 1 atm and a slow pressure relaxation constant of 50 ps with all other parameters preserved from the previous NVT simulation. The restraint set from the third step was preserved through the fourth and fifth steps. Temperature was maintained at 10 k for the fourth step and then elevated to 300 k for the fifth step. In the final stage of the relaxation protocol, all atoms were set free and the simulation was performed using the NPT ensemble for a simulation time of 24 ps using a pressure relaxation constant of 2.0 ps. This final step resembled the conditions of the actual production runs.
The relaxed systems were subjected to a production phase, using the NPT ensemble and periodic boundary conditions for a period of 5 ns. The Martyna-Tobias-Klein80 pressure control method was used to maintain the pressure of the cell at 1.013 bar using the isotropic coupling method. The Nose-Hoover thermostat method was used to control the temperature at 300K. Heavy atom-hydrogen covalent bonds were constrained using the SHAKE algorithm81 which allowed a 2-fs integration step to be used. The integration of the equation of motion as implemented according to the RESPA82 multiple time step scheme was used in the simulations. Long-range electrostatic forces were computed using the smooth Particle-Mesh Ewald (PME) method83. The cutoff distance for calculating short-range electrostatics and Lennard-Jones interaction was set to 9.0 Å. The trajectories and the energies were recorded at every 25 and 6 ps respectively. The glue option was turned on for proper display of recorded trajectories and convenience in structure analysis. The simulation quality analysis tool was used to analyze the energetics of the simulation trajectories. The simulation event analysis tool was used to monitor RMSD fluctuations, hydrogen bond distances, angles and Vanderwaals interactions over the simulation trajectories.
RESULTS AND DISCUSSION
Virtual screening workflow
An in-silico search (Figure 2) was carried using the Asinex platinum collection database (~130,000 compounds). Compounds were enumerated in the form of a Unity database which uses a form of hashed fingerprint for ligand enumeration that has been found to be very effective for similarity searching purposes84. The database was first filtered based on Molecular Weight, letting those ≤ 500 to pass through. We also wanted to evaluate the effectiveness of the similarity principle (discussed later) in its ability to retrieve active compounds within the constraints of this study. Therefore, the second stage of the filtering involved a similarity search using the two hit molecules PJ07 and PJ169 to generate two smaller subsets of ~64,000 (PJ07) and ~27,000 (PJ169) molecules with Tc ≥ 0.55 to the respective query compounds. The third stage of the filtering involved substructure searching. For both scaffolds, a core and two substitution points R1 and R2 were defined (Figure 2) and all unique available substitutions at the two points were identified from the two curtailed subsets obtained from stage two. A subset of substitutions was chosen for each substitution point, based on the likely nature of interactions observed for that position. A set of systemic searches were carried out wherein, substitutions to either the R1 or the R2 positions were assigned from the identified list of substitutions while variations at the other substitution point were explored. Additionally, a few molecules were also chosen that preserved the substitutions to the R1 and R2 positions but had a modified core scaffold. The selections from all the individual searches were combined and checked for redundant entries which resulted in two subsets of ~500 and ~ 1500 molecules for the two queries, PJ07 and PJ169 respectively. Following this, the curtailed database was docked into the crystal structure of SARS-3CLpro (PDB code 2AMD) using the docking protocol described in the methods section. While docking scores may lose resolution in terms of rank ordering of similarly active compounds, it can still be effective in eliminating obvious inactive compounds for which the poses lack shape/interaction complementarity with the binding pocket or are in a highly strained state. Additionally, we have previously observed67, 85 that the use of docking pose based descriptors can complement the docking score by rank ordering poses in terms of their agreement with a desired pharmacophore. The top n % (20 % – PJ07, 7 % – PJ169) of the docked poses corresponding to ~100 molecules, were selected in each case using a consensus ranking based on docking score and docking pose based descriptors (satisfying at least 3 descriptors) generated in Silver. Finally, based on (a) overlap of the core scaffolds in the docking poses to those of the query compounds, (b) diversity of substitutions at the R1 and R2 positions and the corresponding docking pose based descriptors, (c) diversity of similarity to the query compounds (Tc to PJ07 or PJ169) and (d) commercial availability, two selections of compounds comprising of 24 and 20 compounds were made for PJ07 and PJ169 respectively.
Figure 2.
A flowchart showing the protocol utilized for conducting the screening
Biological evaluation of the selected compounds
The selected compounds were put through a sequential biological evaluation funnel to test their enzyme inhibition potential and antiviral activity. The SARS-3CLpro inhibition assay (Supplementary Information) of the compounds was carried out in a two stage procedure. The first screening round involved evaluation of compounds at a single concentration of 10 µM. Compounds showing a significant inhibition (> 25%) at this concentration were then carried forward to the next stage wherein each compound was evaluated using multiple concentrations to obtain an IC50 value from the dose-response curve. Amongst the evaluated compounds (Figure 3), three from the PJ07 series and one from the PJ169 series showed significant inhibition at 10 µM and were carried forward for dose-response studies. The compound PJ207 showed an inhibition of ~25 % at 10 µM and a SARS-3CLpro IC50 of 39.4 µM. The two other actives from the PJ07 series were PJ224 and PJ225. PJ224 and PJ225 showed inhibitions of ~45% and ~61% at 10 µM and demonstrated IC50’s of 15 µM and 5.8 µM respectively. In the PJ169 series, one compound PJ316 showed ~56% inhibition at 10 µM and was found to have an IC50 of 10.3 µM. These four compounds were carried forward and evaluated in the SARS-CoV viral CPE assay (Supplementary Information). PJ207 showed an EC50 of 30 µM in virus infected cells, which is close to its enzymatic IC50, with a corresponding IC50 for cytotoxicity in normal cells at 250 µM. The lack of cellular activity for the other actives maybe due to the relatively higher Polar Surface Area86, 87 (PJ224/PJ225) or higher molecular weight (PJ225/PJ316) in reference to PJ207 which might prevent permeation in amounts required for cellular activity in this particular cellular assay system.
Figure 3.
Compounds from the PJ07 and PJ169 series which were found to demonstrate SARS-3CLpro activity.
Binding pose analysis of identified hits
Based on the peptidic ligand bound to the 2AMD crystal structure, (Figure 1a) the S1’, S2 and S4 all prefer aliphatic hydrophobic groups while the S1 site prefers the cyclized glutamine derivative with the ligand carbonyl forming the critical hydrogen bonding interaction with His163. The predicted binding poses of the identified hits against SARS-3CLpro were indicative of occupying some of these key sub sites. The molecule PJ207 (Figure 4. a, b) binds in a manner similar to PJ07 with the quinazoline core placed over the catalytic cysteine sulfur and the phenyl ring protruding into the S1’ pocket forming hydrophobic interactions with Thr25 and Leu27. The S2 pocket is occupied by the ethyl cyclopentyl group (cyclohexyl in case of PJ07), interacting with Met49 and Met165, supplementing the Leucine residue (P2) of the natural substrate. The amide side chain vectors into the S1 pocket wherein the carbonyl oxygen appears to form a hydrogen bond with His163. The thiazole moiety (methyl furan in case of PJ07) at the end of the linker may undergo weak hydrophobic interactions with the side chain of Leu 141. A similar observation is made in case of the two other hits PJ224 and PJ225. In PJ224 (Figure 4 c, d) the phenyl ring is seen to occupy the S2 pocket (cyclohexyl in case of PJ07) while the phenyl ring of the quinazoline scaffold appears to protrude into the S1’ pocket. The amide side chain is involved in the critical hydrogen bonding interaction with His163. The phenyl ring at the terminus of the amide side chain (methyl furan in case of PJ07) may interact with the side chain of Phe140, His172 and Leu141. It’s also possible for the terminal carboxylate moiety to form a hydrogen bond with Asn142 through a side chain amide flip. The docking pose of PJ225 (Figure 4. e, f) is predicted to be very similar to that of PJ224 with the addition of the ethyl moiety of the ester group which protrudes towards the Leu141 residue in the S1 pocket and appears to form a stronger hydrophobic interaction in comparison to the other PJ07 like hits. The ligand PJ316 (Figure 4 g, h) occupies the S1’, S1 and the S2 pockets with an overall binding pose very similar to the original hit PJ169 in terms of placement of the hydrophobic groups. The carbonyl oxygen from the benzpyridone moiety forms the critical hydrogen bonding interaction with His163 while the two phenyl side chains appear to occupy the S1 and the S1’ sites of the enzymes substrate binding site. The di-methyl substituted phenyl ring could form a hydrophobic interaction with Leu141 in the S1 subsite. An additional hydrogen bond is seen in the S1 subsite between the NH group of the bicyclic ring and the backbone carbonyl oxygen of Phe140. A bifurcated hydrogen bond was observed between the amide carbonyl of the linker (protruding into the S1’ pocket) and the backbone NHs of Gly143 and Ser144 residues which form the oxyanion hole. In PJ169, the non amide side chain vectors into the S1’ pocket. The chloro substitution on the phenyl ring occupying the S2 site is seen to protrude into the S4 subsite and may provide some additional binding affinity through hydrophobic interactions. In case of PJ169, this region is occupied by the methyl phenyl side chain at the end of the amide linker, with the amide carbonyl interacting with the backbone NH of Glu166.
Figure 4.
Interaction profile and surface representation of the poses of the identified hits (a)(b) PJ207, (c)(d) PJ224, (e)(f) PJ225 and (g)(h) PJ316. The yellow and blue surface coloring refers to the position of Cys145 and His163 respectively.
Analysis of the active and inactive (< 25% inhibition at 10 µM) structural analogues (Supplementary Figure 1) of PJ07 were suggestive of certain substitution trends that correlate with activity (Figure 5). At the R2 position, PJ209, PJ210, PJ213–217 and PJ236 have smaller hydrophobic groups (isobutyl, isopropyl or cyclopropyl) while PJ220/221 have a large (adamantyl) group compared to the cyclohexyl (PJ07), ethyl cyclopentyl (PJ207) and phenyl (PJ224, PJ225) groups present in the active compounds. The R1 position seems to prefer an amphiphilic aryl group (Furan-PJ07, Thiazole-PJ207) or an aryl group with a polar substitution (carboxy phenyl- PJ224, PJ225), with the polar portion pointing towards the solvent and the hydrophobic portion pointing downwards into the pocket, possibly interacting with His172. The R1 group of PJ201 (Oxadiazole), PJ203 (Isooxazole), PJ204 (Thiadiazole) and PJ206 (Hydrazine) are likely to be too polar, where as PJ205 (Cyclopropyl), PJ212 (Cyclopentyl) and PJ223 (Methyl Thiophene) are too hydrophobic to meet this requirement. The overall length of the R1 and R2 substitutions and subsequent placement of their terminal groups need to be compatible with each other for binding to occur. In case of PJ208 and PJ231 the overall length is greater (compared to PJ207), while in case of PJ202 its likely lower (compared to PJ07) than ideal, leading to inactivity. The S1’ modification in PJ237 (dimethyl Thiophene) was also not tolerated.
Figure 5.
A summary of the substitution preferences for the two scaffolds which are required for activity.
A similar analysis (Figure 5) of the active and inactive compounds (Supplementary Figure 1) for the PJ169 series suggested that for the R1 group, absence of the one carbon linker as well as polar substitutions on the terminal group (PJ308–315, PJ318–320) were not preferred, leading to inactivity. Similarly, the R2 substitutions also needed to be hydrophobic and PJ305–307, PJ317 had polar substitutions at this position, possibly leading to their lack of activity. Finally, PJ300–303 which met the two top criteria but lacked the specific dimethyl substitution on the bicyclic ring (present in the actives PJ169/PJ316) likely led to their inactivity. This conclusion was further supported by the MD simulation studies of PJ316 (discussed later) which suggested of its role in the stabilization of the benzpyridone moiety through a hydrophobic interaction.
Effectiveness of similarity searching
Similarity searching88, 89 has been utilized as a screening tool in drug discovery applications for over two decades. The basic premise behind this methodology is the “similar property principle”90 which states that structurally similar molecules share similar properties. Thus, given a target molecule with biological activity, its close neighbors in chemical space are likely to exhibit a similar activity profile. This characteristic of structurally related molecules has been termed as the “neighborhood behavior”91. It can be used efficiently92–99 to explore the chemical space around a specific molecule to attain a better understanding of the structure activity/property relationships. The query molecules as well as the searchable chemical database is enumerated using specific techniques100–102 such as 2D fingerprints, descriptors, graphs, pharmacophoric triplets etc. The similarity between the target and the query molecule is usually evaluated using a similarity metric such as the Tc or a distance metric such as the Euclidean distance. Classification of compounds using Tc based chemical similarity and their utility in identifying biologically active compounds have been evaluated extensively. A widely publicized study by Martin et al. 103 showed that selection of compounds based on a Tc ≥ 0.85 enumerated based on Unity fingerprints (used in this study) had an 80% probability of being active in similar assays. A follow up study by Martin et al. 104, however recalibrated this probability to ~30%, citing technical shortcomings of the methodology as well as large changes in activity profiles introduced through small structural perturbations. During our study design, we had reasons to believe (discussed later) that within the limits of this particular application, similarity searching as a ligand based virtual screening method would not be successful. Nonetheless, we wanted to build in provisions (enumeration of compounds based on their Tc similarity) within the screening workflow that would allow us to do a retrospective evaluation of its effectiveness.
Therefore, a further analysis was carried out to analyze hit counts from the biologically evaluated compounds, binned based on their Tc similarity to the query compounds utilized in the selection process. The 44 compounds (24 PJ07 like and 20 PJ169 like) were combined and ranked based on the Tc similarity to their respective query compounds and then binned into 5 categories ranging from 100 to 50% similarity. As seen in Figure 6, the actives were recovered at 3 of the binned categories 80–90 % (1 hit, 8.3% hit rate), 60–70 % (1 hit, 10% hit rate) and the 50–60 % (2 hits, 40% hit rate). Thus, using the 85% similarity cutoff103 we will not recover the three hits in the 60–70% and 50–60% bins. From the 12 compounds which share a common core and are highly similar (Tc > 85) to the queries, only one is found to be active, corresponding to ~8 % hit rate. There could be a number of reasons for this behavior. First, the nature of the target104 (solvent exposed and induced fit effects around the S2 pocket and the dimeric interface) might make the structure activity landscape more complex and very sensitive to structural changes that cannot be captured using a standard structural similarity. For example, in the PJ169 series both actives present a specific dimethyl substitution in addition to the other requirements and the MD simulation study also pointed towards a specific role for this substitution. A similar example is given in the follow up paper on similarity searching by Martin et al.104 where they showed that analogues of an active compound (Pargyline), with differences in a single methyl group could lead to complete loss of activity. While, similarity compares the structure of two molecules as a whole, it doesn’t adjust for conservation of specific areas in the molecule which might weigh more towards activity than others for a given target. Therefore molecules with the same core scaffold which would be considered similar from the perspectives of chemical similarity, synthetic chemistry (accessible from the same synthetic route) or markush definitions from patent literature might actually be quite dissimilar or novel from the perspective of a given target by virtue of specific substitution differences. A secondary reason for this behavior of the similarity search could stem from the fact that the queries used for this similarity search are not highly potent104. They might not have all the pharmacophoric features required for potent activity, thereby making it more difficult to find actives based on the similarity principle.
Figure 6.
A binned histogram showing the counts of the biologically evaluated compounds (active – green, inactive - red) binned based on their similarity to their respective query molecules (PJ07 and PJ169) ranging from 100 to 50% similarity.
Dynamics simulation of identified hits
MD simulations were carried out to investigate the overall stability of the predicted docking poses. They helped in studying the conformational changes of the inhibitors relative to the active site resulting in altered interaction profiles, as well as movements of amino acid side chains lining the binding site, in a physiologically relevant setting. The overall stability of the individual simulations was evaluated by plotting the total energy of the system as a function of time (Figure 7). All the complexes were energetically stable with maximum fluctuations at < 1% of the total energy of the system over the course of the simulation.
Figure 7.
Energy plot for the production phase of dynamic simulation of ligand/SARS-3CLpro complex.
The superposition of the trajectories onto the system conformations at the start of the production phase helped in the analysis of the RMSD progression over the time of the simulation. The average heavy atom RMSD of the binding site (BS) side chain residues (Thr25, Leu27, His41, Met49, Phe140, Leu141, Asn142, Cys145, His 163, Met 165, Glu166, His172, Asp187, Gln189 and Gln 192) of SARS-3CLpro in complex with PJ207, PJ224, PJ225 and PJ316 were 1.14 Å, 1.41 Å, 1.44 Å and 1.49Å respectively for the 5 ns production phase, suggesting a stable binding site. The ligand poses were highly stable with average heavy atom RMSD of 0.72 Å, 1.05 Å, 1.0 Å, and 1.48 Å for PJ207, PJ224, PJ225 and PJ316 respectively (Figure 8b, Figure 9b, Figure 10b and Figure 11b) over the 5 ns production phase. The average heavy atom RMSD of the inhibitors were at the same level (PJ316) or lower (PJ207, PJ224, PJ225) than the corresponding average heavy atom RMSD of the binding site (BS) side chain residues, suggesting that the ligand movements are relatively smaller compared to the overall conformational changes of the SARS-3CLpro binding site. The most noticeable side chain movement was shown by Gln189. This residue is solvent exposed and is located in the opening of the catalytic cleft located between domain I and II105 which might explain its positional variability due to inter domain movements.
Figure 8.
(a) Docking pose of PJ207 showing the hydrogen bond (red) and hydrophobic (green) monitors used during the simulation; (b) Heavy atom RMSD of the side chain residues of enzyme’s binding site (BS), PJ207 and side chain of four residues of BS that showed maximum fluctuations during the simulation; (c) Hydrogen bond (H-A) distances and (d) D-H-A angle of His163[-NH]-[O]PJ207 and Glu166[-NH]-[O]PJ207; (e) Hydrophobic [CH-CH] interactions of PJ207 with Thr25, Leu27, Met49 and M165 and a cation(pi)-pi interaction with His172.
Figure 9.
(a) Docking pose of PJ224 showing the hydrogen bond (red) and hydrophobic (green) monitors used during the simulation; (b) Heavy atom RMSD of the side chain residues of enzyme’s binding site (BS), PJ224 and side chain of four residues of BS that showed maximum fluctuations during the simulation; (c) Hydrogen bond (H-A) distances and (d) D-H-A angle of Asn142[O]-PJ224[NH], His163[-NH]-[O]PJ224 and Gly1[NH]-PJ224[O]; (e) Hydrophobic [CH-CH] interactions of PJ224 with Thr25, Met49, and Met165 and a cation(pi)-pi interaction with His172.
Figure 10.
(a) Docking pose of PJ225 showing the hydrogen bond (red) and hydrophobic (green) monitors used during the simulation; (b) Heavy atom RMSD of the side chain residues of enzyme’s binding site (BS), PJ225 and side chain of four residues of BS that showed maximum fluctuations during the simulation; (c) Hydrogen bond (H-A) distances and (d) D-H-A angle of Asn142[NH]-PJ225[O], Ser144[OH]-PJ225[O] and His163[NH]-[O]PJ225; (e) Hydrophobic [CH-CH] interactions of PJ225 with Thr25, Met49, Phe140, Leu141 and Met165 and a cation(pi)-pi interaction with His172.
Figure 11.
(a) Docking pose of PJ316 showing the hydrogen bond (red) and hydrophobic (green) monitors used during the simulation; (b) Heavy atom RMSD of the side chain residues of enzyme’s binding site (BS), PJ316 and side chain of four residues of BS that showed maximum fluctuations during the simulation; (c) Hydrogen bond (H-A) distances and (d) D-H-A angle of Phe140 [O]-[NH]PJ316, G143[-NH]-[CO]PJ316, Ser144 [NH]-[O]PJ316 and H163[-NH]-[O]PJ316; (e) Hydrophobic [CH-CH] interactions of PJ316 with Thr25, Met49, Leu141 and Met165 and a cation(pi)-pi interaction with His172.
The MD trajectories of the representative inhibitors were analyzed for the presence of hydrogen bonds as well as hydrophobic interactions between the inhibitors and important residues of the enzyme binding site predicted by the docking studies. The analysis indicated the presence of several hydrogen bonds and hydrophobic interactions with moderate to high frequency of occurrence. Several new interactions were observed over the simulation time course (in addition to what was observed from docking), suggesting the possibility of considering these for structure based design. The interactions of each inhibitor with the active site residues of SARS-3CLpro are discussed below in detail. The plots of the MD simulation of all inhibitors are graphically shown in Figure 7–10. The distance monitors (Figure 8a, 9a, 10a and 11a) used for hydrogen bonds and hydrophobic interactions are shown using red and green dotted lines respectively.
The results from the simulation of the PJ207/SARS-3CLpro complex are shown in Figure 8. A very interesting observation was made in respect to the critical hydrogen bonding interaction in the S1 sub pocket. In 0–3 ns of the simulation, the hydrogen bond interaction between the NE2 of His 163 from the S1 subsite of the enzyme and the amide side chain carbonyl of PJ207 was retained with an average D-A distance of 2.69 Å. Following this, the hydrogen bond is lost and is compensated by a new hydrogen bond between the carbonyl side chain of PJ207 and backbone –NH of Glu 166 located close to the S1 pocket with an average D-A distance of 2.85 Å and D-H-A angle of 135° for 4–5 ns of the simulation time (Figure 8 c, d). The hydrophobic interaction profile (Figure 8e) showed interactions of PJ207 with Thr25, Leu27 of the S1’ site as well as Met49 from S2 and Met165 where the corresponding hydrophobic heavy atom distances were retained at ≤ 4.5 Å for 42%, 70.5%, 50%, and 64.5% of the simulation time respectively. A cation (pi)-pi interaction could be observed between the thiazole group and His172. It’s likely, that the relatively polar nature of the aromatic group (compared to the substituents from the other actives) at the end of the S1 substitution could also contribute to the observed higher movement of the amide side chain. It’s also interesting to note that the RMSD fluctuation of Met165 (Figure 8b) shows a biphasic behavior between 0–3 and 3–5 ns of the simulation showing larger fluctuations in the second phase which corresponds with the hydrogen bond compensation event observed between the ligand’s amide side chain carbonyl and the His163 and Glu166 residues (discussed above).
The results from the simulation of the PJ224/SARS-3CLpro complex are shown in Figure. 9. As predicted from docking studies, the amide carbonyl side chain of PJ224 appeared to form a strong hydrogen bond with the NE2 of His163 with an average D-A distance of 2.61 Å and average D-H-A angle of 124.2° (Figure 9 c, d). Another hydrogen bond was observed between the side chain -NH of Asn142 and the side chain carbonyl oxygen of PJ224 with an average D-A distance of 2.9 Å and average D-H-A angle of 120.9° for 0.55–5.0 ns of the simulation time. This interaction was not observed in the static docking pose and suggests the importance of MD simulation in detecting a possible additional interaction. MD simulation also revealed a new hydrogen bond between the carboxylate moiety of PJ224 and N-terminal Gly-1 (of subunit B) with an average D-A distance of 2.69 Å for the first 2.2 ns of the simulation time which was not observed in the docking pose. The additional hydrogen bond interactions in PJ224 vis-a-vis PJ207 may contribute to its higher affinity. Thr25 from the S1’ sub site exhibited strong hydrophobic interactions with the benzpyrimidine scaffold of PJ224 with the corresponding hydrophobic heavy atom distance at ≤ 4.5 Å for 88% of the simulation time with an average distance of 4.03 Å. Met165 and Met49 from the S2 sub site were found to interact with the phenyl side chain of PJ224 with corresponding hydrophobic heavy atom distance at ≤ 4.5 Å for 87% and 71% of the simulation time respectively (Figure 9 e). A transient cation (pi)-pi interaction was observed between the phenyl group on the amide side chain and His172.
The results of the MD studies for SARS-3CLpro in complex with PJ225 are shown in Figure 10. Similar to PJ224, a stable hydrogen bond was observed between the side chain amide carbonyl oxygen of PJ225 with the NE2 of His 163 with an average D-A distance of 2.69 Å and average D-H-A angle of 125.6° (Figure 10 c, d). Also, a transient hydrogen bond was observed between the side chain OH of Ser144 and amide carbonyl oxygen of PJ225 (D-A distance ≤ 3.5 43.5% of the simulation time). This residue is located on the loop (residues 140–146) involved in the oxyanion hole formation and stabilizes the covalent tetrahedral intermediate generated during substrate hydrolysis by the enzyme. Coincidentally, this interaction was not observed in the docking pose of PJ224. However, the Gly-1 hydrogen bond is not possible (ethyl ester in PJ225, carboxylate in PJ224) in case of PJ225 and the Asn142 interaction had a lower residence time in comparison to PJ224. During the course of the simulation, several strong hydrophobic interactions (Figure 10d) were observed. These included interactions of PJ225 with the side chains of Thr25, Leu141, Phe140, Met165 and Met49 with hydrophobic heavy atom distances at ≤ 4.5 Å for 82.5%, 30.5%, 85.5%, 72% and 59% of the simulation time respectively. As predicted from the docking study, PJ225 in comparison to PJ224 and PJ207 demonstrated additional hydrophobic interactions (Figure 10 e) between the ethyl moiety of the ligand with Phe140 and Leu141. A highly stable cation (pi)-pi interaction was observed between the phenyl ring and His 172 with the distance at ≤ 4.5 Å, 100% of the simulation time. These additional interactions in the S1 pocket as well as the transient Ser144 hydrogen bond interaction observed for PJ225 could account for its higher inhibitory activity (IC50 = 5.8 µM) against SARS-3CLpro in comparison to the other active compounds.
The simulation plots for the PJ316/SARS-3CLpro complex are shown in Figure 11. Multiple hydrogen bonds were predicted for PJ316 based on the docking simulations (Figure 11 c, d). The amide carbonyl oxygen of the benzpyridone moiety of PJ316 appeared to form a stable hydrogen bond with NE2 of His 163 (Avg. D-A distance = 2.47Å and Avg. D-H-A angle = 125°) of the S1 pocket. The amide nitrogen of the bicyclic ring of PJ316 also formed a hydrogen bond with the backbone carbonyl oxygen of Phe140 with an average D-A distance of 2.22 Å and D-H-A angle of 139°. In addition, the amide side chain of PJ316 formed a bifurcated hydrogen bond with the backbone amide of Gly143 (Avg. D-A distance = 1.83Å and Avg. D-H-A angle = 164°) and Ser144 which forms the oxyanion hole to stabilize the covalent tetrahedral intermediate resulting from substrate hydrolysis by the enzyme. The hydrophobic interactions profile for PJ316-enzyme complex is shown in Figure 10 e. The side chain of Met49 interacts with PJ316 with the hydrophobic distance at ≤ 4.5 Å, 97.5% of the simulation time. Thr25 also exhibited a strong interaction with the benzyl side chain of PJ316; hydrophobic heavy atom distance at ≤ 4.5 Å for 91.5% of the simulation time and an average hydrophobic heavy atom distance of 4.03 Å. In addition, hydrophobic interactions were observed between (a) Met165 and the phenyl side chain and (b) Leu141 with the benzpyridone moiety of PJ316, with the hydrophobic heavy atom distances at ≤ 4.5 Å for 40% and 51% of the simulation time respectively. Amongst the current hits, only PJ316 can explore the S4 pocket through the chlorine substitution on the phenyl ring. The hydrophobic heavy atom distance progression (Figure 11e) for Leu141 also showed a biphasic behavior (0–1.5 ns and 1.5–5 ns) suggesting the possible stabilization of the interaction with the dimethyl moiety of the ligand in the second phase which would in turn also stabilize the two hydrogen bonds in the S1 pocket formed by the benzpyridone moiety. This would also explain why both the active compounds from this series, PJ169 & PJ316 have this unique dimethyl substitution. The phenyl ring of the benzpyridone moiety also appeared to engage in a cation (pi)-pi interaction with His172.
Based on the overall analysis, certain inferences could be made regarding substitution preferences for binding to this protease. These may be used in structure guided synthesis to evaluate modifications which are currently not available commercially and may also be used for potential scaffold hopping applications. First, the S2 sub site doesn’t appear to tolerate any polar substitutions and appears to be highly specific for hydrophobic groups that may be utilized to occupy this subsite. While the smaller/bulkier aliphatic groups didn’t provide adequate hydrophobic interaction/caused steric clash, longer groups require a corresponding S1 group of compatible size for proper fit. A phenyl group at the S2 position (present in 5 out of 6 actives, including PJ07 and PJ169) is advantageous because it allows a vector into the S4 packet which could be occupied with a hydrophobic group, potentially leading to an increase in overall affinity. In case of PJ316, we observed the hydrophobic chlorine group vectored into the S4 cavity adding to the overall stability of the binding pose and suggest that, such structure-based modifications in both active chemotypes could be beneficial for potency gain. Furthermore, additional smaller hydrophobic substituents may be useful in probing the depths of the S2 pocket. Likewise, the S1’ site also prefers the phenyl group for activity.
The S1 hydrogen bonding interaction with His163 was found to be moderate to highly stable in all the simulations and is one of the most critical binding site requirements. This is also supported by the biological studies on the substrate preference for this site. The alternate hydrogen bonding demonstrated by PJ207 with the backbone of Glu166 in lieu of His163 and the Ser144 (from the oxyanion hole) interaction observed in case of PJ225 suggests that possible modifications vectoring two hydrogen acceptors (e.g. sulfonamide for Ser144) could be utilized to harness additional binding affinity. Also of interest, would be the hydrogen bonding interaction with Asn142 as observed in the MD simulations of PJ224 and PJ225. Additionally, as shown in case of PJ316, the hydrogen bonding interactions with the backbone of Gly143 and Ser144 (residues forming the oxyanion hole) could form a strong anchor point for ligand stability. The Gly-1 residue from the second monomer was found to participate in a transient hydrogen bonding interaction during the MD simulation and may be pursued as part of the structure-based design strategy. It would also appear that, the interaction with Leu141, observed in the docking pose and MD simulations of the two more potent hits PJ225 and PJ316 through aliphatic hydrophobic groups may add to the overall stability by forming additional hydrophobic interactions. These in turn helps in the stabilization of the hydrogen bonds formed in the S1 pocket. Finally, the binding site appears to prefer an aryl group extending into the S1 pocket and is critical towards the stability of the interactions of the linker and the end group located in this region.
CONCLUSIONS
A combined ligand/structure based screening was carried out against the Asinex platinum collection database to identify inhibitors of the SARS-3CLpro enzyme. Two previously identified hits were used for carrying out a ligand based screening which was followed by a structure based screening to generate a final selection of compounds. Forty-four molecules sharing structural similarities to the original hits, but varying substitutions were selected for purchase and subsequent biological evaluation. The compounds were screened against the SARS-3CLpro enzyme using a two stage assay. Amongst these compounds, four were identified as inhibitors of the enzyme with IC50s ranging from 5.8 µM to 39.4 µM. One of the actives, PJ207 also showed activity in a SARS-CoV viral CPE assay with an EC50 of 30 µM. This is significant since, till date very few small molecule SARS-3CLpro inhibitors with antiviral activity have been identified through in-silico approaches. We are aware of only one other case51 where Cinanserin (a known serotonin antagonist) was identified through an in-silico screening approach and had a reported SARS-3CLpro IC50 of 5 µM and EC50 of 31 µM in a viral RNA replication assay. However, in a follow up publication by the authors, the SARS-3CLpro IC50 of Cinanserin was recalibrated to 323 µM which is > 2 log order higher than the previously reported value.
A retrospective analysis of the identified hits based on their Tc similarity to their respective query molecules used in the ligand based screening step also suggested that within the constraints of this screening setup a simple similarity search and selection of compounds using a high similarity cutoff (Tc ≥ 0.85) would not have succeeded in retrieving the majority of the active compounds which originated from a wider similarity window (Tc ≥ 0.55). Bemis & Murcko106 has suggested the use of molecular framework variations as a means of exploring the variance in ligand-protein interaction profiles and based on their definitions the tested compounds belonged to 14 (24 compounds) and 8 (20 molecules) molecular frameworks respectively for PJ07 and PJ169 series and the current hits corresponded to three distinct frameworks which are unique from the framework of the two original query molecules used to identify them.
Comparison of the identified hits spanning ~7 folds in terms of IC50 with the compounds found inactive at the single point test concentration of 10 µM helped in the identification of certain substitution trends that are required for potency against this enzyme. Additionally, the docking poses from the identified hits were utilized for a series of multi nanosecond simulations to study the overall stability of the interaction profiles and novel interactions not observed during the docking study. The information from these analyses would be utilized for further structure guided synthesis of analogues that might help in the identification of potent antiviral leads targeted against SARS-CoV.
Supplementary Material
ACKNOWLEDGEMENTS
Biological evaluations were supported by contracts NO1-AI-30048 (D.S.) from the National Institute of Allergy and Infectious Diseases (NIAID). We would also like to thank Dr. István Kolossváry from D.E.Shaw Research for his help with the technical aspects of the trajectory analysis of the simulations carried out using Desmond.
Footnotes
SUPPORTING INFORMATION
Docking constraints, structures of inactive compounds, spectras and details of bioassays. This material is available free of charge from http://pubs.acs.org.
REFERENCES
- 1.Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF, Lui SF, Szeto CC, Chung S, Sung JJ. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 2.Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, Green K, Tellier R, Draker R, Adachi D, Ayers M, Chan AK, Skowronski DM, Salit I, Simor AE, Slutsky AS, Doyle PW, Krajden M, Petric M, Brunham RC, McGeer AJ. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348(20):1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 3.Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stohr K, Osterhaus AD. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423(6937):240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsang Kenneth W, Ho Pak L, Ooi Gaik C, Yee Wilson K, Wang T, Chan-Yeung M, Lam Wah K, Seto Wing H, Yam Loretta Y, Cheung Thomas M, Wong Poon C, Lam B, Ip Mary S, Chan J, Yuen Kwok Y, Lai Kar N. A cluster of cases of severe acute respiratory syndrome in Hong Kong. The New England journal of medicine. 2003;348(20):1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 5.Drosten C, Preiser W, Gunther S, Schmitz H, Doerr Hans W. Severe acute respiratory syndrome: identification of the etiological agent. Trends Mol. Med. 2003;9(8):325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Medicine. 2006;3(9):1525–1530. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knowles Sandra R, Phillips Elizabeth J, Dresser L, Matukas L. Common adverse events associated with the use of ribavirin for severe acute respiratory syndrome in Canada. Clin. Infect. Dis. 2003;37(8):1139–1142. doi: 10.1086/378304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau EHY, Cowling BJ, Muller MP, Ho L-M, Tsang T, Lo S-V, Louie M, Leung GM. Effectiveness of Ribavirin and Corticosteroids for Severe Acute Respiratory Syndrome. Am. J. Med. 2009;122(12):1150e1111–1150e1121. doi: 10.1016/j.amjmed.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang L-F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 10.Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi H-W, Wong BHL, Wong SSY, Leung S-Y, Chan K-H, Yuen K-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JSM, Poon LLM. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science. 2003;302(5643):276–279. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 12.Tong TR. Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections. Infectious Disorders: Drug Targets. 2009;9(2):223–245. doi: 10.2174/187152609787847659. [DOI] [PubMed] [Google Scholar]
- 13.Patick AK, Potts KE. Protease inhibitors as antiviral agents. Clin. Microbiol. Rev. 1998;11(4):614–627. doi: 10.1128/cmr.11.4.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong L. Viral Proteases. Chem. Rev. 2002;102(12):4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- 15.Bianchi E, Pessi A. Inhibiting viral proteases: challenges and opportunities. Biopolymers. 2002;66(2):101–114. doi: 10.1002/bip.10230. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Montero JV, Barreiro P, Soriano V. HIV protease inhibitors: recent clinical trials and recommendations on use. Expert Opinion on Pharmacotherapy. 2009;10(10):1615–1629. doi: 10.1517/14656560902980202. [DOI] [PubMed] [Google Scholar]
- 17.Judd DA, Nettles JH, Nevins N, Snyder JP, Liotta DC, Tang J, Ermolieff J, Schinazi RF, Hill CL. Polyoxometalate HIV-1 protease inhibitors. A new mode of protease inhibition. J. Am. Chem. Soc. 2001;123(5):886–897. doi: 10.1021/ja001809e. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Labrador F-X. Hepatitis C virus NS3/4A protease inhibitors. Recent Patents on Anti-Infective Drug Discovery. 2008;3(3):157–167. doi: 10.2174/157489108786242369. [DOI] [PubMed] [Google Scholar]
- 19.Waxman L, Darke PL. The herpesvirus proteases as targets for antiviral chemotherapy. Antivir. Chem. Chemother. 2000;11(1):1–22. doi: 10.1177/095632020001100101. [DOI] [PubMed] [Google Scholar]
- 20.Wang QM, Chen S-H. Human rhinovirus 3C protease as a potential target for the development of antiviral agents. Current Protein and Peptide Science. 2007;8(1):19–27. doi: 10.2174/138920307779941523. [DOI] [PubMed] [Google Scholar]
- 21.Ziebuhr J, Heusipp G, Siddell SG. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J. Virol. 1997;71(5):3992–3997. doi: 10.1128/jvi.71.5.3992-3997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dougherty WG, Semler BL. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 1993;57(4):781–822. doi: 10.1128/mr.57.4.781-822.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziebuhr J, Snijder EJ, Gorbalenya AE. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81((Pt 4)):853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 24.Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 2006;103(15):5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Chen L, Tan J, Chen J, Du L, Sun T, Shen J, Chen K, Jiang H, Shen X. Severe acute respiratory syndrome coronavirus 3C-like proteinase N terminus is indispensable for proteolytic activity but not for enzyme dimerization. Biochemical and thermodynamic investigation in conjunction with molecular dynamics simulations. J. Biol. Chem. 2005;280(1):164–173. doi: 10.1074/jbc.M408211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Wei P, Fan K, Liu Y, Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43(15):4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, Wei Z, Song J. Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. J. Biol. Chem. 2004;279(23):24765–24773. doi: 10.1074/jbc.M311744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan K, Wei P, Feng Q, Chen S, Huang C, Ma L, Lai B, Pei J, Liu Y, Chen J, Lai L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279(3):1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu MF, Kuo CJ, Chang KT, Chang HC, Chou CC, Ko TP, Shr HL, Chang GG, Wang AH, Liang PH. Mechanism of the maturation process of SARS-CoV 3CL protease. J. Biol. Chem. 2005;280(35):31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Xie W, Xue X, Yang K, Ma J, Liang W, Zhao Q, Zhou Z, Pei D, Ziebuhr J, Hilgenfeld R, Yuen KY, Wong L, Gao G, Chen S, Chen Z, Ma D, Bartlam M, Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu T, Ooi A, Lee HC, Wilmouth R, Liu DX, Lescar J. Structure of the SARS coronavirus main proteinase as an active C2 crystallographic dimer. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61((Pt 11)):964–966. doi: 10.1107/S1744309105033257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee TW, Cherney MM, Huitema C, Liu J, James KE, Powers JC, Eltis LD, James MN. Crystal structures of the main peptidase from the SARS coronavirus inhibited by a substrate-like aza-peptide epoxide. J. Mol. Biol. 2005;353(5):1137–1151. doi: 10.1016/j.jmb.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh AK, Xi K, Ratia K, Santarsiero BD, Fu W, Harcourt BH, Rota PA, Baker SC, Johnson ME, Mesecar AD. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease inhibitors. J. Med. Chem. 2005;48(22):6767–6771. doi: 10.1021/jm050548m. [DOI] [PubMed] [Google Scholar]
- 36.Lu IL, Mahindroo N, Liang PH, Peng YH, Kuo CJ, Tsai KC, Hsieh HP, Chao YS, Wu SY. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J. Med. Chem. 2006;49(17):5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- 37.Xue X, Yu H, Yang H, Xue F, Wu Z, Shen W, Li J, Zhou Z, Ding Y, Zhao Q, Zhang XC, Liao M, Bartlam M, Rao Z. Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. J. Virol. 2008;82(5):2515–2527. doi: 10.1128/JVI.02114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin J, Niu C, Cherney MM, Zhang J, Huitema C, Eltis LD, Vederas JC, James MN. A mechanistic view of enzyme inhibition and peptide hydrolysis in the active site of the SARS-CoV 3C-like peptidase. J. Mol. Biol. 2007;371(4):1060–1074. doi: 10.1016/j.jmb.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee TW, Cherney MM, Liu J, James KE, Powers JC, Eltis LD, James MN. Crystal structures reveal an induced-fit binding of a substrate-like Aza-peptide epoxide to SARS coronavirus main peptidase. J. Mol. Biol. 2007;366(3):916–932. doi: 10.1016/j.jmb.2006.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan J, Verschueren KH, Anand K, Shen J, Yang M, Xu Y, Rao Z, Bigalke J, Heisen B, Mesters JR, Chen K, Shen X, Jiang H, Hilgenfeld R. pH-dependent conformational flexibility of the SARS-CoV main proteinase (M(pro)) dimer: molecular dynamics simulations and multiple X-ray structure analyses. J. Mol. Biol. 2005;354(1):25–40. doi: 10.1016/j.jmb.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S, Chen SJ, Hsu MF, Wu JD, Tseng CT, Liu YF, Chen HC, Kuo CW, Wu CS, Chang LW, Chen WC, Liao SY, Chang TY, Hung HH, Shr HL, Liu CY, Huang YA, Chang LY, Hsu JC, Peters CJ, Wang AH, Hsu MC. Synthesis, crystal structure, structure-activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J. Med. Chem. 2006;49(16):4971–4980. doi: 10.1021/jm0603926. [DOI] [PubMed] [Google Scholar]
- 42.Verschueren KH, Pumpor K, Anemuller S, Chen S, Mesters JR, Hilgenfeld R. A structural view of the inactivation of the SARS coronavirus main proteinase by benzotriazole esters. Chem. Biol. 2008;15(6):597–606. doi: 10.1016/j.chembiol.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H-M, Liang P-H. Pharmacophores and biological activities of severe acute respiratory syndrome viral protease inhibitors. Expert Opinion on Therapeutic Patents. 2007;17(5):533–546. [Google Scholar]
- 44.Barnard DL, Kumaki Y. Developments in the search for small-molecule inhibitors for treatment of severe acute respiratory syndrome coronavirus. Antiviral Res. 2009:209–222. [Google Scholar]
- 45.Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem. Rev. 2002;102(12):4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh AK, Xi K, Grum-Tokars V, Xu X, Ratia K, Fu W, Houser KV, Baker SC, Johnson ME, Mesecar AD. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2007;17(21):5876–5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goetz DH, Choe Y, Hansell E, Chen YT, McDowell M, Jonsson CB, Roush WR, McKerrow J, Craik CS. Substrate specificity profiling and identification of a new class of inhibitor for the major protease of the SARS coronavirus. Biochemistry. 2007;46(30):8744–8752. doi: 10.1021/bi0621415. [DOI] [PubMed] [Google Scholar]
- 48.Zhang HZ, Zhang H, Kemnitzer W, Tseng B, Cinatl J, Jr, Michaelis M, Doerr HW, Cai SX. Design and synthesis of dipeptidyl glutaminyl fluoromethyl ketones as potent severe acute respiratory syndrome coronovirus (SARS-CoV) inhibitors. J. Med. Chem. 2006;49(3):1198–1201. doi: 10.1021/jm0507678. [DOI] [PubMed] [Google Scholar]
- 49.Liu Z, Huang C, Fan K, Wei P, Chen H, Liu S, Pei J, Shi L, Li B, Yang K, Liu Y, Lai L. Virtual screening of novel noncovalent inhibitors for SARS-CoV 3C-like proteinase. J. Chem. Inf. Model. 2005;45(1):10–17. doi: 10.1021/ci049809b. [DOI] [PubMed] [Google Scholar]
- 50.Chen L, Chen S, Gui C, Shen J, Shen X, Jiang H. Discovering severe acute respiratory syndrome coronavirus 3CL protease inhibitors: virtual screening, surface plasmon resonance, and fluorescence resonance energy transfer assays. J. Biomol. Screen. 2006;11(8):915–921. doi: 10.1177/1087057106293295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L, Gui C, Luo X, Yang Q, Gunther S, Scandella E, Drosten C, Bai D, He X, Ludewig B, Chen J, Luo H, Yang Y, Yang Y, Zou J, Thiel V, Chen K, Shen J, Shen X, Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79(11):7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bacha U, Barrila J, Velazquez-Campoy A, Leavitt SA, Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43(17):4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 53.Jain RP, Pettersson HI, Zhang J, Aull KD, Fortin PD, Huitema C, Eltis LD, Parrish JC, James MN, Wishart DS, Vederas JC. Synthesis and evaluation of keto-glutamine analogues as potent inhibitors of severe acute respiratory syndrome 3CLpro. J. Med. Chem. 2004;47(25):6113–6116. doi: 10.1021/jm0494873. [DOI] [PubMed] [Google Scholar]
- 54.Shie JJ, Fang JM, Kuo CJ, Kuo TH, Liang PH, Huang HJ, Yang WB, Lin CH, Chen JL, Wu YT, Wong CH. Discovery of potent anilide inhibitors against the severe acute respiratory syndrome 3CL protease. J. Med. Chem. 2005;48(13):4469–4473. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- 55.Blanchard JE, Elowe NH, Huitema C, Fortin PD, Cechetto JD, Eltis LD, Brown ED. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11(10):1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuo CJ, Liu HG, Lo YK, Seong CM, Lee KI, Jung YS, Liang PH. Individual and common inhibitors of coronavirus and picornavirus main proteases. FEBS Lett. 2009;583(3):549–555. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai L, Han X, Chen H, Wei P, Huang C, Liu S, Fan K, Zhou L, Liu Z, Pei J, Liu Y. Quaternary structure, substrate selectivity and inhibitor design for SARS 3C-like proteinase. Curr. Pharm. Des. 2006;12(35):4555–4564. doi: 10.2174/138161206779010396. [DOI] [PubMed] [Google Scholar]
- 58.Yeung KS, Meanwell NA. Recent developments in the virology and antiviral research of severe acute respiratory syndrome coronavirus. Infect Disord Drug Targets. 2007;7(1):29–41. doi: 10.2174/187152607780090739. [DOI] [PubMed] [Google Scholar]
- 59.Shah F, Mukherjee P, Desai P, Avery M. Computational approaches for the discovery of cysteine protease inhibitors against malaria and SARS. Current Computer-Aided Drug Design. doi: 10.2174/157340910790980142. In Press. [DOI] [PubMed] [Google Scholar]
- 60.Yang Q, Chen L, He X, Gao Z, Shen X, Bai D. Design and synthesis of cinanserin analogs as severe acute respiratory syndrome coronavirus 3CL protease inhibitors. Chem. Pharm. Bull. (Tokyo) 2008;56(10):1400–1405. doi: 10.1248/cpb.56.1400. [DOI] [PubMed] [Google Scholar]
- 61.Bacha U, Barrila J, Gabelli SB, Kiso Y, Mario Amzel L, Freire E. Development of broad-spectrum halomethyl ketone inhibitors against coronavirus main protease 3CL(pro) Chem. Biol. Drug. Des. 2008;72(1):34–49. doi: 10.1111/j.1747-0285.2008.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghosh AK, Gong G, Grum-Tokars V, Mulhearn DC, Baker SC, Coughlin M, Prabhakar BS, Sleeman K, Johnson ME, Mesecar AD. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2008;18(20):5684–5688. doi: 10.1016/j.bmcl.2008.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shao YM, Yang WB, Kuo TH, Tsai KC, Lin CH, Yang AS, Liang PH, Wong CH. Design, synthesis, and evaluation of trifluoromethyl ketones as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. 2008;16(8):4652–4660. doi: 10.1016/j.bmc.2008.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niu C, Yin J, Zhang J, Vederas JC, James MN. Molecular docking identifies the binding of 3-chloropyridine moieties specifically to the S1 pocket of SARS-CoV Mpro. Bioorg. Med. Chem. 2008;16(1):293–302. doi: 10.1016/j.bmc.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Huitema C, Niu C, Yin J, James MN, Eltis LD, Vederas JC. Aryl methylene ketones and fluorinated methylene ketones as reversible inhibitors for severe acute respiratory syndrome (SARS) 3C-like proteinase. Bioorg. Chem. 2008;36(5):229–240. doi: 10.1016/j.bioorg.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidt MF, Isidro-Llobet A, Lisurek M, El-Dahshan A, Tan J, Hilgenfeld R, Rademann J. Sensitized detection of inhibitory fragments and iterative development of non-peptidic protease inhibitors by dynamic ligation screening. Angew. Chem. Int. Ed. Engl. 2008;47(17):3275–3278. doi: 10.1002/anie.200704594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukherjee P, Desai P, Ross L, White EL, Avery MA. Structure-based virtual screening against SARS-3CL(pro) to identify novel non-peptidic hits. Bioorg. Med. Chem. 2008;16(7):4138–4149. doi: 10.1016/j.bmc.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furet P, Meyer T, Strauss A, Raccuglia S, Rondeau JM. Structure-based design and protein X-ray analysis of a protein kinase inhibitor. Bioorg. Med. Chem. Lett. 2002;12(2):221–224. doi: 10.1016/s0960-894x(01)00715-6. [DOI] [PubMed] [Google Scholar]
- 69.Richardson CM, Gillespie RJ, Williamson DS, Jordan AM, Fink A, Knight AR, Sellwood DM, Misra A. Identification of non-furan containing A2A antagonists using database mining and molecular similarity approaches. Bioorg. Med. Chem. Lett. 2006;16(23):5993–5997. doi: 10.1016/j.bmcl.2006.08.116. [DOI] [PubMed] [Google Scholar]
- 70.Hofmann B, Franke L, Proschak E, Tanrikulu Y, Schneider P, Steinhilber D, Schneider G. Scaffold-hopping cascade yields potent inhibitors of 5-lipoxygenase. ChemMedChem. 2008;3(10):1535–1538. doi: 10.1002/cmdc.200800153. [DOI] [PubMed] [Google Scholar]
- 71.Huang D, Luthi U, Kolb P, Edler K, Cecchini M, Audetat S, Barberis A, Caflisch A. Discovery of cell-permeable non-peptide inhibitors of beta-secretase by high-throughput docking and continuum electrostatics calculations. J. Med. Chem. 2005;48(16):5108–5111. doi: 10.1021/jm050499d. [DOI] [PubMed] [Google Scholar]
- 72.Friedman R, Caflisch A. Discovery of plasmepsin inhibitors by fragment-based docking and consensus scoring. ChemMedChem. 2009;4(8):1317–1326. doi: 10.1002/cmdc.200900078. [DOI] [PubMed] [Google Scholar]
- 73.Novellino E, Cosimelli B, Ehlardo M, Greco G, Iadanza M, Lavecchia A, Rimoli MG, Sala A, Da Settimo A, Primofiore G, Da Settimo F, Taliani S, La Motta C, Klotz KN, Tuscano D, Trincavelli ML, Martini C. 2-(Benzimidazol-2-yl)quinoxalines: a novel class of selective antagonists at human A(1) and A(3) adenosine receptors designed by 3D database searching. J. Med. Chem. 2005;48(26):8253–8260. doi: 10.1021/jm050792d. [DOI] [PubMed] [Google Scholar]
- 74.Ando M, Sekino E, Haga Y, Moriya M, Ito M, Ito J, Iwaasa H, Ishihara A, Kanatani A, Ohtake N. Discovery of novel phenethylpyridone derivatives as potent melanin-concentrating hormone 1 receptor antagonists. Bioorg. Med. Chem. Lett. 2009;19(17):5186–5190. doi: 10.1016/j.bmcl.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 75.Mascarenhas Nahren M, Bhattacharyya D, Ghoshal N. Why pyridine containing pyrido[2,3-d]pyrimidin-7-ones selectively inhibit CDK4 than CDK2: insights from molecular dynamics simulation. J. Mol. Graph. Model. 28(7):695–706. doi: 10.1016/j.jmgm.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Chandrasekaran V, Lee CJ, Lin P, Duke RE, Pedersen LG. A computational modeling and molecular dynamics study of the Michaelis complex of human protein Z-dependent protease inhibitor (ZPI) and factor Xa (FXa) J Mol Model. 2009;15(8):897–911. doi: 10.1007/s00894-008-0444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu S-Y, Jiang Y-J, Lv J, Wu T-X, Yu Q-S, Zhu W-L. Molecular docking and molecular dynamics simulation studies of GPR40 receptor-agonist interactions. J. Mol. Graph. Model. 28(8):766–774. doi: 10.1016/j.jmgm.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Mukherjee P, Pradhan A, Shah F, Tekwani BL, Avery MA. Structural insights into the Plasmodium falciparum histone deacetylase 1 (PfHDAC-1): A novel target for the development of antimalarial therapy. Bioorg. Med. Chem. 2008;16(9):5254–5265. doi: 10.1016/j.bmc.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 79.Berendsen HJC, Postma JPM, Van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81(8):3684–3690. [Google Scholar]
- 80.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101(5):4177–4189. [Google Scholar]
- 81.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23(3):327–341. [Google Scholar]
- 82.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97(3):1990–2001. [Google Scholar]
- 83.Darden T, York D, Pedersen L. Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- 84.Hert J, Willett P, Wilton DJ, Acklin P, Azzaoui K, Jacoby E, Schuffenhauer A. Comparison of fingerprint-based methods for virtual screening using multiple bioactive reference structures. J. Chem. Inf. Comput. Sci. 2004;44(3):1177–1185. doi: 10.1021/ci034231b. [DOI] [PubMed] [Google Scholar]
- 85.Mukherjee P, Desai P, Zhou YD, Avery M. Targeting the BH3 domain mediated protein-protein interaction of Bcl-xL through virtual screening. J. Chem. Inf. Model. 50(5):906–923. doi: 10.1021/ci1000373. [DOI] [PubMed] [Google Scholar]
- 86.Martin YC. A bioavailability score. J. Med. Chem. 2005;48(9):3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
- 87.Refsgaard HH, Jensen BF, Brockhoff PB, Padkjaer SB, Guldbrandt M, Christensen MS. In silico prediction of membrane permeability from calculated molecular parameters. J. Med. Chem. 2005;48(3):805–811. doi: 10.1021/jm049661n. [DOI] [PubMed] [Google Scholar]
- 88.Willett P, Barnard JM, Downs GM. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998;38(6):983–996. [Google Scholar]
- 89.Downs GM, Willett P. Similarity searching in databases of chemical structures. Reviews in Computational Chemistry. 1996;7:1–66. [Google Scholar]
- 90.Johnson MA, Maggiora GM, Lajiness MS, Moon JB, Petke JD, Rohrer DC. Molecular similarity analysis: applications in drug discovery. Methods and Principles in Medicinal Chemistry. 1995;3:89–110. (Advanced Computer-Assisted Techniques in Drug Discovery) [Google Scholar]
- 91.Patterson DE, Cramer RD, Ferguson AM, Clark RD, Weinberger LE. Neighborhood behavior: a useful concept for validation of "molecular diversity" descriptors. J. Med. Chem. 1996;39(16):3049–3059. doi: 10.1021/jm960290n. [DOI] [PubMed] [Google Scholar]
- 92.Mochalkin I, Miller JR, Narasimhan L, Thanabal V, Erdman P, Cox PB, Prasad JV, Lightle S, Huband MD, Stover CK. Discovery of antibacterial biotin carboxylase inhibitors by virtual screening and fragment-based approaches. ACS Chem Biol. 2009;4(6):473–483. doi: 10.1021/cb9000102. [DOI] [PubMed] [Google Scholar]
- 93.Luzhkov VB, Selisko B, Nordqvist A, Peyrane F, Decroly E, Alvarez K, Karlen A, Canard B, Qvist J. Virtual screening and bioassay study of novel inhibitors for dengue virus mRNA cap (nucleoside-2'O)-methyltransferase. Bioorg. Med. Chem. 2007;15(24):7795–7802. doi: 10.1016/j.bmc.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 94.Naylor-Olsen AM, Ponticello GS, Lewis SD, Mulichak AM, Chen Z, Habecker CN, Phillips BT, Sanders WM, Tucker TJ, Shafer JA, Vacca JP. Identification and SAR for a selective, nonpeptidyl thrombin inhibitor. Bioorg. Med. Chem. Lett. 1998;8(13):1697–1702. doi: 10.1016/s0960-894x(98)00296-0. [DOI] [PubMed] [Google Scholar]
- 95.McMasters DR, Garcia-Calvo M, Maiorov V, McCann ME, Meurer RD, Bull HG, Lisnock J, Howell KL, Devita RJ. Spiroimidazolidinone NPC1L1 inhibitors. 1: Discovery by 3D-similarity-based virtual screening. Bioorg. Med. Chem. Lett. 2009;19(11):2965–2968. doi: 10.1016/j.bmcl.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 96.Macias AT, Mia MY, Xia G, Hayashi J, MacKerell AD., Jr Lead validation and SAR development via chemical similarity searching; application to compounds targeting the pY+3 site of the SH2 domain of p56lck. J. Chem. Inf. Model. 2005;45(6):1759–1766. doi: 10.1021/ci050225z. [DOI] [PubMed] [Google Scholar]
- 97.Franke L, Schwarz O, Muller-Kuhrt L, Hoernig C, Fischer L, George S, Tanrikulu Y, Schneider P, Werz O, Steinhilber D, Schneider G. Identification of natural-product-derived inhibitors of 5-lipoxygenase activity by ligand-based virtual screening. J. Med. Chem. 2007;50(11):2640–2646. doi: 10.1021/jm060655w. [DOI] [PubMed] [Google Scholar]
- 98.Edwards PD, Albert JS, Sylvester M, Aharony D, Andisik D, Callaghan O, Campbell JB, Carr RA, Chessari G, Congreve M, Frederickson M, Folmer RH, Geschwindner S, Koether G, Kolmodin K, Krumrine J, Mauger RC, Murray CW, Olsson LL, Patel S, Spear N, Tian G. Application of fragment-based lead generation to the discovery of novel, cyclic amidine beta-secretase inhibitors with nanomolar potency, cellular activity, and high ligand efficiency. J. Med. Chem. 2007;50(24):5912–5925. doi: 10.1021/jm070829p. [DOI] [PubMed] [Google Scholar]
- 99.Shanmugasundaram V, Maggiora GM, Lajiness MS. Hit-directed nearest-neighbor searching. J. Med. Chem. 2005;48(1):240–248. doi: 10.1021/jm0493515. [DOI] [PubMed] [Google Scholar]
- 100.Matter H. Selecting optimally diverse compounds from structure databases: a validation study of two-dimensional and three-dimensional molecular descriptors. J. Med. Chem. 1997;40(8):1219–1229. doi: 10.1021/jm960352+. [DOI] [PubMed] [Google Scholar]
- 101.Bajorath J. Selected concepts and investigations in compound classification, molecular descriptor analysis, and virtual screening. J. Chem. Inf. Comput. Sci. 2001;41(2):233–245. doi: 10.1021/ci0001482. [DOI] [PubMed] [Google Scholar]
- 102.Bender A, Glen RC. Molecular similarity: a key technique in molecular informatics. Org Biomol Chem. 2004;2(22):3204–3218. doi: 10.1039/B409813G. [DOI] [PubMed] [Google Scholar]
- 103.Brown RD, Martin YC. An evaluation of structural descriptors and clustering methods for use in diversity selection. SAR QSAR Environ. Res. 1998;8(1–2):23–39. doi: 10.1080/10629369808033260. [DOI] [PubMed] [Google Scholar]
- 104.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002;45(19):4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 105.Liu H-L, Lin J-C, Ho Y, Chen C-W. Homology models of main proteinase from coronavirus associated with SARS. Chem. Phys. Lett. 2005;401(1–3):24–29. doi: 10.1016/j.cplett.2004.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996;39(15):2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.